Abstract
The present study reports on fibrinogen (Fg) binding of Staphylococcus epidermidis. Adhesion of different S. epidermidis strains to immobilized Fg was found to vary significantly between different strains, and the component responsible was found to be proteinaceous in nature. To further characterize the Fg-binding activity, a shotgun phage display library covering the S. epidermidis chromosome was constructed. By affinity selection (panning) against immobilized Fg, a phagemid clone, pSEFG1, was isolated, which harbors an insert with an open reading frame of ∼1.7 kilobases. Results from binding and inhibition experiments demonstrated that the insert of pSEFG1 encodes a specific Fg-binding protein. Furthermore, affinity-purified protein encoded by pSEFG1 completely inhibited adhesion of S. epidermidis to immobilized Fg. By additional cloning and DNA sequence analyses, the complete gene, termed fbe, was found to consist of an open reading frame of 3,276 nucleotides encoding a protein, called Fbe, with a deduced molecular mass of ∼119 kDa. With a second phage display library made from another clinical isolate of S. epidermidis, it was possible to localize the Fg-binding region to a 331-amino-acid-long fragment. PCR analysis showed that the fbe gene was found in 40 of 43 clinical isolates of S. epidermidis. The overall organization of Fbe resembles those of other extracellular surface proteins of staphylococci and streptococci. Sequence comparisons with earlier known proteins revealed that this protein is related to an Fg-binding protein of Staphylococcus aureus called clumping factor.
Staphylococcus epidermidis and other coagulase-negative staphylococci have been found to be among the most common etiological agents for infections associated with foreign bodies. In a study of incidences of surgical site infections (ranging between 1 and 2.5%), staphylococci accounted for 30 to 40% (36). S. epidermidis is also a common cause of peritonitis among patients undergoing peritoneal dialysis (34) and is often found in neonatal infections (23).
It has been hypothesized that adherence of S. epidermidis is a two-step reaction, in which initial attachment is mainly mediated by hydrophobicity, whereas slime production is important as a secondary step (6). The hydrophobicity seems to be correlated to cell surface proteins, since both adherence to biomaterial and hydrophobicity are reduced by protease treatment. Timmerman et al. (31) and Veenstra et al. (33) have described a cell surface protein that mediates adherence to polystyrene. Also a correlation between the production of a polysaccharide/adhesin and adherence to plastic biomaterial has been reported (21, 32). Another adhesion mechanism used by staphylococci involves their interaction with plasma proteins. Precoating of surfaces in vitro with various plasma proteins, such as albumin, fibrinogen (Fg), and fibronectin, had a blocking effect on early adhesion for most of the S. epidermidis strains tested (6). A decreased binding of S. epidermidis to Fg-coated Dacron was also found by Zdanowski et al. (38). However, in contrast to this finding, it has been clearly shown that several strains of S. epidermidis have a capacity to adhere to immobilized Fg (1, 5, 8, 19, 25, 37).
In this report, we study the Fg-binding activity of S. epidermidis. The binding was found to be dependent on a surface-located protease-sensitive component(s). Therefore, a shotgun phage display library containing chromosomal DNA from a clinical S. epidermidis strain was constructed. The library was affinity selected (panned) against immobilized Fg, which resulted in a specific enrichment of Fg-binding phagemid particles. The inserts of the phagemids were analyzed and found to be identical. By using this insert as a probe, the complete gene, termed fbe, encoding an Fg-binding protein was isolated and characterized. Interestingly, the encoded protein shows similarities to a cell wall-bound Fg-binding protein of Staphylococcus aureus called clumping factor (ClfA), a protein considered to be involved in the virulence of this species (16–18, 20).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. epidermidis HB was obtained from Åsa Ljung, Lund University, Lund, Sweden. This strain was isolated from a human patient with osteomyelitis. S. epidermidis strains 2, 19, 269, and 333 were isolated from patients with peritonitis. The S. epidermidis strains were typed with the API-Staph system (BioMerieux, Lyon, France). S. aureus Newman was used as a control in adhesion experiments. The phagemid pG8H6 (10) was used to construct the phage display library. For additional cloning, the plasmid pUC18 was used. As hosts, the Escherichia coli strains MC1061 [hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi] and TG1 [supE hsdΔ5 thi Δ(lac-proAB) F′(traD36 proAB+ lacIq lacZΔM15] were used. S. epidermidis HB was grown on blood agar plates or in broth culture with tryptone soya broth (Oxoid, Basingstoke, Hampshire, United Kingdom). The E. coli strains were grown in Luria-Bertani (LB) medium supplemented when appropriate with 100 μg of ampicillin per ml or alternatively on LA plates (LB medium supplemented with 1.5% agar and 50 μg of ampicillin per ml). All incubations were at 37°C.
Proteins and reagents.
Human Fg was obtained from IMCO Corporation, Ltd. (Stockholm, Sweden), and anti-human Fg rabbit immunoglobulin G (IgG) conjugated to horseradish peroxidase (HRP) was purchased from Dakopatts A/S, Denmark. Bovine serum albumin (BSA; fraction V, radioimmunoassay grade) was from U.S. Biochemicals (Cleveland, Ohio). Bovine collagen type I and proteinase K were obtained from Boehringer GmbH, Mannheim, Germany. Human serum albumin (HSA), bovine fibronectin, and human transferrin were purchased from Sigma (St. Louis). Human IgG was obtained from Kabi Vitrum (Stockholm, Sweden). Molecular weight markers used in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were obtained from Bio-Rad (Richmond, Calif.). Nitrocellulose (NC) filters (BA-S 85; 0.45-μm pore size) used for Southern and Western blots were from Schleicher and Schuell (Dassel, Germany). Sterile filters (Minisart N; 0.45-μm pore size) were obtained from Sartorius AG (Göttingen, Germany).
Adherence of S. epidermidis to immobilized Fg.
Strains of S. epidermidis were grown on blood agar plates overnight. The bacteria from one plate were harvested with 5 ml of phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.4 mM KH2PO4 [pH 7.4]) and washed once, and the optical density at 600 nm (OD600) was adjusted to 1.0. The adherence was measured as follows. Microtiter wells (Nunc, Copenhagen, Denmark) were coated with Fg in PBS overnight at concentrations ranging from 0.005 to 10 μg/ml. Blocking was done with 2% BSA in PBS for 1 h at 37°C. After washing, bacteria (100 μl) were added and allowed to adhere for 2 h at 37°C. Following additional washing, the microtiter plates were air dried, and bacterial adherence was determined by optical reading with a microtiter plate reader at A405. The reading range was 0.00 to 0.20, with negligible background.
Reduction of binding after protease treatment of bacteria.
Bacteria were treated for 30 min at 37°C with protease K at concentrations ranging from 0.1 ng/ml to 1 mg/ml. After protease treatment, the cells were extensively washed, and 100 μl (OD600 of 1) was transferred to wells coated with Fg (coating concentration, 10 μg/ml) as described above. Four strains of S. epidermidis (2, 19, 269, and HB) and one strain of S. aureus (Newman) were used in this experiment.
Construction of phagemid libraries of S. epidermidis.
All DNA manipulations were performed by standard methods (27). The shotgun phage display library was constructed essentially as described by Jacobsson and Frykberg (9, 10). In short, chromosomal DNA of S. epidermidis HB was prepared and fragmented by sonication. After different time intervals, aliquots were analyzed on an agarose gel. A sample with the majority of the DNA fragments in the size of ∼500 bp was, without prior size fractionation, made blunt ended by using T4 DNA polymerase. The DNA fragments were ligated with the Ready-To-Go T4 DNA ligase kit (Pharmacia Biotech, Uppsala, Sweden) into SmaI-digested and dephosphorylated pG8H6 vector (10). Electrotransformation of the ligated material into E. coli TG1 cells gave ∼4 × 107 ampicillin-resistant transformants. Part of an overnight culture (5 ml) of the electroporated bacteria was infected with helper phage R408 at a multiplicity of infection (MOI) of 20 and poured together with 0.5% soft agar onto 20 LA plates. After incubation overnight, the phage particles were eluted from the soft agar by vigorous shaking. The suspension was centrifuged (15,000 × g), followed by sterile filtration, and the titer of the phage display library was determined to be 1.3 × 1010 CFU/ml.
A phage display library of S. epidermidis 19 was constructed in principle as described for strain HB, except that instead of using the vector pG8H6, a newly developed phagemid vector called pG8SAET modified from pG8SPA1 (11) was used. The number of transformants after ligation and electrotransformation was calculated to be ∼5 × 107. Infection with the helper phage resulted in a library with a titer of 3 × 1010 CFU/ml.
Panning of the phagemid library and identification of an Fg-binding clone.
Microtiter wells (Maxisorp; Nunc, Copenhagen, Denmark) were coated overnight at 4°C with 200 μl of human Fg at a concentration of 100 μg/ml in 0.05 M NaHCO3 (pH 9.7). The wells were blocked with PBS–0.05% Tween 20 (PBST) containing BSA (final concentration, 1 mg/ml) for 1 h at room temperature. After washing with PBST, 600 μl of the phagemid library was added to three coated wells, and the wells were incubated overnight at 4°C. Before elution, the wells were extensively washed with PBST at room temperature and then stepwise eluted with 200-μl buffer solutions consisting of 50 mM Na-citrate and 150 mM NaCl with decreasing pH (5.4, 3.4, and 1.9). The eluates were neutralized by the addition of 75 μl of 2 M Tris-HCl (pH 8.6). From the eluates, 50 μl was used to infect 20 μl of E. coli TG1 cells (overnight culture) supplemented with approximately 100 μl of LB medium. After 20 min of incubation at 37°C, the cells were spread on LA plates containing 2% glucose. The plates were incubated overnight, and colonies corresponding to the two lowest pH elutions were resuspended in LB medium and pooled together. After infection with helper phage R408 at an MOI of 20, the sample was mixed with 5 ml of 0.5% LB soft agar and poured on an LA plate. After incubation overnight, the phagemid particles were eluted and subjected to another round of panning as previously described (9, 10). Finally, after the second panning, individual clones were grown on a small scale for preparation of phagemid DNA in order to sequence the inserts and one such clone, called pSEFG1, was chosen for further studies.
Activity of phagemid particles of pSEFG1.
A phagemid stock of pSEFG1 was prepared as follows. Five hundred microliters of E. coli TG1 cells harboring the phagemid was infected with helper phage R408 (MOI of 20). After propagation in soft agar on an LA plate, the phagemid particles were eluted as described above. The phage stock generated (2 × 109 CFU/ml) was used in an inhibition experiment and to analyze the binding specificity of the phagemid particles. In the binding specificity experiment, the phage stock was panned in duplicates (200 μl/microtiter well) against immobilized Fg, transferrin, fibronectin, collagen type I, IgG, HSA, and plastic (untreated wells) for 3 h at room temperature. The wells were extensively washed with PBST and subsequently eluted by lowering the pH to 1.9. Following neutralization with 2 M Tris-HCl (pH 8.6), aliquots of the eluted phagemid particles were used to infect E. coli TG1 cells and plated on LA plates supplemented with 2% glucose.
In the inhibition experiment, various concentrations of Fg and HSA were separately mixed with 9 × 105 phagemid particles of pSEFG1. After 1 h of incubation at room temperature, the samples (200 μl) were transferred to Fg-coated microtiter wells (100 μg/ml), followed by 3 h of incubation at room temperature. The wells were washed and phagemid particles eluted as described for the binding specificity experiment. E. coli TG1 cells were infected in duplicates corresponding to each concentration of Fg and HSA and plated on LA plates supplemented with 2% glucose.
SDS-PAGE and Western blot analysis.
E. coli MC1061 harboring the phagemid pSEFG1 was grown overnight, diluted 1:10 in LB medium supplemented with ampicillin, and grown to an OD600 of 1.0. As controls, the E. coli host cells, with or without pG8H6, were used. The cultures were induced by the addition of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and incubated for an additional 3 h, whereupon the cells were pelleted, washed twice with 0.01 M Tris-HCl (pH 8.1), and resuspended in a buffer containing 0.03 M Tris-HCl (pH 8.1), 20% (wt/vol) sucrose, and 1 mM EDTA. After 10 min at 37°C, cells were collected and resuspended in ice-cold 0.5 mM MgCl2 for 10 min. After centrifugation, the supernatants were collected and sterile filtered. The released proteins were acetone precipitated, dissolved in PBS, and boiled in an equal amount of sample buffer containing 5% β-mercaptoethanol and 2.5% SDS before being applied to an 8 to 25% gradient SDS-PAGE gel with the PHAST system (Pharmacia Biotech). To transfer the separated proteins, an NC filter was placed on top of the gel at 65°C. After 30 min, the filter was soaked in PBST for 1 h at 37°C. The filter was then incubated for 2 h in PBST containing human Fg (20 μg/ml) at room temperature. After being washed with PBST, the filter was incubated with HRP-labelled rabbit anti-human Fg antibodies (dilution of 1:1,000 in PBST). After 1 h at room temperature, the filter was washed in PBST, and the bound HRP-labelled antibodies were detected with 4-chloro-1-naphthol (Serva, Heidelberg, Germany) as a substrate.
Inhibition of S. epidermidis adherence to Fg by the encoded polypeptide of pSEFG1.
The fact that the insert of pSEFG1 N terminally is fused to a histidine tag (six residues) of the vector was used to purify the encoded polypeptide of pSEFG1 by using the HisTrap kit obtained from Pharmacia Biotech. One hundred microliters of the affinity-purified protein at various concentrations was added to Fg (10 μg/ml)-coated microtiter wells. An unrelated affinity-purified histidine fusion protein was separately added as a control. Binding was allowed for 1 h at 37°C before addition of 106 radiolabelled cells of S. epidermidis 19. After further incubation for 2 h, the wells were washed to remove nonadherent bacteria. Bound bacteria were released from the wells by addition of 50 μl of 3% SDS and quantified by scintillation counting. Radiolabelling was done by growing the bacteria for 5 h in 10 μCi of [3H]thymidine per ml (specific activity, 80 Ci/mol) (Amersham, Buckinghamshire, United Kingdom).
DNA sequencing and homology studies.
The nucleotide sequence of the fbe gene was determined with an ABI PRISM dye terminator cycle sequencing ready reaction kit and the ABI Model 373A DNA sequencer. Alternatively, the Thermo Sequenase fluorescence-labelled primer cycle sequencing kit and ALFexpress DNA sequencer (Pharmacia Biotech) were used. Specific synthetic primers were purchased from Pharmacia Biotech. Computer programs from the PCGENE, DNA, and protein sequence analysis software package (Intelligenetics, Inc., Mountain View, Calif.) were used to record and analyze the sequence data. The databases GenBank, EMBL, Swissprot, and PIR were screened for sequence homologies. The program PALIGN in PCGENE was used for homology studies between the Fbe and ClfA proteins using the structure-genetic matrix with an open gap cost and unit gap cost of 10, respectively.
Detection of fbe in strains of S. epidermidis.
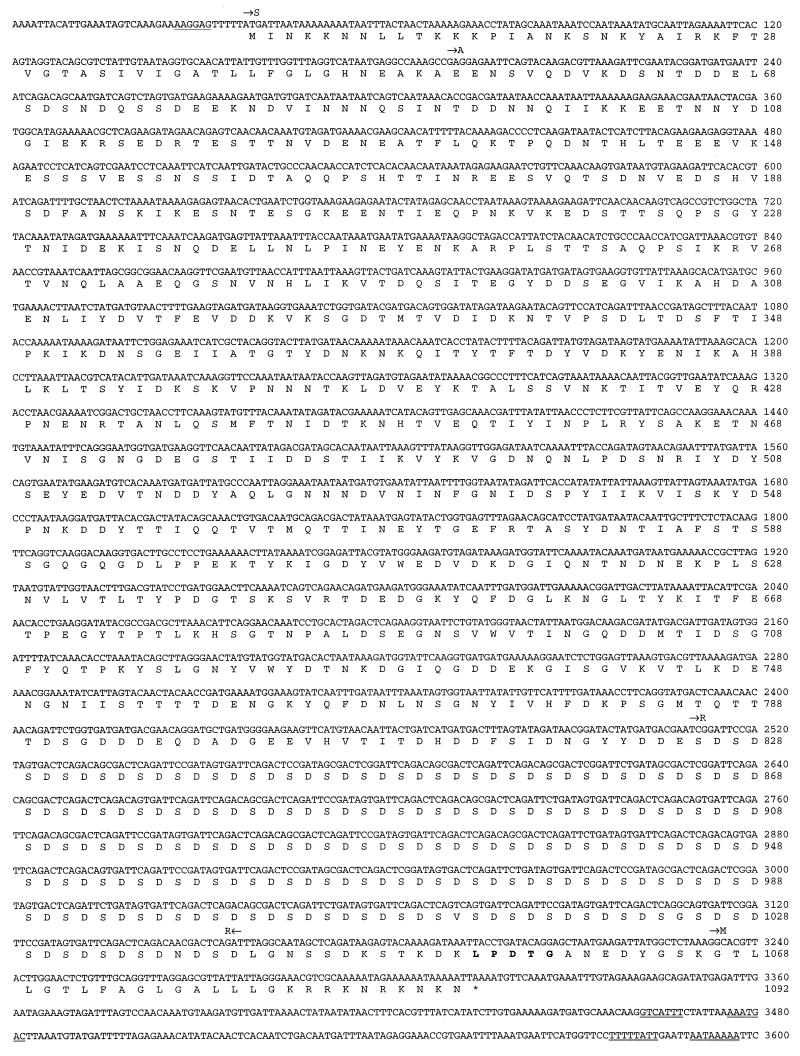
Genomic DNA was prepared from clinical strains of S. epidermidis with a QIA amp tissue kit obtained from Qiagen (Hilden, Germany), with the modification that lysostaphin was used in solution 1. The presence of the fbe gene was detected by PCR with 5′-TAAACACCGACGATAATAACCAAA-3′ as the upstream primer (corresponding to nucleotides 306 to 329 in Fig. 4) and 5′-GGTCTAGCCTTATTTTCATATTCA-3′ as the downstream primer (corresponding to nucleotides 801 to 778 in the complementary strand in Fig. 4). The reactions were amplified for 30 cycles consisting of a 30-s denaturation period at 94°C, a 1-min annealing period at 60°C, and a 1-min extension period at 72°C. After amplification, the samples were analyzed on an agarose gel.
FIG. 4.
Complete nucleotide sequence of the fbe gene from S. epidermidis HB and the deduced amino acid sequence of the encoded protein. A putative ribosomal-binding site (RBS) is underlined, and possible transcription termination hairpin loops are double underlined. The putative signal sequence (S) is followed by the nonrepetitive N-terminal region (A), which harbors the Fg-binding activity. R indicates the highly repetitive region. The amino acid sequence LPDTG, assumed to be involved in cell wall anchoring, is printed in boldface. M indicates the membrane-spanning region, and the translational stop codon is marked with an asterisk.
Nucleotide sequence accession number.
The novel nucleotide sequence of the fbe gene has been deposited in the EMBL sequence data bank and is available under accession no. Y17116.
RESULTS
Adherence of S. epidermidis to Fg.
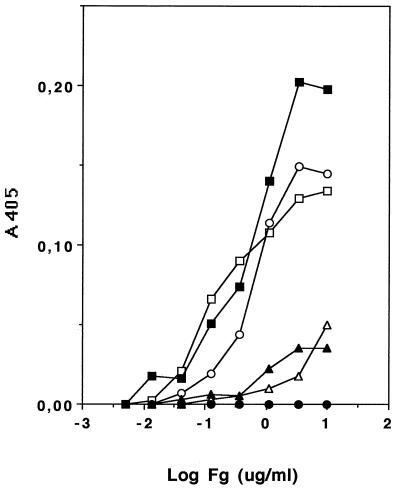
A collection of S. epidermidis strains were screened for their ability to bind immobilized Fg. The protein was immobilized at various concentrations in microtiter wells, and after binding and washing, the bacteria adhering to the wells were measured by the turbidity and light scattering caused by bound bacteria. The result showed a great variation between strains, a finding that can be used to group the strains into three categories: non-, medium-, or high-binders. The adherence values for five strains of S. epidermidis (2, 19, 269, 333, and HB) representing the three categories and S. aureus Newman are presented in Fig. 1, where the adherence values, ranging from 0.00 to 0.20, as a function of coating concentration of Fg are shown. In a separate test, bacteria were treated with protease K and washed prior to addition to immobilized Fg. Four different strains of S. epidermidis (2, 19, 269, and HB) and one strain of S. aureus (Newman) were used in this experiment. All strains tested showed complete loss of Fg binding as a result of the protease treatment (data not shown).
FIG. 1.
Bacterial binding to immobilized Fg. Microtiter wells were coated with fibrinogen at the concentration indicated and blocked with BSA. Adherence was allowed for 2 h, microtiter plates were washed and dried, and relative bacterial adherence was determined spectrophotometrically (A405). □, S. aureus Newman; ○, S. epidermidis 2; ▪, S. epidermidis 19; ▵, S. epidermidis 269; •, S. epidermidis 333; ▴ S. epidermidis HB.
Identification of a phagemid clone displaying specific Fg-binding activity.
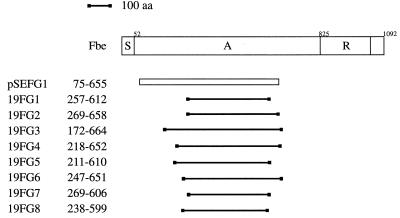
A shotgun phage display library was made with fragmented chromosomal DNA from strain HB. The phagemid library was affinity selected against Fg. The phage stock obtained after the first panning was panned against both Fg and the unrelated protein, BSA. Approximately 20 times more phage were bound to Fg than to BSA, suggesting that the binding was specific (data not shown). From the second panning, eight phagemid clones were chosen for further studies. DNA sequence analysis of the junction between the insert and vector showed that seven of the eight clones examined had an identical insert with an open reading frame in both ends of the inserted fragment. Restriction enzyme cleavage revealed an insert of ∼1.7 kb. One phagemid clone, called pSEFG1, was chosen for further studies (Fig. 2).
FIG. 2.
Schematic presentation of the Fbe protein and alignment of inserts from nine phagemid clones obtained after panning against Fg. The different regions are indicated by S (the signal sequence), A (the Fg-binding region), and R (the highly repetitive region). The insert of the single clone (pSEFG1) originated from strain HB is shown as an open bar, while the eight clones derived from strain 19 are presented by solid lines. The numbers indicate the positions of amino acids (aa) in the Fbe protein as defined in the legend to Fig. 4.
Characterization of pSEFG1.
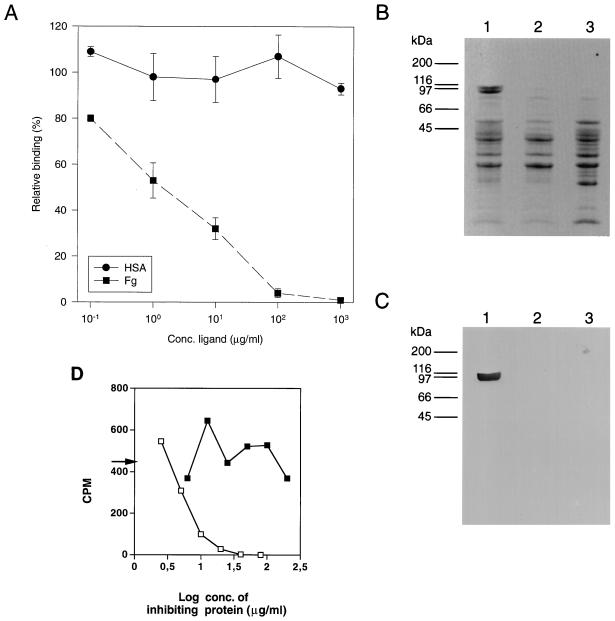
To investigate the binding activity encoded by pSEFG1, E. coli TG1 cells harboring the phagemid were infected with helper phage R408. The generated phage stock was separately panned against six different host proteins and against plastic (uncoated microtiter wells). The proteins used in the assay were collagen type I, Fg, fibronectin, HSA, IgG, and transferrin (Table 1). The binding of phagemid particles was more than 1,000 times higher when panned against Fg than when panned against any of the other proteins or plastic. In addition, an inhibition experiment was performed. Samples of the phage stock were separately preincubated with various concentrations (100 ng/ml to 1 mg/ml) of Fg or HSA. After incubation, the samples were transferred to microtiter wells coated with Fg. The result showed that pretreatment with soluble Fg completely inhibited the binding of the phagemid particles to immobilized Fg (Fig. 3A). The Fg-binding activity of the polypeptide encoded by pSEFG1 was also studied in a Western blot. The phagemid (pSEFG1) was transformed into the nonsuppressive E. coli host MC1061, which results in expression of the insert without fusion to the phage coat protein VIII. After induction with IPTG, the E. coli cells were harvested and treated by an osmotic shock procedure described in Materials and Methods. As shown in Fig. 3B and C, the result confirms the expression of a specific Fg-binding protein in MC1061 harboring pSEFG1. Since the insert of pSEFG1 is a fusion with six histidine residues originating from the vector, the expressed protein was affinity purified by immobilized metal ion adsorption chromatography. The purified protein was allowed to bind at various concentrations to Fg immobilized in microtiter wells prior to addition of radiolabelled cells of S. epidermidis 19. After incubation, the wells were washed, and bound bacterial cells were released by addition of SDS. The result showed that the purified protein of pSEFG1 completely inhibited the binding in contrast to an unrelated histidine fusion protein used as a control (Fig. 3D).
TABLE 1.
Results from panning of a phage stock (pSEFG1) against immobilized ligands
| Ligand | No. of phagemid particles/ml of eluate (pH 1.9)a |
|---|---|
| Fibrinogen | 2.2 × 107 ± 2.2 × 106 |
| Transferrin | 2.1 × 103 ± 1.4 × 102 |
| Fibronectin | 1.0 × 104 ± 7.8 × 102 |
| Collagen (type I) | 1.6 × 103 ± 3.5 × 102 |
| IgG | 2.8 × 103 ± 1.4 × 103 |
| HSA | 2.6 × 103 ± 6.3 × 102 |
| Plastic | 6.7 × 103 ± 1.4 × 103 |
Determined after infection of E. coli TG1 cells as CFU on LA plates supplemented with ampicillin. Values are means ± standard deviations (two samples from two separate microtiter wells).
FIG. 3.
(A) Inhibition of binding of phagemid (pSEFG1) particles to microtiter wells coated with Fg. Various concentrations of Fg and HSA were separately mixed with 9 × 105 phagemid particles of pSEFG1. After 1 h of incubation, the samples (200 μl) were transferred to Fg-coated microtiter wells after 3 h of incubation at room temperature. The wells were washed with PBST and subsequently eluted by lowering the pH to 1.9. Aliquots of the eluted phagemid particles were used to infect E. coli TG1 cells and plated on LA plates supplemented with 2% glucose. The resulting number of CFU per milliliter is shown as a function (percentage) of different concentrations of the two soluble plasma proteins added. Points representing the means of duplicates and standard deviations are indicated. (B) SDS-PAGE. Material released by an osmotic shock procedure from an overnight culture of E. coli MC1061 harboring the phagemid pSEFG1 was concentrated by acetone precipitation. E. coli MC1061 cells, with or without the phagemid vector pG8H6, were used as negative controls. After centrifugation, the pellets were resuspended in PBS and boiled in a sample buffer containing SDS and β-mercaptoethanol. Lanes: 1, E. coli MC1061(pSEFG1); 2, E. coli MC1061(pG8H6); 3, E. coli MC1061. Molecular mass markers are indicated. (C) Western blot analysis. After separation by SDS-PAGE, the samples were transferred to NC filters and analyzed for binding of Fg with HRP-labelled anti-Fg antibodies. (D) Inhibition of the adherence of S. epidermidis (strain 19) to immobilized Fg by purified protein of pSEFG1. Microtiter wells coated with Fg were incubated with the indicated amounts of affinity-purified proteins prior to addition of radiolabelled bacteria. After washing, bacterial adherence was determined by scintillation counting. An arrow shows the value in the absence of fusion proteins. □, purified protein expressed by pSEFG1; ▪, purified unrelated histidine fusion protein.
Characterization of fbe.
To isolate the complete gene encoding the Fg-binding protein of S. epidermidis, a Southern blot analysis was performed with chromosomal DNA from strain HB. An ∼1.3-kb radioactively labelled PCR product of the insert in pSEFG1 was used as a probe. The probe hybridized to an ∼6-kb XbaI fragment (data not shown). This fragment was subsequently ligated into pUC18, and the insert of this plasmid, called pSEFG2, was characterized. Sequence analysis revealed an open reading frame of 3,276 nucleotides starting with an ATG codon at nucleotide position 38 and ending with a TAA at position 3314 (Fig. 4). The open reading frame is preceded by a sequence typical for a ribosome-binding site of gram-positive cocci and is followed by sequences resembling transcriptional termination. The gene, termed fbe, encodes a protein of 1,092 amino acid residues, called Fbe. The deduced protein has a calculated molecular mass of ∼119 kDa. Analysis by the method of von Heijne (35) identified a possible signal cleavage site between amino acids 51 and 52, resulting in a mature protein of 1,041 amino acids with a calculated molecular mass of ∼114 kDa. Following the signal sequence, there is a region, called A, of 773 amino acids. The insert in pSEFG1 contains the sequence corresponding to residues 75 to 655 of the A region (Fig. 2 and 4). The A region is followed by a highly repetitive region of 216 amino acid residues composed of tandemly repeated aspartic acid and serine residues, called R (Fig. 4). The dipeptide region consists of an 18-bp sequence unit (consensus of TCX GAX TCX GAX AGX GAX) repeated 36 times. The 18-bp sequence unit is maintained almost perfectly throughout the whole R region, except for the second unit, which is truncated, consisting of only 12 of the 18 bp and the 3′ end of the R region, in which the consensus sequence is slightly disrupted (units 32, 34, and 36). The changes in these units also result in amino acid exchanges. The C-terminal part of the protein contains several of the features found in gram-positive cell surface-bound proteins (13, 24, 30). The motif LPXTG, shown to be involved in cell wall anchoring (28, 29), is found at position 1053 as LPDTG (Fig. 4). This sequence is followed by a stretch of 17 hydrophobic amino acids, called M, which is proposed to span the cell membrane. The deduced protein ends in a stretch of charged amino acid residues.
Protein Fbe shows sequence similarities to an Fg-binding protein of S. aureus.
With the deduced amino acid sequence of protein Fbe (except for the R region), several protein databases were screened for sequence similarities. Interestingly, the search showed that by far the highest score obtained was for the clumping factor (ClfA), an extracellular protein of S. aureus (16–18). This cell wall-bound protein binds Fg and has been shown to promote aggregation of bacteria in the presence of Fg. Various alignments of ClfA and Fbe were done with the computer program PALIGN (22). The signal sequence and the C-terminal part, including the cell membrane-spanning region of Fbe, show similarity to the corresponding regions in ClfA of 64 and 44%, respectively. In the A regions of Fbe and ClfA, the highest similarity (45%) is located between amino acid positions 373 to 516 and 317 to 460, respectively (Fig. 5). In addition, the most obvious similarity to the clumping factor is the repetitive R region. In both ClfA and Fbe, the R repeat regions are encoded by the same 18-bp consensus unit. A comparison of the nucleotide sequences of fbe and clfA shows that the R regions have approximately 80% homology.
FIG. 5.
Alignment of the deduced amino acid sequence of the parts in the Fg-binding regions of Fbe and ClfA with the highest similarity. The numbering indicates the amino acid position in Fbe according to Fig. 4 and reference 17 for ClfA. Vertical lines indicate identical amino acids, and dots show similar amino acids. Gaps (indicated by dashes) were introduced to obtain optimal alignment.
Occurrence of fbe in strains of S. epidermidis.
A collection of 43 strains of S. epidermidis, including the strains used in the Fg-binding experiment shown in Fig. 1, was screened by PCR for the presence of the fbe gene. The reaction was designed to amplify a region corresponding to a 496-bp-long fragment of the 5′ end of the fbe gene (Fig. 4). The result showed that this fragment was amplified from 40 of the 43 strains tested.
Mapping the Fg-binding region in Fbe by phage display.
An additional phage display library was constructed based on fragmented DNA of S. epidermidis 19. The phage display library of strain 19 was approximately the same size as the library of strain HB. This library gave a much higher enrichment of Fg-binding phagemid particles than the HB library when it was panned against immobilized Fg. Sequence analysis revealed that the inserts of the isolated clones were derived from the fbe gene and that several clones had overlapping inserts. After two separate pannings against Fg, eight different clones covering amino acids 172 to 664 in Fbe were isolated (Fig. 2 and 4). From the polypeptides encoded by the inserts of clones 19FG2, 19FG7, and 19FG8, an Fg-binding domain of 331 amino acids was deduced that covered amino acids 269 to 599. Alignment of the inserts also showed that the nucleotide sequence in the region encoding the Fg-binding domain between strains HB and 19 differed by only one silent nucleotide exchange.
DISCUSSION
Implant biomaterials are instantly covered by circulating plasma components, like Fg (37), promoting adhesion of host cells. One complication that may arise is when contaminating bacteria adhere to the same components on the biomaterial surfaces, leading to infection.
In contrast to S. epidermidis, adherence of S. aureus to Fg has been well characterized. A surface-associated Fg-binding protein, termed clumping factor, mediates S. aureus adherence to immobilized Fg (17) and contributes to virulence in an experimental endocarditis model (20). Also, another surface-located Fg-binding protein in S. aureus has been demonstrated (4). In addition to these, there are no less than three extracellular Fg-binding proteins released into the growth medium, one of which is a coagulase (2, 3). To investigate the Fg-binding nature of S. epidermidis, different strains were tested for their ability to bind to immobilized Fg. The result showed a great variation between strains from non-binders to high-binders, in which the high-binders adhere to immobilized Fg in the same order as S. aureus Newman (Fig. 1). The heterogeneity in binding is in agreement with earlier findings (8) and might reflect different expression levels of Fbe or might be due to production of interfering substances, such as slime (1). The Fg-binding activity of S. epidermidis was found to be protein mediated, since protease treatment destroyed the binding. Thus, to isolate the gene(s) encoding Fg-binding activity, a phage display library of chromosomal DNA from a clinical isolate of S. epidermidis was constructed. These types of libraries have earlier been used successfully to isolate and characterize cell surface proteins from other gram-positive cocci (9–12, 14). Panning of the phage library against Fg resulted in an enrichment of clones. Further analysis revealed that seven of eight clones were identical, harboring an insert of 1,743 nucleotides with one open reading frame.
The Fg-binding activity expressed by pSEFG1 was studied with a specificity test. The pSEFG1 phagemid particles showed no binding activity to the various plasma and extracellular matrix proteins tested (except Fg) or to plastic (Table 1). Furthermore, it was possible to completely inhibit the binding of phagemid (pSEFG1) particles to immobilized Fg in the presence of soluble Fg (Fig. 3A). As seen by SDS-PAGE (Fig. 3B), expression of pSEFG1 in E. coli results in new protein fragments, and a corresponding Western blot indicates that the Fg-binding activity resides in a fraction with a size of around 100 kDa (Fig. 3C). This does not correlate with the calculated molecular mass of the protein encoded by pSEFG1, which is ∼70 kDa. However similar discrepancies have earlier been reported for other cell surface proteins of staphylococci and streptococci (13, 14, 26, 30). Furthermore, affinity-purified protein encoded by pSEFG1 can, in an adhesion experiment, completely inhibit the binding of S. epidermidis to immobilized Fg (Fig. 3D).
Shotgun phage display has proven to be an effective technique in mapping binding domains, since one can rapidly identify many overlapping clones (9–12, 14). With regard to the size of the library of strain HB, it was unexpected to identify only one clone (pSEFG1). The explanation can be that the binding of the S. epidermidis protein to Fg requires a substantial part of the protein and the majority of the ligated chromosomal DNA fragments used to construct the library were only ∼500 bp. Therefore, another phage display library was constructed with chromosomal DNA from strain 19. This strain was chosen because it was grouped into the category of strains that were high in Fg binding (Fig. 1). This time, the fragmentation conditions of the chromosomal DNA were milder, and fragments with the mean size of 1 kb were used for ligation. This had a dramatic effect on the enrichment of phage particles when the library was panned against Fg and resulted in the isolation of several overlapping clones (Fig. 2). By sequence alignment, it was possible to localize the Fg-binding region to a 331-residue-long part in the A region of Fbe located between amino acids 269 and 599 (Fig. 2 and 4).
Since the Fbe protein is thought to be a cell wall-bound protein, it is assumed that a structure mediating this feature would be found in the C-terminal part. Although the C terminus of the protein has the characteristic membrane-spanning region and the LPXTG motif, the predicted charged wall region, rich in proline residues, commonly found among staphylococcal cell surface proteins (7, 24, 30) is not present.
By computer search, it was found that the Fbe protein is related to the S. aureus clumping factor. Comparisons show that both proteins have the same overall organization and partially display a high degree of similarity. Using an inhibition assay and a Western blot analysis, McDevitt et al. (18) located the Fg-binding activity in ClfA at a 329-residue-long fragment in the A region of ClfA. Alignment of the complete A regions shows limited similarity, but the similarities between the proteins increase in their respective Fg-binding domains. The highest similarity in the A region between Fbe and ClfA is found in a stretch of 144 residues located on the fragments mediating Fg binding in both ClfA and Fbe (Fig. 5). The A region of the ClfA protein has, in addition to binding Fg, been shown to be involved in clumping and adherence of S. aureus (18). However, cells of S. epidermidis HB and 19 do not show a positive clumping reaction. Based on the quite moderate homology between Fbe and ClfA, it cannot be ruled out that Fbe binds Fg by a mechanism other than ClfA, which might explain the lack of clumping in S. epidermidis. The most pronounced similarity is found in the highly repetitive R region. The function of the DS repeat region in ClfA is still not clear, but it has been shown that the region is involved in neither Fg binding nor clumping (18). In S. aureus, this region is reported to vary in size between different isolates (16). Furthermore, Southern blot experiments performed by McDevitt et al. (17) showed that the R region had at least four homologous loci in the S. aureus chromosome. In the same way, chromosomal DNA from S. epidermidis HB was analyzed under stringent conditions for the presence of the DS repeat with a probe covering the R region of fbe. The result showed, in contrast to S. aureus, that only a single locus was present in strain HB. Furthermore, it was found by PCR analysis that the occurrence of the fbe gene is common among clinical isolates of S. epidermidis.
It has been suggested that S. epidermidis colonizes biomaterial in a two-step procedure, in which adherence is the primary event followed by biofilm formation (15, 25). The importance of the Fbe protein in virulence of S. epidermidis remains to be demonstrated. We intend to clarify this issue in animal models by using mutants of S. epidermidis in which the fbe gene has been inactivated.
ACKNOWLEDGMENTS
We thank Karl-Erik Johansson and Anja Persson at the National Veterinary Institute, Uppsala, Sweden, for help with the DNA sequencing performed with the ALFexpress DNA sequencer.
This investigation was supported by grants from the Swedish Medical Research Council (B94-16X-03778 and K97-16X-12218-01A9), the Swedish Council for Forestry and Agricultural Research (32.0370/96), and Swedish Research for Engineering Science (96-759).
REFERENCES
- 1.Baldassarri L, Donelli G, Gelosia A, Simpson A W, Christensen G D. Expression of slime interferes with in vitro detection of host protein receptors of Staphylococcus epidermidis. Infect Immun. 1997;65:1522–1526. doi: 10.1128/iai.65.4.1522-1526.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodén M, Flock J I. Evidence for three different fibrinogen-binding proteins with unique properties from Staphylococcus aureus strain Newman. Microb Pathog. 1992;12:289–298. doi: 10.1016/0882-4010(92)90047-r. [DOI] [PubMed] [Google Scholar]
- 3.Bodén M, Flock J I. Cloning and characterization of a gene for a 19 kDa fibrinogen binding protein from Staphylococcus aureus. Mol Microbiol. 1994;12:599–606. doi: 10.1111/j.1365-2958.1994.tb01046.x. [DOI] [PubMed] [Google Scholar]
- 4.Cheung A I, Projan S J, Edelstein R E, Fischetti V A. Cloning, expression, and nucleotide sequence of a Staphylococcus aureus gene (fbpA) encoding a fibrinogen-binding protein. Infect Immun. 1995;63:1914–1920. doi: 10.1128/iai.63.5.1914-1920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai N P, Hossainy S F, Hubbell J A. Surface-immobilized polyethylene oxide for bacterial repellence. Biomaterials. 1992;13:417–420. doi: 10.1016/0142-9612(92)90160-p. [DOI] [PubMed] [Google Scholar]
- 6.Galliani S, Viot M, Cremieux A, Van der Auwera P. Early adhesion of bacteremic strains of Staphylococcus epidermidis to polystyrene: influence of hydrophobicity, slime production, plasma, albumin, fibrinogen, and fibronectin. J Lab Clin Med. 1994;123:685–692. [PubMed] [Google Scholar]
- 7.Guss B, Uhlén M, Nilsson B, Lindberg M, Sjöquist J, Sjödahl J. Region X, the cell-wall-attachment part of staphylococcal protein A. Eur J Biochem. 1984;138:413–420. doi: 10.1111/j.1432-1033.1984.tb07931.x. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann M, Vaudaux P E, Pittet D, Auckenthalter R, Lew P D, Schumacher-Perdreau F, Peters G, Waldvogel F A. Fibronectin, fibrinogen, and laminin act as mediators of adherence of clinical staphylococcal isolates to foreign material. J Infect Dis. 1988;158:693–701. doi: 10.1093/infdis/158.4.693. [DOI] [PubMed] [Google Scholar]
- 9.Jacobsson K, Frykberg L. Cloning of ligand-binding domains of bacterial receptors by phage display. BioTechniques. 1995;18:878–885. [PubMed] [Google Scholar]
- 10.Jacobsson K, Frykberg L. Phage display shot-gun cloning of ligand-binding domains of prokaryotic receptors approaches 100% correct clones. BioTechniques. 1996;20:1070–1081. doi: 10.2144/96206rr04. [DOI] [PubMed] [Google Scholar]
- 11.Jacobsson K, Frykberg L. Gene VIII-based, phage-display vectors for selection against complex mixtures of ligands. BioTechniques. 1998;24:294–301. doi: 10.2144/98242rr01. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsson K, Jonsson H, Lindmark H, Guss B, Lindberg M, Frykberg L. Shot-gun phage display mapping of two streptococcal cell-surface proteins. Microbiol Res. 1997;152:121–128. doi: 10.1016/S0944-5013(97)80002-X. [DOI] [PubMed] [Google Scholar]
- 13.Lindgren P E, McGavin M J, Signäs C, Guss B, Gurusiddappa S, Höök M, Lindberg M. Two different genes coding for fibronectin-binding proteins from Streptococcus dysgalactiae. The complete nucleotide sequences and characterization of the binding domains. Eur J Biochem. 1993;214:819–827. doi: 10.1111/j.1432-1033.1993.tb17985.x. [DOI] [PubMed] [Google Scholar]
- 14.Lindmark H, Jacobsson K, Frykberg L, Guss B. Fibronectin-binding protein of Streptococcus equi subsp. zooepidemicus. Infect Immun. 1996;64:3993–3999. doi: 10.1128/iai.64.10.3993-3999.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mack D, Nedelmann M, Krokotsch A, Schwartzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDevitt D, Foster T J. Variation in the size of the repeat region of the fibrinogen receptor (clumping factor) of Staphylococcus aureus strains. Microbiology. 1995;141:937–943. doi: 10.1099/13500872-141-4-937. [DOI] [PubMed] [Google Scholar]
- 17.McDevitt D, Francois P, Vaudaux P, Foster T J. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol Microbiol. 1994;11:237–248. doi: 10.1111/j.1365-2958.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 18.McDevitt D, Francois P, Vaudaux P, Foster T J. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol Microbiol. 1995;16:895–907. doi: 10.1111/j.1365-2958.1995.tb02316.x. [DOI] [PubMed] [Google Scholar]
- 19.Mohammad S F, Topham N S, Burns G L, Olsen D B. Enhanced bacterial adhesion on surfaces pretreated with fibrinogen and fibronectin. ASAIO Trans. 1988;34:573–577. [PubMed] [Google Scholar]
- 20.Moreillon P, Entenza J M, Francioli P, McDevitt D, Foster T J, François P, Vaudaux P. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–4743. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller E, Hübner J, Gutierrez N, Takeda S, Goldmann D A, Pier G B. Isolation and characterization of transposon mutants of Staphylococcus epidermidis deficient in capsular polysaccharide/adhesin and slime. Infect Immun. 1993;61:551–558. doi: 10.1128/iai.61.2.551-558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers E W, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 23.Neumeister B, Kastner S, Conrad S, Klotz G, Bartmann P. Characterization of coagulase-negative staphylococci causing nosocomial infections in preterm infants. Eur J Clin Microbiol Infect Dis. 1995;14:856–863. doi: 10.1007/BF01691491. [DOI] [PubMed] [Google Scholar]
- 24.Patti J M, Jonsson H, Guss B, Switalski L M, Wiberg K, Lindberg M, Höök M. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J Biol Chem. 1992;267:4766–4772. [PubMed] [Google Scholar]
- 25.Paulsson M, Kober M, Freij-Larsson C, Stollenwerk M, Wesslen B, Ljungh Å. Adhesion of staphylococci to chemically modified and native polymers, and the influence of preadsorbed fibronectin, vitronectin, and fibrinogen. Biomaterials. 1993;14:845–853. doi: 10.1016/0142-9612(93)90006-n. [DOI] [PubMed] [Google Scholar]
- 26.Rakonjac J V, Robbins J C, Fischetti V A. DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect Immun. 1995;63:622–631. doi: 10.1128/iai.63.2.622-631.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 29.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 30.Signäs C, Raucci G, Jönsson K, Lindgren P E, Anantharamaiah G M, Höök M, Lindberg M. Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc Natl Acad Sci USA. 1989;86:699–703. doi: 10.1073/pnas.86.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timmerman C P, Fleer A, Besnier J M, De Graaf L, Cremers F, Verhoef J. Characterization of a proteinaceous adhesin of Staphylococcus epidermidis which mediates attachment to polystyrene. Infect Immun. 1991;59:4187–4192. doi: 10.1128/iai.59.11.4187-4192.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tojo M, Yamashita N, Goldmann D A, Pier G B. Isolation and characterization of a capsular polysaccharide adhesin from Staphylococcus epidermidis. J Infect Dis. 1988;157:713–722. doi: 10.1093/infdis/157.4.713. [DOI] [PubMed] [Google Scholar]
- 33.Veenstra G J C, Cremers F F M, van Dijk H, Fleer A. Ultrastructural organization and regulation of a biomaterial adhesin of Staphylococcus epidermidis. J Bacteriol. 1996;178:537–541. doi: 10.1128/jb.178.2.537-541.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.von Graevenitz A, Amsterdam D. Microbiological aspects of peritonitis associated with continuous ambulatory peritoneal dialysis. Clin Microbiol Rev. 1992;5:36–48. doi: 10.1128/cmr.5.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong E S. Surgical site infections. In: Mayhall C G, editor. Hospital epidemiology and infection control. Baltimore, Md: Williams & Wilkins; 1996. pp. 154–175. [Google Scholar]
- 37.Yu J, Montelius M N, Paulsson M, Gouda I, Larm O, Montelius L, Ljungh Å. Adhesion of coagulase-negative staphylococci and adsorption of plasma proteins to heparinized polymer surfaces. Biomaterials. 1994;15:805–814. doi: 10.1016/0142-9612(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 38.Zdanowski Z, Ribbe E, Schalen C. Influence of some plasma proteins on in vitro bacterial adherence to PTFE and Dacron vascular prosthesis. APMIS. 1993;101:926–932. doi: 10.1111/j.1699-0463.1993.tb00203.x. [DOI] [PubMed] [Google Scholar]