Abstract
An efficient and quantitative method to analyze the transposition of various insertion sequence (IS) elements in Burkholderia multivorans ATCC 17616 was devised. pGEN500, a plasmid carrying a Bacillus subtilis-derived sacB gene, was introduced into ATCC 17616 cells, and 25% of their sucrose-resistant derivatives were found to carry various IS elements on pGEN500. A PCR-based experimental protocol, in which a mixture of several specific primer pairs was used, revealed that pGEN500 captured, in addition to five previously reported IS elements (IS401, IS402, IS406, IS407, and IS408), three novel IS elements, ISBmu1, ISBmu2, and ISBmu3. The global transposition frequency of these IS elements was enhanced more than sevenfold under a high-temperature condition (42°C) but not under oxidative stress or starvation conditions. To our knowledge, this is the first report demonstrating the elevated transposition activities of several IS elements at a high temperature. The efficient experimental protocol developed in this study will be useful in quantitatively and simultaneously investigating various IS elements, as well as in capturing novel functional mobile elements from a wide variety of bacteria.
The transposition of insertion sequence (IS) elements within a genome plays important roles in the evolution of host cells (23, 26). IS transposition usually completely inactivates a gene at a target site but sometimes leads to the constitutive expression of an adjacently located cryptic or regulated gene by delivering IS-loaded promoter sequences (16, 22, 23). Moreover, two or more copies of an identical IS element dispersed over a genome promote various genetic rearrangements, including inversion, deletion, and duplication of the intervening region, and fusion of two replicons residing in the same cell (1, 10, 23, 29). Therefore, IS transposition is one of the most important driving forces that enhance the variability and consequently the adaptive and evolutionary capacities of their hosts.
The Burkholderia cepacia complex (Bcc) consists of several species of closely related bacteria belonging to the β-subgroup of proteobacteria (5, 28). The Bcc species are widely disseminated throughout various environments, including animals, plants, and soil (3, 17, 20, 34). Such wide dissemination of the Bcc has been proposed to be due to their extraordinary metabolic versatility per se and their capability to adapt to new environments as a result of a high degree of IS-mediated genome rearrangements (21). A soil-derived Burkholderia multivorans strain, ATCC 17616 (20), which belongs to the Bcc, has three circular chromosomes and a 170-kb plasmid, pTGL1 (4, 19). This strain has been reported to carry 10 active IS elements (IS401, IS402, IS403, IS404, IS405, IS406, IS407, IS408, IS411, and IS415) (22). IS402, IS403, IS404, and IS405 have been identified based on their ability to allow the expression of a promoterless gene by providing outwardly directed promoter sequences present on the IS elements (8, 32). IS411 has been found to be an active IS element on pTGL1 (1). The remaining IS elements have been identified in a screening protocol, in which their insertion into the λcI repressor gene resulted in the derepression of the cI-repressed genes (1, 30, 35, 36). However, the available sequences of the IS elements in the public databases are limited to the complete sequences of IS401, IS402, IS406, and IS407 as well as the incomplete sequence of IS408.
The transposition of IS elements under certain growth conditions is likely to be beneficial for the survival of host cells and for the expression of new genetic traits. The aim of this study was to test whether any of several growth conditions would affect the transposition activities of the IS elements in B. multivorans ATCC 17616 cells. For this purpose, we employed a sacB-based strategy (11) for the entrapment of active IS elements from the ATCC 17616 genome and a PCR-based strategy, in which several specific primer pairs were simultaneously used to classify the entrapped IS elements. Four growth conditions were examined; we entrapped various IS elements that included three novel IS elements (ISBmu1, ISBmu2, and ISBmu3), and a high-temperature condition was found to enhance the transposition activities of several IS elements. The experimental scheme employed in this study will be generally applied for the quantitative and simultaneous detection of the transposition activities of various IS elements in a wide variety of bacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study were B. multivorans ATCC 17616 (34) and three Escherichia coli strains, JM109, S17-1, and HB101 (24). JM109 was used for the construction and preparation of plasmids. S17-1 had a chromosomal copy of a trimethoprim resistance gene (19). E. coli and B. multivorans cells, unless otherwise stated, were cultivated at 37 and 30°C, respectively. The liquid media used were M9 minimal medium containing 10% sucrose as a carbon source and 1/3 Luria-Bertani (LB) medium (6.66 g of Bacto Tryptone per liter, 3.33 g of yeast extract per liter, and 5 g of NaCl per liter). The solid media were prepared by the addition of 1.5% agar. Selective agents added to the media were as follows: kanamycin at 30 μg/ml, tetracycline (TET) at 20 μg/ml, and trimethoprim at 100 μg/ml.
Basic DNA manipulation.
Established protocols (19) were used for the following procedures: preparation of genomic and plasmid DNA, DNA digestion with restriction endonucleases, ligation, agarose gel electrophoresis, Southern hybridization, and transformation of E. coli and B. multivorans cells. PCR was performed with ExTaq DNA polymerase (TaKaRa, Kyoto, Japan) using the reaction solution provided by the manufacturer.
Entrapment of IS elements from ATCC 17616.
A TET-resistant plasmid, pNIT6012 (15), carries the replication origins of a Pseudomonas plasmid, pVS1, and an E. coli plasmid, p15A. The oriT region from p15A and the mob region from RP4 were also present on pNIT6012, therefore allowing its conjugal mobilization by supplying in trans the RK2-specified transfer genes from, for example, pRK2013 (9). A 2.7-kb SphI-HindIII fragment containing the Bacillus subtilis-derived sacB gene was excised from pMOB3 (31) and inserted into the corresponding sites of pNIT6012 to generate pGEN500 (Fig. 1A). The frozen stock cells of B. multivorans ATCC 17616(pGEN500) were streaked onto a 1/3 LB agar plate containing TET, and a single colony was cultivated in 1/3 LB broth for 12 h with vigorous shaking. A 150-μl portion of the culture was transferred to 5 ml of fresh 1/3 LB broth and incubated under various growth conditions. Viable cells in the resulting culture were counted by plating appropriate dilutions onto 1/3 LB agar plates containing TET. A portion of the same culture was plated onto M9 agar plates containing TET and 10% sucrose, and the colonies thus formed were transferred to and grown on 1/3 LB agar plates containing TET. Each colony was directly transferred by a sterilized toothpick in a microcentrifuge tube containing 20 μl of electrophoresis loading buffer with RNase A (10 μg/ml). After the subsequent addition of 20 μl of phenol-chloroform reagent, the tube was vortexed for 3 min and centrifuged for 5 min at 15,000 rpm (20,400 × g), and the supernatant was used for agarose gel electrophoresis. The colonies that contained plasmids larger than pGEN500 were retained for further analysis.
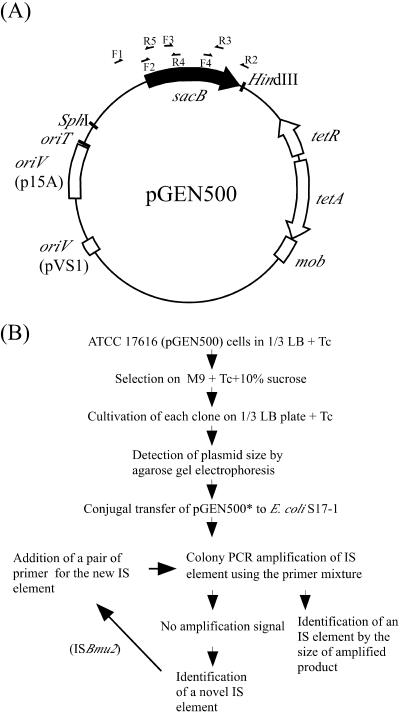
FIG. 1.
Strategy to entrap IS elements from ATCC 17616. (A) Structure of pGEN500. The arrows indicate the primer-annealing regions. F1 to F4 and R2 to R5 denote primers listed in Table 2. (B) The experimental scheme developed and used in this study. See the text for the details of this scheme. The pGEN500::IS derivative is indicated as pGEN500*. Tc, tetracycline.
Conjugal transfer of plasmid from ATCC 17616 into E. coli S17-1.
Individual ATCC 17616 clones containing the pGEN500 derivatives were grown overnight in 50 μl of 1/3 LB broth in a 96-well microtiter plate. Each 50-μl portion of the recipient (S17-1) and the helper [HB101(pRK2013)] cells grown overnight was then added to each well. The 20-μl portions of the mixture were transferred to a new 96-well microtiter plate filled with LB agar. The plate was air dried to facilitate the conjugation and incubated for 24 h at 30°C. The cells in the wells were thereafter suspended in 20 μl of LB and transferred to an LB agar plate containing TET and trimethoprim in order to select the transconjugants that received the pGEN500 derivatives.
Analysis of IS elements entrapped on pGEN500.
Classification of the IS elements entrapped on pGEN500 was carried out by colony PCR. The S17-1 cells carrying the pGEN500 derivative were transferred by a toothpick into a tube containing the PCR solution. The primers listed in Table 1 were used, each at a final concentration of 0.2 pmol/μl of reaction mixture. PCR was carried out under the following conditions: 1 cycle of 94°C for 5 min; 30 cycles of 94°C for 15 s, 60°C for 15 s, and 72°C for 30 s; and 1 cycle of 72°C for 5 min. The PCR products were analyzed by electrophoresis through 2% agarose gel, and the IS elements entrapped on pGEN500 were classified by the size of the amplified products (Table 1). When no PCR amplification of any of the expected sizes was detected, the corresponding plasmid was prepared by a standard protocol from a liquid culture. For the determination of the insertion sites of the novel IS elements on such plasmids, four PCRs that were designed to amplify different parts of the sacB region on pGEN500 were carried out (Table 2 and Fig. 1A). The primers used in these PCRs were also used for DNA sequencing.
TABLE 1.
Primers for classification of entrapped IS elements
| Target | Primer | Sequence (5′ to 3′) | Primer-annealing regiona | Expected length of the product (bp) |
|---|---|---|---|---|
| IS401 | IS401X | GAGGAGGCTCCAACTCTTGAGAGAAT | 19 to 44 | 100 |
| IS401Y | CGGTGCTCTTGCACCATGCGCA | 118 to 97 | ||
| IS402 | IS402X | GCGGTGAGCTGTTAACCTCAGGCA | 41 to 64 | 150 |
| IS402Y | AAACAGGCAGGCGGCCCGGGTTCT | 190 to 167 | ||
| IS406 | IS406X | AAGGAGTGTATCCCTCATGGCGAT | 145 to 168 | 200 |
| IS406Y | CATTGGCGTAATCGCGCCGTGTTT | 344 to 321 | ||
| IS407 | IS407X | ACAGGGCCAGCCGGAGTCTAGTAA | 22 to 45 | 250 |
| IS407Y | TCTTCAGCCGGGCATTCTCCACCT | 271 to 248 | ||
| IS408 | IS408X | GCTGTCGTGCGGCATCTGCTGA | 1552 to 1573 | 300 |
| IS408Y | GACTGCATCAACATCGTGACCTCCTT | 1851 to 1826 | ||
| ISBmu2 | ISBmu2X | GGAACGGACCCACGACGATGAAGA | 70 to 93 | 350 |
| ISBmu2Y | GCGCCGGAACTTCAACAGCGTGGT | 419 to 396 |
The numbers indicate the primer-annealing region on the IS element. The left end of each IS element was defined as position 1.
TABLE 2.
Primers to investigate the IS insertion site on pGEN500
| Primer | Sequence (5′ to 3′) | Primer-annealing regiona |
|---|---|---|
| sacB-F1 | GATTTTTTAGTTCTTTAGGCC | −311 to −291 |
| sacB-R5 | GTAAAGGTTAATACTGTTGC | 47 to 28 |
| sacB-F2 | AACATCAAAAAGTTTGCAAAAC | 4 to 25 |
| sacB-R4 | TCATTTGCATCGAATTTGTC | 458 to 439 |
| sacB-F3 | GCCGCGTCTTTAAAGACAGC | 419 to 438 |
| sacB-R3 | CGTTTAGCTCAATCATACCG | 939 to 958 |
| sacB-F4 | TTAGCAAACGGCGCTCTCGG | 922 to 941 |
| sacB-R2 | TTATTTGTTAACTGTTAATTGTCC | 1422 to 1399 |
The numbers indicate the primer-annealing region on pGEN500. The first A residue of the sacB start codon was defined as position +1.
Nucleotide sequencing and accession numbers of the IS elements.
The nucleotide sequencing was carried out using an ABI310 sequencer (Applied Biosystems Japan, Tokyo, Japan). The DNA sequences of novel IS elements were determined by primer walking. The complete nucleotide sequences of ISBmu1, ISBmu2, ISBmu3, and IS408 have been deposited in the DDBJ/EMBL/GenBank databases under the accession numbers AB159773, AB159774, AB159775, and AB182267, respectively.
RESULTS
Experimental scheme for the entrapment and identification of IS elements.
We have previously observed that the expression of the sacB gene in the presence of 10% sucrose was severely toxic to the growth of B. multivorans ATCC 17616 cells (19). This observation led us to expect that the sacB gene might be suitable to entrap active IS elements from this strain. Since pNIT6012 (15) was found to be stably maintained in ATCC 17616, the sacB gene was inserted into pNIT6012 to obtain pGEN500 (Fig. 1A).
The quantitative analysis of the transposition of various IS elements usually requires the multistep and laborious characterization of a large number of candidate plasmids that might have entrapped the IS elements (13, 14, 18, 25, 30). To overcome these inconveniences, we developed an efficient experimental scheme, depicted in Fig. 1B. The sucrose-resistant derivatives of ATCC 17616(pGEN500) were selected on M9 agar plates containing TET and 10% sucrose. The plasmids residing in these derivatives were directly extracted from the colonies and electrophoresed in agarose gel in order to examine their sizes. The plasmids larger than pGEN500 were thereafter conjugally transferred from B. multivorans ATCC 17616 to E. coli S17-1 by triparental mating using pRK2013 as a helper plasmid. The pGEN500 derivatives transferred were then analyzed by colony PCR. For this PCR analysis, a mixture of six primer pairs was designed based on the nucleotide sequences of five reported IS elements (IS401, IS402, IS406, IS407, and IS408) as well as on that of an IS element newly identified in this study (ISBmu2) (Table 1). Each primer pair was specifically able to amplify a part of one of the six IS elements, with a size unique to each element, thereby allowing the rapid identification of an IS element entrapped on pGEN500. No amplification by the colony PCR suggested the presence of novel IS elements on pGEN500. Each of the plasmids was prepared by the standard protocol, and the approximate insertion site of the IS element in each plasmid was determined by the results of four PCRs, in which four partially overlapping regions in the sacB gene on pGEN500 were targeted for amplification. No PCR amplification in one of the four regions identified the region carrying the IS element (Fig. 1A and Table 2). The primers used for the determination of the insertion site were also used for the determination of the nucleotide sequence of the entrapped IS elements.
Entrapment of IS elements and their analysis.
Since some growth conditions have been reported to affect the transposition of mobile genetic elements (23, 27), the entrapment of IS elements from ATCC 17616 was examined under the normal growth conditions (i.e., overnight growth at 30°C) and under high-temperature, oxidative stress, and starvation growth conditions (overnight growth at 42°C, overnight growth at 30°C in the presence of 0.1 mM paraquat [0.5 mM was lethal to the cells], and growth for 3 days at 30°C [which led to a 10-fold decrease in the number of viable cells], respectively). After the cultivation of ATCC 17616(pGEN500) cells under each of the four conditions, the cells were plated onto M9 medium containing TET and sucrose. For each of the four conditions, three independent experiments were performed, and 32 sucrose- and TET- resistant clones were randomly chosen in each experiment. Consequently, we collected 96 clones for each condition and 384 clones in total (Tables 3 and 4). Among the 384 clones, 97 clones had plasmids larger than pGEN500. The PCR analysis using mixed primers for IS401, IS402, IS406, IS407, and IS408 indicated that 23 plasmids carried one of the five elements (Table 4). The IS elements on the remaining 74 plasmids differed from the four previously reported IS elements, IS403, IS404, IS411, and IS415 (10, 32, 36), in terms of their size and/or restriction pattern (data not shown). Subsequent PCR and sequencing analyses revealed that 71 of the 74 plasmids had an insert of a novel 1.2-kb IS element, ISBmu2 (accession no. AB159774) and that the remaining three plasmids carried two novel IS elements, ISBmu1 (accession no. AB159773) and ISBmu3 (accession no. AB159775) (Table 4).
TABLE 3.
IS transposition frequency under several growth conditions
| Condition and experiment no. | TET-resistant cells/ml of culture (A) | Sucrose-resistant clones/ml of culture (B) | Ratio of IS-mediated-sucrose-resistant clones among 32 sucrose-resistant clones (C) | IS transposition frequencya (B × C/A) | Average | Standard deviation |
|---|---|---|---|---|---|---|
| Normal | ||||||
| 1 | 2.4 × 109 | 2.9 × 104 | 9/32 | 3.4 × 10−6 | 2.1 × 10−6 | 1.3 × 10−6 |
| 2 | 3.4 × 109 | 2.5 × 104 | 9/32 | 2.1 × 10−6 | ||
| 3 | 3.7 × 109 | 2.8 × 104 | 3/32 | 7.0 × 10−7 | ||
| Oxidative | ||||||
| 1 | 2.8 × 109 | 2.1 × 104 | 6/32 | 1.5 × 10−6 | 2.3 × 10−6 | 1.3 × 10−6 |
| 2 | 3.6 × 109 | 2.7 × 104 | 7/32 | 1.6 × 10−6 | ||
| 3 | 2.7 × 109 | 2.5 × 104 | 13/32 | 3.8 × 10−6 | ||
| High temperature | ||||||
| 1 | 3.8 × 109 | 1.1 × 105 | 9/32 | 8.6 × 10−6 | 1.6 × 10−5 | 7.2 × 10−6 |
| 2 | 3.8 × 109 | 1.9 × 105 | 10/32 | 1.5 × 10−5 | ||
| 3 | 2.1 × 109 | 1.3 × 104 | 12/32 | 2.3 × 10−5 | ||
| Starvation | ||||||
| 1 | 2.0 × 109 | 5.2 × 103 | 10/32 | 8.1 × 10−7 | 1.6 × 10−6 | 1.9 × 10−6 |
| 2 | 1.8 × 109 | 4.4 × 103 | 5/32 | 3.8 × 10−6 | ||
| 3 | 2.0 × 109 | 5.5 × 104 | 4/32 | 3.1 × 10−7 |
The number of sucrose- and TET-resistant clones per milliliter of culture (B) was multiplied by the ratio of IS-mediated-sucrose-resistant clones among 32 sucrose-resistant clones (C) and then divided by the number of TET-resistant cells per milliliter of culture (A). For each condition, three independent experiments were performed (experiments 1 to 3), and the average and standard deviation were calculated.
TABLE 4.
Distribution of IS elements entrapped under different growth conditionsa
| Growth condition | No. of plasmids carrying:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| IS401 | IS402 | IS406 | IS407 | IS408 | ISBmu1 | ISBmu2 | ISBmu3 | Total out of 96 clones | |
| Normal | 0 | 0 | 0 | 1 | 2 | 0 | 18 | 0 | 21 |
| Oxidative | 0 | 0 | 0 | 1 | 4 | 0 | 21 | 0 | 26 |
| High temperature | 2 | 1 | 6 | 1 | 4 | 0 | 15 | 2 | 31 |
| Starvation | 0 | 0 | 0 | 0 | 1 | 1 | 17 | 0 | 19 |
IS401 and IS407 belong to the IS3 family, IS402 and ISBmu2 belong to the IS5 family, IS406 belongs to the IS256 family, and IS408, ISBmu1, and ISBmu3 belong to the IS21 family.
The 3′ portion of the IS408 sequence has not yet been reported. The determination in this study of the complete sequence indicated the following characteristics regarding IS408: (i) it was 2,798 bp in size, (ii) it possessed 47-bp terminal inverted repeats (IRs) with 9-bp mismatches, (iii) it encoded the two putative transposition-related proteins, IstA and IstB, with sizes of 518 and 231 amino acids, respectively, and (iv) it generated the 7-bp duplication of its target sequences (Fig. 2). These properties supported the previously proposed classification of IS408 to the IS21 family (2). The less-conserved multiple terminal repeats commonly present at both ends of members of the IS21 family (2) were also found in three and two copies at the left and right ends, respectively (Fig. 2). There existed between the 3′ end of istB and the right end of IS408 three copies of the canonical −35 and −10 sequences of the E. coli promoter, and all of these putative promoters were rightward directed.
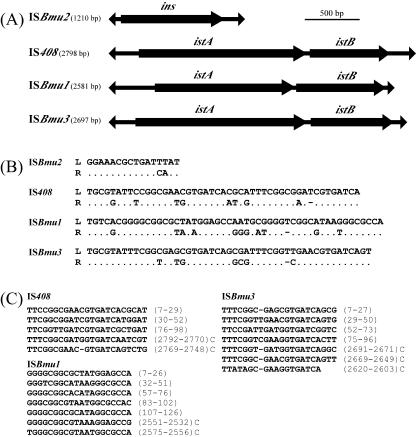
FIG. 2.
IS elements identified in this study. (A) Structures of IS408, ISBmu1, ISBmu2, and ISBmu3. (B) Nucleotide sequences of the terminal IRs. The left (L) and right (R) ends of each IS element are defined as those located upstream and downstream, respectively, of the ins or istAB genes. The same nucleotides are represented by dots. Note that some gaps (indicated by hyphens) were introduced for maximum matching. (C) The alignment of multiple terminal repeats with their coordinates in the IS21 family. Hyphens indicate gaps. C, complementary strand.
The two novel IS elements, ISBmu1 (2,581 bp) and ISBmu3 (2,697 bp), generated 5-bp target duplication, and the ends showed the 5′-TG-3′ dinucleotide and seven copies of the multiple terminal repeats that were approximately 20 bases in length (Fig. 2). ISBmu1 and ISBmu3 had long terminal IRs (i.e., 51-bp IRs with 12-bp mismatches and 49-bp IRs with 8-bp mismatches, respectively) and carried two genes (istA and istB) that were separated by 4 and 15 bases, respectively, bearing putative ribosome-binding sequences. The phylogenetic analysis of the IstA and IstB proteins of the two IS elements revealed that their IstA and IstB proteins were transposases and helper proteins, respectively, of the IS21 family. A BLAST search of the databases and subsequent phylogenetic analysis (data not shown) revealed that the IstA and IstB proteins of ISBmu1 showed the highest identities with Rv3428c (34%) and RV3427c (37%), respectively, of Mycobacterium tuberculosis H37Rv (6), and the proteins of ISBmu3 showed the highest identities with Reut5638 (73%) and Reut5637 (77%), respectively, of Ralstonia metallidurans. While the DNA region between the istB gene and the right end of ISBmu3 carried one copy of the rightward directed promoter sequence, no such sequence was found in the corresponding region of ISBmu1. ISBmu3 showed 75% nucleotide identity with IS408.
ISBmu2 was 1,210 bp in size and shared the structural properties commonly associated with the IS5 subgroup in the IS5 family, since this element (i) had the 16-bp terminal IRs with 4-bp mismatches and one orf (ins) encoding a 330-amino-acid transposase with a DDE motif and (ii) generated 4-bp duplication of the target sequences (Fig. 2). This Ins protein showed the highest (82%) identity with that of ISRso3, another member of the IS5 subgroup from Ralstonia solanacearum GMI1000. Since ISBmu2 accounted for more than 75% of the IS elements entrapped in this study, eight randomly chosen pGEN500::ISBmu2 plasmids were analyzed with respect to their insertion sites. Six out of the eight ISBmu2 inserts generated the target duplication of the CTAA sequence at five different positions in the sacB gene. This finding indicated that ISBmu2 had a preference for its target site selection.
To determine the copy numbers of IS408, ISBmu1, ISBmu2, and ISBmu3 in ATCC 17616, its genomic DNA was digested by the appropriate restriction enzymes, separated by agarose gel electrophoresis, and blotted onto a nylon membrane. Use of a portion of each IS element as a probe revealed that the ATCC 17616 genome contained three, four, nine, and two copies of IS408, ISBmu1, ISBmu2, and ISBmu3, respectively (data not shown).
Effect of growth conditions on IS transposition frequency.
In this study, the global IS transposition frequency was calculated using the following formula: (number of sucrose- and TET-resistant clones per milliliter of culture × ratio of IS-mediated-sucrose-resistant clones among 32 sucrose-resistant clones)/number of TET-resistant cells per milliliter of culture. The IS transposition frequency under the high-temperature condition (1.6 × 10−5/viable cell) was more than sevenfold higher than that observed under the three other growth conditions. Three additional experiments were further carried out to determine the global IS transposition frequencies under the high-temperature and normal growth conditions, and the transposition under the former condition was 16-fold higher than that in the latter condition (data not shown). As is shown in Table 4, the transposition of four IS elements, IS401, IS402, IS406, and ISBmu3, was observed only under the high-temperature condition, showing their preferential transposition under high-temperature conditions. This result again emphasized the induction of IS transposition at the high-temperature condition.
DISCUSSION
The experimental scheme developed in this study allowed us to do the following things: (i) screen a large number of sucrose-resistant clones without their cultivation in liquid medium and (ii) identify and classify the previously characterized IS elements by colony PCR with mixed primer pairs. Employment of this scheme led to the successful entrapment of the five previously reported IS elements as well as three novel IS elements. Our scheme was also effective for quantitatively analyzing the transposition of these eight IS elements. The scheme employed in this study will be useful for the quantitative analysis of IS transposition in other bacterial strains.
With respect to the genomes of strains belonging to the Bcc, the complete sequence of a Burkholderia cenocepacia strain (J2315) and the draft sequences of two B. cepacia strains (R1808 and R18194) are available in public databases. We surveyed the distribution of the eight ATCC 17616-derived IS elements in the three strains. IS401, IS406, IS408, and their highly homologous sequences were not found among these three strains. IS402, IS407, and their isoforms are abundant in J2315 and R1808, and the latter strain additionally carries the isoforms of ISBmu2 and ISBmu3. In contrast to R1808, R18194 did not carry any of the eight IS elements or their isoforms, suggesting that R18194 is a unique strain from a standpoint of the distribution of IS elements. Various isoforms of IS407 were also distributed in pathogenic Burkholderia species, such as Burkholderia mallei and Burkholderia pseudomallei, and in other various environmental Burkholderia strains (25), thus indicating that IS407 or its highly homologous element must have been residing in a common ancestral strain prior to diversification to establish the Bcc.
Extensive studies of prokaryotic mobile DNA elements revealed that their transposition activities were modulated by several growth conditions and that two alternative, and in some cases mutually related, mechanisms govern transposition activity (27). According to one of these mechanisms, the growth conditions control the expression of transposition-related genes and/or the activities of the gene products. In the other type of mechanism, the growth conditions regulate the activities of various fundamental host factors that subsequently modulate transposition activity. The following candidate host factors have thus far been suggested: (i) the so-called DNA chaperones IHF, HU, H-NS, and FIS, which are important for the maintenance of proper DNA and genome architectures; (ii) cold- and heat shock protein chaperone systems; and (iii) recombination systems, including the SOS system. However, it has been reported that the effects of both the growth conditions and the host factors were in general specific for each mobile element (23). Taking these effects into consideration, we investigated in this study the transposition of the B. multivorans ATCC 17616 IS elements under normal conditions as well as under the following three conditions. The first of these three conditions was starvation, since this strain was isolated from a soil environment that was poor in nutrients (33). The two other conditions were high-temperature and oxidative stress conditions. The main reason why we employed the two latter conditions was that the Bcc strains, including B. multivorans, are often associated with animals. Upon infection of animals, Bcc cells are likely to be subjected to attack by the host defense system, which generates high temperatures as well as superoxide, a typical reactive oxygen species that disrupts the bacterial DNA molecules and therefore induces the host-cell SOS system (7). Among the four growth conditions examined in this study, only the high-temperature condition led to more than a sevenfold increase in the transposition activity of all of the IS elements except for ISBmu1, and transposition of IS401, IS402, IS406, and ISBmu3 was detected only under this condition (Table 4). The higher transposition activities of these IS elements at 42°C in the ATCC 17616 cells contrasted with the decreased transposition activities of various mobile DNA elements (e.g., Tn3, IS1, IS30, and IS911) in E. coli cells at this temperature (27). The temperature sensitivities of transposition in E. coli have been considered to be the intrinsic properties of the transposases (12). The observation of a simultaneous increase in ATCC 17616 cells of the transposition activities of the seven IS elements belonging to four different IS families (Table 4) suggested that some common host factors affect transposition at high temperatures. Some unknown factors that inhibit transposition might be depleted at higher temperatures. Alternatively, other unknown factors might contribute to changing the donor and/or target DNA architecture into a form that enables the IS transposition machinery to work more effectively. Whatever the mechanism might be, the observed increase in IS transposition frequency indicated that the IS elements examined here play an important role in cells exposed to high temperatures, thus generating genetic diversity in a population of B. multivorans ATCC 17616; therefore, these IS elements also appear to be important for adaptation. To the best of our knowledge, this is the first report demonstrating an elevation in the transposition activity of several IS elements at a high temperature.
Acknowledgments
This work was supported by the Research for the Future Programs of the Japan Society for the Promotion of Science and by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The complete and preliminary sequence data for various Burkholderia genomes were obtained from the DOE Joint Genome Institute at http://www.jgi.doe.gov/JGI_microbial/html/ and from the Sanger Institute at ftp://ftp.sanger.ac.uk/pub/pathogens/.
REFERENCES
- 1.Barsomian, G., and T. G. Lessie. 1986. Replicon fusions promoted by insertion sequences on Pseudomonas cepacia plasmid pTGL6. Mol. Gen. Genet. 204:273-280. [DOI] [PubMed] [Google Scholar]
- 2.Berger, B., and D. Haas. 2001. Transposase and cointegrase: specialized transposition proteins of the bacterial insertion sequence IS21 and related elements. Cell. Mol. Life Sci. 58:403-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkholder, W. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-118. [Google Scholar]
- 4.Cheng, H. P., and T. G. Lessie. 1994. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J. Bacteriol. 176:4034-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrante, A. A., and T. G. Lessie. 1991. Nucleotide sequence of IS402 from Pseudomonas cepacia. Gene 102:143-144. [DOI] [PubMed] [Google Scholar]
- 9.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaffney, T. D., and T. G. Lessie. 1987. Insertion-sequence-dependent rearrangements of Pseudomonas cepacia plasmid pTGL1. J. Bacteriol. 169:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gay, P., D. Le Coq, M. Steinmetz, T. Berkelman, and C. I. Kado. 1985. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 164:918-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haren, L., M. Betermier, P. Polard, and M. Chandler. 1997. IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol. Microbiol. 25:531-540. [DOI] [PubMed] [Google Scholar]
- 13.Hasebe, A., and S. Iida. 2000. The novel insertion sequences IS1417, IS1418, and IS1419 from Burkholderia glumae and their strain distribution. Plasmid 44:44-53. [DOI] [PubMed] [Google Scholar]
- 14.Hasebe, A., S. Tsushima, and S. Iida. 1998. Isolation and characterization of IS1416 from Pseudomonas glumae, a new member of the IS3 family. Plasmid 39:196-204. [DOI] [PubMed] [Google Scholar]
- 15.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 16.Hübner, A., and W. Hendrickson. 1997. A fusion promoter created by a new insertion sequence, IS1490, activates transcription of 2,4,5-trichlorophenoxyacetic acid catabolic genes in Burkholderia cepacia AC1100. J. Bacteriol. 179:2717-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 18.Jäger, W., A. Schäfer, J. Kalinowski, and A. Pühler. 1995. Isolation of insertion elements from gram-positive Brevibacterium, Corynebacterium and Rhodococcus strains using the Bacillus subtilis sacB gene as a positive selection marker. FEMS Microbiol. Lett. 126:1-6. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu, H., Y. Imura, A. Ohori, Y. Nagata, and M. Tsuda. 2003. Distribution and organization of auxotrophic genes on the multichromosomal genome of Burkholderia multivorans ATCC 17616. J. Bacteriol. 185:3333-3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessie, T. G., and T. D. Gaffney. 1986. Catabolic potential of Pseudomonas cepacia, vol. 10. AcademicPress, Inc., Orlando, Fla.
- 21.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 22.Lessie, T. G., M. S. Wood, A. Byrne, and A. Ferrante. 1990. Transposable gene-activating elements in Pseudomonas cepacia, p. 279-291. In S. Silver, A. M. Chakrabarty, B. Iglewski, and S. Kaplan (ed.), Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. American Society for Microbiology, Washington, D.C.
- 23.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Miche, L., D. Faure, M. Blot, E. Cabanne-Giuli, and J. Balandreau. 2001. Detection and activity of insertion sequences in environmental strains of Burkholderia. Environ. Microbiol. 3:766-773. [DOI] [PubMed] [Google Scholar]
- 26.Mira, A., L. Klasson, and S. G. Andersson. 2002. Microbial genome evolution: sources of variability. Curr. Opin. Microbiol. 5:506-512. [DOI] [PubMed] [Google Scholar]
- 27.Nagy, Z., and M. Chandler. 2004. Regulation of transposition in bacteria. Res. Microbiol. 155:387-398. [DOI] [PubMed] [Google Scholar]
- 28.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 29.Riehle, M. M., A. F. Bennett, and A. D. Long. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider, D., D. Faure, M. Noirclerc-Savoye, A. C. Barriere, E. Coursange, and M. Blot. 2000. A broad-host-range plasmid for isolating mobile genetic elements in gram-negative bacteria. Plasmid 44:201-207. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer, H. P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195-1204. [DOI] [PubMed] [Google Scholar]
- 32.Scordilis, G. E., H. Ree, and T. G. Lessie. 1987. Identification of transposable elements which activate gene expression in Pseudomonas cepacia. J. Bacteriol. 169:8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic Pseudomonas: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 34.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 35.Wood, M. S., A. Byrne, and T. G. Lessie. 1991. IS406 and IS407, two gene-activating insertion sequences for Pseudomonas cepacia. Gene 105:101-105. [DOI] [PubMed] [Google Scholar]
- 36.Wood, M. S., C. Lory, and T. G. Lessie. 1990. Activation of the lac genes of Tn951 by insertion sequences from Pseudomonas cepacia. J. Bacteriol. 172:1719-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]