Abstract
Rapid population growth and urban development along waterways and coastal areas have led to decreasing water quality. To examine the effects of upstream anthropogenic activities on microbiological water quality, methods for source-specific testing are required. In this study, molecular assays targeting human enteroviruses (HEV), bovine enteroviruses (BEV), and human adenoviruses (HAdV) were developed and used to identify major sources of fecal contamination in the lower Altamaha River, Georgia. Two-liter grab samples were collected monthly from five tidally influenced stations between July and December 2002. Samples were analyzed by reverse transcription- and nested-PCR. PCR results were confirmed by dot blot hybridization. Eleven and 17 of the 30 surface water samples tested positive for HAdV and HEV, respectively. Two-thirds of the samples tested positive for either HEV or HAdV, and the viruses occurred simultaneously in 26% of samples. BEV were detected in 11 of 30 surface water samples. Binary logistic regression analysis showed that the presence of both human and bovine enteric viruses was not significantly related to either fecal coliform or total coliform levels. The presence of these viruses was directly related to dissolved oxygen and streamflow but inversely related to water temperature, rainfall in the 30 days preceding sampling, and chlorophyll-a concentrations. The stringent host specificity of enteric viruses makes them good library-independent indicators for identification of water pollution sources. Viral pathogen detection by PCR is a highly sensitive and easy-to-use tool for rapid assessment of water quality and fecal contamination when public health risk characterization is not necessary.
Fecal coliform bacteria and other bacterial indicators have been used by most water quality regulators in the United States for over a century as standard tools to measure fecal contamination and determine if a body of water is suitable for its designated use (e.g., fishing, drinking, recreational, industrial, wildlife preserve, etc.). These standards have helped to improve water sanitation and protect public health (29); however, there are several drawbacks to these indicators that make them unreliable for predicting the occurrence of many waterborne pathogens and identifying fecal contamination sources. Fecal coliform bacteria may be found in both human and animal feces; therefore, tracking and monitoring the source of contamination is impossible without sophisticated microbial source tracking techniques, such as multiple antibiotic resistance profiling, ribotyping, and pulsed-field gel electrophoresis, which require an extensive strain database and can be laborious and costly (13, 49, 56). Coliform standards often fail to predict the occurrence of many waterborne human pathogens, such as pathogenic bacteria, the protozoan parasites Cryptosporidium and Giardia, and enteric viruses, which are most often the cause of disease from recreational exposure (23, 29, 60). Furthermore, traditional bacterial indicators generally die off quickly in marine water compared to viruses and protozoa (34, 40, 41, 46, 48, 67). Studies have shown that human pathogenic viruses have been isolated from sites with no violation of coliform standards (40, 41), and outbreaks of gastroenteritis have been associated with water supplies with acceptable fecal coliform counts (for examples, see references 8, 11, and 31).
Over 100 types of pathogenic viruses have been found in sewage-contaminated aquatic environments, such as ground water, coastal marine water, coastal river water, aerosols emitted from sewage treatment plants, insufficiently treated drinking water, and private wells that received treated or untreated wastewater either directly or indirectly (26, 38, 54, 57, 68). These viruses, collectively known as enteric viruses, are transmitted via the fecal-oral route, and they infect and replicate in the gastrointestinal tract of the hosts. Enteric viruses are excreted in high concentrations in human and animal feces and, in certain cases, urine (43, 50). Infected individuals suffering from viral gastroenteritis and hepatitis may excrete 105 to 1011 viral particles per gram of stool, with average levels between 106 and 108 viral particles per gram of stool for enteroviruses and hepatitis A virus, respectively (17, 18, 22). Enteric virus concentrations in raw sewage and polluted surface water have been estimated at around 102 viral particles 100 ml−1 and 1 to 10 viral particles 100 ml−1, respectively (25, 58, 64). Enteric viruses also are more resistant than many other sewage-associated pathogens and bacterial indicators to extreme environmental conditions and conventional wastewater treatment, such as chlorination, UV radiation, and filtration (61, 62). These viruses can also remain infective for long periods in the environment; they have been reported to survive for up to 130 days in seawater, up to 120 days in freshwater and sewage, and up to 100 days in soil at 20 to 30°C (1, 7, 65, 66). These survival periods surpass those reported for fecal coliform and other indicator bacteria in similar environments (34, 44). Therefore, the traditional bacterial indicators are poor proxies to monitor the presence of pathogenic viruses.
The host specificity of enteric viruses and their prevalence in sewage- and fecal-contaminated waters suggest that they can be promising library-independent microbial source tracking tools for polluted environmental waters (35, 39, 42, 49). Human enteroviruses, which consist of poliovirus, coxsackieviruses A and B, echoviruses, and the numbered enteroviruses, have been included by the European Union regulations governing water quality as a parameter for evaluating viral pollution of a water body, because they can easily be isolated and quantified as plaque-forming units in cell culture (50, 53). Recent studies conducted in Europe have also suggested using adenoviruses as an index of pollution of human origin in waters, given their high numbers in sewage and contaminated aquatic environments (32, 34, 37, 50). Human adenoviruses (HAdV) are the only human enteric viruses that contain double-stranded DNA instead of RNA, potentially are more stable in various environments, and are more resistant to UV irradiation and other water purification treatments than other human enteric viruses, because they are able to use the host cell DNA repair mechanism to repair damage in their DNA caused by UV irradiation (24, 32, 45). Adenoviruses have been found to survive three to five times longer than poliovirus in seawater, wastewater, and tap water (15). Both human enteroviruses (HEV) and HAdV are readily detected in surface water as well as coastal waters that have received anthropogenic inputs (6, 34, 40, 50, 52). Recently, animal-specific enteroviruses and adenoviruses have been identified and may be used as indicators to identify and monitor fecal contamination originating from cattle farms, swine farms, or other animal sources (35, 39, 42).
The use of PCR-based viral pathogen detection assays to identify the source categories of fecal pollution in coastal rivers has not been previously evaluated. In this study, we examined the extent and relative importance of fecal contamination from agricultural activities (cattle), anthropogenic activities, and development upstream in coastal reaches of the lower Altamaha River, Georgia, by detecting three groups of host-specific enteric viruses: human enteroviruses, human adenoviruses, and bovine enteroviruses (BEV). We also compared the findings from PCR-based assays to concurrently collected bacterial indicator data and other environmental variables, such as rainfall, streamflow, and water temperature, to evaluate the use of this assay in defining estuarine water quality in a mixed-use watershed.
MATERIALS AND METHODS
Sampling sites.
The Altamaha River is the largest river of the Georgia coast and the second largest basin in the eastern United States (21). It drains more than one-fourth of the state, with a drainage area of approximately 7,107 km2 (21). The Altamaha supports more than 30% of Georgia's $80 million commercial fishery and about one-third of Georgia's $350 million recreational fishery, according to the Altamaha Riverkeeper (3). The lower Altamaha River acts as a conduit for discharging the combined flow from two major rivers in Georgia: the Ocmulgee River and the Oconee River. Urban development, population growth, and a growing ecotourism industry in the coastal areas as well as upstream urban, agricultural, and industrial discharges have degraded the quality and productivity of the lower Altamaha River markedly, and there is evidence of increasing coastal salinity, harmful algal populations, and declining fishery stocks in the river (21).
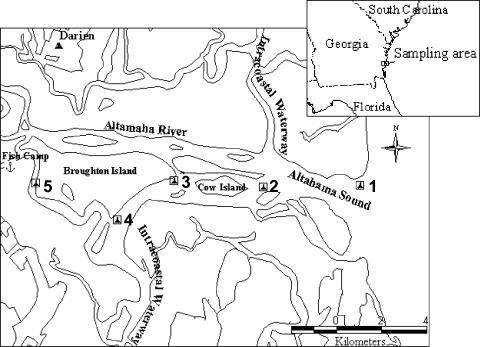
Samples were collected from five stations along a 15-km stretch of the lower Altamaha River, located between Glynn and McIntosh counties, in conjunction with the Georgia Marine Extension Service, Brunswick (Fig. 1). Sampling stations were located approximately 11 and 23 km from regulated commercial shellfish harvesting areas in McIntosh and Glynn counties, respectively. Although not regulated, all sampling sites were within an area considered to be shellfish supporting by the state (20). The sampling stations are surrounded by marsh islands and wetlands that are mainly inhabited by cattle, feral hogs, and waterfowl, and they are within the waterfowl management area. Samples were collected monthly from July to December 2002 to observe and track the changes in enteric virus loading in the river between wet and dry seasons. All samples were collected on an outgoing tide, starting with station 1, which is located at the Altamaha Sound (mouth of the river) and ending with station 5, which is located farther inland, west of Broughton Island and downstream from a commercial fish camp (Fig. 1). At each station, a grab sample was collected from just below water surface. Water samples were collected from the bottom of the water column in July, but because of processing difficulties due to high turbidity, no additional samples were collected. The 2-liter samples were kept on ice (∼4°C) and processed within 24 h of collection. Salinity, water temperature, pH, chlorophyll-a levels, turbidity, dissolved oxygen (DO) levels, and weather conditions (i.e., ambient temperature, rainfall, wind, etc.) were noted for each sample at collection. Streamflow data for the area were obtained from the U.S. Geological Survey (USGS) stream gauge at Doctortown, Ga. (USGS station 02226000). Rainfall data for Glynn County were obtained from the Georgia Automated Environmental Monitoring Network (www.georgiaweather.net). The Georgia Marine Extension Service provided fecal coliform and total coliform counts for each sample.
FIG. 1.
Sampling stations (1 through 5) in the lower Altahama River. The Altamaha River and South Altamaha River are the main sources of flow towards the coast. The Atlantic Intracoastal Waterway provides flow from coastal areas primarily to stations 1 (on the outgoing tide), 2, and 4; station 3 is outside of the main flow.
Concentration of viruses.
Water samples were concentrated according to the method described by Katayama et al. (36) with the following modifications. Samples were acidified to pH 3.5 to 4.0 by adding 10% acetic acid prior to filtration. Acidified water samples were filtered through a type HA, negatively charged membrane (Millipore, Billerica, Mass.) with a 47 mm diameter and a 0.45 μm pore size. Because of the high turbidity, the volumes of water filtered ranged between 0.5 and 2.0 liters. A volume of 100 ml of 0.5 mM H2SO4 was then passed through the membrane, and viral particles were finally eluted with 10 ml of 1 mM NaOH. Eluate was recovered in a tube containing 0.1 ml of 50 mM H2SO4 and 0.1 ml of 100× Tris-EDTA (TE) buffer for neutralization. All 10-ml eluates were stored at −20°C. Eluates were further purified, concentrated, and desalted with Centriprep YM-50 concentrator columns (Millipore). The final volume of concentrated eluate recovered was about 2 ml. Concentrates were split in half and stored at −80°C.
Extraction of viral RNA and DNA.
Concentrated samples were extracted for viral nucleic acids and purified through commercial spin columns based on the method of Boom et al. (5), using an RNeasy Mini kit or DNeasy Tissue kit (QIAGEN, Valencia, Calif.) following the manufacturer's protocol. Purified viral RNA was then eluted and resuspended in 50 μl of RNase-free water. Extracted DNA was eluted and resuspended in 50 μl of AE buffer (elution buffer) provided in the kit. Each concentrated and purified sample was serially diluted to a concentration of 10−3.
Oligonucleotides.
Primers and probes were selected from highly conserved regions of the HEV, HAdV, and BEV genomes, which allowed for detection of multiple members from each group of viruses. For HEV detection, the pan-enterovirus primer set (ENT-up-2 and ENT-down-1) developed by De Leon et al. (12) was used in conjunction with another HEV primer set (ENT-up-1 and ENT-down-2; developed by J. H. Paul at the University of South Florida) to develop a reverse transcription (RT)-nested-PCR assay for the pan-enterovirus group (Table 1). Primers were selected from the 5′-untranslated region (UTR) of HEV genomes by aligning with previously published sequences available on the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). HEV primer sets were able to pick up at least 25 different HEV; echovirus 22 was not detected. A nested primer set designed by Allard et al. (2) was used to amplify HAdV in this study (Table 1). The primers are able to identify 47 HAdV serotypes, including the more common HAdV types 2, 40, and 41 (2, 52). BEV primers were designed by aligning and evaluating the 5′ UTR of BEV genomes available in the NCBI database (Table 1). The sequences amplified by the BEV primer set were BLAST searched in GenBank and showed exact matches with seven different strains of bovine enteroviruses (strains PS87, RM2, SL305, K2577, BOT/209/67, BEV261, and VG527) and one sheep enterovirus isolate (strain 82Sh2R). The primers were tested against poliovirus 1 (strain Lsc) and HAdV 2 and showed no cross-reaction with either of the virus groups. The internal probe used for HEV dot blot hybridization was described by De Leon et al. (12) and is able to pick up 25 different enteroviruses (28). Internal probes for HAdV and BEV were developed in this study (Table 1).
TABLE 1.
Nucleotide sequences for primers and probes used for PCR amplification and dot blot hybridization of HEV, BEV, and HAdV
| Virus | Primer(s) and Probe(s) | Sequence (5′ to 3′)d | Targeta | Amplicon (bp) | Reference or source |
|---|---|---|---|---|---|
| HEV | ENT-up-1 | GTAGATCAGGTCGATGAGTC | This study | ||
| ENT-down-1 | ACYGGRTGGCCAATC | 5′ UTR | 330 | 12 | |
| ENT-up-2b | CCTCCGGCCCCTGAATG | 12 | |||
| ENT-down-2b | ATTGTCACCATAAGCAGCC | 5′ UTR | 154 | This study | |
| EV-probe | TACTTTGGGTGTCCGTGTTTCc | 12 | |||
| BEV | BEV-up | GAGTAGTCCGACTCCGCWCC | This study | ||
| BEV-down | CAGAGCTACCACTGGGGT | 5′ UTR | 270 | This study | |
| BEV-probe | AAYCGACCAATAKVNGCc | This study | |||
| HAdV | AV-A1 | GCCGCAGTGGTCTTACATGCACATC | 2 | ||
| AV-A2 | CAGCACGCCGCGGATGTCAAAGT | Hexon gene | 300 | 2 | |
| AV-B1b | GCCACCGAGACGTACTTCAGCCTG | 2 | |||
| AV-B2b | TTGTACGAGTACGCGGTATCCTCGCGGTC | Hexon gene | 143 | 2 | |
| AV-probe | ACGCACGACGTAACCACAGACc | This study |
UTR, untranslated region.
Internal primer set.
5′ end is biotin labeled.
Y = G + T; R = A + G; W = A + T; K = G + T; V = A + C + G; N = A + T + C + G.
RT-nested-PCR for HEV.
HEV were amplified with RT-nested-PCR. RT-nested-PCR was performed with the RNA-PCR core kit by Applied Biosystems (Foster City, Calif.). For reverse transcription, 2.5 μl of concentrated and purified sample RNA was added to 7.5 μl of reaction mixture containing 5.0 mM MgCl2, 1× PCR buffer, 0.75 mM deoxynucleoside triphosphates (dNTPs), 2.5 μM random hexamer (as provided in the kit), 2.5 U of reverse transcriptase μl−1, and 1.0 U of RNase inhibitor μl−1. The temperature cycle for RT was 22°C for 10 min, 42°C for 15 min, and 99°C for 5 min. Samples were cooled to 4°C before the addition of master mix for the first round of PCR (PCR I). All reactions were performed in a DNA Engine PTC-0200 thermal cycler (MJ Research, Inc., Waltham, Mass.).
To optimize the detection sensitivity by PCR, we tested several concentrations of primers, MgCl2, and dNTPs as well as annealing temperatures and numbers of cycles. During PCR I, the 5′ UTR of the virus cDNA was amplified with primers ENT-up-1 and ENT-down-1, yielding amplicons of 333 bp in size. PCR I was carried out by adding 20 μl of PCR master mix to the entire reaction mixture from RT. The PCR I reaction mixture had a final concentration of 2.9 mM MgCl2, 1× PCR buffer, 0.4 mM dNTPs, 0.2 μM each primer, 2.1 U of Taq polymerase enzyme 100 μl−1, and 1× Eppendorf TaqMaster (Brinkmann Instruments, Inc. Westbury, N.Y.). PCR I consisted of 40 cycles of denaturing at 95°C for 30 s, annealing at 57.7°C for 30 s, and extension at 72°C for 45 s. During the last cycle of amplification, an extra 5 min at 72°C for extension was included.
One microliter of amplified PCR product from PCR I was transferred into the master mix for the second round of PCR (PCR II). The reaction mixture for PCR II had a final volume of 50 μl, containing 2.5 mM MgCl2, 1× PCR buffer, 0.2 mM dNTPs, 0.2 μM each primer (ENT-up-2 and ENT-down-2), and 2.5 U of Taq polymerase enzyme 100 μl−1. PCR II consisted of 40 cycles of denaturing at 95°C for 30 s, annealing at 56.5°C for 30 s, and extension at 72°C for 30 s. As with PCR I, a final extension of 5 min was included during the last cycle. Amplicons were 154 bp. Poliovirus 1 (vaccine strain Lsc, courteously provided by C. P. Gerba, University of Arizona) was used as a positive control, and molecular-grade nuclease-free water was used as a no-template negative control. The equivalent original volume of water analyzed by this HEV RT-nested-PCR for each sample ranged between 2.5 and 10 ml, depending on the volume filtered for adsorption-elution.
RT-PCR for BEV.
RT-PCR for BEV was performed under the same conditions and concentrations as RT and PCR I for HEV. In PCR, the cDNA was amplified for 40 cycles, which consisted of denaturing at 95°C for 30 s, annealing at 56°C for 30 s, and an extension at 72°C for 1 min followed by a final 5-min extension. The resulting amplicons were ∼270 bp. BEV type 1 (ATCC VR-248) was used as the positive control; human poliovirus 1 (strain LSc), HAdV type 2, and molecular-grade nuclease-free water were used as negative controls to test for cross-reaction and contamination, respectively. As with HEV, the equivalent original volume of water analyzed for each sample ranged between 2.5 and 10 ml.
Nested-PCR for HAdV.
Nested-PCR for HAdV was performed by amplifying the open reading frame of the hexon gene of adenoviruses following the protocol of Pina et al. (50) with modifications in the concentration of primers and dNTPs, annealing temperature, and the addition of Eppendorf TaqMaster (Brinkmann Instruments, Inc., Westbury, N.Y.) to the PCR mixtures. Two rounds of PCR consisting of 40 cycles each were performed. In the first round of PCR (PCR I), 1.5 μl of sample DNA was added to a 23.5-μl reaction mixture. The reaction mixture contained 1.5 mM MgCl2, 1× PCR buffer, 0.5 mM dNTPs, 0.8 μM each primer, 1× TaqMaster, and 2.5 U of Taq polymerase enzyme 100 μl−1. Three microliters of PCR I product were used as template for PCR II. Both rounds of PCR were performed under the same conditions: initial DNA denaturation at 94°C for 4 min followed by 40 cycles of denaturation at 92°C for 30 s, annealing at 60°C for 30 s, and an extension at 72°C for 1 min. A final extension at 72°C for 5 min was added during the last cycle of amplification. Amplicons of 300 and 142 bp were produced from PCR I and PCR II, respectively. HAdV type 2 was used as the positive control (courteously provided by C. P. Gerba), and molecular-grade nuclease-free water was used as a no-template negative control. The equivalent original volume of water analyzed for each sample ranged between 1.5 and 6 ml.
Visualization and confirmation of PCR products.
PCR products (12.5 ml) were analyzed by gel electrophoresis on a 2.2% average strength Omnipur agarose gel (EM Science, Darmstadt, Germany). The gel was stained in ethidium bromide and viewed under UV light. PCR products were then confirmed by dot blot hybridization with biotin-labeled probes (Table 1) internal to the amplified viral regions and detected by chemiluminescence following the protocol for Southern light chemiluminescent detection system for biotin-labeled DNA from Applied Biosystems (Bedford, Mass.) (12, 28, 40). All blots were hybridized overnight at 37°C. Stringency wash temperatures for HEV, HAdV, and BEV probes were 47, 65, and 51°C, respectively.
Detection efficiency.
The sensitivities of all PCR assays were determined by limiting dilution experiments of pure virus stocks in cell culture lysates (∼108 viral particles ml−1 for HEV and HAdV and ∼107 ml−1 for BEV). Viral particles were counted using the method described by Noble and Fuhrman (47), with the exception that viruses were stained with SYBR Gold rather than SYBR Green I nucleic acid gel stain (Molecular Probes, Eugene, Oreg.). Viral RNA (enteroviruses) and DNA (adenoviruses) extracts were serially diluted to 10−8, reversed transcribed (HEV and BEV), and amplified by PCR methods described previously.
The efficiency of viral detection after concentration by adsorption-elution was also evaluated using a seeded study. MilliQ water was used as a control. A fresh estuarine water sample (with 50/00 salinity and pH 7.9) was filtered through 0.45-μm-pore-size membrane filters to remove particles. Prior to inoculation, the filtered estuarine water sample was exposed to UV light for at least 24 h to reduce or eliminate any background viruses. For quality control, two samples were taken from each type of water before inoculation as preseed controls. One liter of each of the two water types was inoculated with known amounts of HEV, BEV, and HAdV, serially diluted to 1 virus particle ml−1, and mixed at room temperature for at least an hour before processing. Seeded samples were concentrated, extracted, detected by PCR, and confirmed by dot blot hybridization following the protocols described previously.
Statistical analysis.
The Pearson correlation test was used to evaluate relationships among water quality and environmental variables. Binary logistic regression was used to analyze the relationship between the occurrence of viruses and levels of bacterial indicators and other environmental variables collected in this study. Minitab Release 12.2 (State College, Pa.) was used for logistic regression and correlation analyses. The analysis of variance (ANOVA) and subsequent comparisons to determine differences in mean levels of bacterial indicators and other environmental variables were performed using GraphPad Prism (GraphPad Software, Inc., San Diego, Calif.). In all cases, significance was determined at the 95% confidence level.
RESULTS
Physical and chemical parameters related to water quality.
A total of 35 samples were collected (including 5 samples that were collected from the bottom of the water column in July). Water temperature ranged between 10.5°C (December) and 31.0°C (July). Average water temperatures were similar for July, August, September, and October (means ranged between 27.7 and 29.5°C) and dropped significantly as winter approached (November mean, 20.2°C; December mean, 11.0°C) (P < 0.001). Increases in dissolved oxygen levels corresponded to the decrease in water temperature (r = −0.791, P < 0.001) and were lowest in August, with an average of 2.86 mg liter−1, and highest in December, with an average of 9.35 mg liter−1. Salinity varied between sites and sampling dates and ranged between 0.10/00 (station 5 in December) and 23.10/00 (station 1 in September). Station 5 (located farthest inland) consistently had the lowest salinity (mean, 1.00/00). Salinity levels were significantly lower at all stations in November and December (mean, 2.9 in each month) (P = 0.01) and were moderately influenced by increased streamflow (r = −0.5446, P = 0.0019).
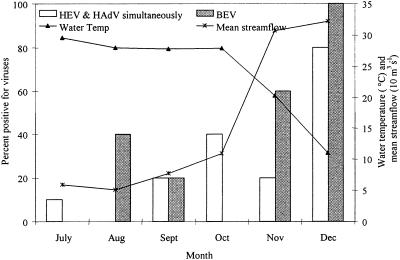
Total monthly precipitation ranged from a seasonal high of 16.5 cm in August to a winter-time low of 5.6 cm in December, which was significantly lower than precipitation levels in any other month. However, streamflow was lowest in August, at 51 m3 s−1, and highest in December, at 322 m3 s−1 (Fig. 2).
FIG. 2.
Percentage of samples positive for human enteroviruses and human adenoviruses (detected simultaneously) as well as bovine enteroviruses by month versus the mean monthly water temperature (°C) and stream flow (10 m3 s−1) along the lower Altahama River between July and December 2002 (n = 5).
Biological water quality indicators.
Chlorophyll-a measurements did not vary significantly among stations, and the averages ranged from 7.52 μg liter−1 (station 5) to 8.54 μg liter−1 (station 3). Monthly averages were more variable and ranged from 10.69 μg liter−1 in August to 4.86 μg liter−1 in December, which was significantly lower than that in other months (P < 0.001).
Over the course of the study, fecal coliform levels ranged between a most probable number (MPN) of 2 100 ml−1 and an MPN of 170 100 ml−1 (Table 2). Increases in fecal coliform levels corresponded to decreasing salinity (r = −0.648, P < 0.001). Fecal coliform levels were the lowest at station 1 (located at the mouth of the river) and increased up the river; station 5 consistently had the highest fecal coliform levels (Table 2). Total coliform levels ranged between 11 and 2,400 MPN 100 ml−1, following the same trend as fecal coliform levels.
TABLE 2.
The range and geometric mean MPN fecal and total coliform bacterial counts at each sampling station in the lower Altamaha River between July and December 2002a
| Station | Fecal coliform bacteria
|
Total coliform bacteria
|
||
|---|---|---|---|---|
| Range | Geometric mean | Range | Geometric mean | |
| 1 | 2-23 | 7 | 11-170 | 51 |
| 2 | 8-34 | 22 | 30-700 | 91 |
| 3 | 13-70 | 34 | 50-1,600 | 238 |
| 4 | 13-130 | 36 | 30-900 | 162 |
| 5 | 30-170 | 74 | 110-2,400 | 786 |
Fecal coliform bacteria and total coliform bacteria are measured in MPN 100 ml−1.
Viral detection efficiency.
Pure cultures of HEV and HAdV could each be detected to a 10−8 dilution (∼4 viral particles ml−1) with RT-nested-PCR and nested-PCR, respectively. BEV was detected by RT-PCR to a 10−3 dilution (∼40,000 viral particles ml−1). The efficiency of viral detection in MilliQ water after concentration by adsorption-elution was 4 HAdV particles ml−1 and 40 HEV particles ml−1. Both HAdV and HEV were detected to a level of 40 particles ml−1 in filtered estuarine water. BEV were detected to a concentration of 4 × 105 viral particles ml−1 both in MilliQ and filtered estuarine water with RT-PCR.
Dot blot hybridization showed no significant improvement in detection efficiency for seeded HAdV (detection limits remain 4 viral particles ml−1 in MilliQ water and 40 viral particles ml−1 in filtered estuarine water). However, the additional hybridization step did improve the detection of HEV by an order of magnitude (to 4 viruses ml−1 in both water types). Detection efficiency for BEV also improved by 10- to 100-fold after dot blot hybridization in which they were detected to a concentration of 4 × 104 and 4 × 103 viral particles ml−1 in filtered estuarine water and MilliQ water, respectively.
Detection of human enteric viruses from environmental samples.
HEV were detected in 17 out of 30 (∼57%) surface water samples and two out of five (40%) bottom water samples (collected only in July). HAdV were detected in 11 (∼37%) surface samples and 1 (20%) bottom sample. Sixty-seven percent of surface water samples and 40% of bottom water samples were positive for either HEV or HAdV. The viruses were detected simultaneously in nine (∼26%) samples (including one of the bottom water samples); this consisted of 41% of those samples in which either HEV or HAdV was detected.
Human enteric viruses were most frequently detected at stations 1 and 4, followed by station 5, station 2, and station 3 (Table 3). Six out of seven (86%) samples taken from station 1 were positive for either HEV or HAdV. HEV were detected from four (67%) surface samples and from the bottom sample collected in July, and HAdV was detected from three (50%) surface samples at station 1. At station 4, HEV and HAdV were each detected from four out of six (67%) surface samples; HEV and HAdV were detected concurrently from three (50%) surface samples. For station 5, HEV and HAdV were each detected from three out of six (50%) surface samples, and HEV and HAdV were detected simultaneously from the bottom sample collected in July. Four HEV-positive samples and only one HAdV-positive sample were detected from station 2. Human enteric viruses were only detected at station 2 from July to October. Station 3 was the least contaminated station throughout the study period, with only two HEV-positive (October and November) and no HAdV-positive samples.
TABLE 3.
Occurrence of HEV, HAdV, and BEV at each sampling station in the lower Altamaha River between July and December 2002a
| Station | Human Impact
|
Bovine Impact
|
|||
|---|---|---|---|---|---|
| Month of HEV- positive sample | Month of HAdV-positive sample | Rank | Month of BEV- positive sample | Rank | |
| 1 | July, Aug, Oct, Nov, Dec | Sept, Oct, Dec | 1 | Nov, Dec | 2 |
| 2 | July, Aug, Sept, Oct | Sept | 3 | Dec | 3 |
| 3 | Oct, Nov | 4 | Sept, Dec | 2 | |
| 4 | Aug, Oct, Nov, Dec | Sept, Oct, Nov, Dec | 1 | Sept, Oct, Nov, Dec | 1 |
| 5 | July, Aug, Dec | July,b Sept, Dec | 2 | Nov, Dec | 2 |
Sites are ranked by occurrence of viruses, with 1 being the most contaminated.
Bottom-water sample.
The occurrence of human enteric viruses demonstrated a clear seasonal trend in which the frequency of detection increased at lower water temperatures and increased streamflow (Fig. 2). Both of these conditions reached their greatest extent in December, at which time HEV and HAdV were detected simultaneously in 80% of the samples. Conversely, only three samples (from stations 1, 2, and 5) were positive for any human virus in July (average temperature, 31°C), and two of these samples (from stations 1 and 5) were collected from the bottom of the water column.
The occurrence of human enteric viruses was significantly related to streamflow (in cubic meters per second) on the day samples were collected, mean daily rainfall for the 30 days preceding the sample collection, and other water quality variables (i.e., water temperature and DO and chlorophyll-a levels) but was not related to rainfall on the sample collection day and mean daily rainfall for up to 7 days preceding the sample collection, salinity, pH, and nutrient levels. Binary logistic regression models using streamflow (on sampling days), water temperature, 30-day mean rainfall, and DO and chlorophyll-a concentrations as independent variables were able to predict the presence or absence of HEV and HAdV together in a sample 79 to 85% of the time (P < 0.05) (Table 4). The presence or absence of human enteric viruses was directly (positively) related to streamflow and DO and inversely related to temperature, rainfall, and chlorophyll-a concentrations. Fecal coliform and total coliform levels in a sampling area were generally low and did not show any correlation to viral detection (Table 4).
TABLE 4.
Binary logistic regression analyses for the occurrence of HEV, HAdV, and BEV using water quality or environmental variables
| Parameter | HAdV
|
HEV
|
HEV and HAdVa
|
BEV
|
||||
|---|---|---|---|---|---|---|---|---|
| Concordance (%) | P valued | Concordance (%) | P value | Concordance (%) | P value | Concordance (%) | P value | |
| DO | 75.6 | 0.031* | 59.3 | 0.625 | 85.8 | 0.012* | 88.5 | 0.018* |
| Water temp | 75.6 | 0.062 | 63.8 | 0.183 | 81.8 | 0.012* | 86.6 | 0.006* |
| Total coliform | 51.2 | 0.431 | 44.3 | 0.429 | 48.9 | 0.233 | 42.6 | 0.567 |
| Fecal coliform | 50.7 | 0.545 | 52.9 | 0.643 | 57.4 | 0.377 | 44 | 0.811 |
| Streamflowb | 70.8 | 0.040* | 54.3 | 0.246 | 79 | 0.009* | 85.6 | 0.005* |
| Rainfallc | 70.8 | 0.08 | 54.3 | 0.204 | 79 | 0.012* | 85.6 | 0.003* |
| Chlorophyll-a | 74.2 | 0.034* | 42.5 | 0.956 | 80.1 | 0.019* | 80.9 | 0.009* |
HEV and HAdV were detected in the same sample simultaneously.
Streamflow value on sampling days was used.
Mean rainfall 30 days preceding sampling was used.
An asterisk indicates the value is significant (P < 0.05).
Detection of bovine enteric viruses from environmental samples.
BEV were detected from 11 out of 30 (∼37%) surface water samples and none of the bottom water samples. Over the course of study, station 4 was the most contaminated site for BEV, where it was detected from four out of six (∼67%) surface samples. Station 1, station 3, and station 5 each had two BEV-positive samples, while only one BEV-positive sample was discovered at station 2 (Table 3).
The occurrence of BEV demonstrated a seasonal trend similar to those of human enteric viruses, with frequency of detection increasing at lower water temperatures and increased streamflow (Fig. 2). BEV were detected from three out of five (60%) stations in November and all stations in December. Neither July nor August had any positive BEV samples. Binary logistic regression analysis for the occurrence of BEV using streamflow on sampling days, 30-day mean rainfall, water temperature, and other water variables (i.e., DO and chlorophyll-a levels) as independent variables showed relationships for BEV similar to those for human enteric viruses (Table 4). There was no significant relationship between the presence of BEV and total or fecal coliform counts (Table 4).
DISCUSSION
Freshwater demand in the 24 counties of Georgia's coast has increased tremendously from 1980 to 1997 because of population growth and increased water use by industry and agriculture (16). Furthermore, exponential population growth and rapid development along coastal rivers in general has generated many concerns about decreasing water quality (21), including increased pumpage of groundwater and freshwater, conversion of open lands into nonpermeable surfaces, changes in hydrologic conduits, and increased wastewater discharge (16). These changes have affected water quality through saltwater intrusion and the addition of sediment, toxic chemicals, pathogenic microorganisms, and nutrients into the coastal rivers and estuaries.
Water quality issues in the Altamaha River.
In 1999, water quality data collected by the United States Geological Survey in the Altamaha River Basin indicated DO and fecal coliform impairments in many segments of the Altamaha tributaries (19, 21). The state of Georgia has conducted several studies, in accordance with Section 303(d) of the Clean Water Act and the U.S. Environmental Protection Agency (EPA) Water Quality Planning and Management Regulations (40 CFR Part 130), to develop fecal coliform bacteria and DO total maximum daily loads (TMDLs) for water bodies in the Altamaha River basin that are listed as either not supporting or partially supporting designated use classifications, due to exceedence of water quality standards (19, 21). One of the main purposes of the TMDL analysis is to identify the source categories or individual sources of fecal pollution in a watershed and the amount of loading contributed by each of these sources so that the long-term effects of anthropogenic activities and development upstream on water quality can be better monitored (21). Presently, there are more than 100 permits allowing treated sewage, discharge from paper and pulp operations, and other pollutants to be discharged into the Altahama River (4). Typical non-point sources of fecal pollution in the Altahama River include urban development (storm water runoff and leaking sewer collection lines), leaking septic systems, land application of agricultural manure, livestock grazing, and wildlife (21). Based on an EPA survey in 2001, 5% of the septic systems in the watershed leak (21). In the Altamaha River Basin, animal waste might be one of the main contributors to non-point source contamination via land application of manure, runoff from confined animal feeding operations, and unconfined animals (i.e., deer, hogs, and other wildlife) (21). The marsh islands surrounding our sampling stations in the lower Altamaha River are inhabited primarily by cattle, a limited number of feral hogs, and waterfowl (Katy Austin, Georgia Marine Extension Services, personal communication).
Fecal indicator bacteria.
Several studies have suggested that fecal coliform levels cannot be used to predict the occurrence of human viruses, and this is consistent with the results of our study (28, 50, 67). While human and bovine enteric viruses were frequently detected at our sampling stations, fecal coliform readings at sampling stations were generally low and never exceeded Georgia's recreational water quality control one-time sampling limit of 500 MPN 100 ml−1 during our study period (20). Fecal coliform levels ranged between 13 and 130 MPN 100 ml−1 at station 4, which was ranked the highest in human and bovine enteric virus contamination, combined. Fecal coliform bacteria, therefore, may not be a reliable indicator to assess risks associated with enteric viruses in the coastal rivers of Georgia, especially given that symptomatic infections can be caused by less than 1 PFU of enteric virus (66). Furthermore, without additional strain typing of these bacteria the source is unknown.
Enteric viruses as water quality assessment tools.
The levels of enteric viruses in natural aquatic environments usually are low, and detection methods with high sensitivity and specificity are needed to study the occurrence of these viruses. Molecular detection of viruses offers several advantages over traditional viral assays, such as cell culture. PCR viral detection is less laborious and time-consuming but is more specific and sensitive than cell culture (10, 30, 34). PCR is capable of detecting viruses that are either difficult to grow in cultured cells or replicate without producing cytopathogenic effects in cells (9, 41, 51). For example, adenoviruses, one of the most important human pathogens present in polluted water, are slow growing and often do not produce cytopathogenic effects in cells, therefore they are consistently underestimated when fast-growing enteroviruses are present (32, 59). Pina et al. (50) suggested that the highly sensitive PCR detection method had led to higher detection rate of adenoviruses in environmental samples. Furthermore, the specificity of PCR combined with the host specificity of enteric viruses allows a contaminant source to be identified immediately without the need for an extensive database of strains, as is required with bacterial source tracking (49).
The development and application of RT-nested-PCR protocols to detect HEV and HAdV in our study increased the detection limit compared to that of conventional PCR protocols (12). Nested-PCR is generally more specific and has shown a higher level of sensitivity in detecting enteric viruses from environmental samples (52). The detection limit for HAdV by nested-PCR as used in this study was ∼4 viral particles ml−1, which is consistent with previously reported sensitivities as high as one purified viral particle (2). This is about 100 to 1,000 times more sensitive than cell culture assays (52). The RT-nested-PCR protocol for HEV detection developed in this study was also shown to have a sensitivity of ∼4 viral particles ml−1. Previous unnested PCR assays had a reported sensitivity of 103 viral particles ml−1 (28).
The addition of dot blot hybridization after PCR further increased detection sensitivity, especially in natural water samples, and prevented false-negative results as well as confirmed PCR positives by gel electrophoresis. In our seeded experiment, dot blot hybridization improved detection sensitivity of HEV by about one order of magnitude in filtered estuarine water and one to two orders of magnitude for BEV in MilliQ water and filtered estuarine water. While there was no significant improvement for HAdV in MilliQ water or filtered estuarine water, in environmental samples our rate of detection was higher after dot blot hybridization in all cases. This finding is consistent with the results from previous studies in which dot blot hybridization increases detection sensitivity by at least an order of magnitude (12, 28, 63). For our environmental samples, dot blot hybridization increased the number of HAdV-, HEV-, and BEV-positive samples compared to gel electrophoresis by 71, 111, and 100%, respectively.
Although direct PCR and dot blot hybridization offer quick, highly specific, and sensitive ways to detect enteric viruses from environmental samples, it does not necessarily represent a public health risk, because little is known about the infectivity of the viruses. Because PCR can detect nucleic acids from both infectious and noninfectious viruses, data derived from direct PCR is most useful when the infectivity of these viruses is not an important issue or not of public health concern (i.e., solely to track the source of water pollution).
Human and animal fecal loading in the lower Altamaha.
The prevalence of positive human enteric virus samples detected in this study suggests that the water quality at the lower Altamaha River is affected by contaminants of human origin, such as wastewater (including leaking septic systems) and urban runoff. BEV also were frequently detected in the study area, despite a reduced detection efficiency compared to that of the human enteric viruses. This suggests that bovine species also contribute to fecal loading in this watershed and indicates potential for transmission of zoonotic pathogens.
The Altamaha River drains one-fourth of the state of Georgia, and therefore contamination may reflect both upstream as well as local pollution. The most human viral loading was noted at station 1, which is located near the Altamaha Sound. Contamination at this station may be higher, because in addition to the lower Altamaha River, it also receives flow from the Atlantic Intracoastal Waterway (which flows through populated areas of coastal Georgia). Station 4, which ranked the highest in both bovine and human enteric virus contamination, is also influenced by the Atlantic Intracoastal Waterway (which flows south of a marsh island near station 3 inhabited primarily by cattle, locally known as Cow Island) and the south Altamaha tributary (Fig. 1). Station 5, our most upstream site and located on the south Altamaha tributary, ranked second in human enteric virus load. A commercial fish camp with restaurant and lodging upstream from the station might have contributed to human fecal contamination.
In our study, all viruses were detected at a higher frequency in December, reflecting the importance of low temperature and high streamflow in viral stability and loading, respectively. This finding is consistent with previous reports that the stability of viral particles is highly influenced by water temperature (46, 66). The important effect of streamflow in the loading of viruses in coastal waters has also been demonstrated by Goyal et al. (27) and Lipp et al. (40). Increase in streamflow may have caused more remote influx of contaminants as well as more widespread viral loading. Our results showed no immediate response in viral detection to rainfall events and an inverse response with mean rainfall in the 30 days preceding sampling, which may reflect a complex hydrology in this watershed. Seasonal cycles in viral infection and excretion in the population also might have played a role in the elevated detection of enteric viruses in winter months (33).
The ability to identify the major source categories of fecal contamination is important in managing both water quality and human health risk. Several studies have evaluated the prevalence of animal-specific enteric viruses (i.e., bovine enteroviruses, porcine teschoviruses, and bovine and porcine adenoviruses) in animal feces and the environment (35, 39, 42, 49). Findings of this study along with those of previous works suggest that molecular detection of animal-specific viruses can be highly sensitive and specific markers for tracing sources of fecal contamination; however, more testing will be needed to study the geographical and seasonal distribution of these viruses.
In conclusion, the stringent host specificity of enteric viruses makes them good library-independent indicators for identifying sources of water pollution. PCR can be used to detect and differentiate specific groups of viruses when infectivity data are unnecessary in decision-making. Detection assays specific for other groups of animal enteric viruses, such as avian and porcine enteric viruses, could be developed to better characterize sources of contamination.
Acknowledgments
This study was funded by the Georgia Sea Grant College program development funds (UGA) and USDA Hatch Project no. GEO00978; additional partial support was provided by USEPA grant no. X7-97480102-0.
Special thanks to Patricia Smith and Ethell Vereen, Jr., for help with sample processing. Katy L. Austin and the sampling team from the Marine Extension Service of the University of Georgia in Brunswick provided valuable support in sample collection and transportation, and they provided water quality data and coliform readings. Charles P. Gerba (University of Arizona) provided strains of poliovirus and adenovirus for use as positive controls; George Lukasik provided cultures of bovine enteroviruses; and John H. Paul assisted with primer development for human enteroviruses.
REFERENCES
- 1.Abad, F., R. Pinto, C. Villena, R. Gajardo, and A. Bosch. 1997. Astrovirus survival in drinking water. Appl. Environ. Microbiol. 63:3119-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allard, A., B. Albinsson, and G. Wadell. 1992. Detection of adenoviruses in stools from healthy persons and patients with diarrhea by two-step polymerase chain reaction. J. Med. Virol. 37:149-157. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 2004, posting date. The Altamaha River. [Online.] http://www.altamahariverkeeper.org/river_news/altamaha/altamaha_river.asp.
- 4.Anonymous. 2004, posting date. James Holland and the Altamaha Riverkeeper. [Online.] http://www.altamahariverkeeper.org/river_news/altamaha/altamaha_river_keeper.asp.
- 5.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-Dillen, and J. Van Der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch, A. 1998. Hum. enteric viruses in the water environment: a minireview. Int. Microbiol. 1:191-196. [PubMed] [Google Scholar]
- 7.Bosch, A. 1995. The survival of enteric viruses in the water environment. Microbiologia SEM 11:393-396. [PubMed] [Google Scholar]
- 8.Bosch, A., F. Lucena, J. M. Diez, R. Gajardo, M. Blasi, J. Jofre. 1991. Waterborne viruses associated with hepatitis outbreak. J. Am. Water Works Assoc. 83:80-83. [Google Scholar]
- 9.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, H., L. Jaykus, and M. Sobsey. 1996. Detection of human enteric viruses in oysters by in vivo and in vitro amplification of nucleic acids. Appl. Environ. Microbiol. 62:3772-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craun, G. F. 1991. Causes of waterborne outbreaks in the United States. Water Sci. Technol. 24:17-20. [Google Scholar]
- 12.De Leon, R., C. Shieh, R. S. Baric, and M. D. Sobsey. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction, p. 833-853. In Proceedings of the 1990 Water Quality Technology Conference, Denver, Colorado, American Water Works Association, Washington, D.C.
- 13.Dombek, P. E., L. K. Johnson, S. T. Zimmerley, and M. J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Appl. Environ. Microbiol. 66:2572-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donaldson, K. A., D. W. Griffin, and J. H. Paul. 2002. Detection, quantitation and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Res. 36:2505-2514. [DOI] [PubMed] [Google Scholar]
- 15.Enriquez, C. E., C. J. Hurst, and C. P. Gerba. 1995. Survival of the enteric adenoviruses 40 and 41 in tap, sea, and waste water. Water Res. 29:2548-2553. [Google Scholar]
- 16.Fanning, J. L. 1999. Water use in coastal Georgia by county and source, 1997; and water-use trends, 1980-97. U.S. Geological Survey, Washington, D.C.
- 17.Farthing, M. J. G. 1989. Viruses and the gut. Smith Kline & French, Walwyn Garden City, Hertfordshire, United Kingdom.
- 18.Feachem, R. G., D. J. Bradley, H. Garelick, and D. D. Mara. 1983. Sanitation and disease: health aspects of excreta and wastewater management. John Wiley & Sons, Inc., New York, N.Y.
- 19.Georgia Department of Natural Resources. 2002. Altamaha River basin dissolved oxygen TMDLs. U.S. Environmental Protection Agency Region 4. Georgia Department of Natural Resources, Atlanta, Ga.
- 20.Georgia Department of Natural Resources. 2002. Total maximum daily loads (TMDLs) for fecal coliform in 303(d) Listed Streams in The Altamaha River Basin. U.S. Environmental Protection Agency, Region 4. Georgia Department of Natural Resources, Atlanta, Ga.
- 21.Georgia Department of Natural Resources. 2004. State of Georgia rules and regulations for water quality control chapter 391-3-6. Georgia Department of Natural Resources, Atlanta, Ga.
- 22.Gerba, C. P. 2000. Assessment of enteric pathogen shedding during recreational activity and its impact on water quality. Quant. Microbiol. 2:55-68. [Google Scholar]
- 23.Gerba, C. P., S. M. Goyal, R. L. Labelle, I. Cech, and G. F. Bodgan. 1979. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am. J. Pub. Health. 69:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerba, C. P., D. M. Gramos, and N. Nwachuku. 2002. Comparative inactivation of enteroviruses and adenovirus 2 by UV light. Appl. Environ. Microbiol. 68:5167-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerba, C. P., J. B. Rose, C. N. Haas, and K. D. Crabtree. 1996. Waterborne rotavirus: a risk assessment. Water Res. 30:2929-2940. [Google Scholar]
- 26.Goyal, S. M., and C. P. Gerba. 1983. Viradel method for detection of rotavirus from seawater. J. Virol. Methods 7:279-285. [DOI] [PubMed] [Google Scholar]
- 27.Goyal, S. M., C. P. Gerba, and J. L. Melnick. 1979. Human enteroviruses in oysters and their overlying waters. Appl. Environ. Microbiol. 37:572-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffin, D. W., C. J. Gibson, E. K. Lipp, K. Riley, J. H. Paul, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin, D. W., E. K. Lipp, M. R. McLaughlin, and J. B. Rose. 2001. Marine recreation and public health microbiology: quest for the ideal indicator. BioScience 51:817-825. [Google Scholar]
- 30.Hafliger, D., M. Gligen, J. Luthy, and P. Hubner. 1997. Seminested RT-PCR systems for small round structured viruses and enteric viruses detection in seafood. Int. J. Food Microbiol. 37:27-36. [DOI] [PubMed] [Google Scholar]
- 31.Hejkal, T. W., B. Keswick, R. L. LaBelle, C. P. Gerba, Y. Sanchez, G. Dreesman, B. Hafkin, and J. L. Melnick. 1982. Viruses in a community water supply associated with an outbreak of gastroenteritis and infectious hepatitis. J. Am. Water Works Assoc. 150:318-321. [Google Scholar]
- 32.Irving, L. G., and F. A. Smith. 1981. One-year survey of enteroviruses, adenoviruses, and reoviruses isolated from effluent at an activated-sludge purification plant. Appl. Environ. Microbiol. 41:51-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaykus, L. A., M. T. Hemard, and M. D. Sobsey. 1994. Human enteric pathogenic viruses, p. 92-153. In H. a. M. D. Pierson (ed.), Environmental indicators and shellfish safety. Chapman and Hall, New York, N.Y.
- 34.Jiang, S., R. Noble, and W. P. Chui. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiménez-Clavero, M. A., C. Fernandez, J. A. Ortiz, J. Pro, G. Carbonell, J. V. Tarazona, N. Roblas, and V. Ley. 2003. Teschoviruses as indicators of porcine fecal contamination of surface water. Appl. Environ. Microbiol. 69:6311-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama, H., A. Shimasaki, and S. Ohgaki. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krikelis, V., P. Markoulatos, N. Spyrou, and C. Serie. 1985. Detection of indigenous enteric viruses in raw sewage effluents of the city of Athens, Greece, during a two year survey. Water Sci. Technol. 17:159-164. [Google Scholar]
- 38.Lee, S. H., and S. J. Kim. 2002. Detection of infectious enteroviruses and adenoviruses in tap water in urban areas in Korea. Water Res. 36:248-256. [DOI] [PubMed] [Google Scholar]
- 39.Ley, V., J. Higgins, and R. Fayer. 2002. Bovine enteroviruses as indicators of fecal contamination. Appl. Environ. Microbiol. 68:3455-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipp, E. K., R. Kurz, R. Vincent, C. Rodriguez-Palacios, S. R. Farrah, and J. B. Rose. 2001. The effects of seasonal variability and weather on microbial fecal pollution and enteric pathogens in a subtropical estuary. Estuaries 24:266-276. [Google Scholar]
- 41.Lipp, E. K., J. Lukasik, and J. B. Rose. 2001. Human enteric viruses and parasites in the marine environment, p. 559-588. In J. H. Paul (ed.), Methods in microbiology, vol. 30. Academic Press, London, United Kingdom. [Google Scholar]
- 42.Maluquer de Motes, C., P. Clemente-Casares, A. Hundesa, M. Martin, and R. Girones. 2004. Detection of bovine and porcine adenoviruses for tracing the source of fecal contamination. Appl. Environ. Microbiol. 70:1448-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnick, J. L. 1984. Etiologic agents and their potential for causing waterborne virus diseases, p. 1-16. In J. L. Melnick (ed.), Enteric viruses in water, vol. 15. Karger, Basel, Switzerland. [Google Scholar]
- 44.Melnick, J. L., and C. P. Gerba. 1989. The ecology of enteroviruses in natural waters. Crit. Rev. Environ. Control 10:65. [Google Scholar]
- 45.Meng, Q. S., and C. P. Gerba. 1996. Comparative inactivation of enteric adenovirus, poliovirus and coliphages by ultraviolet irradiation. Water Res. 30:2665-2668. [Google Scholar]
- 46.Nasser, A. M., N. Zaruk, L. Tenenbaum, and Y. Netzan. 2003. Comparative survival of Cryptosporidium, coxsackievirus A9 and Escherichia coli in stream, brackish and sea waters. Water Sci. Technol. 47:91-96. [PubMed] [Google Scholar]
- 47.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 48.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 49.Noble, R. T., S. M. Allen, A. D. Blackwood, W. Chu, S. C. Jiang, G. L. Lovelace, M. D. Sobsey, J. R. Stewart, and D. A. Wait. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial course tracking comparison study. J. Water Health 1:195-207. [PubMed] [Google Scholar]
- 50.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pommepuy, M., and F. Le Guyader. 1998. Molecular approaches to measuring microbial marine pollution. Curr. Opin. Biotechnol. 9:292-299. [DOI] [PubMed] [Google Scholar]
- 52.Puig, M., J. Jofre, F. Lucena, A. Allard, G. Wadell, and R. Girones. 1994. Detection of adenoviruses and enteroviruses in polluted waters by nested-PCR amplification. Appl. Environ. Microbiol. 60:2963-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao, V. C., and J. L. Melnick. 1986. Environmental biology. In J. A. Cole, C. J. Knowles, and D. Schlessinger (ed.), Aspects of microbiology, vol. 13. American Society of Microbiology, Washington, D.C.
- 54.Rose, J. B. 1986. Microbial aspects of wastewater reuse for irrigation. Crit. Rev. Environ. Control 16:231-256. [Google Scholar]
- 55.Schiff, G. M., G. M. Stefanovic, B. Young, and J. K. Pennekamp. 1984. Minimum human infectious dose of enteric viruses (echovirus-12) in drinking water, p. 222-228. In J. L. Melnick (ed.), Enteric viruses in water, vol. 15. Karger, Basel, Switzerland. [Google Scholar]
- 56.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sobsey, M. D., P. A. Shields, F. S. Hauchman, R. L. Hazard, and L. W. Caton. 1986. Survival and transport of hepatitis A virus in soils, groundwater and wastewater. Water Sci. Technol. 18:97-106. [Google Scholar]
- 58.Straub, T. M., and D. P. Chandler. 2003. Towards a unified system for detecting waterborne pathogens. J. Microbiol. Methods 53:185-197. [DOI] [PubMed] [Google Scholar]
- 59.Tani, N., Y. Dohi, N. Jurumatani, and K. Yonemasu. 1995. Seasonal distribution of adenoviruses, enteroviruses and reoviruses in urban river water. Microbiol. Immunol. 39:577-580. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, M. B., P. J. Becker, E. J. Van-Rensburg, B. N. Harris, I. W. Bailey, and W. O. Grabow. 1995. A serosurvey of water-borne pathogens amongst canoeists in South Africa. Epidemiol. Infect. 115:299-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, and C. P. Gerba. 2003. Chlorine inactivation of adenovirus type 40 and feline calicivirus. Appl. Environ. Microbiol. 69:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thurston-Enriquez, J. A., C. N. Haas, J. Jacangelo, K. Riley, and C. P. Gerba. 2003. Inactivation of feline calicivirus and adenovirus type 40 by UV radiation. Appl. Environ. Microbiol. 69:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsai, Y. L., M. D. Sobsey, L. R. Sangermano, and C. J. Palmer. 1993. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl. Environ. Microbiol. 59:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.U.S. Environmental Protection Agency. 1988. Comparative health effects assessment of drinking water. U.S. Environmental Protection Agency, Washington, D.C.
- 65.U.S. Environmental Protection Agency and U. S. Agency for International Development. 1992. Manual on guidelines for water reuse. EPA/625/R-92/004. Center for Environmental Reservation Information, Cincinnati, Ohio.
- 66.Wetz, J. J., E. K. Lipp, D. W. Griffin, J. Lukasik, D. Wait, M. D. Sobsey, T. M. Scott, and J. B. Rose. 2004. Presence, infectivity, and stability of enteric viruses in seawater: relationship to marine water quality in the Florida Keys. Mar. Pollut. Bull. 48:698-704. [DOI] [PubMed] [Google Scholar]
- 67.Wyer, M. D., J. M. Fleisher, J. Gough, D. Kay, and H. Merrett. 1995. An investigation into parametric relationships between enterovirus and faecal indicator organisms in the coastal waters of England and Wales. Water Res. 29:1863-1868. [Google Scholar]
- 68.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]