Abstract
Genetic manipulation of antibiotic producers, such as Streptomyces species, is a rational approach to improve the properties of biologically active molecules. However, this can be a slow and sometimes problematic process. Red/ET recombination in an Escherichia coli host has permitted rapid and more versatile engineering of geldanamycin biosynthetic genes in a complementation plasmid, which can then be readily transferred into the Streptomyces host from which the corresponding wild type gene(s) has been removed. With this rapid Red/ET recombination and gene complementation approach, efficient gene disruptions and gene replacements in the geldanamycin biosynthetic gene cluster have been successfully achieved. As an example, we describe here the creation of a ketoreductase 6 null mutation in an E. coli high-copy-number plasmid carrying gdmA2A3 from Streptomyces hygroscopicus NRRL3602 and the subsequent complementation of a gdmA2A3 deletion host with this plasmid to generate a novel geldanamycin analog.
Polyketides represent a large class of structurally diverse natural products which have widespread use as therapeutics. They are structurally complex compounds that are difficult to make or modify chemically (18). The polyketide geldanamycin is a potential anticancer agent. It and its derivatives bind to heat shock protein 90 (Hsp90) and destabilize its client proteins (which include v-Src, Bcr-Abl, Raf-1, ErbB2, mutated p53, some growth factor receptors, and steroid receptors) that are often involved in human cancers (17). Many geldanamycin analogs (13, 23, 24), including 17-allylamino-17-demethoxygeldanamycin, which is in multiple clinical trials, have been produced through chemical modification of the parent molecule.
The polyketide synthase (PKS) responsible for geldanamycin biosynthesis, like other modular PKSs, consists of a set of multifunctional enzymes encoded by a large gene cluster. We are interested in making geldanamycin analogs that are not easily accessible by chemical synthesis through genetic engineering of the geldanamycin PKS. For this purpose, the gene cluster (gdm) for geldanamycin production has been isolated and sequenced (20). This cluster consists of three genes, gdmA1, gdmA2, and gdmA3, which encode a seven-module PKS, in addition to other genes for oxidative modification and carbamoyl attachment and genes for methoxymalonyl-ACP precursor biosynthesis.
Since the discovery of the eryA gene cluster for the erythromycin PKS (5, 6), methodologies have been developed for genetically engineering PKS gene clusters. The conventional gene replacement procedure, which can be slow and tedious for DNA manipulation in the producing actinomycete strains, employs direct selection for a first crossover that integrates a vector and then screening for a second crossover that exchanges the wild-type allele with the mutated allele and eliminates the remaining vector sequences. In an improvement over the double-reciprocal recombination approach, PKS genes have been cloned in shuttle vectors that are capable of replication in Streptomyces, as well as in Escherichia coli, downstream of promoters that permit expression in several Streptomyces hosts (10, 27, 30). This system allows for the engineering, in E. coli, of domains for alternative substrate specificities and β-carbon processing activities, followed by efficient transfer to the Streptomyces hosts. A multiple-plasmid system that facilitates combinatorial biosynthesis of type I PKSs can speed up the process by combining several plasmids encoding functional mutants of PKS subunits with one or more additional mutants (26).
Our approach to make geldanamycin analogs through genetic engineering of the PKS was based on recombinogenic engineering by using Red/ET recombination and its recent adaptation to Streptomyces (8, 12, 14). Red/ET recombination is based on the discovery that allelic exchanges on the E. coli chromosome can be achieved by recombination with a selectable marker flanked by only short stretches of homology to the desired region in the chromosome, when either Redα/Redβ from λ phage or RecE/RecT from the Rac phage is present in the targeted strain (14, 15, 28, 29). By using Red/ET recombination technology, we were able to circumvent many of the present limitations in the engineering of polyketide systems. Current methods for combinatorial biosynthesis are limited by the need to find or create appropriate restriction endonuclease sites and by the recently introduced idea that testing a number of alternative splice sites, although laborious, can be an important factor in the success of experiments (19). In addition, the inefficient in vitro methods in which restriction endonuclease digestion and ligation with the usually large shuttle plasmids used for the expression of PKS genes are used makes the process even more troublesome.
Genetic engineering of polyketide biosynthetic genes has been used successfully to create many novel unnatural natural compounds that are not readily accessible via direct chemical modification (16, 25). In order to create novel geldanamycin analogs, gene knockouts and gene replacements in the geldanamycin biosynthetic gene cluster have been obtained (McDaniel, unpublished results). One desired modification is the introduction of a keto group at the C-5 position of geldanamycin, which can serve as a handle for further chemical modification. Initially, traditional genetic approaches, such as streptomycete phage- or plasmid pKC1139 (2)-mediated gene replacement, were found to be ineffective and tedious. To accomplish this goal, an approach using Red/ET recombination with gene complementation was developed.
Since the geldanamycin biosynthetic gene cluster spans over 60 kb, it would be difficult to work on a plasmid carrying such a large amount of DNA (20). Therefore, a segment containing gdmA2A3 and some of the downstream tailoring genes was subcloned into a ΦC31-derived attB-integrating vector to facilitate engineering of the desired region of the gene cluster and to reduce the chances of intramolecular recombination between similar active site domains, such as ketosynthase domains. After Red/ET recombination was performed in an E. coli host, the resulting plasmid containing the genetic modification, a point mutation that inactivated the ketoreductase 6 (KR6) domain, was introduced by conjugation into a Streptomyces hygroscopicus gdmA2A3 deletion mutant strain, where it complemented the resident gdmA1 gene to produce a novel compound. In our strategy, instead of deletion of the entire KR6 domain, a conserved tyrosine (Y1888) in the ketoreductase catalytic domain was mutated to phenylalanine because this was reported to be superior to ketoreductase deletion (21).
MATERIALS AND METHODS
Bacterial strains and growth condition.
The geldanamycin-producing strain S. hygroscopicus NRRL3602 has been described previously (20). Geldanamycin production medium (GPM) (4) with 40 g of XAD-16 (Rohm-Haas) per liter was used as the fermentation medium. When necessary, apramycin was added to a final concentration of 60 μg/ml. Cultures were grown at 30°C.
Plasmid pSC101-BAD-gbaA (Tetr) bearing genes encoding Redα and Redβ was obtained from Gene Bridges (Dresden, Germany) and was introduced by transformation into E. coli DH5α. The resulting strain was used as the host for Red/ET recombination. E. coli DH5α was also used as the host for routine DNA cloning.
Plasmid construction.
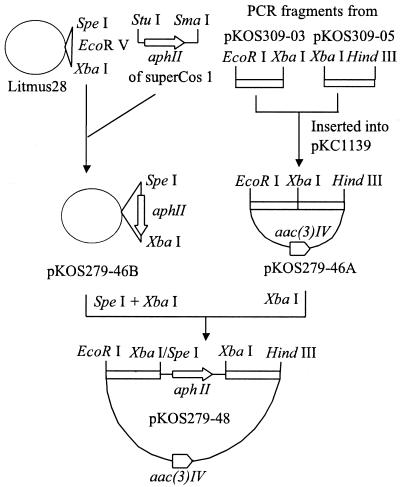
pKOS279-48 is a pKC1139 (2)-based plasmid and contains the aphII gene of Tn5 between the ketosynthase 4 (KS4) and dehydratase 7 (DH7) domains of a 7.8-kb gdmA fragment. This plasmid was used in construction of the gdmA2A3 deletion host as described below. Construction of pKOS279-48 (Fig. 1) was accomplished by inserting the SpeI-XbaI fragment of pKOS279-46B into the XbaI site of pKOS279-46A. pKOS279-46B resulted from insertion of the StuI-SmaI fragment (carrying the aphII gene) of SuperCos 1 (Stratagene) into the unique EcoRV site of Litmus 28 (New England Biolabs). pKOS279-46A was made by cloning two 1.5-kb fragments from the gdm PKS cluster between the EcoRI and HindIII sites of pKC1139 in the same orientation as in the natural gdmA2A3 gene. The left fragment consisted of DNA immediately upstream of the acyltransferase 4 (AT4) domain, and the right fragment consisted of DNA immediately downstream of the AT7 domain, cloned in pKOS309-03 and pKOS309-05, respectively (K. Patel, unpublished results).
FIG. 1.
Construction of pKOS279-48. The experiments are described in the text. aac(3)IV, apramycin resistance gene (3).
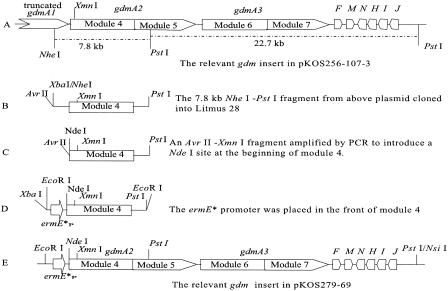
pKOS279-69 carries gdmA2A3, gdmF, gdmM, and several additional downstream genes. In this plasmid, the ermE* promoter is placed before gdmA2A3. This plasmid carries elements such as the integrase gene (int) and the attB sequences from streptomycete phage ΦC31, so it can be integrated into the chromosomes of strains of interest. Construction of pKOS279-69 (Fig. 2) was started by subcloning the 7.8-kb NheI-PstI fragment (carrying module 4 and part of module 5) from pKOS256-107-3 (20) into Litmus 28 to obtain pKOS313-57. At the same time, an AvrII-XmnI fragment was generated by PCR with primers M4F (5′-TCCTAGGACATATGGCGAATGACGAGC) and M4R (5′-GCGTCGAAGAGGTTCTCCAG) (AvrII, NdeI, and XmnI restriction sites in the primers are underlined) by using pKOS256-107-3 as the template. The amplified AvrII-XmnI fragment was used to replace the AvrII-XmnI fragment of pKOS313-57 to make pKOS279-68. Then the NdeI-PstI fragment carrying module 4 of pKOS279-68 and the XbaI-NdeI fragment (carrying the ermE* promoter) of pKOS159-8 (22) were ligated and inserted between XbaI and PstI sites of Litmus 28 to produce pKOS279-68B. Plasmid pKOS279-69 finally resulted from insertion of the EcoRI-PstI fragment from pKOS279-68B and a 22.7-kb PstI fragment from pKOS256-107-3 into pKOS159-8 linearized with EcoRI and NsiI.
FIG. 2.
Relevant genes and restriction endonuclease enzyme sites of the gdm inserts (and the ermE* promoter) in pKOS256-107-3 (A), pKOS313-57 (B), pKOS279-68 (C), pKOS279-68B (D), and pKOS279-69 (E). ermE*P, mutated ermE gene promoter (1).
pKOS272-134 and pKOS272-139 were made to construct a KR6 null version of pKOS279-69. To construct the inactivated KR6 (KR6°) cassette, four primers were used to produce by PCR two overlapping fragments (1.5 kb each). The 5′ half of the KR6° cassette was generated with primers KR6-1 (5′-GCGGAGAAGTTGCCCTGGCCGGGCCCGCCTAGGACTCCGGCGGCGGACGAGTACA) and KR6-3 (5′-CGGGATCCGAGCCCCAACTGGCGGTGCGCGGT), and the 3′ half was generated with primers KR6-2 (5′-GAGTCCTAGGCGGGCCCGGCCAGGGCAACTTCTC CGCCGCCAACGCCTATCTGGA) and KR6-4 (5′-GCTCTAGAGGGTCCGTTGGGCGCGGTGAGGCC) (Y1888F is in boldface type). Overlapping oligonucleotides KR6-1 and KR6-2 contained a point mutation that changed tyrosine (Y1888) in the active site to a phenylalanine, an approach previously used to inactivate the KR6 domain in deoxyerythronolide B synthase (21). These oligonucleotides also included a single base change that resulted in a silent mutation to introduce an AvrII site (underlined). The PCR products were assembled into Litmus 28, and then the cassette was subcloned into Litmus 38 (New England Biolabs), resulting in pKOS272-134. Then the subcloned aphII gene from pKOS279-46B was inserted at the AvrII site of pKOS272-134 to obtain pKOS272-139.
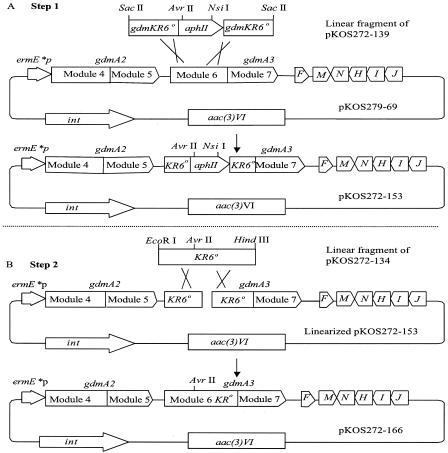
pKOS272-166 was made by two steps of Red/ET recombination to introduce the KR6° mutation into pKOS279-69 (the gdmA2A3 expression plasmid) (Fig. 3), as follows. In the first step, 1 μg of a 2.3-kb linear SacII fragment from pKOS272-139 containing approximately 0.4 kb of the KR6° cassette flanking the kanamycin gene marker (aphII) was coelectroporated with 3 μg of pKOS279-69. The total volume of DNA added never exceeded 4 μl. Analysis of all the kanamycin-resistant (Kmr) clones indicated that there was a mixed population of the original plasmid and the desired KR6° plasmid. The plasmid preparation was then diluted and used for transformation. All the Kmr clones resulting from latter transformation were the expected expression plasmid (pKOS272-153). In the second step, the KR6° cassette fragment linearized with EcoRI and HindIII from pKOS272-134 was coelectroporated with pKOS272-153 completely digested with AvrII and NsiI into the Red/ET recombination host. Recombinants were selected with 60 μg of apramycin per ml. Different sets of restriction endonucleases were used to verify the desired product. Approximately 30 to 40% of the clones contained a mixed population of the desired KR6° expression plasmid and the parent wild-type plasmid. The KR6° expression plasmid, pKOS272-166, was purified by passage through DH5α and was verified by restriction endonuclease analysis and loss of Kmr.
FIG. 3.
Schematic diagram of the Red/ET recombination approach for construction of pKOS272-166 (gdmA2A3-KR6° expression plasmid). In step 1, a linear DNA fragment containing the aphII gene and approximately 0.4 kb of the KR6° cassette flanking the gene marker was cotransformed with plasmid pKOS279-69 into competent Red/ET recombination cells. Kmr clones were selected. In step 2, a linear DNA fragment containing the KR6° cassette was coelectroporated with linearized pKOS272-153 cut by AvrII and NsiI. Aprr transformants were selected. ermE*P, mutated ermE gene promoter (1); int, integrase gene from streptomycete phage ΦC31.
Fermentation of the recombinant strain and isolation of the novel compound.
Plasmid pKOS272-166 carrying the KR6° mutation was introduced into the gdmA2A3 deletion host K279-48 by conjugation, using the procedure reported by Flett et al. (7). The resulting strain, S. hygroscopicus K272-166, was fermented in 40 250-ml baffled flasks containing 40 ml of GPM and 1.6 g of XAD-16. At the end of day 4, the contents of the flasks were pooled and centrifuged, and the supernatant was decanted. To separate cells from XAD-16, the mixture was resuspended in water, and the XAD-16 was allowed to settle. The supernatant with suspended cells was then decanted. This procedure was repeated two or three times until most of the mycelia were removed. The XAD-16 was filtered by using a separation funnel with Whatman filter paper and washed briefly with water.
The XAD-16 was extracted with 200 ml of methanol three times. The aqueous methanolic extract was concentrated on a rotary evaporator and freeze-dried. The resultant brown residue was resuspended in 120 ml of methanol and filtered. The filtrate was evaporated, which resulted in a brown solid, which was redissolved in dichloromethane-methanol. About 10 g of silica gel was added to the solution, and the mixture was evaporated to obtain a free-flowing powder. This powder was loaded on a silica gel column and eluted with a 0 to 10% methanol gradient in dichloromethane. The fractions showing mass spectrometric (MS) peaks at 506 and 528 were pooled and evaporated, which resulted in a light brown solid. The crude sample was further purified by high-performance liquid chromatography (HPLC) using a C18 column (length, 150 mm; inside diameter, 21.2 mm; MetaChem Polaris 5u C18-A) that was eluted with a gradient of acetonitrile in water. Fractions were pooled based on liquid chromatography-MS analysis. KOSN-1869 was obtained as a white solid after lyophilization. 1H nuclear magnetic resonance (NMR) (CD3OD, 400 MHz): δ (relative to CHD2OD at 3.30 ppm) 0.74 (d, 3, J = 6.4 Hz), 0.99 (d, 3, J = 6.8 Hz), 1.05 (m, 1), 1.60 (m, 1), 1.61 (s, 3), 1.83 (s, 3), 1.83 (m, 1), 2.22 (dd, 1, J = 8.0, 13.2 Hz), 2.40 (m, 1), 2.41 (dd, 1, J = 6.4, 13.2 Hz), 3.03 (d, 1, J = 10.4 Hz), 3.22 (s, 3), 3.34 (s, 3), 3.43 (dd, 1, J = 3.0, 8.2 Hz), 3.88 (d, 1, J = 6.4 Hz), 4.22 (d, 1, J = 6.4 Hz), 5.34 (d, 1, J = 10.0 Hz), 6.05 (s, 1), 6.07 (s, 1), 6.10 (s, 1), 6.18 (s, 1). 13C NMR (CD3OD, 100 MHz): δ (relative to CD3OD at 49.0 ppm) 8.6, 13.0, 17.2, 19.7, 31.8, 35.4, 36.3, 45.5, 56.7, 58.3, 75.1, 78.3, 81.3, 83.8, 100.3, 101.7, 104.0, 108.3, 110.0, 133.1, 133.6, 144.2, 148.0, 158.4, 159.4, 168.1, 168.4. Electrospray ionization time of flight MS m/z 506.2768, calculated for C27H40NO8 ([M+H]+) 506.2748. UV λmax 211, 240 (sh), 285 nm.
RESULTS
Construction of a gdmA2A3 deletion host and complementation of this host.
Our approach was based on complementation of an S. hygroscopicus gdmA2A3 deletion host by integration of a Streptomyces expression plasmid carrying engineered gdmA2A3 genes. This strategy required generation of a strain in which a gene(s) was knocked out or deleted and also construction of an expression plasmid to express the corresponding gene(s) or its mutant(s). Thus, strain K279-48, the deletion host, was constructed for this purpose. In this strain, gdmA2 and gdmA3 were disrupted by inserting the aphII gene between the gdm KS4 and DH7 domains via a double crossover by using pKOS279-48. The genotype of the deletion host was verified by Southern hybridization analysis by using the aphII gene as the probe (data not shown). Fermentation of K279-48 showed no trace of geldanamycin production as determined by HPLC-MS. The deletion of part of gdmA2 and gdmA3 might have affected the transcription of gdmF and gdmM, which were located downstream of gdmA3 (Fig. 2 and 3). These two genes might be under the control of the promoter which directed the transcription of gdmA1 through gdmA3. To construct the expression plasmid for gene complementation, gdmF and gdmM were cloned together with gdmA2A3 into pKOS279-69 in which the ermE* promoter was placed in front of gdmA2. To facilitate cloning, gdmNHIJ, which obviously were transcribed in the direction opposite gdmA1 to gdmA3 and would not be affected by the deletion, were also included in pKOS279-69 (Fig. 2). When pKOS279-69 was introduced by conjugation into the deletion host strain, K279-48, the resulting recombinant strain, K279-48/pKOS279-69, restored geldanamycin production at a level comparable to the 1-g/liter level produced by the NRRL3602 strain (Fig. 4A).
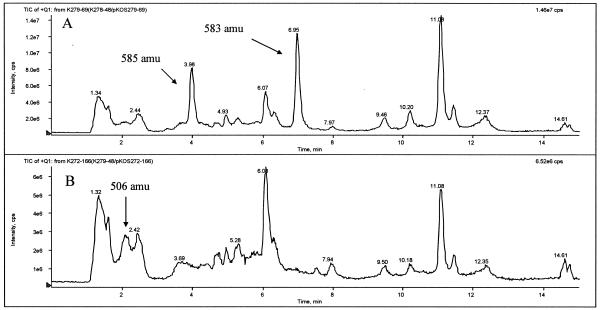
FIG. 4.
Total ion chromatograms (TIC) from HPLC-MS analysis of extracts from fermentation of strains K279-48/pKOS279-69 (A) and K272-166 (B). The positions of geldanamycin and related compounds (as determined by their fragmentation patterns) are indicated. Geldanamycin and its reduced form, 18,21-dihydro-geldanamycin, appeared at about 6.95 and 3.98 min on the chromatogram of strain K279-48/pKOS279-69 with MS peaks at 583 and 585 AMU ([M+Na]+), respectively. KOSN-1869 appeared at about 2.1 min on the chromatogram of strain K272-166 with a base MS peak at 506 AMU ([M+H]+).
Construction of a gdmA3 KR6 knockout in the gdmA2A3 expression plasmid by Red/ET recombination.
For construction of gdmA3 KR6°, a Y1888F mutation was introduced to inactivate the catalytic tyrosine residue of the ketoreductase domain since this strategy was reported to be more effective in generating the cognate keto derivatives than the introduction of a ketoreductase deletion (21). The Y1888F mutation was introduced into the complementation plasmid, pKOS279-69, by two-step Red/ET recombination cloning. First, the KR6 domain in pKOS279-69 was replaced with a KR6°-aphII cassette from pKOS272-139. The replacement was accomplished by coelectroporation of a linear KR6°-aphII cassette with the recipient plasmid pKOS279-69 into a Red/ET recombination-competent E. coli strain (Fig. 3). Approximately 200 apramycin-resistant (Aprr) Kmr colonies were obtained from the transformation described above. Restriction nuclease digestion analysis of plasmids from representative transformants revealed mixtures of the recipient plasmid and the expected KR6°-aphII mutant. The plasmid mixtures were diluted and used for further transformation. All the Kmr clones resulting from the latter round of transformation carried only the desired plasmid (pKOS272-153).
The next step was a second round of Red/ET recombination to replace the selectable marker with the KR6° cassette (Fig. 3). In this step, the linear KR6° cassette fragment from pKOS272-134 was used as a donor, and linearized pKOS272-153 was used as the recipient, which was cut in the region homologous to the donor. The use of a linearized recipient plasmid favored selection of the recombinogenic event, which circularized the target plasmid in the absence of a counterselection gene marker. Restriction endonuclease digestion analysis of 10 Aprr colonies indicated that 40% of the clones obtained were mixtures of the recipient plasmid and the expected KR6° mutants, and the rest appeared to be rearranged recipient plasmid. The desired expression plasmid, pKOS272-166 carrying gdmA2A3 with a KR6° mutation, was purified from the mixture as described above. All the Aprr and kanamycin-sensitive clones harbored plasmid pKOS272-166, showing the expected DNA banding patterns on agarose gels after restriction endonuclease digestion and carrying gdmA2A3 with a KR6° mutation.
Novel compound resulting from a KR6 null mutation and characterization of this compound.
Plasmid pKOS272-166 containing the KR6° modification was introduced into strain K279-48 by conjugation. Exconjugants were fermented in GPM for 4 days. Analysis by HPLC-MS of the major metabolites produced did not indicate any compound with the expected mass of 5-keto geldanamycin (575 atomic mass units [amu]). However, a major product (KOSN-1869) (Fig. 5B) of strain K272-166 with a nominal mass of 505 amu was found (Fig. 4B). Sixteen milligrams of KOSN-1869 was recovered from 1.6 liters of culture. HR-electrospray ionization-time of flight-MS analysis confirmed that the molecular formula was C27H39NO8, indicating that the carbamoyl group was missing. 1H, 13C, two-dimensional correlation spectroscopy (COSY), heteronuclear single quantum correlation (HSQC), and heteronuclear multiple bond correlation (HMBC) NMR data for this compound in methanol-d4 clearly established that the structure of the fragment for C-6 to C-15 was identical to that of 7-O-decarbamoylgeldanamycin. The only peaks on the 1H NMR spectrum downfield of the doublet for C-9 H at δ 5.33 ppm were four singlets at 6.0 to 6.2 ppm, which belonged to protons attached to carbons at δ 101 to 110 ppm. There was one quaternary carbon at 100.3 ppm that showed a strong cross peak with the C-2 Me (1H δ 1.82 ppm) in the HMBC spectrum. This indicated that the aryl ring existed in a phenolic form and C-2 to C-5 belonged to enol systems. The UV spectrum showed a λmax at 285 nm due to the extended conjugated enol system, while it lacked the λmax at 305 nm, a characteristic of the quinone system observed in geldanamycin. KOSN-1869 was also differentiated physicochemically from geldanamycin by its increased polarity, as indicated by a much shorter retention time in reversed-phase HPLC (Fig. 4B) and relative insolubility in chloroform. Overall, the analytical data for KOSN-1869 are consistent with data for 3,5-dihydroxy-4,5-dehydroprogeldanamycin, which is the di(enol) form of a 3,5-diketo derivative of geldanamycin (Fig. 5B).
FIG. 5.
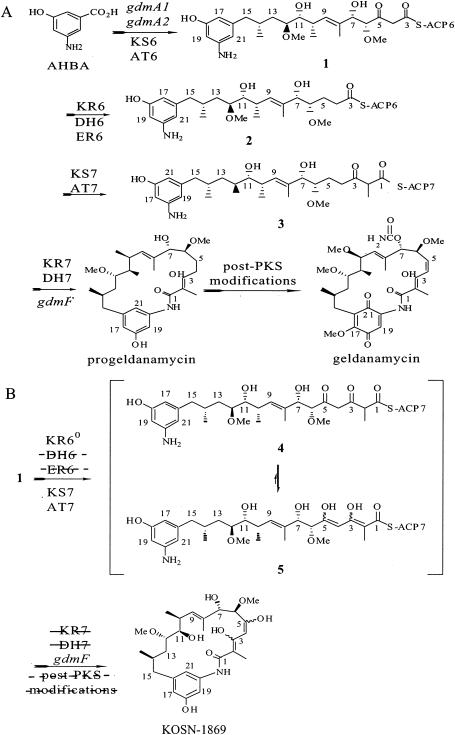
(A) Proposed biosynthetic pathway for geldanamycin in its native producer. (B) Proposed biosynthetic pathway for KOSN-1869 in the KR6 null strain. The dashed lines through DH6, ER6 (enoyl reductase 6), KR7, DH7, and post-PKS modifications indicate that the reactions did not occur. AHBA, 3-hydroxy-5-aminobenzoic acid; ACP, acyl carrier protein.
Although polyketide synthesis might be viewed as a series of discrete enzymatic steps, where events in one cycle do not affect those that take place in later cycles, an altered structure that results from an early step genetic change may present novel acyl chains to be processed by downstream PKS domains. A central question in combinatorial biosynthesis is whether such modified chains would be processed, even though the changes would not be at subsequent processing centers (11).
In the biosynthesis of geldanamycin from 3-hydroxy-5-aminobenzoic acid (Fig. 5A), the 5-keto intermediate 1 is reduced by the KR6 domain, resulting in a 5-hydroxy intermediate, which is further processed by the DH6 and enoyl reductase 6 domains. The next polyketide chain elongation via module 7 ketosynthase (KS7) and acyltransferase (AT7) results in the linear polyketide chain 3, which undergoes 3-keto reduction (KR7) and 2,3-dehydration (DH7), followed by cyclization via the lactam synthase (GdmF), forming progeldanamycin. Modifications by post-PKS enzymes lead to geldanamycin (20).
In the KR6° strain, the 5-keto intermediate 1 directly underwent chain elongation via KS7 and AT7, forming the 3,5-diketo intermediate 4 (Fig. 5B). Because of the dipolar repulsion between the carbonyl groups at C-1, C-3, and C-5, this intermediate underwent tautomerization, resulting in the much more stable 3,5-dien-3,5-diol tautomer 5. Interestingly, such di(enol) structures were observed in the tetraketide shunt products synthesized by a mutant with a mutation in the rifamycin pathway (9). Since intermediate 5 was not a substrate for KR7, it underwent cyclization via the lactam synthase (GdmF), resulting in KOSN-1869. The fact that only KOSN-1869 was isolated suggested that it was not a good substrate for the post-PKS enzymes, such as the monooxygenase(s) to transform it into a benzoquinone form or the carbamoyl transferase to form the 7-carbamate.
DISCUSSION
Gust et al. (8) described an efficient gene replacement method for Streptomyces that involved engineering conjugative cosmid clones by using Red/ET-mediated recombination in an E. coli host. This approach demonstrated that recombinogenic engineering rapidly generated Streptomyces coelicolor mutants. In other work, Kim et al. (12) reported the use of Red/ET recombination to generate a large number of shuttle plasmids from a diverse library of chimeric domains for directed evolution of the pikromycin PKS.
Because of the need to boost the flow of new nontraditional drug leads into preclinical studies, we adopted the Red/ET recombination technology for generating new polyketides because it overcame some of the limitations of traditional approaches for PKS engineering. Streptomyces genetic manipulation presents many challenges. In addition to the slow growth of the majority of Streptomyces strains, unknown restriction-modification systems, low frequencies of homologous recombination in polyketide-producing hosts, and a lack of efficient vectors often further slow the process of DNA engineering. Using Red/ET recombination in E. coli, we engineered the geldanamycin gdmA3 KR6 domain to eliminate its activity. This approach obviated the need for introducing appropriate restriction endonuclease sites for the engineering, as well as restriction endonuclease digestion and ligation of large plasmids used to complement the gdmA2A3 deletion strain. Red/ET recombination could also facilitate the testing of alternative splice sites and donor domains when acyltransferase domain swaps are objects, although it was not necessary in this work. By using the Red/ET recombination and gene complementation approach, novel geldanamycin analogues resulting from different acyltransferase domain swaps in the geldanamycin gene cluster have recently been generated rapidly by our colleagues.
Acknowledgments
We thank David A. Hopwood and Leonard Katz for critical reading of the manuscript. We also thank Gary Ashley and John Carney for helpful discussions in solving the structure of KOSN-1869.
REFERENCES
- 1.Bibb, M. J., J. White, J. M. Ward, and G. R. Janssen. 1994. The mRNA for the 23S rRNA methylase encoded by the ermE gene of Saccharopolyspora erythraea is translated in the absence of a conventional ribosome-binding site. Mol. Microbiol. 14:533-545. [DOI] [PubMed] [Google Scholar]
- 2.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 3.Davies, J., and S. O'Connor. 1978. Enzymatic modification of aminoglycoside antibiotics: 3-N-acetyltransferase with broad specificity that determines resistance to the novel aminoglycoside apramycin. Antimicrob. Agents Chemother. 14:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBoer, C., P. A. Meulman, R. J. Wnuk, and D. H. Peterson. 1970. Geldanamycin, a new antibiotic. J. Antibiot. (Tokyo) 23:442-447. [DOI] [PubMed] [Google Scholar]
- 5.Donadio, S., and L. Katz. 1992. Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111:51-60. [DOI] [PubMed] [Google Scholar]
- 6.Donadio, S., M. J. Staver, J. B. McAlpine, S. J. Swanson, and L. Katz. 1992. Biosynthesis of the erythromycin macrolactone and a rational approach for producing hybrid macrolides. Gene 115:97-103. [DOI] [PubMed] [Google Scholar]
- 7.Flett, F., V. Mersinias, and C. P. Smith. 1997. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155:223-229. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunziker, D., T.-W. Yu, C. R. Hutchinson, H. G. Floss, and C. Khosla. 1998. Primer unit specificity in rifamycin biosynthesis principally resides in the later stages of the biosynthetic pathway. J. Am. Chem. Soc. 120:1092-1093. [Google Scholar]
- 10.Kao, C. M., L. Katz, and C. Khosla. 1994. Engineered biosynthesis of a complete macrolactone in a heterologous host. Science 265:509-512. [DOI] [PubMed] [Google Scholar]
- 11.Katz, L., and R. McDaniel. 1999. Novel macrolides through genetic engineering. Med. Res. Rev. 19:543-558. [DOI] [PubMed] [Google Scholar]
- 12.Kim, B. S., D. H. Sherman, and K. A. Reynolds. 2004. An efficient method for creation and functional analysis of libraries of hybrid type I polyketide synthases. Protein Eng. Des. Sel. 17:277-284. [DOI] [PubMed] [Google Scholar]
- 13.Le Brazidec, J. Y., A. Kamal, D. Busch, L. Thao, L. Zhang, G. Timony, R. Grecko, K. Trent, R. Lough, T. Salazar, S. Khan, F. Burrows, and M. F. Boehm. 2004. Synthesis and biological evaluation of a new class of geldanamycin derivatives as potent inhibitors of Hsp90. J. Med. Chem. 47:3865-3873. [DOI] [PubMed] [Google Scholar]
- 14.Muyrers, J. P., Y. Zhang, V. Benes, G. Testa, W. Ansorge, and A. F. Stewart. 2000. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep. 1:239-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyrers, J. P., Y. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myles, D. C. 2003. Novel biologically active natural and unnatural products. Curr. Opin. Biotechnol. 14:627-633. [DOI] [PubMed] [Google Scholar]
- 17.Ochel, H. J., K. Eichhorn, and G. Gademann. 2001. Geldanamycin: the prototype of a class of antitumor drugs targeting the heat shock protein 90 family of molecular chaperones. Cell Stress Chaperones 6:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Hagan, D. 1991. The polyketide metabolites. Ellis Howood, Chichester, United Kingdom.
- 19.Petkovic, H., R. E. Lill, R. M. Sheridan, B. Wilkinson, E. L. McCormick, H. A. McArthur, J. Staunton, P. F. Leadlay, and S. G. Kendrew. 2003. A novel erythromycin, 6-desmethyl erythromycin D, made by substituting an acyltransferase domain of the erythromycin polyketide synthase. J. Antibiot. (Tokyo) 56:543-551. [DOI] [PubMed] [Google Scholar]
- 20.Rascher, A., Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis, and C. R. Hutchinson. 2003. Cloning and characterization of a gene cluster for geldanamycin production in Streptomyces hygroscopicus NRRL 3602. FEMS Microbiol. Lett. 218:223-230. [DOI] [PubMed] [Google Scholar]
- 21.Reid, R., M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. R. Hutchinson, and R. McDaniel. 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42:72-79. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez, E., Z. Hu, S. Ou, Y. Volchegursky, C. R. Hutchinson, and R. McDaniel. 2003. Rapid engineering of polyketide overproduction by gene transfer to industrially optimized strains. J. Ind. Microbiol. Biotechnol. 30:480-488. [DOI] [PubMed] [Google Scholar]
- 23.Schnur, R. C., M. L. Corman, R. J. Gallaschun, B. A. Cooper, M. F. Dee, J. L. Doty, M. L. Muzzi, C. I. DiOrio, E. G. Barbacci, P. E. Miller, V. A. Pollack, D. M. Savage, D. E. Sloan, L. R. Pustilnik, J. D. Moyer, and M. P. Moyer. 1995. erbB-2 Oncogene inhibition by geldanamycin derivatives: synthesis, mechanism of action, and structure-activity relationships. J. Med. Chem. 38:3813-3820. [DOI] [PubMed] [Google Scholar]
- 24.Tian, Z.-Q., Y. Liu, D. Zhang, Z. Wang, S. D. Dong, C. W. Carreras, Y. Zhou, G. Rastelli, D. V. Santi, and D. C. Myles. 2004. Synthesis and biological activities of novel 17-aminogeldanamycin derivatives. Bioorg. Med. Chem. 12:5317-5329. [DOI] [PubMed] [Google Scholar]
- 25.Tsoi, C. J., and C. Khosla. 1995. Combinatorial biosynthesis of ‘unnatural’ natural products: the polyketide example. Chem. Biol. 2:355-362. [DOI] [PubMed] [Google Scholar]
- 26.Xue, Q., G. Ashley, C. R. Hutchinson, and D. V. Santi. 1999. A multiplasmid approach to preparing large libraries of polyketides. Proc. Natl. Acad. Sci. USA 96:11740-11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon, Y. J., B. J. Beck, B. S. Kim, H. Y. Kang, K. A. Reynolds, and D. H. Sherman. 2002. Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem. Biol. 9:203-214. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, Y., F. Buchholz, J. P. Muyrers, and A. F. Stewart. 1998. A new logic for DNA engineering using recombination in Escherichia coli. Nat. Genet. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, Y., J. P. Muyrers, J. Rientjes, and A. F. Stewart. 16 January 2003, posting date. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. [Online.] http://www.biomedcentral.com/1471-2199/4/1. [DOI] [PMC free article] [PubMed]
- 30.Ziermann, R., and M. C. Betlach. 1999. Recombinant polyketide synthesis in Streptomyces: engineering of improved host strains. BioTechniques 26:106-110. [DOI] [PubMed] [Google Scholar]