Abstract
Osteosarcoma is a differentiation-deficient disease, and despite the unique advantages and great potential of differentiation therapy, there are only a few known differentiation inducers, and little research has been done on their targets. Cell differentiation is associated with an increase in mitochondrial content and activity. The metabolism of some tumor cells is characterized by impaired oxidative phosphorylation, as well as up-regulation of aerobic glycolysis and pentose phosphate pathways. Leucine-containing zipper and EF-hand transmembrane protein 1 (LETM1) is involved in the maintenance of mitochondrial morphology and is closely associated with tumorigenesis and progression, as well as cancer cell stemness. We found that MG63 and 143B osteosarcoma cells overexpress LETM1 and exhibit abnormalities in mitochondrial structure and function. Knockdown of LETM1 partially restored the mitochondrial structure and function, inhibited the pentose phosphate pathway, promoted oxidative phosphorylation, and led to osteogenic differentiation. It also inhibited spheroid cell formation, proliferation, migration, and invasion in an in vitro model. When LETM1 was knocked down in vivo, there was reduced tumor formation and lung metastasis. These data suggest that mitochondria are aberrant in LETM1-overexpressing osteosarcoma cells, and knockdown of LETM1 partially restores the mitochondrial structure and function, inhibits the pentose phosphate pathway, promotes oxidative phosphorylation, and increases osteogenic differentiation, thereby reducing malignant biological behavior of the cells.
Keywords: Cancer metabolism, Differentiation, LETM1, Osteosarcoma, Stemness
Introduction
Osteosarcoma (OS) is a common non-hematological bone malignancy in both children and adults. There is increasing evidence that OS may be a differentiation-associated disease.1,2 Osteogenic differentiation is a precisely regulated process that may manifest as OS when the process is disturbed or deregulated.3,4 Therapies that promote differentiation or circumvent differentiation defects may be attractive new strategies for chemotherapy or chemoprevention.2
Otto Warburg’s theory on the origin of cancer postulates that tumor cells are defective in mitochondrial oxidative phosphorylation and rely on high levels of aerobic glycolysis as a significant source of ATP during cell proliferation.5, 6, 7 An attractive model for tumor cell metabolism is that oncogene-induced transformation depends on elevated glycolysis for ATP synthesis and the transfer of glycolytic intermediates to the pentose phosphate pathway (PPP).8,9 In support of this, recent studies have shown that in cancer cells, oncogenic mutations typically increase glycolysis and PPP flux to support cell biogenesis and the production of macromolecules required for rapid growth and division.8,10 Mitochondrial disorders have been identified in OS cells and have demonstrated the importance of the “Warburg effect” in OS, in which various mitochondrial proteins are thought to play a role.11
Cell differentiation is associated with increased mitochondrial content and activity, as well as growth and a shift toward oxidative phosphorylation in metabolism.12,13 Mesenchymal stem cells (MSCs) depend more on glycolysis, whereas differentiated cells are more dependent on oxidative phosphorylation.14,15 Mitochondria maintain low activity in MSCs, and mitochondrial function is up-regulated after the induction of osteogenesis. Similarly, inhibition of mitochondrial oxidative phosphorylation (OXPHOS) leads to defective differentiation and instead supports the maintenance of pluripotency.12,16,17 Therefore, we speculate that disruption of the normal osteogenic differentiation process during OS formation is related to metabolic reprogramming.
Leucine-containing zipper and EF-hand transmembrane protein 1 (LETM1) is a mitochondrial inner membrane protein.18 The LETM1 protein is required to maintain the tubular shape of mitochondria, mitochondrial cristae, for the assembly of respiratory chain supercomplexes, and helps regulate cell viability.18,19 Recent studies have shown that the formation of a LETM1/GRP75/GRP78 complex regulates mitochondrial quality control in lung cancer.20 High LETM1 expression has been detected in multiple human malignancies and is associated with a poor prognosis, poor survival, up-regulation of cancer stemness genes, and enhanced angiogenesis.21, 22, 23 In the current study, we demonstrate that LETM1 is highly expressed in OS and is associated with changes in the mitochondrial structure and mitochondrial dysfunction in OS cells. Restoring the mitochondrial structure and function can promote the osteogenic differentiation of OS cells.
Materials and methods
The use of TNMplot, Kaplan–Meier plotter, and GEPIA 2 databases
The TNMplot database utilizes gene array data from the NCBI-GEO, RNA-seq data from The Cancer Genome Atlas (TCGA), the TARGET database for pediatric tumors, and the Genotype-Tissue Expression (GTEx) repository.24 In brief, we logged on to “https://tnmplot.com/analysis/”, selected “TN: compare Tumor and Normal”, then selected” RNA-Seq data”, and entered the gene “LETM1" to analyze the differences in gene expression between normal (no malignancy) (n = 564) and OS (n = 88) samples. To generate Kaplan–Meier curves, we logged in to “http://kmplot.com/analysis/index.php?p=service”, selected “mRNA RNA-seq” in “Pan-Cancer”, entered the gene “LETM1”, selected “Auto select best cutoff”, chose the tumor type “Sarcoma”, and clicked on “Draw Kaplan–Meier Plot” to plot the overall survival curve. We logged in to “http://gepia2.cancer-pku.cn”, entered the gene “LETM1”, selected “SARC”, and clicked on “Plot” to plot the disease-free survival curve. The HR represents the risk factor for the high-expression group relative to the low-expression group, and an HR > 1 means that the gene is a risk factor (the higher the expression, the worse the prognosis).
Cell culture
The TE85, MG63, and 143B human OS cell lines, and normal human bone marrow mesenchymal stem cells (BMSCs) were stored in our laboratory. The normal human osteoblast cell line, hFOB1.19, was obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 1% penicillin-streptomycin in a 37 °C incubator with 5% CO2.
Adenovirus infection
Three recombinant adenoviruses expressing target shRNA (sh-LETM1-1: 5′-GCAGCAAATGATCGGGCAGAT-3′, sh-LETM1-2: 5′-CCAGAGATTGTGGCAAAGGAA-3′, sh-LETM1-3: 5′-GCTATGGATCGACACCAAGAT-3′) and negative control virus (expressing 5′-TTCTCCGAACGTGTCACGT-3′) were designed and synthesized by GeneChem, and all expressed green fluorescent protein. The infection of cells was performed for 48–72 h. Fluorescence expression was observed under a microscope and the fluorescence rate reached nearly 90%. The cells were therefore considered to be in good growth condition for subsequent experiments. The knockdown efficiency was verified by quantitative real-time PCR (qPCR) and western blotting.
RNA isolation and qPCR analysis
Total RNA was extracted using a Simply P Total RNA Extraction Kit (BSC52S1, BioFlux, Beijing, China). The PrimeScript RT Reagent kit (TaKaRa, Japan) was used to reverse-transcribe one microgram of total RNA into single-stranded complementary DNA (cDNA). Then, the cDNA samples, gene-specific primers, and SYBR Green Master Mix kit (MCE, USA) were used for amplification in a CFX-96 real-time system (Bio-Rad). Results were calculated using the 2−ΔΔCT method. The primer sequences are shown in Table S1.
Western blot
Total proteins were extracted according to the manufacturer’s instructions (P0033, Beyotime). The total protein concentration was determined using a BCA kit (P0010, Beyotime). The proteins were then dissolved in 5 × SDS-PAGE sample buffer (P0015, Beyotime) and heated to 99 °C for 10 min. Equal amounts of proteins were loaded into each well of polyacrylamide gels (6%–15%, Epizyme Biotechnology) for electrophoresis, and then transblotted onto a 0.2-μm polyvinylidene fluoride (PVDF) membrane (ISEQ00010, MerckMillipore). After being washed three times, the membranes were blocked with QuickBlockTM blocking buffer for Western blotting (P0252, Beyotime) at room temperature for 30 min. The blots were then incubated with anti-LETM1 (1:4000, Proteintech), anti-G6PD (1:1000, PTM BIO), anti-PKM2 (1:2000, PTM BIO), anti-OPN (1:2000, ZEN BIO), anti-Osteocalcin (1:500, Biorbyt) or anti-β-actin (1:4000, ZSGB BIO) antibodies at 4 °C overnight, and then with horseradish peroxidase-linked anti-mouse or anti-rabbit secondary antibodies (1:10,000, ZEN BIO) at room temperature for 2 h. The bands were detected with a chemiluminescent horseradish peroxidase substrate (NCM Biotech). The expression of β-actin was used as a loading control.
Transmission electron microscopy
Cells were washed with phosphate-buffered saline solution (PBS) twice and then digested with 0.25% trypsin–EDTA solution (C0201, Beyotime). Cells were loaded into 1.5-mL centrifuge tubes and centrifuged at 1200 rpm for 10 min; 4% glutaraldehyde solution was added along the walls of the tubes. The cell samples were subsequently rinsed with distilled water, dehydrated with a gradient of ethanol and methanol, and then soaked and embedded in epoxy resin. Finally, the ultrathin sections were randomly observed using a transmission electron microscope (H-7500).
Mitochondrial staining
The MitoTracker® red CMXRos (40741ES50, Yeasen Biotechnology) intracellular dye was used for mitochondrial imaging. The cells were incubated with 20 nM MitoTracker red for 25 min and then the nuclei were stained with Hoechst 33,342 (C1028, Beyotime) for 10 min. After being washed twice with PBS, the cells were imaged using a confocal microscope (Nikon, Japan).
Detection of mitochondrial membrane potential (MMP)
MMP was measured using the MMP assay kit with JC-1 (C2006, Beyotime) or tetramethylrhodamine ethyl ester perchlorate (TMRE) (HY-D0985A, MedChemExpress). Cells were inoculated into confocal dishes. After being washed twice with PBS, the cells were incubated at 37 °C in a 5% CO2 incubator for 30 min following the addition of the JC-1 working solution or TMRE working solution. Photographs were taken under a confocal microscope (Nikon). The ratio between the monomeric (Em = 590 nm) and aggregate (Em = 525 nm) forms of JC-1 represents the MMP. A high ratio indicates a high MMP. For the TMRE method, the ΔΨm of cells was immediately analyzed by confocal microscopy or flow cytometry (BD FACSCanto II). The NIS-Elements Viewer software was used to analyze the average fluorescence intensity.
ATP assay
The total intracellular ATP levels were determined by applying the ATP assay kit (Beyotime, S0026) according to the manufacturer’s protocol.25 Briefly, after the cells were lysed with ATP lysate, the supernatant was removed by centrifugation at 4 °C, and 100 μL of ATP assay working solution was added to each well of a black 96-well plate. After 5 min at room temperature, 20 μL of cell supernatant was added to the wells, and chemiluminescence was detected by a multifunctional enzyme marker. Protein concentration was determined as before. The ATP concentration was calculated from the standard curve and converted to nmol/mg protein.
Correlation score
The RNA-sequencing expression (level 3) profiles and corresponding clinical information for sarcoma were downloaded from the TCGA dataset (https://portal.gdc.com). The R software GSVA package and the ssGSEA method were used to analyze the data. We focused on groups of genes included in related pathways.26 The correlation between genes and pathway scores was analyzed by Spearman’s correlation. A one-class logistic regression algorithm was used to calculate mRNAsi which was constructed by Malta.27 Based on the mRNA expression signature, the gene expression profile contained 11,774 genes. The minimum value was subtracted, and the result was divided by the maximum maps of the dryness index to the range [0, 1]. All the analysis methods and R packages were implemented in R version 4.0.3. A P-value <0.05 was considered statistically significant.
Detection of α-KGDH and ICDHm activities
Seventy-two hours after adenovirus infection, the activity levels of α-ketoglutarate dehydrogenase (α-KGDH) and mitochondrial isocitrate dehydrogenase (ICDHm) were measured and calculated using the α-KGDH assay kit and the ICDHm assay kit (Solebro, Beijing), respectively, according to the manufacturer’s instructions.28 Three separate independent replicate experiments were performed.
Detection of the lactic acid content
Cells were collected, and lactate levels were measured and calculated using the Lactic Acid Assay Kit (Nanjing Jiancheng Bioengineering Institute) according to the manufacturer’s instructions.28
Measurement of cellular glucose content and uptake
The culture medium was collected from cells in the logarithmic growth phase. The cells were then washed twice with PBS, then the cell lysate for the glucose assay was added, and cells were scraped off with a cell scraper. The collected cell lysates were centrifuged at 12,000 g for 5 min, then the supernatants were collected, and the absorbance at 630 nm was measured according to the manufacturer’s instructions for the glucose assay kit (S0201S, Beyotime). The glucose concentration in the sample was calculated from the standard curve, and the protein concentration was measured to calculate the glucose content in the sample per unit of protein. The amount of glucose in the old media and cell lysates was determined, and the amount of glucose in the lysates was subtracted from the amount in the old medium and divided by the protein concentration to calculate the glucose uptake.
Alkaline phosphatase (ALP) staining and activity assay and alizarin red S (ARS) staining
Cells were induced to differentiate by an osteogenic induction medium. Early osteogenic differentiation was detected using the ALP assay and late osteogenic differentiation was detected based on ARS staining. When the cells were induced for two weeks, ALP staining was performed with a BCIP/NBT ALP color development kit (C3206, Beyotime) according to the manufacturer’s instructions.29 The supernatant from cell lysis was used to determine the ALP activity using the ALP assay kit (P0321S, Beyotime).30 When the cells were induced for three weeks, intracellular calcium salt nodules were visualized by 0.2% ARS (G1450, Solarbio) staining after the cells were fixed with 4% paraformaldehyde.31
Colony-formation assay
After 48-h adenovirus infection, cells were inoculated into six-well plates. MG63 cells (2000 cells/well) and 143B cells (1000 cells/well) were cultured for 12 days. After 12 days, the medium in the well plates was discarded, cells were washed three times with PBS, and then fixed in 4% paraformaldehyde for 10 min. Subsequently, the plates were stained with crystal violet staining solution (Beyotime, C0121) for 10 min and photographed. Colonies that were more prominent than 50 cells were counted under a microscope.
EdU with Alexa Fluor 555 proliferation assay
The proliferation of MG63 and 143B cells was determined using the BeyoClick™ Edu-555 kit (C0075S, Beyotime) according to the manufacturer’s protocol. Both cell lines were seeded at 10,000 cells per well into 24-well plate cell slides after 72-h adenovirus infection. An anti-fluorescence quencher was used to seal the slices, which were then photographed during laser confocal microscopy.
Wound healing assay
Cells were inoculated into six-well plates and infected with adenovirus for 72 h. After the cells reached 100% confluency in the monolayer, a linear wound was created by scraping on the cell surface using a 200-μL pipette tip. Cells were cultured in a serum-free medium. Photographs of the exact locations were taken at 0 and 24 h to calculate the wound healing rate.
Transwell assay for cell migration and invasion
Cell migration and invasion were assayed using Transwell chambers with and without Matrigel (Corning). After 72-h adenovirus infection, cells from each group were resuspended with serum-free Dulbecco’s modified Eagle’s medium, then 100 μL of cell suspension (1 × 104 per well of 143B cells or 5 × 104 per well of MG63 cells for migration, and 2 × 104 per well of 143B cells and 1 × 105 per well of MG63 cells for invasion) was added to the upper chambers, and Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum was added to the lower chambers. After 36-h incubation, the chambers were carefully removed. The lower cells were fixed, and the upper cells were wiped off and stained with crystal violet (Beyotime) for 20 min. Images of the cells were obtained using a microscope (Nikon).
Cell cycle analysis
Cells were fixed with 70% cold ethanol at 4 °C for 24 h. After the fixative was washed off with PBS, 500 μL of PI/RNase A staining solution (keygentec) was added and cells were incubated in the dark at room temperature for 60 min. The cells were then read by a machine that recorded red fluorescence at an excitation wavelength of 488 nm.
In vivo tumorigenesis and metastasis model
All procedures for animal experiments were approved by the Ethics Committee of the Children’s Hospital of Chongqing Medical University. Female BALB/c nude mice (6 weeks old, n = 10) were purchased from Chongqing Tengxin Biotechnology Co., Ltd. and housed at 25 °C under specific pathogen-free conditions with a 12-h/12-h light/dark cycle, with free access to food and water. A total of 2 × 106 143B cells transfected with sh-LETM1 or ADCON were injected into the tibial periosteum via the knee joint while nude mice were anesthetized. Each group contains five mice. Animals were monitored daily for health and behavior, and mice exhibiting tibial were photographed weekly using IVIS Spectrum Imaging, and the results were analyzed by the Living Image software. Mice were euthanized if the maximum length of any tumor reached 15 mm or when there was a significant decrease in body weight. The remaining animals were sacrificed after 4 weeks. Lungs were harvested, fixed, sectioned, and subjected to HE staining (4-μm thickness).
Statistical analysis
All data were expressed as mean ± standard deviation and were analyzed using GraphPad Prism 9.0 software (GraphPad Software, Inc., USA). Statistical analyses were performed using a two-tailed Student’s t-test to determine the significance of differences between two groups. The one-way ANOVA was used to measure the significance of differences among more than two groups. A P-value < 0.05 was considered statistically significant.
Results
LETM1 was overexpressed and associated with an abnormal mitochondrial structure in MG63 and 143B cells
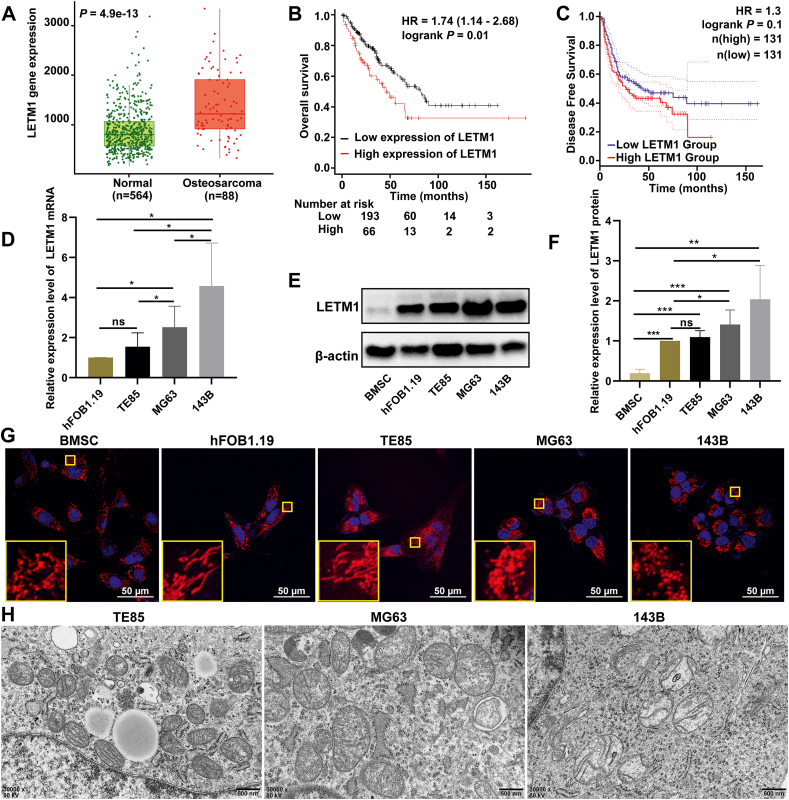
The TNMplot database was used to analyze the expression levels of LETM1 in OS tissues compared to tissues from adult patients without malignancy and normal pediatric tissues. The results showed that the expression levels of LETM1 were significantly higher in OS tissues compared to normal tissues (Fig. 1A). The impact of LETM1 expression on the prognosis of sarcoma patients was assessed using the Kaplan–Meier plotter and GEPIA 2 database, and the results showed that the overall survival and the disease-free survival of patients in the high LETM1 expression group was shorter than that of the patients in the low LETM1 expression group (Fig. 1B, C).
Figure 1.
Overexpression of LETM1 in OS tissue and cells and abnormal mitochondrial structures in MG63 and 143B cells. (A) The expression of LETM1 in OS tissue, normal samples from patients without malignancy, and other (non-malignant) pediatric tissues was assessed using the TNMplot database. (B) The relationship between the expression level of LETM1 in sarcomas and overall survival. (C) The relationship between the expression level of LETM1 in sarcomas and disease-free survival. (D) Analysis of the mRNA expression of the LETM1 gene in normal osteoblasts (hFOB1.19) and OS cells (TE85, MG63, and 143B) by real-time PCR. (E, F) The expression level of the LETM1 protein was analyzed by Western blotting. (G) The structures of cellular mitochondria were observed under a laser confocal microscope by staining with MitoTracker red probe. (H) The mitochondria were also measured by transmission electron microscopy. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
We next investigated the expression of LETM1 in OS cell lines. LETM1 expression was increased in MG63 and 143B OS cells compared to normal osteoblasts (hFOB1.19; Fig. 1D–F). The LETM1 expression was also increased in TE85, MG63, and 143B OS cells compared to normal BMSCs (Fig. 1E, F).
LETM1 is an anchoring protein that forms complexes with mitochondrial ribosomes and regulates mitochondrial biogenesis.32 Studies have reported mitochondrial dysfunction and an abnormal permeability transition in OS cells showing the Warburg effect.33 Therefore, we evaluated the mitochondrial morphology of OS cells in vitro. We used laser confocal microscopy for live cell imaging after staining with the mitochondrial red fluorescent probe and Hoechst 33,342. The mitochondria of hFOB1.19 and TE85 cells were mallet-shaped and had a network of highly connected filamentous mitochondria, while the mitochondria of MG63 and 143B cells were round and had limited branching, and the mitochondria of BMSCs were a mixture of punctate and longer tubular mitochondria (Fig. 1G). To further examine the mitochondrial structure of MG63 and 143B OS cells, we used transmission electron microscopy to observe the organelles. Compared with the less malignant TE85 cells, the MG63 and 143B cells had enlarged mitochondria with a lower electron density matrix, an indication of swollen mitochondria, and the swelling was more pronounced in 143B cells than in MG63 cells (Fig. 1H). Thus, the expression of LETM1 was increased in OS cells, and the mitochondrial structure appeared to correspond to the LETM1 expression.
Mitochondrial dysfunction was present in MG63 and 143B cells
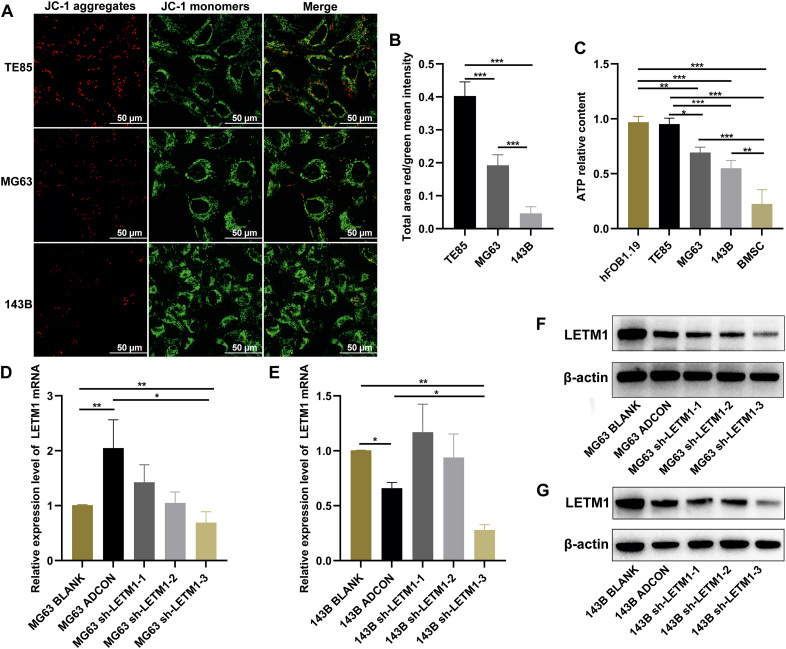
The MMP plays a critical role in mitochondrial functions, and its dissipation signifies mitochondrial dysfunction. Since we observed abnormal mitochondrial structures in MG63 and 143B OS cells (Fig. 1G, H), we assessed the mitochondrial function by monitoring changes in the MMP. MG63 and 143B cells had lower red-green fluorescence ratios than TE85 cells, and the ratio for 143B cells was lower than that for MG63 cells (Fig. 2A, B). Mitochondria are known for their role in the production of ATP through aerobic respiration. A decrease in ATP levels usually indicates impaired or decreased mitochondrial function. An analysis of the total cellular ATP content revealed that hFOB1.19 cells had a higher ATP content than BMSCs, and the ATP content of the three OS cell lines was between that of the BMSCs and hFOB1.19 cells. The MG63 and 143B cells had a lower ATP content than TE85 cells, and 143B cells had a lower ATP content than MG63 cells (Fig. 2C). These findings suggested the presence of mitochondrial dysfunction in MG63 and 143B cells.
Figure 2.
Mitochondrial dysfunction in MG63 and 143B cells. (A, B) Cells were stained with the JC-1 fluorescent probe and photographed under a laser confocal microscope, and the ratio of red fluorescence to green fluorescence was determined. (C) The total cellular ATP levels were detected by an ATP assay kit. (D, E) Cells were infected with adenovirus for 48 h, and then RNA was extracted and the mRNA expression levels of the LETM1 gene were detected by real-time PCR. (F, G) Cells were infected with adenovirus for 72 h, and then the total protein was extracted and the expression levels of the LETM1 protein were detected by Western blotting. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
To better understand how LETM1 protein plays a role in OS progression, its expression was disrupted using targeted LETM1 shRNA. Three potential LETM1-targeting shRNAs were used, and the results showed that sh-LETM1-3 had the most substantial inhibitory effect on LETM1 expression compared with the ADCON group (Fig. 2D–G). Therefore, sh-LETM1-3 was used for further experiments.
Correlation analysis
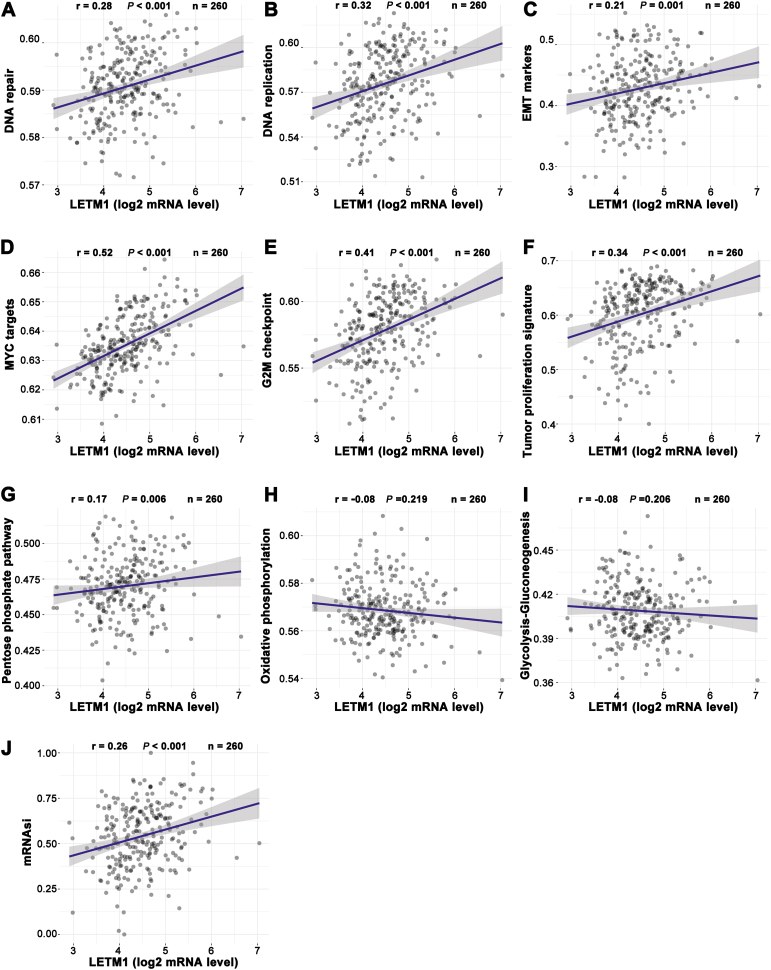
Based on the Spearman correlation analysis of 260 sarcoma cases included in the TCGA database, we predicted that LETM1 plays a role in tumor development, metabolism, and cancer stemness. It was noted that the expression of LETM1 was positively correlated with DNA repair, DNA replication, epithelial-to-mesenchymal transition makers, MYC targets, the G2M checkpoint, and the tumor proliferation signature (Fig. 3A–F). The expression of LETM1 was also positively correlated with the PPP (Fig. 3G). The OXPHOS and glycolysis-gluconeogenesis were not strongly correlated with the expression of LETMT (Fig. 3H, I). However, the expression of LETM1 was positively correlated with the mRNA expression-based stemness index (si; Fig. 3J).
Figure 3.
The correlations between LETM1 and the pathway score were analyzed with a Spearman analysis. The abscissa represents the distribution of the gene expression, and the ordinate represents the distribution of the pathway score; “r" represents the correlation coefficient, where a positive number indicates a positive correlation and a negative number indicates a negative correlation. The larger the absolute value, the stronger the correlation. “P" represents the correlation P value.
Knockdown of LETM1 partially restored the structure of mitochondria
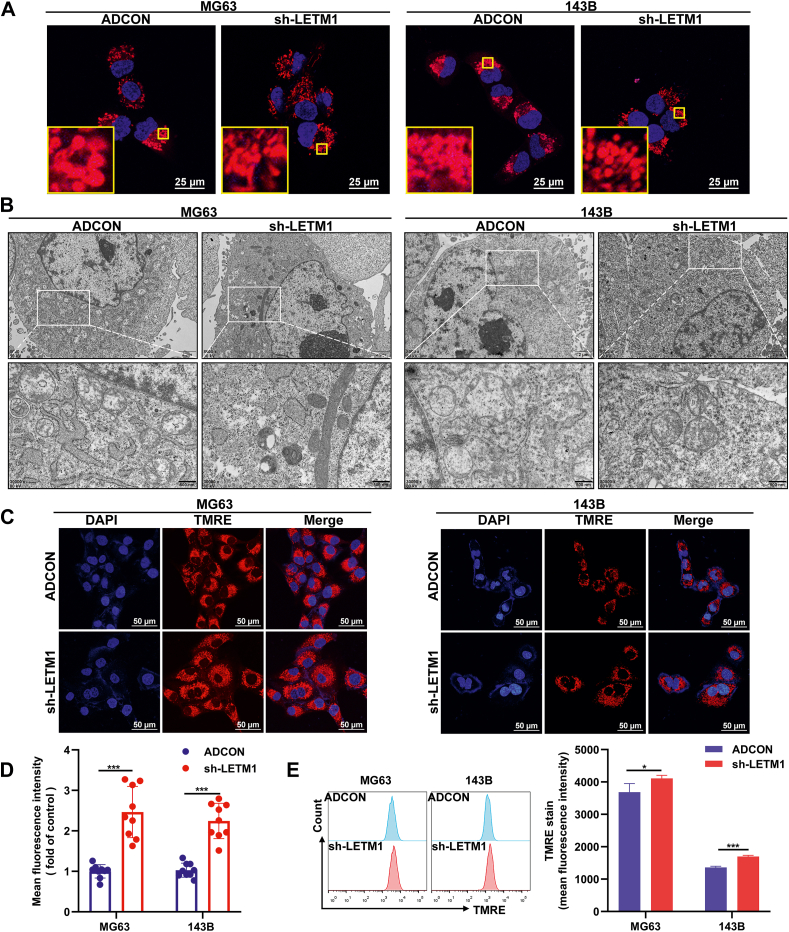
LETM1 is a mitochondrial inner membrane protein involved in regulating mitochondrial ion channels, maintaining mitochondrial morphology, and respiratory chain formation,19,32,34 so we examined the changes in mitochondrial structure in cells with LETM1 knockdown. We observed by MitoTracker red staining that both MG63 and 143B sh-LETM1 groups had longer rod-shaped mitochondria and more mitochondrial branches compared to the ADCON group (Fig. 4A). Subsequent transmission electron microscopy showed that mitochondria in both the MG63 and 143B sh-LETM1 groups had increased electron-dense substrates, clearer cristae structures, and varying degrees of improvement in the mitochondrial swelling compared to the ADCON group (Fig. 4B). Taken together, these findings suggest that the knockdown of LETM1 can partially restore the structure of mitochondria.
Figure 4.
Knocking down LETM1 partially restored the structure of mitochondria and increased the MMP. (A) The structure of the cellular mitochondria was observed under a laser confocal microscope after staining with a MitoTracker red probe. (B) Mitochondria were measured by transmission electron microscopy. (C, D) Cells were stained with the TMRE fluorescent probe and photographed under the same parameters using a laser confocal microscope, and the red fluorescence intensity of individual cells was counted. (E) The MMP was compared by TMRE flow detection, with the mean fluorescence intensity compared between the ADCON and sh-LETM1 groups. ∗P < 0.05, ∗∗∗P < 0.001.
Knocking down LETM1 improved the function of mitochondria
It has been reported in the literature that LETM1 reduces mitochondrial biogenesis and ATP production,35 so we examined the changes in mitochondrial function following LETM1 knockdown in our cells. Because our virus carried green fluorescent protein, the MMP was detected by TMRE. TMRE fluorescent probe staining and an analysis of the average fluorescence intensity of individual cells revealed an increased MMP in the sh-LETM1 group compared to the ADCON group in both the MG63 and 143B cells (Fig. 4C, D). A TMRE flow assay showed that the MG63 sh-LETM1 group and 143B sh-LETM1 group had increased MMP compared to the ADCON groups (Fig. 4E). Both α-KGDH and ICDHm are critical enzymes of the tricarboxylic acid cycle. The sh-LETM1 groups had increased α-KGDH and ICDHm activities compared to the ADCON group in both the MG63 and 143B cells (Fig. 5A, B). The ATP content assay revealed that the sh-LETM1 group had increased ATP content compared to the ADCON group for both the MG63 and 143B cell lines (Fig. 5C). Together, these observations indicate that knockdown of LETM1 can improve the mitochondrial function.
Figure 5.
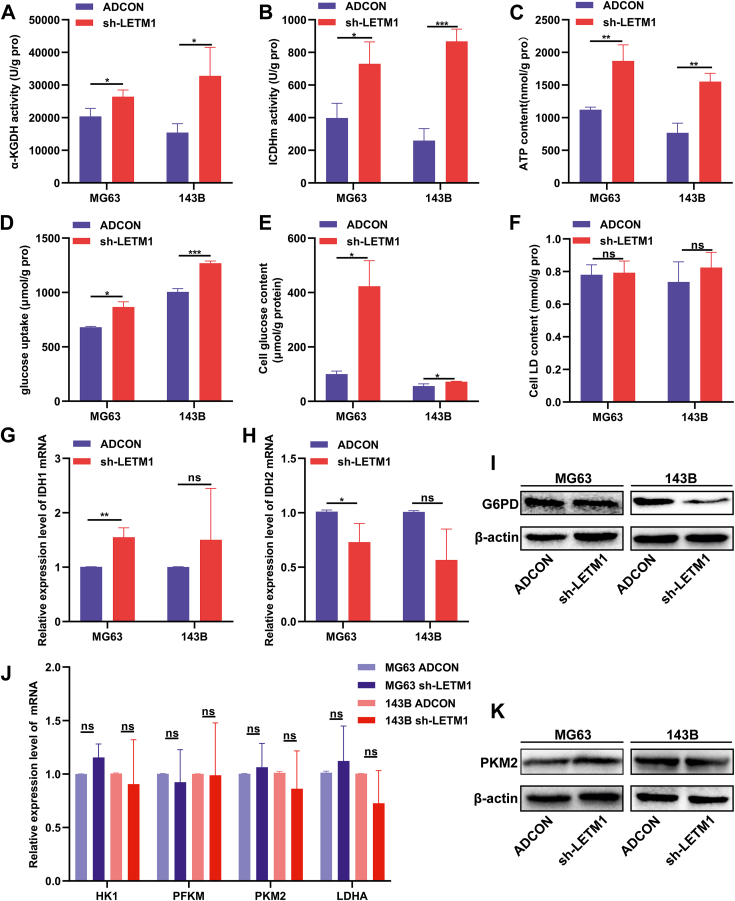
Knockdown of LETM1 improved the function of mitochondria, inhibited the PPP, and promoted the OXPHOS in MG63 and 143B cells. Cells were infected with adenovirus for 72 h, and then (A) the α-KGDH activity assay kit was used to detect and compare the α-KGDH activity in the ADCON and sh-LETM1 groups. (B) The ICDHm activity assay was used to detect the ICDHm activity, which was compared between the ADCON and sh-LETM1 groups. (C) The total ATP content in the ADCON and sh-LETM1 groups was determined using an ATP assay kit. (D) The glucose uptake was determined by subtracting the amount of glucose present from the amount originally contained in the culture medium, divided by the protein level. (E) Detection of the intracellular glucose content was determined after cell lysis. (F) The lactic acid levels were measured and calculated using a lactic acid assay kit. (G, H, J) The mRNA expression levels of IDH1, IDH2, HK1, PFKM, PKM2, and LDHA were analyzed by real-time PCR. (I, K) The expression levels of the G6PD and PKM2 proteins were detected by Western blotting. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
LETM1 knockdown inhibited the PPP and promoted the OXPHOS
The sh-LETM1 MG63 and 143B cells showed significant increases in the cellular glucose content and glucose uptake compared to the ADCON groups (Fig. 5D, E). No significant differences were noted in the lactic acid content (Fig. 5F). After LETM1 knockdown, the isocitrate dehydrogenase 1 (IDH1) gene expression was increased, and the isocitrate dehydrogenase 2 (IDH2) gene expression was decreased in MG63 cells (Fig. 5G, H). Knockdown of LETM1 also reduced the expression of G6PD, the first rate-limiting enzyme of the PPP (Fig. 5I). No differences were detected in the expression of the HK1, PFKM, PKM2, and LDHA genes related to glycolysis (Fig. 5J, K). These findings suggested that the knockdown of LETM1 inhibited the PPP, promoted the OXPHOS, and had no significant effect on glycolysis.
Knockdown of LETM1 promoted osteogenic differentiation
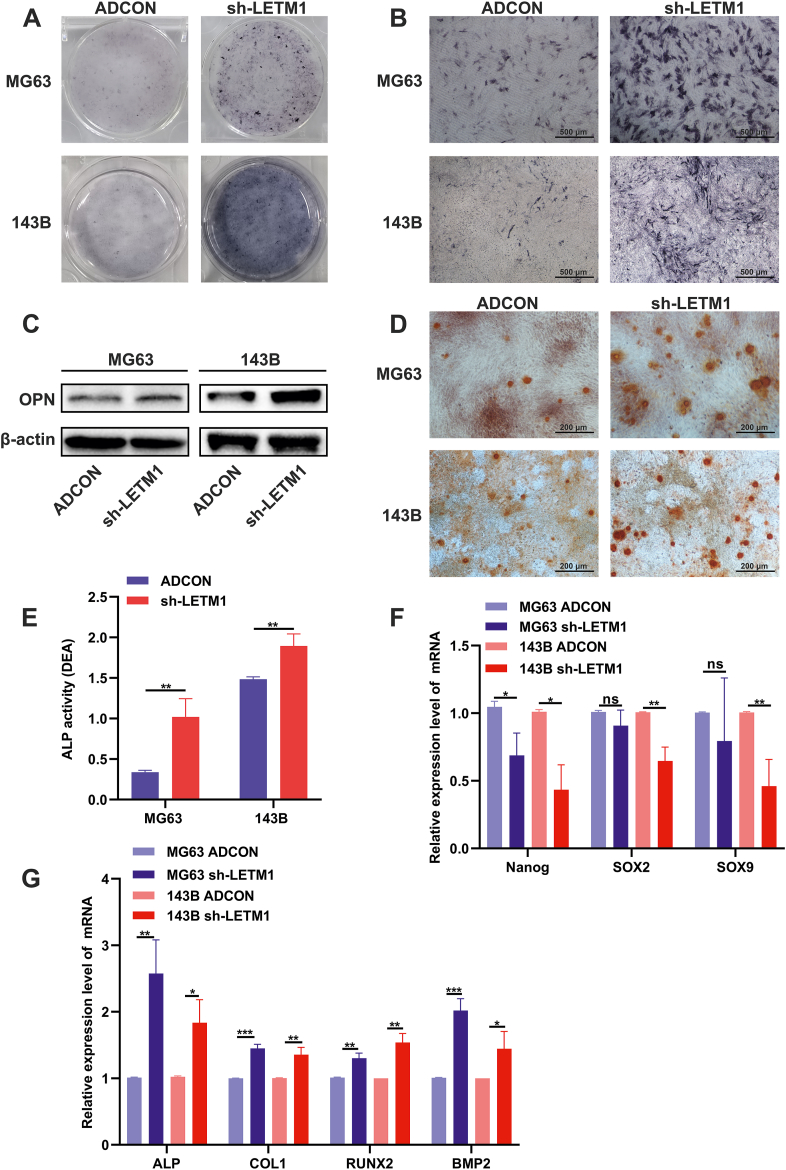
Cell differentiation is associated with increased mitochondrial content and activity and a metabolic shift toward increased OXPHOS. Osteogenic differentiation is a precisely regulated process that may manifest as OS if disturbed or uncontrolled.1,3,36 As shown in Figure 6A and B, the sh-LETM1 group had darker ALP staining than the ADCON group in both the MG63 and 143B cells. The ALP activity assay also found increased ALP activity in the sh-LETM1 group compared to the ADCON group in both OS cell lines (Fig. 6E).
Figure 6.
LETM1 knockdown promoted the osteogenic differentiation of MG63 and 143B cells. (A, B) After adenovirus infection for 48 h, cells were cultured in an osteogenic medium for an additional 14 days and then were fixed and stained for ALP. (C) The expression level of the OPN protein was detected by Western blotting. (D) After adenovirus infection for 48 h, the cells were cultured in an osteogenic medium for another 21 days, and then were fixed and stained with Alizarin red S, and observed for calcium salt nodules. (E) Cells were assayed for ALP activity. (F) The mRNA expression levels of cancer stemness-associated genes including Nanog, SOX2, and SOX9 were analyzed by real-time PCR. (G) The mRNA expression levels of osteogenic differentiation-related genes including ALP, COL1, RUNX2, and BMP2 were analyzed by real-time PCR. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
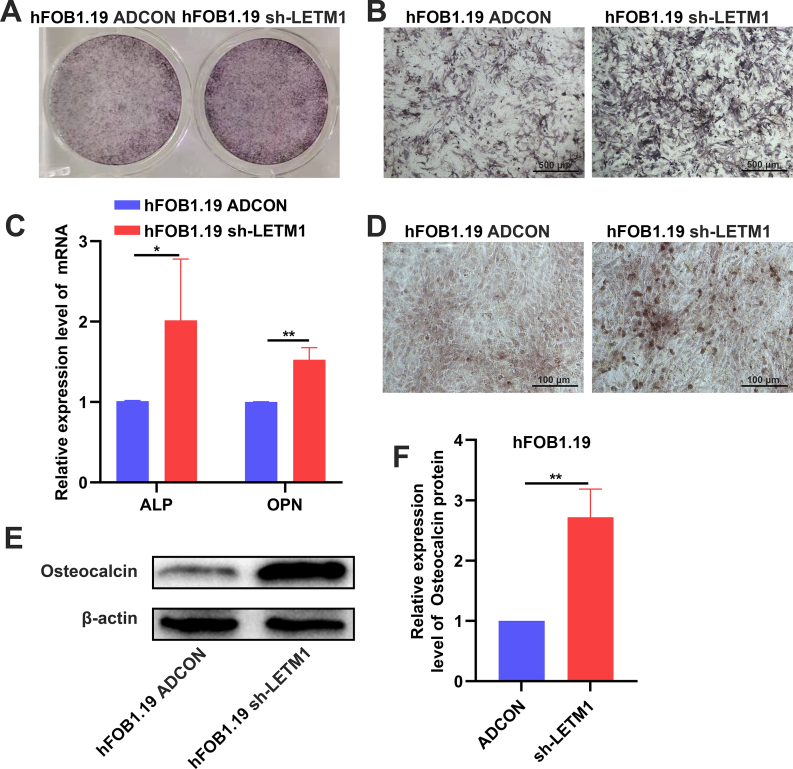
We detected mineralized nodules by alizarin red staining, and we found increased calcium salt nodules in the sh-LETM1 group compared to the ADCON group in both the MG63 and 143B cell lines (Fig. 6D). We then measured the expression of genes related to osteogenic differentiation. Knocking down LETM1 increased the expression of ALP, COL1, RUNX2, and BMP2 in both the MG63 and 143B cells (Fig. 6G). As shown in Figure 6C, LETM1 knockdown led to increased expression of OPN. We repeated the above experiments in normal osteoblasts (hFOB1.19 cells). The sh-LETM1 group had darker ALP staining and increased calcium salt nodules than the ADCON group in the hFOB1.19 cells (Fig. S1A, B, D). Knocking down LETM1 increased the expression of ALP and OPN in the hFOB1.19 cells (Fig. S1C). As shown in Figure S1E and F, LETM1 knockdown led to increased expression of osteocalcin. We found that inhibiting the expression of LETM1 also promoted the osteogenic differentiation of these osteoblast cells. We also found that LETM1 knockdown decreased the expression of Nanog, SOX2, and SOX9, stemness-related genes, in the OS cell lines (Fig. 6F). In summary, LETM1 knockdown reduced the stemness and promoted the osteogenic differentiation of MG63 and 143B cells.
LETM1 knockdown inhibited the malignant biological behavior of MG63 and 143B cells
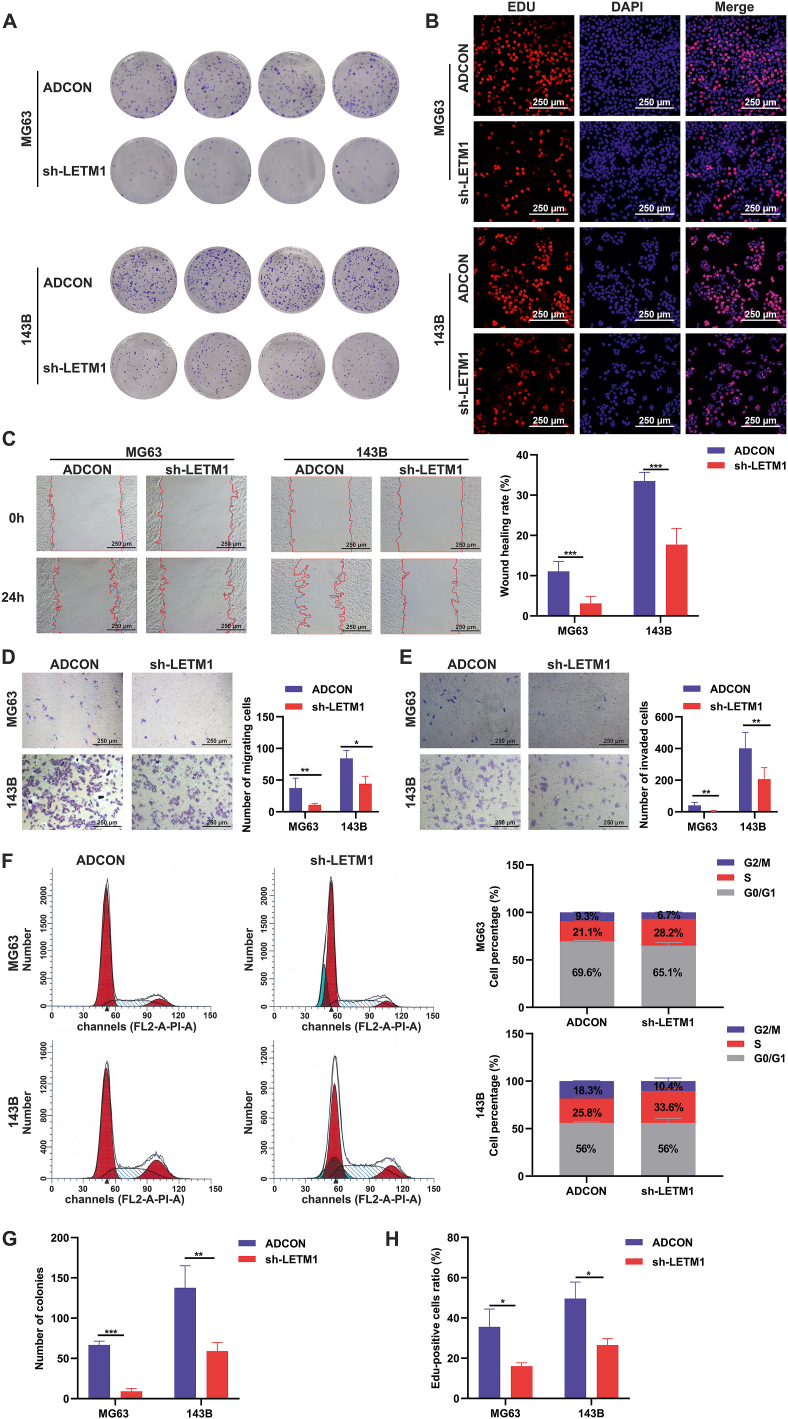
Cellular metabolic shifts and cell differentiation affect biological behavior. Impaired mitochondrial function in OS is associated with a more aggressive phenotype.11 The clone formation assay and EDU-555 cell proliferation assay revealed that knocking down LETM1 inhibited the clone formation and proliferation of the MG63 and 143B cells (Fig. 7A, B, G, H). The MG63 and 143B cells with LETM1 knockdown also exhibited reduced horizontal and vertical migration and invasion in scratch and Transwell assays (Fig. 7C–E). Studies of the cell cycle distribution showed that the proportion of cells in the S phase was significantly increased in the sh-LETM1 group compared to the ADCON group in both MG63 and 143B cells (Fig. 7F). The data collectively suggest that knocking down LETM1 inhibited the proliferation, migration, and invasion of MG63 and 143B OS cells, and affected the cell cycle distribution.
Figure 7.
Knockdown of LETM1 inhibited the malignant biological behavior of MG63 and 143B cells. (A, G) Colony formation assays were performed to detect the effects of LETM1 knockdown on OS cell colony formation. (B, H) Cells in the ADCON and sh-LETM1 groups were stained with EDU-555 and the results were quantified to evaluate the proliferation of the cells. (C) The wound healing assay was used to detect cell migration and the wound healing rate. (D, E) Transwell migration and invasion assays were performed for 24 h, and then the cells were fixed and stained with crystal violet, and the number of migrated cells was counted. (F) The cell cycle distribution in the MG63 and 143B cells was analyzed in the ADCON and sh-LETM1 groups. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Knocking down LETM1 reduced the distant metastasis of OS in vivo
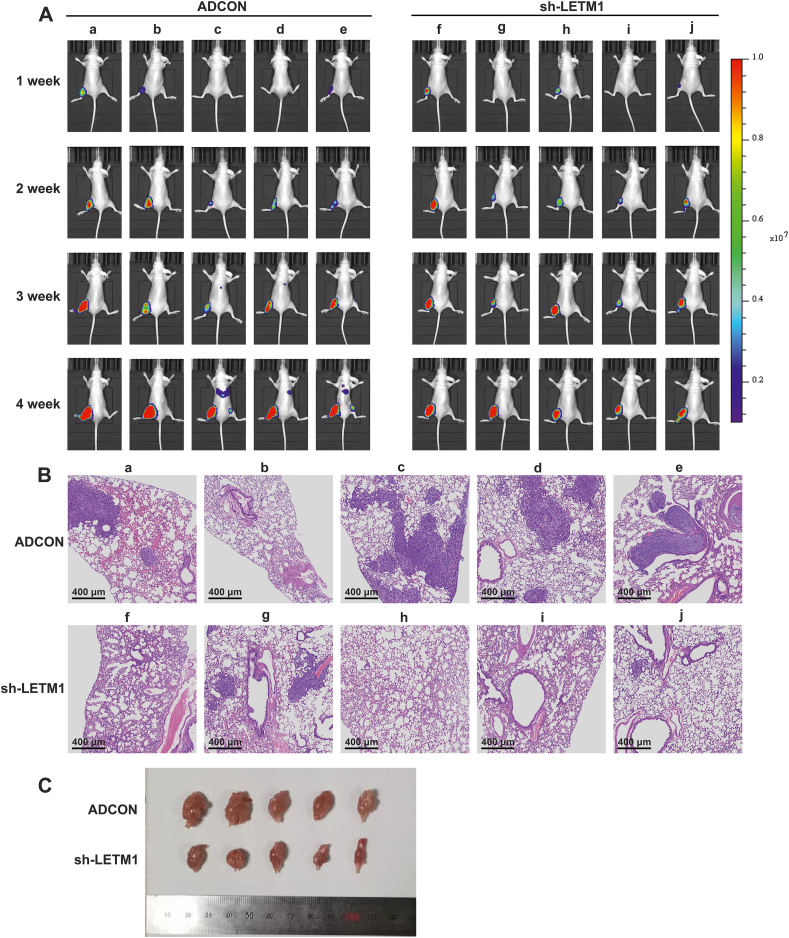
To evaluate the in vivo effects of LETM1, a clinically relevant intra-tibial injection model of OS was established using 143B cells. This model leads to the formation of lung metastases in nude mice. The results of imaging studies showed that mice numbered c, d, and e in the ADCON group had pulmonary metastases (Fig. 8A). HE staining of the lungs revealed that there were more lung metastases in the ADCON group than in the sh-LETM1 group (Fig. 8B). Moreover, knocking down LETM1 reduced the ability of 143B cells to form tumors in nude mice (Fig. 8C). Thus, LETM1 knockdown reduced the tumorigenic capacity and lung metastasis of 143B OS cells in vivo.
Figure 8.
Knocking down LETM1 reduced OS tumor growth and distant metastasis in vivo. (A) Live imaging of nude mice. (B) HE staining of lungs from nude mice with tibial tumors. (C) Specimens of tibial tumors formed in nude mice.
Discussion
Warburg initially observed increased aerobic glycolysis and reduced mitochondrial function in cancer cells.7,37 The Warburg effect is a natural phenomenon that has been observed both in vitro and in vivo in several mouse models of cancer and in human cancer patients.38, 39, 40, 41 Previous studies indicated that LM7 and 143B OS cells exhibit the Warburg effect and have many swollen mitochondria.11,33 Dysfunctional mitochondria not only contribute to the metabolic reprogramming of cancer cells but also regulate several cellular processes involved in tumorigenesis.42, 43, 44 In this study, we also found that MG63 and 143B OS cells overexpress LETM1 compared to normal osteoblasts (hFOB1.19), and these OS cells also had fragmented mitochondria, whereas the less malignant TE85 cells had typical mitochondrial structures. Fragmented mitochondria were also found in lung cancer cells with LETM1 overexpression.20
LETM1 has been shown to play an essential role in maintaining mitochondrial ion homeostasis, volume, and morphology, as well as the respiratory electron transport chain.18,19,45,46 Several studies have shown that LETM1 depletion leads to punctate mitochondrial fragmentation, cristae swelling, and the absence of tubular mitochondrial networks.47,48 Other studies have found that LETM1 overexpression inhibits mitochondrial biogenesis, leading to a significant reduction in the total cellular mitochondrial mass and ATP production, and that prolonged overexpression of LETM1 leads to a shift in cellular metabolism toward glycolysis.20,35,49 Our study found that knocking down LETM1 improved mitochondrial quality and function and increased the ATP levels in MG63 and 143B cells. The knockdown of LETM1 also reduced the mitochondrial membrane permeability transition pores, as indicated by TMRE staining. Inhibition of the mitochondrial membrane permeability transition has been shown to improve mitochondrial function and reverse metabolic reprogramming and the Warburg effect in OS cells.33 LETM1 is known to play an essential role in maintaining mitochondrial ion homeostasis, but the mechanism of action of LETM1 regarding the mitochondrial membrane permeability transition needs to be investigated in further studies.
Warburg hypothesized that a metabolic shift from OXPHOS to glycolysis converts differentiated cells into undifferentiated cells.50,51 Although high levels of aerobic glycolysis have been confirmed in tumor cells, the role of mitochondria in tumor cells has been controversial. Recent studies have shown that the PPP works with glycolysis to coordinate glucose flux and support the cellular biogenesis of macromolecules and energy production.8 The PPP plays a key role in helping glycolytic cancer cells meet their anabolic demands and counteract oxidative stress.52, 53, 54 G6PD is the rate-limiting enzyme for the oxidation of the PPP and determines the flux of G6P directly into the pathway. The mitochondria in MSCs are maintained at relatively low levels of activity, and after induction of differentiation, the mtDNA copy number, respiratory enzyme protein levels, and intracellular ATP content all increase.12,55,56
OS is a clinically and molecularly heterogeneous group of malignancies characterized by varying degrees of mesenchymal differentiation. Bone formation is a carefully regulated process in which pluripotent MSCs undergo successive stages of differentiation requiring the activation of many osteogenic transcription factors (including Runx2, Osterix, and β-linked proteins) and repression of adipogenic transcription factors (PPARγ and CEBPα), which results in a reduced propensity to proliferate.57 Disruption or deregulation of this process may manifest as the development of OS, and the earlier stage at which this deregulation occurs, the less differentiated, more aggressive, and more malignant the OS that develops.3,58, 59, 60 In our study, we used TE85, MG63, and 143B cells, three types of OS cells with different degrees of malignancy, with 143B being the most malignant and MG63 the second most malignant. We observed that 143B cells had the most severely disordered mitochondrial structures and functions, followed by MG63 cells, while the TE85 cells were relatively normal. This may suggest that the three cell lines became abnormal at different stages of differentiation during the process of bone formation. If this is the case, it may then be possible to change the metabolism of cells to allow them to continue to differentiate towards osteogenesis and thus reduce their malignancy characteristics.
Our study found that knocking down LETM1 partially restored the mitochondrial structure and function, inhibited the PPP, promoted OXPHOS and osteogenic differentiation, and reduced malignant biological behavior. LETM1 knockdown also promoted osteogenic differentiation in normal osteoblasts (hFOB1.19). A previous study found that silencing LETM1 inhibited preadipocyte differentiation and reduced the potential of these cells for adipogenesis.61 This suggests that inhibition of LETM1 expression may favor osteogenic differentiation by inhibiting lipogenic differentiation.
It is also necessary to know whether mitochondrial inhibitors can reverse these alterations. Osteogenic differentiation is a dynamic process, and the changes that occur in the mitochondria may both underlie and result from this process. LETM1 has been considered a cancer stemness-related gene based on previous cancer studies.23,62,63 It was also found to be associated with cancer stemness-related genes in our study. Hypoxia affects the differentiation of MSCs, and the environment in which OS grows is hypoxic. Our study was performed under normoxia, so further studies should be performed under hypoxia. Our study also found that the cells tended to become larger after LETM1 was knocked down. The genes controlling cell size are also involved in controlling mitochondrial respiration and mitochondrial mass,64 so the mechanism responsible for the observed cell enlargement needs further investigation.
In summary, we demonstrated that OS cells and tumors overexpressing LETM1 had mitochondrial structural abnormalities and mitochondrial dysfunction. Knocking down LETM1 in these cells improved the mitochondrial quality, increased mitochondrial OXPHOS, decreased the PPP, and promoted the differentiation of MG63 and 143B cells toward osteogenesis, thus inhibiting cell proliferation, migration, and invasion. This implies that LETM1 may play a role in the development and/or progression of OS. Our study identified a relationship between metabolic reprogramming and OS differentiation, which provides new opportunities for drug development for OS-inducing differentiation therapy.
Author contributions
Yulu Shi: methodology, software, validation, data curation, writing–original draft, visualization, conceptualization, and formal analysis. Quan Kang: project administration, and funding acquisition. Hong Zhou: validation, and data curation. Xiaohan Yue: validation. Yang Bi: conceptualization. Qing Luo: conceptualization, writing–review & editing, supervision, and funding acquisition.
Conflict of interests
Authors declare no conflict of interests.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81172545) and the Chongqing Science and Technology Commission, China (No. cstc2020jcyj-msxmX0113).
Acknowledgements
We are grateful to the teachers of the Pediatric Institute of Chongqing Medical University public platform for their technical help.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2023.05.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
References
- 1.Mortus J.R., Zhang Y., Hughes D.P.M. Developmental pathways hijacked by osteosarcoma. Adv Exp Med Biol. 2014;804:93–118. doi: 10.1007/978-3-319-04843-7_5. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Cao J., Zhang N., et al. Advances in differentiation therapy for osteosarcoma. Drug Discov Today. 2020;25(3):497–504. doi: 10.1016/j.drudis.2019.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Tang N., Song W.X., Luo J., Haydon R.C., He T.C. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466(9):2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubio R., Abarrategi A., Garcia-Castro J., et al. Bone environment is essential for osteosarcoma development from transformed mesenchymal stem cells. Stem Cell. 2014;32(5):1136–1148. doi: 10.1002/stem.1647. [DOI] [PubMed] [Google Scholar]
- 5.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 6.Feng Z., Ou Y., Hao L. The roles of glycolysis in osteosarcoma. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.950886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pascale R.M., Calvisi D.F., Simile M.M., Feo C.F., Feo F. The Warburg effect 97 years after its discovery. Cancers. 2020;12(10):2819. doi: 10.3390/cancers12102819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang P., Du W., Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell. 2014;5(8):592–602. doi: 10.1007/s13238-014-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghanem N., El-Baba C., Araji K., El-Khoury R., Usta J., Darwiche N. The pentose phosphate pathway in cancer: regulation and therapeutic opportunities. Chemotherapy. 2021;66(5–6):179–191. doi: 10.1159/000519784. [DOI] [PubMed] [Google Scholar]
- 10.Stincone A., Prigione A., Cramer T., et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev. 2015;90(3):927–963. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapovalov Y., Hoffman D., Zuch D., de Mesy Bentley K.L., Eliseev R.A. Mitochondrial dysfunction in cancer cells due to aberrant mitochondrial replication. J Biol Chem. 2011;286(25):22331–22338. doi: 10.1074/jbc.M111.250092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q., Gao Z., Chen Y., Guan M.X. The role of mitochondria in osteogenic, adipogenic and chondrogenic differentiation of mesenchymal stem cells. Protein Cell. 2017;8(6):439–445. doi: 10.1007/s13238-017-0385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khacho M., Slack R.S. Mitochondrial activity in the regulation of stem cell self-renewal and differentiation. Curr Opin Cell Biol. 2017;49:1–8. doi: 10.1016/j.ceb.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann A.D., Beyer M., Krause-Buchholz U., Wobus M., Bornhäuser M., Rödel G. OXPHOS supercomplexes as a hallmark of the mitochondrial phenotype of adipogenic differentiated human MSCs. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu Y.C., Wu Y.T., Yu T.H., Wei Y.H. Mitochondria in mesenchymal stem cell biology and cell therapy: from cellular differentiation to mitochondrial transfer. Semin Cell Dev Biol. 2016;52:119–131. doi: 10.1016/j.semcdb.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Guo Y., Chi X., Wang Y., et al. Mitochondria transfer enhances proliferation, migration, and osteogenic differentiation of bone marrow mesenchymal stem cell and promotes bone defect healing. Stem Cell Res Ther. 2020;11(1):245. doi: 10.1186/s13287-020-01704-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S.Y., An H.J., Kim J.M., et al. PINK1 deficiency impairs osteoblast differentiation through aberrant mitochondrial homeostasis. Stem Cell Res Ther. 2021;12(1):589. doi: 10.1186/s13287-021-02656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura S., Matsui A., Akabane S., et al. The mitochondrial inner membrane protein LETM1 modulates cristae organization through its LETM domain. Commun Biol. 2020;3(1):99. doi: 10.1038/s42003-020-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natarajan G.K., Mishra J., Camara A.K.S., Kwok W.M. LETM1: a single entity with diverse impact on mitochondrial metabolism and cellular signaling. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.637852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran Q., Lee H., Jung J.H., et al. Emerging role of LETM1/GRP78 axis in lung cancer. Cell Death Dis. 2022;13(6):543. doi: 10.1038/s41419-022-04993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piao L., Yang Z., Feng Y., Zhang C., Cui C., Xuan Y. LETM1 is a potential biomarker of prognosis in lung non-small cell carcinoma. BMC Cancer. 2019;19(1):898. doi: 10.1186/s12885-019-6128-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q., Chen S., Chen L. LETM1 (leucine zipper-EF-hand-containing transmembrane protein 1) silence reduces the proliferation, invasion, migration and angiogenesis in esophageal squamous cell carcinoma via KIF14 (kinesin family member 14) Bioengineered. 2021;12(1):7656–7665. doi: 10.1080/21655979.2021.1982275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piao L., Feng Y., Yang Z., et al. LETM1 is a potential cancer stem-like cell marker and predicts poor prognosis in colorectal adenocarcinoma. Pathol Res Pract. 2019;215(7) doi: 10.1016/j.prp.2019.152437. [DOI] [PubMed] [Google Scholar]
- 24.Á Bartha, Győrffy B. TNMplot.com: a web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int J Mol Sci. 2021;22(5):2622. doi: 10.3390/ijms22052622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X., Jiang B., Zou Y., Zhang J., Fu Y.Y., Zhai X.Y. Roxadustat (FG-4592) facilitates recovery from renal damage by ameliorating mitochondrial dysfunction induced by folic acid. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.788977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei J., Huang K., Chen Z., et al. Characterization of glycolysis-associated molecules in the tumor microenvironment revealed by pan-cancer tissues and lung cancer single cell data. Cancers. 2020;12(7):1788. doi: 10.3390/cancers12071788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malta T.M., Sokolov A., Gentles A.J., et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173(2):338–354.e15. doi: 10.1016/j.cell.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X., Zhao Q., Qi J., et al. lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARγ pathway in hepatocellular carcinoma. Int J Oncol. 2018;53(2):551–566. doi: 10.3892/ijo.2018.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., Xu L., Nie H., Lei L. Dexamethasone-loaded β-cyclodextrin for osteogenic induction of mesenchymal stem/progenitor cells and bone regeneration. J Biomed Mater Res. 2021;109(7):1125–1135. doi: 10.1002/jbm.a.37104. [DOI] [PubMed] [Google Scholar]
- 30.Sun M., Hu L., Wang S., et al. Circulating microRNA-19b identified from osteoporotic vertebral compression fracture patients increases bone formation. J Bone Miner Res. 2020;35(2):306–316. doi: 10.1002/jbmr.3892. [DOI] [PubMed] [Google Scholar]
- 31.Jiang T., Zhao J., Yu S., et al. Untangling the response of bone tumor cells and bone forming cells to matrix stiffness and adhesion ligand density by means of hydrogels. Biomaterials. 2019;188:130–143. doi: 10.1016/j.biomaterials.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y., Tran Q., Shrestha R., et al. LETM1 is required for mitochondrial homeostasis and cellular viability. Mol Med Rep. 2019;19(5):3367–3375. doi: 10.3892/mmr.2019.10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giang A.H., Raymond T., Brookes P., et al. Mitochondrial dysfunction and permeability transition in osteosarcoma cells showing the Warburg effect. J Biol Chem. 2013;288(46):33303–33311. doi: 10.1074/jbc.M113.507129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao J., Fu Z., Ji Y., et al. Leucine zipper-EF-hand containing transmembrane protein 1 (LETM1) forms a Ca2+/H+ antiporter. Sci Rep. 2016;6 doi: 10.1038/srep34174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piao L., Li Y., Kim S.J., et al. Association of LETM1 and MRPL36 contributes to the regulation of mitochondrial ATP production and necrotic cell death. Cancer Res. 2009;69(8):3397–3404. doi: 10.1158/0008-5472.CAN-08-3235. [DOI] [PubMed] [Google Scholar]
- 36.Haydon R.C., Luu H.H., He T.C. Osteosarcoma and osteoblastic differentiation. Clin Orthop Relat Res. 2007;454:237–246. doi: 10.1097/BLO.0b013e31802b683c. [DOI] [PubMed] [Google Scholar]
- 37.Hamanaka R.B., Chandel N.S. Cell biology. Warburg effect and redox balance. Science. 2011;334(6060):1219–1220. doi: 10.1126/science.1215637. [DOI] [PubMed] [Google Scholar]
- 38.Gatenby R.A., Gillies R.J. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4(11):891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 39.Martínez-Reyes I., Chandel N.S. Cancer metabolism: looking forward. Nat Rev Cancer. 2021;21(10):669–680. doi: 10.1038/s41568-021-00378-6. [DOI] [PubMed] [Google Scholar]
- 40.Schreiber J.R., Balcavage W.X., Morris H.P., Pedersen P.L. Enzymatic and spectral analysis of cytochrome oxidase in adult and fetal rat liver and Morris hepatoma 3924A. Cancer Res. 1970;30(10):2497–2501. [PubMed] [Google Scholar]
- 41.Jiang F., Ryan M.T., Schlame M., et al. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275(29):22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- 42.Zong W.X., Rabinowitz J.D., White E. Mitochondria and cancer. Mol Cell. 2016;61(5):667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Missiroli S., Perrone M., Genovese I., Pinton P., Giorgi C. Cancer metabolism and mitochondria: finding novel mechanisms to fight tumours. eBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace D.C. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Doonan P.J., Chandramoorthy H.C., Hoffman N.E., et al. LETM1-dependent mitochondrial Ca2+ flux modulates cellular bioenergetics and proliferation. FASEB J. 2014;28(11):4936–4949. doi: 10.1096/fj.14-256453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Austin S., Nowikovsky K. LETM1: essential for mitochondrial biology and cation homeostasis? Trends Biochem Sci. 2019;44(8):648–658. doi: 10.1016/j.tibs.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Dimmer K.S., Navoni F., Casarin A., et al. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum Mol Genet. 2008;17(2):201–214. doi: 10.1093/hmg/ddm297. [DOI] [PubMed] [Google Scholar]
- 48.Tamai S., Iida H., Yokota S., et al. Characterization of the mitochondrial protein LETM1, which maintains the mitochondrial tubular shapes and interacts with the AAA-ATPase BCS1L. J Cell Sci. 2008;121(Pt 15):2588–2600. doi: 10.1242/jcs.026625. [DOI] [PubMed] [Google Scholar]
- 49.Piao L., Li Y., Kim S.J., et al. Regulation of OPA1-mediated mitochondrial fusion by leucine zipper/EF-hand-containing transmembrane protein-1 plays a role in apoptosis. Cell Signal. 2009;21(5):767–777. doi: 10.1016/j.cellsig.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 50.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 51.Weinberg F., Hamanaka R., Wheaton W.W., et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107(19):8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patra K.C., Hay N. The pentose phosphate pathway and cancer. Trends Biochem Sci. 2014;39(8):347–354. doi: 10.1016/j.tibs.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghashghaeinia M., Köberle M., Mrowietz U., Bernhardt I. Proliferating tumor cells mimick glucose metabolism of mature human erythrocytes. Cell Cycle. 2019;18(12):1316–1334. doi: 10.1080/15384101.2019.1618125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang H.C., Stern A., Chiu D.T.Y. G6PD: a hub for metabolic reprogramming and redox signaling in cancer. Biomed J. 2021;44(3):285–292. doi: 10.1016/j.bj.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rafalski V.A., Mancini E., Brunet A. Energy metabolism and energy-sensing pathways in mammalian embryonic and adult stem cell fate. J Cell Sci. 2012;125(Pt 23):5597–5608. doi: 10.1242/jcs.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shum L.C., White N.S., Mills B.N., Bentley K.L., Eliseev R.A. Energy metabolism in mesenchymal stem cells during osteogenic differentiation. Stem Cell Dev. 2016;25(2):114–122. doi: 10.1089/scd.2015.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Almalki S.G., Agrawal D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation. 2016;92(1–2):41–51. doi: 10.1016/j.diff.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruh M., Stemmler M.P., Frisch I., et al. The EMT transcription factor ZEB1 blocks osteoblastic differentiation in bone development and osteosarcoma. J Pathol. 2021;254(2):199–211. doi: 10.1002/path.5659. [DOI] [PubMed] [Google Scholar]
- 59.Thomas D., Kansara M. Epigenetic modifications in osteogenic differentiation and transformation. J Cell Biochem. 2006;98(4):757–769. doi: 10.1002/jcb.20850. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W., Zhuang Y., Zhang Y., et al. Uev1A facilitates osteosarcoma differentiation by promoting Smurf1-mediated Smad1 ubiquitination and degradation. Cell Death Dis. 2017;8(8) doi: 10.1038/cddis.2017.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xue H., Wang Z., Hua Y., et al. Molecular signatures and functional analysis of beige adipocytes induced from in vivo intra-abdominal adipocytes. Sci Adv. 2018;4(7):eaar5319. doi: 10.1126/sciadv.aar5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Che N., Yang Z., Liu X., et al. Suppression of LETM1 inhibits the proliferation and stemness of colorectal cancer cells through reactive oxygen species-induced autophagy. J Cell Mol Med. 2021;25(4):2110–2120. doi: 10.1111/jcmm.16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Z., Ni W., Cui C., Qi W., Piao L., Xuan Y. Identification of LETM1 as a marker of cancer stem-like cells and predictor of poor prognosis in esophageal squamous cell carcinoma. Hum Pathol. 2018;81:148–156. doi: 10.1016/j.humpath.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 64.Yamamoto K., Gandin V., Sasaki M., et al. Largen: a molecular regulator of mammalian cell size control. Mol Cell. 2014;53(6):904–915. doi: 10.1016/j.molcel.2014.02.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.