Abstract
A simple and highly efficient method was developed to produce a library of Escherichia coli clones that express a particular chromosomal gene at a wide range of expression levels. The basic strategy was to replace all or part of the upstream region of a coding sequence containing the elements involved in its expression (promoter, operator, gene coding for a regulator, ribosome binding site, and start codon) with a PCR-generated library of expression cassettes.
Escherichia coli is widely used in the biotechnology industry for the production of pharmaceuticals, fine chemicals, and feed. In most of these applications, genetic manipulations of the production strain have been considered in order to improve the overall performances of the bioprocess. Particularly, it is often necessary to tune the level of many enzymes to achieve the expected flux and yield of production of the desired product (10, 13).
A new approach for modulating chromosomal gene expression has recently been described by Jensen and collaborators (11, 12, 20). The two limitations of this method are that (i) to achieve a promoter replacement on the chromosome, a cloning step is needed, and (ii) it allows only promoter replacement which limits the maximal level of expression of a gene present in only one copy.
Datsenko and Wanner (6) have recently developed a fast and efficient method to disrupt chromosomal genes in E. coli, in which PCR primers provide the homology to the targeted gene(s). In the work presented here, we have extended the use of the tools developed by Datsenko and Wanner to create a simple and highly efficient method of producing a library of E. coli clones that express a particular gene located on the chromosome at a very wide range of expression levels. The main advantages of this approach are that (i) without a cloning step into a plasmid, a library of clones with different levels of expression of a particular chromosomal gene can be obtained, (ii) it is possible to get both very high and very low levels of expression of a chromosomal gene, (iii) the expression cassette can be moved from strain to strain by P1 transduction (17), and (iv) if needed, markerless strains can be made (4).
Description of the strategy.
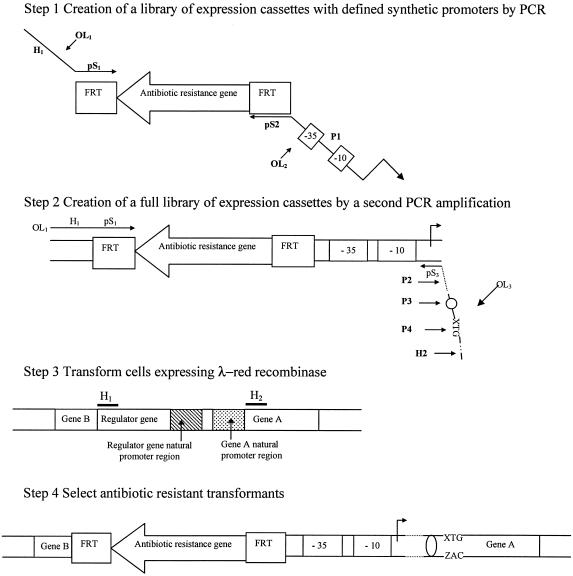
The construction of a library of clones that express a chromosomal gene at different levels was done in three steps. The first step was the creation of a library of expression cassettes with defined artificial promoters. This step included the design of two 100-mer oligonucleotides (OL1 and OL2) containing various specific regions (called pS1, pS2, H1, and P1) (Fig. 1). The reverse oligonucleotide was degenerated on specific bases of the sequence coding for a very short artificial promoter (P1) to create a library of promoters. The template plasmid pKD3 or pKD4 (6) and the two oligonucleotides (OL1 and OL2) were then used in a PCR (Fig. 1) that produced a first set of expression cassettes.
FIG. 1.
Strategy for the creation of a library of clones with different levels of expression of a chromosomal gene. pS1 and pS2 are the priming sites for pKD3 or pKD4. H1 and H2 are the homology extensions. P1 is the degenerated part of the reverse oligonucleotide OL2 containing synthetic short GI promoters. OL3 can contain up to three degenerated sequences: P2 for mRNA-stabilizing sequences, P3 for RBSs, and P4 for start codons (see Results for a description). FRT is the FLP recombinase recognition target (4).
The second step was the extension of the initial promoter library by adding modified and defined ribosomal binding site (RBS) and/or mRNA-stabilizing sequences and/or start codons, depending on the range of expression level needed. This PCR step used the first set of expression cassettes as a template. The forward oligonucleotide was the same as that used for the first step (OL1). The reverse oligonucleotide (OL3) contained (i) a priming sequence (pS3) for the artificial promoter; (ii) defined and degenerated sequences that encoded mRNA-stabilizing sequences (P2), RBSs (P3), and start codons (P4); and (iii) a flanking sequence (H2) that was homologous to the chromosomal DNA downstream of the start codon (Fig. 1). After the second PCR, a full library of expression cassettes was obtained.
The third step of the method was the in vivo recombination in E. coli. This last step was achieved by the transformation of competent cells (expressing the Gam, Bet, and Exo proteins from the plasmid pKD46) with the library of expression cassettes (6, 7, 16). The result of the in vivo recombination was the replacement of all or part of the upstream region of a coding sequence containing the elements involved in its expression by a PCR-generated library of expression cassettes.
Evaluation of the feasibility of the method with the lacZ gene as a model.
The initial experiment was designed to evaluate the feasibility of the method for a small library and the range of expression levels that could be obtained from the very short promoters included in an oligonucleotide. The LacZF and LacZR oligonucleotides (Table 1) were then designed to delete the lacI repressor and to replace the natural promoter region of the lactose operon by three very short artificial glucose isomerase (GI) promoters of different strengths (Fig. 2) and by an antibiotic resistance gene. The different artificial GI promoters were derived from the GI promoter of Streptomyces spp. (9), a promoter that was shown to be relatively strong in E. coli. The corresponding set of expression cassettes obtained by PCR was used to transform Escherichia coli K-12 MG1655 (F− LAM rph-1; Genetic Stock Center, New Haven, Conn.) carrying pKD46 competent cells with selection on agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and chloramphenicol. Ten transformants were checked for recombination at the lacZ locus (by PCR using the LacseqF and LacseqR oligonucleotides [Table 1] located outside the recombination region) and for β-galactosidase activity (15). The sequence analysis of the promoter region of the different types of recombinant clones showed, as expected, that they differed by only one base pair in their −35 region. This experiment also demonstrated that, with very short artificial promoters, it was possible to achieve specific β-galactosidase activities ranging from two times higher (2.3 ± 1.05 U/mg of proteins) to four times lower (0.3 ± 0.11 U/mg of proteins) than those produced with the IPTG (isopropyl-β-d-thiogalactopyranoside)-induced (1 mM) natural lacZ promoter (1.2 ± 0.54 U/mg of proteins).
TABLE 1.
Structures of the different oligonucleotides used in this study
| Oligonucleotide | Structure or sequencea |
|---|---|
| LacZF | 79-nt homology extension (from positions 366734 to 366655) at the 5′ end of the lacI gene and the 20-nt priming sequence (pS1) for pKD3 (6) |
| GapZF | 59-nt homology extension (positions 1860478 to 1860536) from the yeaA-gapA intergenic region and the same 20-nt pS1 priming sequence as that in LacZF |
| LacZR | 39-nt homology extension from +1 of the transcription start site to the ATG of lacZ (positions 365529 to 365567) and 42 nt of the short GI promoter sequence (9) from 4 bp upstream of the −35 site to 8 bp downstream of the −10 site, degenerated at the last base of the −35 site (TTGACH) (where H is A, C, or T) and the priming sequence (pS2) for pKD3 (6) |
| LacZRT | Identical to LacZR without the degeneration of the last base of the −35 site (TTGACA) |
| LacZRBSR | 60-nt homology extension to lacZ after the start codon (positions 362455 to 362512) and 40-nt homology extension to the lacZR oligonucleotide. This oligonucleotide is degenerated in the RBS sequence (AGYAY, where Y is G or A) and in the start codon sequence (ATG degenerated at the A position to a G or a T) |
| LacZRBS2R | 58-nt homology extension to lacZ after the start codon and 42-nt homology extension to the lacR oligonucleotide, including a degenerated RBS sequence (AAYYAGGAAA) from the 15th to the 5th nt upstream of the start codon (where Y is G or A) (18) |
| LacZmRNA | 43-nt homology extension of lacZ downstream of the RBS site, 34 nt of an mRNA-stabilizing structure, and a 23-nt homology extension to the lacZR oligonucleotide upstream of +1 of the transcription start site. This oligonucleotide is degenerated in the mRNA-stabilizing sequence (1) as follows: in GGTCGAGTTATCTCGAGTGAGATATTGTTGACG, there is degeneration of the 1st G and the 2nd G to a C, of the 4th C to a G, and of the 7th G to a C. |
| GapR | 38-nt homology extension from +3 relative to the transcription start site of P1 (natural gapA promoter [3]) to the ATG start codon of gapA, 42 nt of the short GI promoter sequence (9), from 4 bp upstream of the −35 site to 8 bp downstream of the −10 site, degenerated at the last base of the −35 site (TTGACH) (wherein H is A, C, or T) and the priming sequence (pS2) for pKD3 (6) |
| LacseqF | GGCTGCGCAACTGTTGGGAA(positions 365367 to 365386) |
| LacseqR | CATTGAACAGGCAGCGGAAAAG (positions 366915 to 366934) |
| GapseqR | AGTCATATATTCCACCAGCTATTTGTTAGTG (positions 1860770 to 1860800) |
| GapseqF | GTCGACAAACGCTGGTATACCTCA (positions 1859940 to 1859963) |
nt, nucleotide.
FIG. 2.
Sequences of three very short, artificial GI promoters introduced upstream of the +1 site of lacZ transcription. The −35 and −10 sites are recognition sites for the housekeeping RNA polymerase.
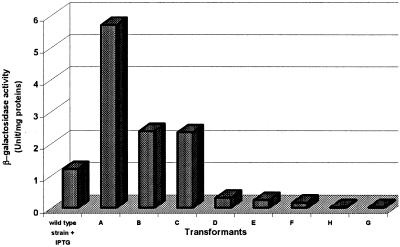
The possibility of using a controlled-degeneracy library of RBS sequences in combination with an artificial promoter was then shown by use of the LacZF, LacZRT, and LacZRBSR oligonucleotides (Table 1). Eight transformants were selected, and their β-galactosidase activities and DNA sequences upstream of lacZ were determined. As expected, the transformants differed only in their RBS sequences, and the β-galactosidase activity ranged between 0.02 ± 0.01 and 5.7 ± 1.2 U/mg of proteins (Fig. 3). These values are 60 times lower and 5 times higher, respectively, than the value produced by the IPTG-induced natural lacZ promoter (1.2 ± 0.54 U/mg of proteins).
FIG. 3.
Influence of different RBS sequences on β-galactosidase activities. (A) CAAGGAGGAAACAGCTATG; (B) CAAGAAGGAAACAGCTATG; (C) CACACAGGAAACAGCTATG; (D) CTCACAGGAGACAGCTATG; (E) CTCACAGGAAACAGCTATG; (F) CACACAGAAAACAGCTATG; (G) CTCACAGAGAACAGCTATG; (H) CTCACAGAAAACAGCTATG. Boldface indicates RBS sequences, and italic indicates start codons.
In order to extend the range of expression of lacZ in the higher values, the possibility of using a controlled-degeneracy library of mRNA-stabilizing sequences with a strong, short artificial promoter was demonstrated by use of the LacZF and the LacZmRNA oligonucleotides (Table 1). Five transformants were selected, and their β-galactosidase activities and the DNA sequences upstream of lacZ were determined (Table 2). For the best clone, the activity was up to 15 times higher than that of the IPTG-induced natural lacZ promoter. Based on a specific activity of 1,000 U/mg of pure β-galactosidase, it can be calculated that in the best case, this enzyme represents approximately 2% of the total protein in the cell.
TABLE 2.
Influence of mRNA-stabilizing sequences on β-galactosidase activities
| Transformant | mRNA-stabilizing sequence | Sp act (U/mg) |
|---|---|---|
| 1 | GGTCGAGTTATCTCGAGTGAGATATTGTTGACG | 5.9 ± 2.7 |
| 2 | GGTGGAGTTATCTCGAGTGAGATATTGTTGACG | 11 ± 5 |
| 3 | CCTCGAGTTATCTCGAGTGAGATATTGTTGACG | 18.5 ± 8.5 |
| 4 | GGTGGACTTATCTCGAGTGAGATATTGTTGACG | 7.3 ± 3.3 |
| 5 | GCTGGACTTATCTCGAGTGAGATATTGTTGACG | 4.1 ± 1.8 |
| Wild-type strain + IPTG | None | 1.2 ± 0.5 |
Finally, to demonstrate that it would be possible to create a library of E. coli clones with different levels of lacZ expression by simultaneously introducing different artificial promoters, modified start codons, and modified RBS sequences, the LacZR, LacZF, and LacZRBS2R oligonucleotides (Table 1) were used in two successive PCRs, as described in Fig. 1 and 2. The results shown in Table 3 indicate that the simultaneous modification of the three elements that affect the expression level of a chromosomal gene was achieved in one step. The expression levels that were found in five representative members of the library showed that both very low (0.03 ± 0.01 U/mg of proteins) and high (5.7 ± 1.2 U/mg of proteins) levels of lacZ expression could be obtained, demonstrating the validity of this approach.
TABLE 3.
Comparison of the β-galactosidase activities of four randomly picked clones from the library and corresponding sequences of the lacZ upstream regions
| Transformant | Artificial promoter | Start codon | RBS sequence | β-Galactosidase activity (U/mg) |
|---|---|---|---|---|
| 1 | 1.6 GI | TTG | TCACAGGAGA | 0.28 ± 0.1 |
| 2 | 1.6 GI | ATG | AAGGAGGAAA | 5.7 ± 1.2 |
| 3 | 1.2 GI | ATG | ACACAGGAAA | 0.7 ± 0.3 |
| 4 | 1.6 GI | TTG | ACACAGAAGA | 0.03 ± 0.01 |
Creation of a library of E. coli clones with different levels of expression of the glyceraldehyde-3-phosphate dehydrogenase and selection of the best level for growth or for glycerol production.
To demonstrate the usefulness of the method for metabolic pathway engineering, we chose gapA (encoding the glyceraldehyde-3-phosphate dehydrogenase) as a target gene because of its importance in the central metabolism of E. coli (5). The GapZF and the GapR oligonucleotides were then designed to replace the natural promoter region of gapA by the three artificial GI promoters that were previously used for lacZ. Ten transformants were selected and analyzed for (i) recombination at the gapA locus (by PCR using the GapseqR and GapseqF oligonucleotides that are located outside the recombination region) and (ii) GapA activity. The transformants were then classified in the three expected categories (1.2 GI, 1.5 GI, and 1.6 GI), based on the GapA activities and the promoter sequence upstream of gapA. One member of each category was then individually grown in a shake flask at 34°C in M9 medium (19) supplemented with 10 g of glucose per liter, and the optical density at 550 nm was measured to estimate the specific growth rate (Fig. 4). The best specific growth rate was obtained for the 1.6 GI gapA clone, which also exhibited the highest GapA activity and an activity close to that of the wild-type strain.
FIG. 4.
Replacement of the natural promoter region of gapA by three different short GI artificial promoters. (A) Influence of the type of synthetic GI promoter on GapA activity and specific growth rate of the E. coli strain; (B) influence of the type of synthetic GI promoter on average glycerol production rate (grams per liter per hour) and on the glycerol molar yield (mole/mole) on the E. coli(pAH48) strains. Fermentations were done in duplicate at 34°C in shake flasks (200 rpm) on M9 medium supplemented with 10 g of glucose per liter. Fermentation product and glucose were quantified as previously described (8). Variation between duplicate experiments was less than 10%.
To evaluate the effect of the gapA level on the production of a chemical compound, the three different modified gapA strains were transformed with the pAH48 plasmid that carries the genes used to convert dihydroacetone-phosphate into glycerol (9). The batch fermentations showed (Fig. 4) that the 1.5 GI gapA strain gave both the best rate and the best molar yield of glycerol production. This result demonstrated that the optimal level of a metabolic enzyme for production of a chemical may not necessarily be the highest achievable expression level and emphasizes the need for the selection of the appropriate expression for the desired application. Moreover, in a single step, it is possible to make use of this library to select the best mutant for growth or for glycerol production. For metabolic engineering, the method has several advantages over existing inducible expression systems (14). One advantage is that after the optimal expression has been determined, the strain is, in principle, ready for use in the industrial fermentation process. Another important feature is the option to modulate, to different extents, the expression of many genes located at different positions of the genome in the same strain. This approach has been extensively and successfully used to develop an industrial E. coli strain that produces 1,3-propanediol at very high yields (2).
REFERENCES
- 1.Carrier, T. A., and J. D. Keasling. 1999. Library of synthetic 5′ secondary structures to manipulate mRNA stability in Escherichia coli. Biotechnol. Prog. 15:58-64. [DOI] [PubMed] [Google Scholar]
- 2.Cervin, M., P. Soucaille, and F. Valle. 2002. Process for the biological production of 1,3-propanediol with high yield. U.S. patent 60/416192.
- 3.Charpentier, B., and C. Branlant. 1994. The Escherichia coli gapA gene is transcribed by the vegetative RNA polymerase holoenzyme Eσ70 and by the heat shock RNA polymerase Eσ32. J. Bacteriol. 176:830-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: Tcr and Kmr cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 5.D'Alessio, G., and J. Josse. 1971. Glyceraldehyde phosphate dehydrogenase of Escherichia coli: structural and catalytic properties. J. Biol. Chem. 246:4326-4333. [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, H. M., D. Yu, T. DiTizio, and D. L. Court. 2001. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. USA 98:6742-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartlep, M., W. Hussmann, N. Prayitno, I. Meynial-Salles, and A. P. Zeng. 2002. Study of two-stage processes for the microbial production of 1,3-propanediol from glucose. Appl. Microbiol. Biotechnol. 60:60-66. [DOI] [PubMed] [Google Scholar]
- 9.Hsu, A. K., D. E. Trimbur, R. Nair, M. Payne, F. Valle, S. K. Picataggio, and P. Soucaille. 2003. Promoter and plasmid system for genetic engineering. Patent Corporation Treaty international application WO 03089621.
- 10.Jensen, P. R., and K. Hammer. 1998. Artificial promoters for metabolic optimization. Biotechnol. Bioeng. 58:191-195. [PubMed] [Google Scholar]
- 11.Jensen, P. R., and K. Hammer. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, P. R., and K. Hammer. 1998. Artificial promoter libraries for selected organisms and promoters derived from such libraries. Patent Corporation Treaty international application WO 9807846.
- 13.Jensen, P. R., H. V. Westerhoff, and O. Michelsen. 1993. Excess capacity of H(+)-ATPase and inverse respiratory control in Escherichia coli. EMBO J. 12:1277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keasling, J. D. 1999. Gene-expression tools for the metabolic engineering of bacteria. Trends Biotechnol. 17:452-460. [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Murphy, K. C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provence, D. L., and R. Curtiss III. 1994. Gene transfer in gram-negative bacteria, p. 317-347. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 18.Ringquist, S., S. Shinedling, D. Barrick, L. Green, J. Binkley, G. D. Stormo, and L. Gold. 1992. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6:1219-1229. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 20.Solem, C., and P. R. Jensen. 2002. Modulation of gene expression made easy. Appl. Environ. Microbiol. 68:2397-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]