Abstract
Propene monooxygenase has been cloned from Mycobacterium sp. strain M156, based on hybridization with the amoABCD genes of Rhodococcus corallinus B276. Sequencing indicated that the mycobacterial enzyme is a member of the binuclear nonheme iron monooxygenase family and, in gene order and sequence, is most similar to that from R. corallinus B-276. Attempts were made to express the pmoABCD operon in Escherichia coli and Mycobacterium smegmatis mc2155. In the former, there appeared to be a problem resolving overlapping reading frames between pmoA and -B and between pmoC and -D, while in the latter, problems were encountered with plasmid instability when the pmoABCD genes were placed under the control of the hsp60 heat shock promoter in the pNBV1 vector. Fortuitously, constructs with the opposite orientation were constitutively expressed at a level sufficient to allow preliminary mutational analysis. Two PMO active-site residues (A94 and V188) were targeted by site-directed mutagenesis to alter their stereoselectivity. The results suggest that changing the volume occupied by the side chain at V188 leads to a systematic alteration in the stereoselectivity of styrene oxidation, presumably by producing different orientations for substrate binding during catalysis. Changing the volume occupied by the side chain at A94 produced a nonsystematic change in stereoselectivity, which may be attributable to the role of this residue in expansion of the binding site during substrate binding. Neither set of mutations changed the enzyme's specificity for epoxidation.
Several bacteria are capable of aerobic growth on short-chain alkene substrates (C2 to C4) as sole carbon and energy sources. These include strains of Rhodococcus spp., Mycobacterium spp., Nocardia spp., and Xanthobacter autotrophicus (10). Mycobacterium sp. strain M156 was isolated on propene as the sole carbon source and was shown to initiate alkene oxidation by an O2- and NAD(P)H-dependent monooxygenase reaction (32). In propene-utilizing organisms that have been characterized thus far, the further metabolism of propene involves carboxylation to acetoacetate (6), and the absolute requirement of carbon dioxide for the growth of M156 on propene in sparged bioreactors (32) suggests that this is also the case for this strain.
Alkene monooxygenases have interesting prospects as biocatalysts for asymmetric synthesis (15). Although they belong to the same family of binuclear nonheme iron monooxygenases as soluble methane monooxygenase (sMMO) (14), they are able to discriminate between nonactivated C-H bonds and double bonds, specifically epoxidizing the latter, often with high enantiomeric excess (20). This discrimination appears to be a fundamental part of the catalytic mechanism, but stereoselectivity is probably a reflection of substrate binding. Therefore, it should be possible to modify stereoselectivity through protein engineering without a loss of reaction specificity, which is not likely to be the case with alkane or methane monooxygenases, for which epoxidation is a fortuitous consequence of the catalytic cycle and competing reactions such as allylic hydroxylation can also occur. However, nonheme iron monooxygenases are relatively complex, placing a considerable synthetic burden on the host cells, and simpler systems would be advantageous. The enzymes from X. autotrophicus Py2 and Rhodococcus corallinus are the most thoroughly characterized alkene monooxygenases. The former (XAMO) is a four-component system comprised of (i) an NADH oxidoreductase containing a noncovalently bound FAD molecule and a [2Fe-2S] cluster, (ii) a Rieske-type ferredoxin, (iii) a hexameric oxygenase (α2β2γ2) containing the binuclear iron active site, and (iv) a small coupling protein which is essential for activity (26). It has a high degree of sequence similarity to aromatic monooxygenases, including toluene 2-, 3-, and 4-monooxygenases (T2/3/4MO) and benzene monooxygenase, and also to isoprene monooxygenase, suggesting that these form a homologous group (35).
The alkene monooxygenase from R. corallinus B-276 (AMO) is a simpler, three-component system encoded by the four-gene operon amoABCD (23). It lacks the ferredoxin and the oxygenase γ subunit (the oxygenase is an α2β2 tetramer [18]), and the gene order differs from that in X. autotrophicus Py2. However, spectroscopic studies and important elements of structure and sequence conservation revealed that the α subunit contains a typical binuclear nonheme iron binding site (8).
The monooxygenase from Mycobacterium sp. strain M156 (PMO) has been more difficult to purify. However, a partial purification (32) indicated that it did not contain a ferredoxin component, suggesting that it is similar to the simpler three-component enzyme from R. corallinus B-276, and we have used this information as a starting point for the cloning, sequencing, and expression of the mycobacterial enzyme. Although the high levels of expression necessary for use as a recombinant biocatalyst have yet to be achieved, expression was sufficient to allow confirmation that by targeting appropriate residues, the stereoselectivity of alkene monooxygenases can be modified without losing reaction specificity.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Mycobacterium sp. strain M156 was originally isolated on propene as the sole carbon source (32). Mycobacterium smegmatis mc2155 is a high-efficiency transformation strain (28) and was obtained from W. R. Jacobs, Jr. Escherichia coli XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIq ZΔM15 Tn10 (Tetr)]} was purchased from Stratagene. E. coli DH5α [supE44 ΔlacY169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1] and E. coli DH10B (F− mcrA Δ[mrr hsdRMS mcrBC] φ80d lacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ[ara leu]7697 galU galK l− rpsL nupG) were obtained from GIBCO BRL.
The E. coli cloning vector pBluescript II was purchased from Stratagene, and pSP72 (Ampr; contains oriE and a multiple cloning site) was purchased from Promega. The E. coli-mycobacterial shuttle vector pNBV1 (Hygr; contains oriE, oriM, and a multiple cloning site; suitable for blue-white colony screening) was obtained from W. R. Bishai (11), and pUS1872 (Kanr; contains oriM) was obtained from J. MacFadden (unpublished data).
Materials.
Hygromycin was obtained as a sterile stock solution (50 mg ml−1) from Roche Diagnostics Ltd. All other chemicals were from Sigma-Aldrich and were analytical grade. Restriction enzymes and DNA-modifying enzymes (calf intestinal alkaline phosphatase and DNA ligase) were purchased from New England Biolabs and Roche Diagnostics Ltd., respectively. High-purity plasmid DNAs were purified from E. coli clones by the use of QIAprep Spin miniprep kits (QIAGEN), while DNAs were recovered from agarose gels by use of a DNA Wizard clean-up kit from Promega or a QIAQuick gel extraction kit from QIAGEN. Chemically competent E. coli DH5α, used for routine subcloning, was either purchased from Invitrogen or prepared as described by Sambrook et al. (24). E. coli DH10B which had been rendered electrocompetent (24) was used for mycobacterium library constructions. DNA primers were ordered from MWG-Biotech, and DNA sequencing was done by use of a BigDye Terminator cycle sequencing kit (Perkin-Elmer) with AmpliTaq FS DNA polymerase and analyzed on an ABI 377 automated sequencer (Applied Biosystems).
Southern blot analysis.
Chromosomal DNA from Mycobacterium sp. strain M156 was extracted as described by Pitcher et al. (19), and Southern blots were done according to the method of Sambrook et al. (24). Probe labeling and membrane hybridizations were performed according to the instructions in an ECL random-prime labeling and detection kit (Amersham). After overnight probe hybridization, Hybond N+ membranes containing chromosomal DNA were washed twice in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 15 min at 60°C. Membranes containing plasmid DNA after the lysis of colonies were treated as described by Sambrook et al. (24), and posthybridization washes were done with 1× SSC-0.1% SDS and 0.2× SSC-0.1% SDS for 15 min each at 60°C.
Sequence analysis.
Open reading frame (ORF) searches were done on the Frameplot server (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl). Codon usage tables were created at http://www.kazusa.or.jp/codon/countcodon.html and compared (M. Furhmann, L. Ferbitz, A. Hausherr, and T. Schodl, unpublished data) at http://gcua.schoedl.de/. BLAST searches were performed at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST) by use of the BLOSUM62 matrix, with a gap open penalty of 11 and a gap extension penalty of 1. Amino acid identities were calculated with the ALIGN program at http://www.infobiogen.fr/services/analyseq/cgi-bin/alignp_in.pl by use of the BLOSUM 50 matrix. Multiple alignments were done with CLUSTAL-X, version 1.8 (31). The Genedoc alignment editor was used (http://www.psc.edu/biomed/genedoc/). Secondary structure predictions were done at http://bioinf.cs.ucl.ac.uk/psipred/ (16), and protein modeling was done at the website of Swiss-Model (http://www.expasy.org/swissmod/SWISS-MODEL.html) in the alignment interface mode (25).
Expression plasmid construction.
An intermediate construct, pNBVSpmo9, was digested with BamHI, and the resulting 4.8-kb fragment containing the pmo cluster was gel purified. After ligation to BamHI-cut pUS1872 and transformation in E. coli DH5α, two types of clone were isolated, depending on the orientation of the insert relative to the hsp60 promoter. pUSpmo11 contained the pmo cluster downstream of hsp60p, and pUSpmo5 contained the pmo cluster upstream of hsp60p.
To obtain pNBVpmo11 and pNBVpmo15, we cut pUSpmo11 and pUSpmo5, respectively, with BglII, treated them with T4 DNA polymerase, and then cut them with HindIII before agarose gel purification. The purified 5-kb DNA fragment (containing the hsp60p and pmo genes) was ligated to pNBV1 after cutting with ClaI, filling in with T4 DNA polymerase, and cutting with HindIII. The selection of DH5α transformants on Luria broth plus hygromycin (0.2 mg/ml) plates yielded pNBVpmo11 and pNBVpmo15, respectively (Fig. 1).
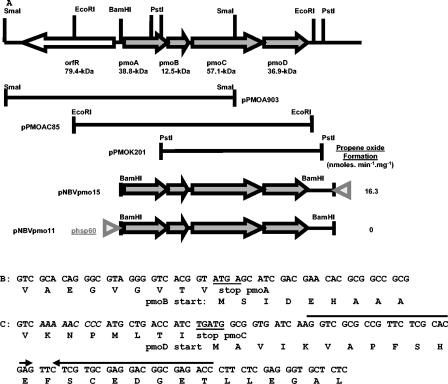
FIG. 1.
Genetic map and features of the alkene monooxygenase genes of Mycobacterium sp. strain M156. (A) pPMOA903, pPMOAC85, and pPMOK201 are constructs created by cloning of the respective SmaI, EcoRI, and PstI fragments, as indicated by the horizontal bars. The E. coli-mycobacterium shuttle constructs for homologous expression are also shown as pNBVpmo11 and pNBVpmo15, with the location of the hsp60 promoter (gray triangles) indicated. Propene oxide formation activities obtained from whole-cell assays are indicated on the right. (B) DNA region showing the end of pmoA overlapping with the start of pmoB. The stop codon and start codon are underlined. (C) Region at the end of pmoC displaying the slippage codons (in italics), the overlapping start and stop codons (underlined), and the plausible stem-loop (inverted arrows).
Site-directed mutagenesis.
Site-directed mutagenesis was performed by use of a QuikChange site-directed mutagenesis kit (Stratagene). The introduction of single specific mutations into pmoC was achieved by the use of pSPpmo2 (pSP72 with a 2.7-kb NheI-NdeI insert containing pmoABC from Mycobacterium sp. strain M156) as template DNA together with the following pairs of mutagenic primers: for isoform A94G, A94Gfwd (5′-CCG ATC CTG ACC AAT GGC GAG TAT CAG GCG GTC-3′) and A94Grev (5′-GAC CGC CTG ATA CTC GCC ATT GGT CAG GAT CGG-3′); for A94V, A94Vfwd (5′-CCG ATC CTG ACC AAT GTC GAG TAT CAG GCG GTC-3′) and A94Vrev (5′-GAC CGC CTG ATA CTC GAC ATT GGT CAG GAT CGG-3′); for V188A, V188Afwd (5′-GAT CTG AAC ATC GTC GCG GAG ACG GCG TTC AC-3′) and V188Arev (5′-GTG AAC GCC GTC TCC GCG ACG ATG TTC AGA TC-3′); and for V188L, V188Lfwd (5′-GAT CTG AAC ATC GTC CTG GAG ACG GCG TTC AC-3′) and V188Lrev (5′-GTG AAC GCC GTC TCC AGG ACG ATG TTC AGA TC-3′) (the nucleotide changes giving the appropriate mutations are shown in bold). After initial denaturation at 95°C for 0.5 min, the cycling parameters were 0.5 min at 95°C followed by 1.0 min at 55°C and 12 min at 68°C (12 cycles).
The reaction mixtures were placed on ice for 2 min, and then the parental, supercoiled double-stranded DNA was digested with 0.5 μl of DpnI at 37°C for 1 h before being transformed into competent E. coli XL1 Blue cells. Mutations were verified by DNA sequencing.
Recombinant M. smegmatis induction experiments.
For expression, the 2.1-kb NdeI-NheI fragment of wild-type pmo in pNBVpmo15 was replaced with the appropriate mutant version, followed by electroporation into M. smegmatis mc2155 (12). The resulting transformants were used as templates for colony PCRs with Accuzyme DNA polymerase (BioLine) and the primers PM3025 (5′-GTT GTC ACC GTT GCG CAC-3′) and SEA3 (5′-GCA AAT CAA CGG ATC GTC-3′). The cycling parameters were an initial melting step at 94°C for 5 min followed by 30 cycles of melting at 94°C for 30 s, annealing at 55°C for 1 min, and extension at 72°C for 2 min. A 1-kb DNA fragment confirmed the presence of the pmo insert, and sequencing of the PCR product verified the presence of the appropriate mutation(s). Recombinant strains of M. smegmatis mc2155 were grown aerobically at 28°C in 500 ml of NMS medium (32) containing 0.4% (wt/vol) sodium succinate, Tween 20 (20% [vol/vol]), and either 0.05 mg of hygromycin/ml or 0.025 mg of kanamycin/ml in a 2.5-liter flask until an optical density at 600 nm (OD600) of 0.1 was reached. One milliliter of acidified FeSO4 solution (1.12% [wt/vol]) was added to the culture, which was then incubated as described above for a further 5 h (final OD600, ∼0.5). The cells were harvested by centrifugation at 5,000 × g at 4°C for 20 min and washed sequentially with 20 ml of NMS medium and then 40 ml of ice-cold MOPS (morpholinepropanesulfonic acid) buffer (pH 7.5). The resulting cell pellet was resuspended in sufficient MOPS buffer (pH 7.5) to give a final OD600 of 20.
Monooxygenase assay. (i) Propene biotransformation.
Cell suspensions (1 ml) were assayed for propene oxidation at 30°C as previously described (33).
(ii) Styrene biotransformation assays.
For styrene biotransformation assays, the assay for propene oxidation was followed except that 10 μl of 1 M styrene in acetone was added instead of propene. After 90 min, the reaction was quenched on ice, the cells were pelleted by centrifugation (15,000 × g, 10 min), and the supernatant was extracted with 200 μl of ethyl acetate. The organic layer was dried over anhydrous sodium sulfate prior to analysis. The initial quantification of styrene oxide was done by gas chromatography-flame ionization detection (GC-FID) as described above on a 1M Tenax TA column at 200°C. For chiral separation, a 50-m by 0.25-mm FS-Lipodex C chiral capillary column (Macherey-Nagel, Duren, Switzerland), maintained isothermally at 70°C, was used, with the injector and detector maintained at 200°C and a split ratio of 1:50. Under these conditions, R- and S-styrene oxide enantiomers eluted at 44.9 and 47.1 min, respectively.
Nucleotide sequence accession number.
The sequences described in this report have been deposited with GenBank at NCBI (http://www.ncbi.nlm.nih.gov/) and are available under accession number AY455999.
RESULTS AND DISCUSSION
Molecular cloning of the mycobacterial pmoABCD gene cluster.
A 6.5-kb BamHI fragment containing the whole R. corallinus amo gene cluster (23) was used as a probe for the chromosomal DNA of Mycobacterium sp. strain M156 (data not shown). The hybridization signals were strong, suggesting significant sequence identity between the two operons. Subsequently, single sharp hybridization bands were obtained for electrophoretically separated chromosomal restriction digests with BamHI (5.2 kb), EcoRI (5.5 kb), and PstI (6.5 kb). SmaI-digested fragments gave a strong band at 6.9 kb and a faint band at 2.7 kb. The chromosomal DNA was therefore cut with EcoRI, PstI, and SmaI and separated in an agarose gel, and fragments of approximately 5.5, 6.5, and 6.9 kb, respectively, were recovered and ligated into pBluescript II. After electroporation into E. coli DH10B, 2,000 colonies from each cloning fragment were screened by colony hybridization, yielding three or four positive clones for each cloning exercise. Representative EcoRI, PstI, and SmaI fragment clones were sequenced, and the results are presented in Fig. 1A. This sequencing revealed a gene cluster with the same order as that of the R. corallinus amoABCD cluster, and the cluster was named pmoABCD. The cluster was cleaved by both PstI and SmaI but not by EcoRI. However, an ORF encoding a probable regulatory protein (OrfR) was cleaved by EcoRI, so a combination of all fragments was required to cover the entire operon.
Sequence analysis of the pmoABCD gene cluster.
Four potential genes were assigned by taking into account the higher G+C contents in their coding regions (2) and also by analogy with the translated sequence of R. corallinus AMO. The start codon of pmoA is GTG instead of the ATG for R. corallinus; it is preceded by a possible ribosomal binding site (AGGA) eight nucleotides upstream. As in the case of R. corallinus and Pseudonocardia K1 (30), the first nucleotide of the initiation codon for the coupling protein immediately precedes the stop codon of the oxygenase small subunit (Fig. 1B). No obvious ribosome binding site was found for pmoB (except an AGGGG sequence nine nucleotides upstream). An intergenic distance of 31 nucleotides separates the stop codon of pmoB and the start codon of pmoC, which is significantly more than the 11 nucleotides in the case of R. corallinus. A possible ribosome binding site (GAAAGG) is present 11 bp before pmoC. The start codon of pmoD and the stop codon of pmoC overlap by 1 bp (TGATG) (Fig. 1B), and as in the case of pmoB, no ribosome binding site was evident for pmoD. However, potential slippage codons are present 15 bp upstream of the pmoD start codon (AAAAACCCC) (Fig. 1B) and a possible 23-bp stem-loop is present 11 bp after the initiation codon of pmoD (−39.5 kcal/mol) (Fig. 1), both of which may contribute to ribosomal frameshifting (7) and therefore enable the translation of pmoD. The intergenic distance between pmoD and the next ORF is 180 bp.
The G+C content of the pmoABCD cluster is 64.0%, which is consistent with values described for other mycobacterial genes (3, 9), and its codon usage is typical of Mycobacterium spp. (data not shown). The G+C contents at silent third positions of sense codons for the pmoABCD genes are 84.1, 80.4, 88.2, and 82.7%, respectively, which were slightly closer to that of M. smegmatis than to that of Mycobacterium tuberculosis (particularly for Asn, His, Ile, Lys, Phe, and Tyr) when compared by use of a graphical codon usage analyzer (http://gcua.schoedl.de/).
Modeling of PmoC.
Similarity searches showed that PmoA, -B, -C, and -D are most similar to their counterparts from AMO of R. corallinus (23) and from ETN of Mycobacterium rhodesiae (4). However, significantly, PmoA and PmoC have 25 and 33% sequence identities with the β and α subunits of sMMO, respectively, which are sufficient to allow realistic modeling of the available structures for this enzyme. The crystal structures of the hexameric oxygenase components of sMMO from Methylococcus capsulatus and Methylosinus trichosporium have shown that the catalytic di-iron center is located at the base of a hydrophobic cavity formed by residues from four helices in the α subunit (5, 21, 22). The six iron binding residues, Glu114 (helix B), Glu144, His147 (helix C), Glu209 (helix E), Glu243, and His 246 (helix F), together with the helical structure and several other residues, are completely conserved in many monooxygenase homologues, including PmoC and AmoC (Fig. 2), and PmoC models closely to the sMMO α subunit in this region. Furthermore, electron paramagnetic resonance studies of the R. corallinus oxygenase indicated the existence of a bridged binuclear iron center (8). Given the extent of amino acid sequence identity between AmoC and PmoC and the conservation of key residues involved in iron binding, we envisage that the PMO cluster also contains a binuclear nonheme iron center.
FIG. 2.
Active-site residues of Mycobacterium sp. strain M156 β subunit (PmoC) aligned with homologs. The sequences aligned with PmoC are as follows: MEMA_METTR from Methylosinus trichosporium OB3b (GenBank accession no. P27353); MEMA_METCA from Methylococcus capsulatus Bath (GenBank accession no. P22869); AmoC from Rhodococcus corallinus B-276 (GenBank accession no. BAA07114); ETNC from M. rhodesiae JS60 (GenBank accession no. AAO48576); the rsph3375 protein, a Rhodococcus sphaeroides hypothetical oxygenase (GenBank accession no. ZP_00007442); the blr3677 protein, a Bradyrhizobium japonicum hypothetical oxygenase (GenBank accession no. BAC48942); PRMA from Gordonia sp. strain TY-5 (GenBank accession no. BAD03956); THMA from Pseudonocardia K1 (GenBank accession no. CAC10506); and TMOA_PSEME from Pseudomonas mendocina (GenBank accession no. Q00456). The alignment shows only some of the amino acids within the active site (Leu110 to Ile239), as previously defined (5, 21, 22). Ligands of the binuclear iron center are joined with black lines. The residues forming the active site are highlighted with white diamonds (⋄).The positions of Ala 94 and Val 188 in PmoC, which were subjected to mutagenesis in this study, are indicated with black triangles. Residues that are known or predicted to form helices (H) are indicated. Secondary structure information was taken from M. capsulatus Bath (pdb code 1mty chain D) and Methylosinus trichosporium (pdb code 1mhy chain D). PmoCpred is the predicted secondary structure of PmoC, as described in Materials and Methods.
Expression of pmoABCD in M. smegmatis mc2155.
Attempts to express the mycobacterial propene monooxygenase in an active form in E. coli have so far been unsuccessful. The main problem appears to be resolving the gene overlaps between pmoA and pmoB and between pmoC and pmoD, as constructs in which the two pairs of genes (pmoA-pmoB and pmoC-pmoD) were placed under inducible control only produced polypeptides of sizes corresponding to PmoA and PmoC, respectively (data not shown). Expression of the complete AMO system was achieved in Streptomyces lividans by cloning the amo operon into a thiostrepton-inducible expression plasmid, which allowed the production of AMO with comparable activity to that of the wild type (27). We therefore followed an analogous strategy of expression in the “homologous host,” M. smegmatis. For this strategy, the pmoABCD DNA fragment was subcloned into the shuttle vector pNBV1. pNBVpmo11 and pNBVpmo15 contain the whole pmoABCD cluster as a 4.6-kb fragment subcloned into pNBV1 (starting with the BamHI site 159 bp upstream of the pmoA start codon and finishing at the PstI site 529 bp downstream of the pmoD stop codon [Fig. 1A]). In construct pNBVpmo11, the entire gene cluster is under the control of the Mycobacterium bovis heat shock promoter, hsp60p. For pNBVpmo15, the orientation with respect to hsp60p was reversed for use as a negative control. Transformants of M. smegmatis mc2155 always showed a high plasmid stability for the negative control pNBVpmo15 (as assessed by restriction digests of plasmid DNAs from 10 randomly picked transformants), but transformants produced with pNBVpmo11 all contained plasmid deletions of various sizes. This was a reproducible phenomenon when equivalent constructs were made with different shuttle vectors, such as pUS1872. Subsequent studies showed that this was not limited to the pmo gene cluster, as the amo gene cluster from R. corallinus under the control of hsp60p also displayed pNBV1-based plasmid construct instabilities (data not shown). The hsp60 promoter has also been reported to induce plasmid deletions in the M. bovis BCG strain (1).
Unsurprisingly, given the high instability of the constructs, no propene oxidation activity was detected with pNBVpmo11 transformants. However, a low activity was seen with pNBVpmo15, suggesting that expression occurred from a weak, uncharacterized promoter. These experiments were done with cells grown on 7H9 medium, which is a relatively rich medium. In a minimal salts medium with supplements of iron, a much higher constitutive activity, 16.3 nmol/min/mg of protein (Fig. 1), was obtained. Cell extracts of active recombinant cells did not show any overexpressed polypeptides by SDS-polyacrylamide gel electrophoresis, unlike the situation with the wild type. This suggests that the low level of activity was the result of poor expression from an uncharacterized promoter situated in the pAL5000 component. Only ∼708 bp separates the end of pAL5000 and the initiation codon of pmoA in pNBVpmo15, and the restriction map of pNBV1 indicates that the short rap gene (29) lies immediately upstream of pmoABCD.
Site-directed mutagenesis of PmoC and stereoselectivity of PmoC isoforms.
Two glycine residues (G113 and G208) lie adjacent to the nonbridging glutamate residues that coordinate the iron center in sMMO (5, 21, 22) The equivalent residues in other nonheme iron monooxygenase enzymes are usually aliphatic, but the length (and hence bulk) of the side chain varies. It seems plausible that the increasing steric bulk of these residues may be at least partly responsible for the increasing stereoselectivity of epoxidation observed upon going from sMMO via PMO and AMO to XAMO. Therefore, we mutated A94 and V188 (the equivalent residues to G113 and G208) in the PMO α subunit to increase and decrease the steric bulk and then investigated the resultant effects on the stereoselectivity of styrene epoxidation reactions.
The A94G, A94V, V188A, and V188L mutants were constructed by site-directed mutagenesis and introduced into the same expression system, as described above. The activity of PMO in these mutant isoforms was verified under standard assay conditions, and all were found to possess propene oxidation activity at a level similar to that of the recombinant wild-type enzyme, demonstrating that A94 and V188 are not essential for catalysis. Furthermore, propene oxide was the only product obtained in these assays, indicating that there had been no loss of reaction selectivity. Additionally, whole cells of M. smegmatis mc2155 expressing the natural and mutant isoforms of pmoC were found to catalyze the oxidation of styrene to styrene oxide, again at similar levels and with styrene oxide as the only product. Whole cells of M. smegmatis mc2155 did not catalyze this reaction, nor did whole cells of M. smegmatis mc2155 transformed with a control plasmid (pNBV1).
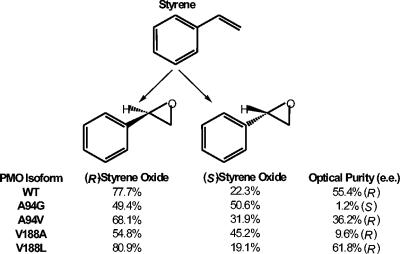
Figure 3 shows the stereoselectivities of styrene epoxidation catalyzed by the natural isoform and the mutant isoforms of PmoC described in this work. The recombinant wild-type enzyme converted styrene to styrene oxide, producing mainly the (R)-enantiomer, with an enantiomeric excess of 55.4%. The epoxidation of styrene by the mutant isoforms also predominantly produced (R)-styrene oxide, except for that of the A94G mutant, which effectively produced racemic styrene oxide [enantiomeric excess of 1.2% for the (S)-enantiomer]. Although three of the four mutations resulted in lower degrees of stereoselectivity, the V188L isoform, in which the steric bulk of the side chain was increased, gave an improved enantiomeric excess of 61.8%.
FIG. 3.
Stereoselectivities of styrene epoxidation catalyzed by the natural isoform (WT) and mutant isoforms of PMO. The results are from capillary GC-FID analysis of the reaction products obtained from resting cell biotransformation assays. Each experiment was performed in triplicate, and the mean data are presented.
The results obtained for the V188A and V188L isoforms appeared to follow a logical pattern. An increase in the volume of the side chain at this position resulted in an increase in the stereoselectivity of the reaction, while introducing a residue with a smaller side chain resulted in the stereoselectivity of the reaction being less rigorously controlled. This suggests that the residue at position 188 in PmoC can influence the positioning of the substrate during catalysis, with a smaller side chain allowing a higher degree of flexibility in substrate binding, allowing the enzyme to accept the substrate in an orientation that is less favored in the natural isoform. Residue V188 lies adjacent to a conserved glutamate residue (E189) that would coordinate with one of the nonheme iron atoms, and therefore this residue is sufficiently close to the enzyme active site to affect the orientation of the substrate with respect to the catalytic iron-bound oxygen species.
The stereoselectivities of styrene oxidation observed for the A94G and A94V isoforms are less straightforward to explain. Although the introduction of a smaller residue at this position led to a virtually complete loss in the optical purity of the product compared to the natural isoform, the converse cannot be said to be true. The A94V isoform, in which valine was substituted for alanine, exhibited a lower stereoselectivity than the wild-type enzyme. As with V188, A94 is adjacent to a conserved glutamate residue (E95) that coordinates with a nonheme iron atom. Primary sequence alignments showed that many nonheme iron monooxygenases (including methane monooxygenase, which is not known to be stereoselective) have a glycine residue that aligns with A94 in PmoC, while the propane monooxygenase from Gordonia sp. strain TY-5 (13) and two hypothetical nonheme iron monooxygenases have a proline at this position (Fig. 2). Both glycine and proline are known to disrupt protein α-helical regions. Although the crystal structures of methane monooxygenase (5, 22) indicate that the equivalent glycine (G113) lies in the middle of an α-helix, this may reflect the fact that in the absence of substrates, the surrounding four-helix bundle provides the rigidity to counteract the disruptive effect of the glycine. However, during the catalytic cycle, expansion of the four-helix bundle might allow the glycine to act as a hinge point, generating some flexibility in helix B. The enzymes with a proline at this position presumably have a permanent bend in the equivalent helix. With the notable exception of a valine in XAMO (34), most of the other oxygenases have an alanine at this position, including all of the three-component alkene monooxygenases. This would serve to reduce the flexibility of the protein backbone, which might be the real reason for the difference at A94 and would explain the lack of correlation between a change in the volume of the side chain at this position and the stereoselectivity. The equivalent residue in T4MO (G103) has already been targeted by site-directed mutagenesis, and it was reported that introducing a leucine residue in place of glycine at this position in T4MO significantly alters the regiospecificity of aromatic hydroxylation compared to the natural isoform of the enzyme (17). This could easily be the result of a change in the backbone structure resulting from a local increase in helix stability rather than a direct effect arising from the increased bulk of the amino acid side chain.
Acknowledgments
We thank W.R. Bishai for the gift of pNBV1 and J. McFadden for the gift of pUS1872.
S.A. was supported by a BBSRC Committee Studentship. C.K.C.K.C. was funded by BBSRC grant 28/T02831.
REFERENCES
- 1.Al Zarouni, M., and J. W. Dale. 2002. Expression of foreign genes in Mycobacterium bovis BCG strains using different promoters reveals instability of the hsp60 promoter for expression of foreign genes in Mycobacterium bovis BCG strains. Tuberculosis (Edinburgh) 82:283-291. [DOI] [PubMed] [Google Scholar]
- 2.Bibb, M. J., P. R. Findlay, and M. W. Johnson. 1984. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene 30:157-166. [DOI] [PubMed] [Google Scholar]
- 3.Clark-Curtiss, J. E., J. E. Thole, M. Sathish, B. A. Bosecker, S. Sela, E. F. de Carvalho, and R. E. Esser. 1990. Protein antigens of Mycobacterium leprae. Res. Microbiol. 141:859-871. [DOI] [PubMed] [Google Scholar]
- 4.Coleman, N. V., and J. C. Spain. 2003. Epoxyalkane: coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J. Bacteriol. 185:5536-5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elango, N., R. Radhakrishnan, W. A. Froland, B. J. Wallar, C. A. Earhart, J. D. Lipscomb, and D. H. Ohlendorf. 1997. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 6:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ensign, S. A., and J. R. Allen. 2003. Aliphatic epoxide carboxylation. Annu. Rev. Biochem. 72:55-76. [DOI] [PubMed] [Google Scholar]
- 7.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60:103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallagher, S. C., R. Cammack, and H. Dalton. 1997. Alkene monooxygenase from Nocardia corallina B-276 is a member of the class of dinuclear iron proteins capable of stereospecific epoxygenation reactions. Eur. J. Biochem. 247:635-641. [DOI] [PubMed] [Google Scholar]
- 9.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartmans, S., J. A. M. de Bont, and W. Harder. 1989. Microbial metabolism of short chain unsaturated hydrocarbons. FEMS Microbiol. Rev. 63:235-264. [DOI] [PubMed] [Google Scholar]
- 11.Howard, N. S., J. E. Gomez, C. Ko, and W. R. Bishai. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181-182. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 13.Kotani, T., T. Yamamoto, H. Yurimoto, Y. Sakai, and N. Kato. 2003. Propane monooxygenase and NAD(+)-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J. Bacteriol. 185:7120-7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leahy, J. G., P. J. Batchelor, and S. M. Morcomb. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27:449-479. [DOI] [PubMed] [Google Scholar]
- 15.Leak, D. J., P. J. Aikens, and M. Seyed-Mahmoudian. 1992. The microbial production of epoxides. Trends Biotechnol. 10:256-261. [Google Scholar]
- 16.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 17.Mitchell, K. H., J. M. Studts, and B. G. Fox. 2002. Combined participation of hydroxylase active site residues and effector protein binding in a para to ortho modulation of toluene 4-monooxygenase regiospecificity. Biochemistry 41:3176-3188. [DOI] [PubMed] [Google Scholar]
- 18.Miura, A., and H. Dalton. 1995. Purification and characterization of the alkene monooxygenase from Nocardia corallina B-276. Biosci. Biotechnol. Biochem. 59:853-859. [Google Scholar]
- 19.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 20.Rigby, S. R., C. S. Matthews, and D. J. Leak. 1994. Epoxidation of styrene and substituted styrenes by whole cells of Mycobacterium sp. M156. Bioorg. Med. Chem. 2:553-556. [DOI] [PubMed] [Google Scholar]
- 21.Rosenzweig, A. C., H. Brandstetter, D. A. Whittington, P. Nordlund, S. J. Lippard, and C. A. Frederick. 1997. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): implications for substrate gating and component interactions. Proteins 29:141-152. [PubMed] [Google Scholar]
- 22.Rosenzweig, A. C., C. A. Frederick, S. J. Lippard, and P. Nordlund. 1993. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 366:537-543. [DOI] [PubMed] [Google Scholar]
- 23.Saeki, H., and K. Furuhashi. 1994. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene monooxygenase. J. Ferment. Bioeng. 78:399-406. [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 25.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Small, F. J., and S. A. Ensign. 1997. Alkene monooxygenase from Xanthobacter strain Py2—purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J. Biol. Chem. 272:24913-24920. [DOI] [PubMed] [Google Scholar]
- 27.Smith, T. J., J. S. Lloyd, S. C. Gallagher, W. L. Fosdike, J. C. Murrell, and H. Dalton. 1999. Heterologous expression of alkene monooxygenase from Rhodococcus rhodochrous B-276. Eur. J. Biochem. 260:446-452. [DOI] [PubMed] [Google Scholar]
- 28.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 29.Stolt, P., Q. Zhang, and S. Ehlers. 1999. Identification of promoter elements in mycobacteria: mutational analysis of a highly symmetric dual promoter directing the expression of replication genes of the Mycobacterium plasmid pAL5000. Nucleic Acids Res. 27:396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiemer, B., J. R. Andreesen, and T. Schrader. 2003. Cloning and characterization of a gene cluster involved in tetrahydrofuran degradation in Pseudonocardia sp. strain K1. Arch. Microbiol. 179:266-277. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woodland, M. P., C. S. Matthews, and D. J. Leak. 1995. Properties of a soluble propene monooxygenase from Mycobacterium sp. (strain M156). Arch. Microbiol. 163:231-234. [Google Scholar]
- 33.Zhou, N. Y., C. K. Chan Kwo Chion, and D. J. Leak. 1996. Cloning and expression of the genes encoding the propene monooxygenase from Xanthobacter Py2. Appl. Microbiol. Biotechnol. 44:582-588. [Google Scholar]
- 34.Zhou, N. Y., A. Jenkins, C. K. Chan Kwo Chion, and D. J. Leak. 1998. The alkene monooxygenase from Xanthobacter Py2 is a binuclear non-haem iron protein closely related to toluene 4-monooxygenase. FEBS Lett. 430:181-185. [DOI] [PubMed] [Google Scholar]
- 35.Zhou, N. Y., A. Jenkins, C. K. Chan Kwo Chion, and D. J. Leak. 1999. The alkene monooxygenase from Xanthobacter strain Py2 is closely related to aromatic monooxygenases and catalyzes aromatic monohydroxylation of benzene, toluene, and phenol. Appl. Environ. Microbiol. 65:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]