Abstract
Pseudoalteromonas tunicata is a biofilm-forming marine bacterium that is often found in association with the surface of eukaryotic organisms. It produces a range of extracellular inhibitory compounds, including an antibacterial protein (AlpP) thought to be beneficial for P. tunicata during competition for space and nutrients on surfaces. As part of our studies on the interactions between P. tunicata and the epiphytic bacterial community on the marine plant Ulva lactuca, we investigated the hypothesis that P. tunicata is a superior competitor compared with other bacteria isolated from the plant. A number of U. lactuca bacterial isolates were (i) identified by 16S rRNA gene sequencing, (ii) characterized for the production of or sensitivity to extracellular antibacterial proteins, and (iii) labeled with a fluorescent color tag (either the red fluorescent protein DsRed or green fluorescent protein). We then grew single- and mixed-species bacterial biofilms containing P. tunicata in glass flow cell reactors. In pure culture, all the marine isolates formed biofilms containing microcolony structures within 72 h. However, in mixed-species biofilms, P. tunicata removed the competing strain unless its competitor was relatively insensitive to AlpP (Pseudoalteromonas gracilis) or produced strong inhibitory activity against P. tunicata (Roseobacter gallaeciensis). Moreover, biofilm studies conducted with an AlpP− mutant of P. tunicata indicated that the mutant was less competitive when it was introduced into preestablished biofilms, suggesting that AlpP has a role during competitive biofilm formation. When single-species biofilms were allowed to form microcolonies before the introduction of a competitor, these microcolonies coexisted with P. tunicata for extended periods of time before they were removed. Two marine bacteria (R. gallaeciensis and P. tunicata) were superior competitors in this study. Our data suggest that this dominance can be attributed to the ability of these organisms to rapidly form microcolonies and their ability to produce extracellular antibacterial compounds.
Biofouling is ubiquitous in the marine environment, and bacteria are among the first organisms to foul surfaces. They form biofilms which serve as a focus for the attachment and growth of other organisms, such as invertebrates, sessile plants, and animals (15). Mature marine biofouling communities are complex, highly dynamic ecosystems and, once established, are extremely difficult to eradicate (28).
Many marine organisms have evolved efficient strategies to combat epibiosis. Seaweeds employ a number of physical and chemical defense systems to prevent fouling, such as the shedding of outer layers of cells (35) or production of inhibitory compounds (17). However, antifouling defense is costly (57), and it has been hypothesized that the seaweed Ulva lactuca, which has neither physical nor chemical defenses, relies on microbial defense (19, 32). It has been demonstrated that bacterial biofilms are present on the surface of U. lactuca (53), and it has been shown that biofilms can be beneficial for their hosts by enhancing their antifouling strategies (2, 7, 16, 24, 30, 46).
One important group of marine bacteria that is found in association with living surfaces is Pseudoalteromonas spp. This genus produces a diverse range of biologically active compounds that specifically target marine fouling organisms. Perhaps the most extensively studied species in the genus is Pseudoalteromonas tunicata. P. tunicata is a green-pigmented gram-negative gamma-proteobacterium. It colonizes living marine surfaces, including U. lactuca (29), and produces at least five novel inhibitory compounds. One of these compounds is a 190-kDa multisubunit antibacterial protein designated AlpP which is effective against both gram-negative and gram-positive bacteria from a range of environments (33). However, this protein was also found to be active against P. tunicata itself, which raises a question about the ecological role of AlpP. It has been speculated that AlpP might provide a competitive advantage to P. tunicata during biofilm growth in the marine environment.
P. tunicata has been shown to form complex differentiated biofilms (39). Biofilm formation appears to typically follow a sequence. Single cells attach to a surface and differentiate into matrix-enclosed microcolonies separated by a network of open water channels. Dispersal of bacteria from the interior regions of the microcolonies has been observed, resulting in formation of hollow voids inside the microcolonies (39). Microcolony development is thought to be a coordinated adaptive response that facilitates continued biofilm development and dispersal (60). Very little is known about the role of microcolonies within biofilms. Recent findings suggest that the formation of microcolonies by Pseudomonas aeruginosa allows it to escape predation by flagellates (41). Generally, microcolonies may be an adaptive strategy for competing under stressful conditions.
The microcolonies that constitute a biofilm can be composed of single-species populations or mixed populations with various degrees of interaction, depending on the environmental conditions under which they were formed. Two species formed mixed microcolonies when they were grown under commensal conditions and formed separate microcolonies under noncommensal conditions (48). Thus, commensal relationships can play a role in determining the spatial distribution of the organisms in microbial communities. Meanwhile, in competitive interactions between bacteriocin-producing and bacteriocin-sensitive strains, the bacteria formed biofilms in which each strain formed its own microcolonies (55). Generally, surprisingly little is known about the factors that govern the establishment and distribution of bacteria within multispecies biofilms in marine environments. A few studies on epiphytic microbial communities present on macroalgae have highlighted the complex spatial distribution of bacterial populations (12), but the strategies that marine bacteria use to colonize surfaces and to compete with other bacteria are poorly understood.
In this study we investigated the hypothesis that P. tunicata competes effectively with other marine bacterial isolates during biofilm formation based on the isolation of this organism from marine plants in repeated sampling and based on the production of the potent antibacterial compound AlpP by this species. We observed that in pure cultures all the marine isolates formed stable biofilms within 72 h. P. tunicata was found to be the dominant isolate and totally removed a competing strain unless the competitor was not sensitive to the P. tunicata antibacterial protein or exhbited strong inhibitory activity against P. tunicata. In competition studies in which microcolonies were allowed to form before the introduction of P. tunicata, biofilms coexisted for greatly extended periods. Studies with a P. tunicata alpP mutant suggested that AlpP provides an advantage during colonization of biofilms formed by other marine bacteria. Our data also suggest that microcolonies may be protective structures during biofilm development that enhance the persistence of an organism during competitive interactions.
MATERIALS AND METHODS
Isolation of marine strains.
P. tunicata and other isolates were originally isolated from the surface of the common marine alga U. lactuca, which was collected from the rocky intertidal zone near Sydney, Australia. The algal thallus was suspended in sterile nine-salt solution, and surface bacteria were removed by vortexing. Aliquots of the samples were then spread on the complex marine medium Vaatanen nine-salt solution (VNSS) (40) containing 1.5% agar and incubated at 23°C for 48 h. Morphologically distinct bacterial colonies were selected. Bacteria were stored at −70°C in 30% glycerol. While over 50 different colony phenotypes were observed, only 20 strains survived repeated subculturing on VNSS. These strains were routinely grown and maintained on VNSS agar at 25°C. Sequencing of these strains was part of another project and was carried out by Tujula et al. (N. Tujula, J. S. Webb, I. Dahllöf, C. Hölmström, and S. Kjelleberg, unpublished data). Four of the 20 culturable strains were chosen in addition to P. tunicata, as these strains are commonly found growing on algae. The strains were Pseudoalteromonas gracilis, Alteromonas sp., Cellulophaga fucicola, and Roseobacter gallaeciensis strains.
Fluorescent labeling of marine isolates.
Marine strains were labeled with a green fluorescent protein (GFP) color tag by transconjugation by using the constitutive GFP expression plasmid pCJS10. This plasmid contains the gfpmut3 gene (11) on an RSF1010 backbone from the broad-host-range vector pHRP304 (3). In addition, strains were labeled with a red fluorescent protein (RFP) color tag by using the pCJS10-derived plasmid pCJS10R. This plasmid contains the RFP gene dsred (Clontech) in place of gfpmut3 on pCJS10. Plasmids pCJS10 and pCJS10R were gifts from Charles Svenson, University of New South Wales, Sydney, Australia. GFP triparental conjugations were carried out as described previously (19), and labeled transconjugants were grown on VNSS agar plates containing 15 μg of chloramphenicol per ml and 100 μg of streptomycin per ml. GFP- and RFP-labeled strains showed bright fluorescence after overnight culture, and we observed no differences in the growth rates or surface attachment properties of the labeled strains compared with the unlabeled parent strain (data not shown).
Detection of inhibitory compounds produced by marine bacterial isolates.
In order to detect inhibitory compounds produced by marine bacterial isolates, concentrated supernatants from P. tunicata and other marine isolates were prepared by the method described previously (33), with some modifications. The strains were grown in 3M medium with 0.2% (wt/vol) trehalose and 0.2% (wt/vol) glucose as the carbon sources for 48 h, harvested by centrifugation (12,000 × g for 20 min), and resuspended in 3M medium (47) containing trehalose at a density of 0.7 g ml−1. Each concentrated cell suspension was incubated without shaking at 28°C for 24 h. Cells were removed by centrifugation (14,000 × g for 1.5 h), and the concentrated supernatant was sterilized by using a 0.2-μm-pore-size sterile filter (Millipore).
Supernatant samples were assayed for inhibitory activity by the drop test assay (33). Briefly, 100 μl of an overnight broth culture of the target strain was spread on a VNSS agar plate, and the plate was dried at 30°C for 30 min. Drops containing 20 μl of the concentrated supernatant, as well as a control (nine-salt solution), were placed on the agar surface and incubated overnight at room temperature to allow formation of inhibition zones.
Biofilm experiments.
Biofilms were grown in continuous-culture flow cells (channel dimensions, 1 by 4 by 40 mm) at room temperature as previously described (44). Channels were inoculated with overnight cultures and incubated with no flow for 1 h at room temperature. Cultures were adjusted so that biofilms were established with a flow rate of 150 μl min−1. R. gallaeciensis, which could not be labeled with a fluorescent color tag, was stained with Syto 59 diluted to a concentration of 3 μl ml−1 in biofilm media. (Biofilms were grown in 3M medium containing 0.01% trehalose, 0.01% glucose, and 0.01% fructose.) Biofilms were visualized with a confocal laser scanning microscope (Olympus) by using fluorescein isothiocyanate and tetramethyl rhodamine isocyanate optical filters.
To cultivate mixed biofilms, flow chambers were inoculated with 500 μl of a mixture of stationary-phase cultures of P. tunicata and a competitive marine strain. In order to ensure that the initial ratio of attached cells of the two competing strains was 1:1, we first monitored the initial attachment of the mixed culture after a 1-h adhesion period. In most cases, the initial levels of attachment to the glass surface were approximately equal, as determined by counting the numbers of red- and green-labeled cells by epifluorescence microscopy by using an eyepiece grid. However, for C. fucicola and Alteromonas it was necessary to increase the ratios of cells to 4:1 and 2:1, respectively, in order to achieve equal levels of attachment for the two competing strains.
To investigate in more detail whether microcolonies improved the competitiveness of strains, marine strains were allowed to preestablish and form microcolonies for 24 h (P. gracilis) and 48 h (C. fucicola and Alteromonas sp.). Each preformed biofilm was inoculated with ∼10 7 cells of wild-type P. tunicata, and the flow was stopped for 1 h. After resumption of the flow, the biofilm was examined for red and green fluorescence. Experiments were repeated in three separate rounds with three independent flow cells running in parallel.
RESULTS AND DISCUSSION
Characterization of marine strains. (i) 16S rRNA gene sequencing.
We first carried out partial 16S rRNA gene sequencing of bacteria isolated from U. lactuca (Table 1). We found that the four isolates studied all exhibited 16S rRNA gene homology with organisms that are commonly isolated from marine eukaryotic surfaces. C. fucicola belongs to the Flavobacterium group of the Bacteroidetes, a diverse group with members commonly found in coastal marine regions (49). C. fucicola is frequently found on the surface of marine benthic macroalgae (6) and has been found to decompose highly polymeric material from the brown alga Fucus serratus (34). The sequenced strains in our collection also contained Pseudoalteromonas and Alteromonas spp. The recently described genus Pseudoalteromonas resulted from division of the genus Alteromonas into two genera, Alteromonas and Pseudoalteromonas, based on the phylogenetic comparison of Gauthier et al. (21). Numerous bacteria belonging to these genera are frequently isolated from marine waters and are found in association with marine invertebrates, algae, plants, and animals (31). They are readily culturable and are also capable of surviving in nutrient-poor environments. P. gracilis is frequently found on algae such as Gracilaria and produces disease symptoms due to its agarolytic activity (52). Members of the genus Roseobacter are often found on the surfaces of algae (9, 43).
TABLE 1.
16S rRNA gene identification of bacteria isolated from the marine alga U. lactuca
| Strain | % Identity to sequence in database | Closest relative |
|---|---|---|
| 2.06 | 97 | Cytophaga fucicola |
| 2.10 | 99 | Roseobacter gallaeciensis |
| 2.14 | 95 | Pseudoalteromonas gracilis |
| 2.19 | 99 | Alteromonas sp. |
(ii) Production of extracellular antibacterial compounds.
We examined the ability of the bacterial isolates from U. lactuca to inhibit the growth of each of the other isolates from the plant. Studies of the antibacterial activity in concentrated supernatants indicated that P. tunicata and R. gallaeciensis were the most inhibitory (Table 2). Each of these strains could inhibit the growth of several of the bacterial strains used in this study. P. tunicata exhibited strong inhibitory activity against itself and C. fucicola and weak inhibitory activity against P. gracilis. The 190-kDa protein (AlpP) responsible for the antibacterial activity of P. tunicata has now been well characterized (33). This protein contains at least two subunits (60 and 80 kDa), which are joined together by noncovalent bonds, and it was shown to be released during the stationary phase (33). P. tunicata possesses a ToxR-like regulon which appears to control determinants for the expression of iron uptake and also regulates expression of AlpP (S. Stelzer, S. Egan, and S. Kjelleberg unpublished data).
TABLE 2.
Drop test activity for the detection of extracellular inhibitory compounds active against P. tunicata and sensitivity of each strain to the P. tunicata antibacterial protein AlpP, as determined by the droplet test assay
| Producer | Scores with the following targetsa:
|
||||
|---|---|---|---|---|---|
| P. tuni- cata | P. gra- cilis | Altero- monas sp. | C. fuci- cola | R. gallae- ciensis | |
| P. tunicata | 3 | 0-1 | 1 | 3 | 1 |
| P. gracilis | 1 | 1 | 0 | 2 | 1 |
| Alteromonas sp. | 1 | 0 | 0 | 2 | 1 |
| C. fucicola | 0 | 0 | 0 | 0 | 0 |
| R. gallaeciensis | 3 | 3 | 1 | 0 | 1 |
A score of 3 indicates a high level of sensitivity (complete inhibition of growth in the region of the droplet), and a score of 1 indicates slight sensitivity (partial inhibition of growth in the region of the droplet, which appears turbid with bacterial growth).
R. gallaeciensis exhibited strong activity against both P. tunicata and P. gracilis. Production of secondary metabolites by members of this group has been reported previously (5, 25, 51). One of the inhibitory compounds (proposed to be a peptide) has been reported to be produced only in the presence of other bacteria (51), although in this study we observed strong inhibitory activity with pure-culture supernatants of this strain. More recent work indicated that R. gallaeciensis also produces a new antibiotic called tropodithietic acid (8). This compound showed strong inhibitory activity against marine bacteria belonging to various taxa and marine algae.
C. fucicola did not exhibit antibacterial activity against the strains used in this study. However, a Cytophaga strain (RB1057) has previously been shown to produce an extracellular inhibitor of expansion of colonies of closely related Cytophaga strains (10). The inhibitor (a glycoprotein) inhibited the competing strain's ability to adhere to and glide on a substrate. The inhibitor had no measurable impact on several other related strains of gliding bacteria. Thus, inhibitors produced by this genus may only be effective against closely related bacteria, which may explain why no activity was detected for the Cellulophaga strain tested in this study. In their studies on antagonistic interactions among marine pelagic bacteria, Long and Azam found that the members of the Bacteroidetes group were the most sensitive to inhibition by other marine bacteria and were also the least inhibitory (37). Similarly, Grossart and coworkers found that the Flavobacterium-Sphingobacterium group had the lowest percentage of inhibitory strains among a diverse group of 51 marine bacterial isolates (27).
In this study Alteromonas sp. showed activity against P. tunicata, C. fucicola, and R. gallaeciensis. Alteromonas spp. have been shown to produce a wide range of inhibitory compounds. Some species, such as Alteromonas citrea and Alteromonas rubra, produce only a macromolecular polyanionic substance, whereas other species, such as Alteromonas luteoviolacea, produce both a diffusible macromolecule and two intracellular low-molecular-weight brominated compounds (22). Barja et al. (4) found that Alteromonas species isolated from seaweeds produced thermolabile low-molecular-weight inhibitors (molecular weights, less than 2,000), whereas strains isolated from seawater produced a high-molecular-weight glycoprotein (molecular weight, 90,000), which displayed a broad spectrum of inhibitory activity against medical and environmental isolates. An Alteromonas strain (SWAT5) derived from particulate organic matter was found to produce 2-alkyl-quinolinols. The antibiotics were produced only on polysaccharide matrices and were found to be hydrophobic (38).
Single-species biofilm development.
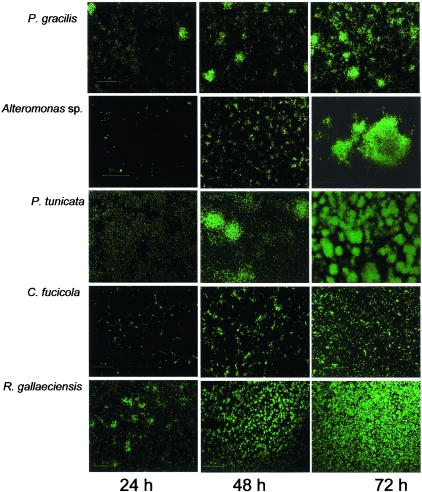
Biofilm development for each of the marine strains in monoculture was monitored in glass flow cells (Fig. 1). For all strains, single cells were observed to attach to the substratum after inoculation. Each strain established a stable biofilm with a characteristic architecture after 72 h. All of the strains exhibited microcolony formation to various extents during the course of biofilm development. P. tunicata and P. gracilis each formed well-defined spherical microcolonies that were up to 50 μm in diameter and 100 μm high within 48 h after inoculation. Alteromonas sp. also formed large, distinct microcolony structures which were frequently over 100 μm in diameter and 50 μm high after 72 h of biofilm development. In contrast, the biofilms formed by R. gallaeciensis and C. fucicola were less structured. The C. fucicola biofilms consisted of both flat unstructured regions and many small microcolonies that were up to 10 μm in diameter and up to 10 μm high. R. gallaeciensis initially formed cell chains which later developed into small microcolonies (10 to 20 μm) during the early stages of biofilm development. These clusters then formed a relatively flat, mat-like biofilm whose thickness approached 10 μm. For each strain, the characteristic architecture formed after 72 h persisted for at least another 2 days within flow cells. However, after 8 days we noted that the majority of the biomass had detached from the flow cells for all of the strains studied, which left only single cells attached to the flow cells (data not shown).
FIG. 1.
Single-species biofilm development for bacteria isolated from the marine alga U. lactuca expressing GFP. Magnification, ×600.
The marine strains studied here comprise both organisms that form microcolonies rapidly and organisms that form less-defined microcolonies. Previously, it has been proposed that microcolonies can result from carbon flux gradients during growth within biofilms (56). In addition, many genetically encoded regulatory and structural determinants of microcolony formation have been revealed over the past few years. Some recent examples include the roles of conjugative plasmids (23, 50) and antigen 43 (13, 50) in enhancing microcolony formation in Escherichia coli biofilms, suggesting that these cell surface structures might act as cellular adhesins in stabilizing microcolony structures. Generally, the formation of multicellular structures, such as microcolonies, is thought to be an adaptive response that mediates survival under unfavorable conditions.
Of the marine organisms studied here, only P. tunicata has previously been characterized for biofilm development under continuous-culture conditions. In agreement with the present study, P. tunicata was observed previously to form prominent microcolony structures during biofilm development (39). Biofilm development has been well characterized in the marine bacterium Vibrio cholerae, which also can form microcolonies (45, 58, 59). To our knowledge, however, there have been no reports that have considered the ecological role of biological structures in mediating interactions between bacteria in mixed-community marine biofilms. In several studies the workers investigated interactions in marine biofilm communities (14, 26, 36). However, these workers focused mainly on succession events or addressed the issue of attachment and detachment of defined pure and mixed cultures on agar beads. In this study, we also examined the role of microcolonies during mixed-species biofilm development.
Mixed-species biofilm development.
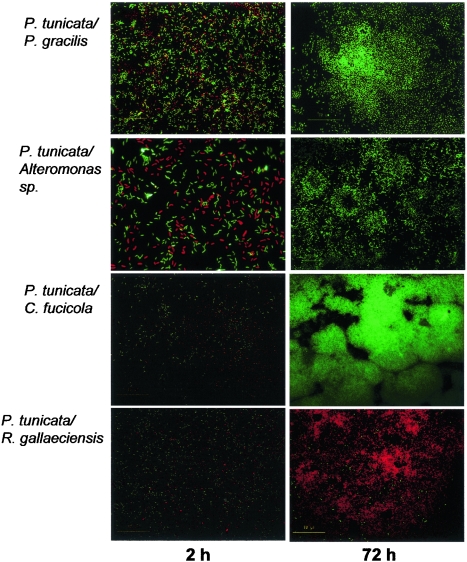
Because of its strong inhibitory activity against a broad range of surface-fouling organisms, we hypothesized that P. tunicata would exhibit an aggressive biofilm-forming lifestyle and would dominate other marine strains during competitive biofilm development. We therefore prepared mixed 1:1 inocula containing P. tunicata (GFP tagged) and a second organism (dsRed tagged) and allowed them to attach in a flow cell. In this experiment, P. tunicata totally outcompeted C. fucicola and Alteromonas sp. (Fig. 2). No cells of the latter competing organisms were visible after 24 h. This is consistent with our observation that P. tunicata produces inhibitory compounds. The results of the drop test assay (Table 2) showed that both C. fucicola and Alteromonas sp. are susceptible to inhibitors from P. tunicata. P. gracilis, which has a lower susceptibility to AlpP, was able to coexist with P. tunicata for up to 72 h before it was removed. The outcome of the competition may be correlated with the results of the drop test assay, which showed that P. gracilis is relatively insensitive to the antibacterial protein.
FIG. 2.
Competitive biofilm development in two-species biofilms containing P. tunicata (expressing GFP) and other marine bacterial isolates (expressing RFP). Magnification, approximately ×600.
In contrast to the dominance of P. tunicata over C. fucicola and Alteromonas, P. tunicata was outcompeted by R. gallaeciensis. While some cells of P. tunicata persisted on the surface throughout the experimental period, these cells did not grow, and R. gallaeciensis rapidly grew and formed a biofilm. This is again consistent with the results of the drop test assay, which showed that P. tunicata is highly susceptible to the inhibitory compounds produced by R. gallaeciensis.
Role of the P. tunicata antibacterial protein AlpP in competitive biofilm development.
To evaluate whether the antibacterial protein AlpP can play a role in competitive biofilm development, a ΔalpP insertional knockout mutant (39) was used in competition studies. We first repeated the mixed-inoculum biofilm experiments described above by using the ΔalpP strain instead of the wild type. In these experiments we observed no difference in the outcome of the biofilm competition experiments when the wild-type and ΔalpP strains were used. When inoculated together with C. fucicola, Alteromonas sp., or P. gracilis, the ΔalpP mutant became dominant after 72 h, similar to the results obtained for the wild-type strain (data not shown). Also, R. gallaeciensis was dominant over the P. tunicata ΔalpP strain in a manner similar to the manner observed with wild-type P. tunicata.
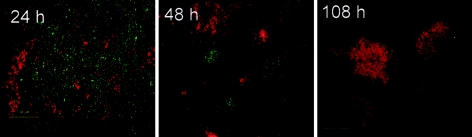
However, we found that AlpP can play an important role in the colonization of preestablished biofilms of other marine strains. As shown in Fig. 3, the ΔalpP strain had a greatly reduced ability to colonize 24-h-old biofilms of P. gracilis. Wild-type P. tunicata could become established within such a biofilm, form microcolonies, and completely remove P. gracilis in a manner similar to the manner observed for the mixed-inoculum biofilm shown in Fig. 2. In contrast, introduction of the ΔalpP strain led to establishment of a mixed-species biofilm containing separate microcolonies of each strain, but after 108 h the ΔalpP mutant strain was completely overtaken by P. gracilis (Fig. 3.). When the P. tunicata ΔalpP strain was introduced into preestablished biofilms of C. fucicola and Alteromonas sp., mixed-species biofilms were formed that persisted for over 9 days. In contrast, the wild-type strain was able to remove these competitors after 120 h (data not shown).
FIG. 3.
A P. tunicata mutant that did not produce the antibacterial protein AlpP had reduced ability to compete against P. gracilis. The images show two-species biofilm development for P. tunicata ΔalpP (green) and P. gracilis (red) strains. After 96 h, microcolonies of P. tunicata and P. gracilis coexisted within the biofilm (compare with Fig. 2, which shows that wild-type P. tunicata could outcompete P. gracilis after 72 h). After 108 h, P. gracilis outcompeted the P. tunicata ΔalpP mutant strain. Magnification, ×600.
These results suggest that AlpP can provide a competitive advantage in certain ecological situations, such as the colonization of established biofilms. The production of inhibitory compounds by marine organisms may be a response to various ecological pressures in the environment. Epiphytic bacteria live in a highly competitive environment in which space and access to nutrients are limited. It is possible that P. tunicata up-regulates production of the antibacterial protein in the presence of competitors; in future studies we will examine AlpP expression in mixed-species biofilms by using a Gfp reporter protein fused to the alpP promoter. Some bacteria exhibit an inducible chemical antagonism when they are grown in the presence of competing marine bacteria (42, 54). It is also possible that the antibacterial protein is a key factor when there is competition between microcolonies. Evans and coworkers (20) compared protease production in planktonic and biofilm cells and found that the production was higher in the latter cells, suggesting that antimicrobial agents were important in biofilms. Furthermore, it has been shown that strains that are normally considered organisms that do not produce inhibitors can express inhibitory activity when they are growing as a biofilm (61). Moreover, there is compelling evidence that mature microcolonies may be the site of inhibitor production in P. tunicata. Production of inhibitors has been linked to pigmentation in P. tunicata (18), and we have observed mature microcolonies become pigmented in biofilms (data not shown).
Role of microcolonies in competitive interactions within biofilms.
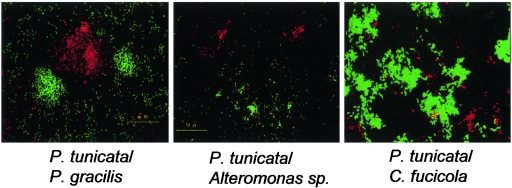
To evaluate the role of microcolonies in competition, microcolonies of Alteromonas sp., P. gracilis, and C. fucicola were allowed to preform before they were colonized by P. tunicata. Our data suggest that microcolony formation may enhance the ability of an organism to compete against P. tunicata and persist within a flow cell reactor. It was found that P. gracilis biofilms containing microcolonies could coexist with P. tunicata for more than 5 days, in contrast to biofilms without microcolonies (Fig. 4). Moreover, preestablished biofilms of C. fucicola and Alteromonas sp. (which normally persist for no more than 24 h in the presence of P. tunicata) were able to persist for up to 72 h before they were dispersed. Thus, high densities of cells within microcolonies may allow enhanced persistence during coculture with the superior competitor P. tunicata. This conclusion is supported by the outcome of competition studies conducted by other researchers. In two-species biofilms, bacteriocin-producing enteric bacteria prevented the colonization of a potential competitor in a preestablished biofilm (55). Al-Bakri et al. (1) also found that preestablished biofilms of P. aeruginosa conferred colonization resistance to Burkholderia cepacia.
FIG. 4.
Microcolonies enhanced the coexistence of strains in competition with P. tunicata. Competing marine strains (red) were allowed to form microcolonies before introduction of the superior competitor P. tunicata (green) into the flow cell. Microcolonies were able to persist for more than 72 h in competition with P. tunicata. In contrast, in mixed-inoculum experiments P. tunicata completely removed inferior competitors after 72 h (see Fig. 2).
Conclusions.
By labeling marine bacteria isolated from the green alga U. lactuca with genetic color tags, we were able to examine the colonization, biofilm formation behavior, and competitive interactions of the strains during mixed-species biofilm development in real time. This study is the first study to demonstrate that inhibitory compounds produced by marine bacteria can provide a competitive advantage during competitive growth within biofilms. We further suggest that microcolony formation may represent one adaptive strategy that increases the ability of bacteria to persist in mixed-species biofilms under competitive biofilm formation conditions.
Our studies showed that the AlpP protein, a potent antibacterial protein produced by P. tunicata, can enhance the ability of P. tunicata to colonize biofilms formed by other bacteria. The wild-type P. tunicata strain aggressively colonized and dominated strains that were sensitive to the AlpP protein. In contrast, a P. tunicata alpP mutant was defective in the ability to colonize and dominate biofilms under the same conditions. Moreover, marine strains that were tolerant to the AlpP protein in drop plate assays were recalcitrant to invasion and displacement by P. tunicata during biofilm development.
Previous studies performed in our laboratory have suggested that bacteria present on the surface of some higher marine eukaryotes may play an important role in the chemical defense against biofouling in the marine environment (30). Many members of the genus Pseudoalteromonas, for example, produce multiple extracellular inhibitory compounds that target different classes of marine fouling organisms (28). Competitive interactions between bacteria, such as those demonstrated in this study, may play an important role in determining the composition of such antifouling communities, because they likely lead to predominance of inhibitory bacteria on the host surface.
We also suggest that specific interactions between inhibitor-producing bacteria and eukaryotic host surfaces may enhance the antifouling defense of the host organism. For example, recent studies in our laboratory have revealed that P. tunicata possesses a mannose-sensitive type IV pilus that promotes attachment to the cellulose-containing surfaces of U. lactuca and Ciona intestinalis, the principal hosts of P. tunicata in the marine environment (D. Saludes, A. Scheffel, S. James, J. Webb, C. Holmstrom, and S. Kjelleberg, unpublished data). Both U. lactuca and C. intestinalis are thought to rely on inhibitor-producing bacteria for defense against biofouling; thus, it may be an advantage to these organisms to become associated with antifouling bacteria such as P. tunicata. The present study provides a platform for further studies of interactions between marine bacteria in surface-associated communities and between biofilms and their eukaryotic host surfaces.
Acknowledgments
This work was supported by a USP-AusAID scholarship, by the Australian Research Council, and by The Leverhulme Trust, United Kingdom.
We thank C. Svenson for providing the plasmids used for labeling strains. We also thank our colleagues at the Centre for Marine Biofouling and Bio-Innovation, University of New South Wales, Sydney, Australia, for their support.
REFERENCES
- 1.Al-Bakri, A. G., P. Gilbert, and D. G. Allison. 2004. Immigration and emigration of Burkholderia cepacia and Pseudomonas aeruginosa between and within mixed biofilm communities. J. Appl. Microbiol. 96:455-463. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, E., L. Yan, K. G. Boyd, P. C. Wright, and J. G. Burgess. 2001. The symbiotic role of marine microbes on living surfaces. Hydrobiologia 461:37-40. [Google Scholar]
- 3.Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. [DOI] [PubMed] [Google Scholar]
- 4.Barja, J. L., M. L. Lemos, and A. E. Toranzo. 1989. Purification and characterization of an antibacterial substance produced by a marine Alteromonas species. Antimicrob. Agents Chemother 33:1674-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettcher, K. J., B. J. Barber, and J. T. Singer. 2000. Additional evidence that juvenile oyster disease is caused by a member of the Roseobacter group and colonization of nonaffected animals by Stappia stellulata-like strains. Appl. Environ. Microbiol. 66:3924-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolinches, J., M. L. Lemos, and J. L. Barja. 1988. Population dynamics of heterotrophic bacterial communities associated with Fucus vesiculosus and Ulva rigida in an estuary. Microb. Ecol. 15:345-357. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, K. G., D. R. Adams, and J. G. Burgess. 1999. Antibacterial and repellent activities of marine bacteria associated with algal surfaces. Biofouling 14:227-236. [Google Scholar]
- 8.Brinkhoff, T., G. Bach, T. Heidorn, L. Liang, A. Schlingloff, and M. Simon. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinkmeyer, R., M. Rappe, S. Gallacher, and L. Medlin. 2000. Development of clade- (Roseobacter and Alteromonas) and taxon-specific oligonucleotide probes to study interactions between toxic dinoflagellates and their associated bacteria. Eur. J. Phycol. 35:315-329. [Google Scholar]
- 10.Burchard, R. P., and M. L. Sorongon. 1998. A gliding bacterium strain inhibits adhesion and motility of another gliding bacterium strain in a marine biofilm. Appl. Environ. Microbiol. 64:4079-4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Corre, S., and D. Prieur. 1996. Density and morphology of epiphytic bacteria on the kelp Laminaria digitata. Bot. Mar. 33:515-523. [Google Scholar]
- 13.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 14.Dang, H., and C. R. Lovell. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of 16S rRNA genes. Appl. Environ. Microbiol. 66:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, A. R., N. M. Targett, O. J. McConnel, and C. M. Young. 1989. Epibiosis of marine algae and benthic invertebrates: natural products chemistry and other mechanisms inhibiting settlement and overgrowth. Bioorg. Mar. Chem. 3:85-114. [Google Scholar]
- 16.Dobretsov, S. V., and P. Y. Qian. 2002. Effect of bacteria associated with the green alga Ulva reticulata on marine micro- and macrofouling. Biofouling 18:217-228. [Google Scholar]
- 17.Dworjanyn, S. A., R. de Nys, and P. D. Steinberg. 1999. Localization and surface quantification of secondary metabolites in the red alga Delisea pulchra. Mar. Biol. 133:727-736. [Google Scholar]
- 18.Egan, S., S. James, C. Holmstrom, and S. Kjelleberg. 2002. Correlation between pigmentation and antifouling compounds produced by Pseudoalteromonas tunicata. Environ. Microbiol. 4:433-442. [DOI] [PubMed] [Google Scholar]
- 19.Egan, S., S. James, and S. Kjelleberg. 2002. Identification and characterization of a putative transcriptional regulator controlling the expression of fouling inhibitors in Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 68:372-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, E., M. R. Brown, and P. Gilbert. 1994. Iron chelator, exopolysaccharide and protease production in Staphylococcus epidermidis: a comparative study of the effects of specific growth rate in biofilm and planktonic culture. Microbiology 140:153-157. [DOI] [PubMed] [Google Scholar]
- 21.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 22.Gauthier, M. J., and G. N. Flatau. 1976. Antibacterial activity of marine violet pigmented Alteromonas with special reference to the production of brominated compounds. Can. J. Microbiol. 22:349-354. [DOI] [PubMed] [Google Scholar]
- 23.Ghigo, J.-M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 24.Gil-Turness, M. S., and W. Fenical. 1992. Embryos of Homarus americanus are protected by epibiotic bacteria. Biol. Bull. 182:105-108. [DOI] [PubMed] [Google Scholar]
- 25.Gram, L., H. P. Grossart, A. Schlingloff, and T. Kiorboe. 2002. Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl. Environ. Microbiol. 68:4111-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossart, H.-P., T. Kiorboe, K. Tang, and H. Ploug. 2003. Bacterial colonization of particles: growth and interactions. Appl. Environ. Microbiol. 69:3500-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grossart, H. P., A. Schlingloff, M. Bernhard, M. Simon, and T. Brinkhoff. 2004. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol. Ecol. 47:387-396. [DOI] [PubMed] [Google Scholar]
- 28.Holmstrom, C., S. Egan, A. Franks, S. McCloy, and S. Kjelleberg. 2002. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 41:47-58. [DOI] [PubMed] [Google Scholar]
- 29.Holmstrom, C., S. James, B. A. Neilan, D. C. White, and S. Kjelleberg. 1998. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 48:1205-1212. [DOI] [PubMed] [Google Scholar]
- 30.Holmstrom, C., and S. Kjelleberg. 1994. The effect of external biological factors on settlement of marine invertebrate and new antifouling technology. Biofouling 8:147-160. [Google Scholar]
- 31.Holmstrom, C., and S. Kjelleberg. 1999. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol. Ecol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 32.Holmstrom, C., D. Rittschof, and S. Kjelleberg. 1992. Inhibition of settlement by larvae of Balanus amphitrite and Ciona intestinalis by a surface-colonizing marine bacterium. Appl. Environ. Microbiol. 58:2111-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.James, S., C. Holmstrom, and S. Kjelleberg. 1996. Purification and characterization of a novel antibacterial protein from the marine bacterium D2. Appl. Environ. Microbiol. 62:2783-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansen, J. E., P. Nielsen, and C. Sjoholm. 1999. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of Cytophaga lytica to Cellulophaga lytica gen. nov., comb. nov. Int. J. Syst. Bacteriol. 49:1231-1240. [DOI] [PubMed] [Google Scholar]
- 35.Keats, D. W., M. A. Knight, and C. M. Pueschel. 1997. Antifouling effects of epithallial shedding in three crustose coralling algae (Rhodophyta, Coralines) on a coral reef. J. Exp. Mar. Biol. Ecol. 213:281-293. [Google Scholar]
- 36.Kiorboe, T., K. Tang, H.-P. Grossart, and H. Ploug. 2003. Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl. Environ. Microbiol. 69:3036-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long, R. A., and F. Azam. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long, R. A., A. Qureshi, D. J. Faulkner, and F. Azam. 2003. 2-n-Pentyl-4-quinolinol produced by a marine Alteromonas sp. and its potential ecological and biogeochemical roles. Appl. Environ. Microbiol. 69:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mai-Prochnow, A., F. Evans, D. Dalisay-Saludes, S. Stelzer, S. Egan, S. James, J. S. Webb, and S. Kjelleberg. 2004. Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70:3232-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marden, P., A. Tunlid, K. Malmcrona-Friberg, G. Odham, and S. Kjelleberg. 1985. Physiological and morphological changes during short term starvation of marine bacteria isolates. Arch. Microbiol. 142:326-332. [Google Scholar]
- 41.Matz, C., T. Bergfeld, S. A. Rice, and S. Kjelleberg. 2004. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ. Microbiol. 6:218-226. [DOI] [PubMed] [Google Scholar]
- 42.Mearns-Spragg, A., M. Bregu, K. G. Boyd, and J. G. Burgess. 1998. Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates after exposure to terrestrial bacteria. Lett. Appl. Microbiol. 27:142-146. [DOI] [PubMed] [Google Scholar]
- 43.Meusnier, I., J. L. Olsen, W. T. Stam, C. Destombe, and M. Valero. 2001. Phylogenetic analysis of Caulerpa taxifolia (Chlorophyta) and its associated bacterial microflora provide clues to the origin of the Mediterranean introduction. Mol. Ecol. 10:931-946. [DOI] [PubMed] [Google Scholar]
- 44.Moller, S., C. Sternberg, J. B. Andersen, B. B. Christensen, J. L. Ramos, M. Givskov, and S. Molin. 1998. In situ gene expression of mixed-culture biofilms: evidence of metabolic interactions between community members. Appl. Environ. Microbiol. 64:721-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moorthy, S., and P. I. Watnick. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakasono, S., J. G. Burgess, K. Takashi, M. Koike, C. Murayama, S. Nakamura, and T. Matsunaga. 1993. Electrochemical prevention of marine biofouling with a carbon-chloroprene sheet. Appl. Environ. Microbiol. 59:3757-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neidhart, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen, A., T. Tolker-Neilsen, K. Barken, and S. Molin. 2000. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ. Microbiol. 2:59-68. [DOI] [PubMed] [Google Scholar]
- 49.Pinhassi, J., U.-L. Zweifel, and A. Hagstrom. 1997. Dominant marine bacterioplankton species found among colony-forming bacteria. Appl. Environ. Microbiol. 63:3359-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reisner, A., J. A. J. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 51.Ruiz-Ponte, C., V. Cilia, C. Lambert, and J. L. Nicolas. 1998. Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int. J. Syst. Bacteriol. 48:537-542. [DOI] [PubMed] [Google Scholar]
- 52.Schroeder, D. C., M. A. Jaffer, and V. E. Coyne. 2003. Investigation of the role of a β(1-4)agarase produced by Pseudoalteromonas gracilis B9 in eliciting disease symptoms in the red alga Gracilaria gracilis. Microbiology 149:2919-2929. [DOI] [PubMed] [Google Scholar]
- 53.Sieburth, J. M. 1975. Microbial seascapes: a pictoral essay on marine microorganisms and their environments. University Park Press, Baltimore, Md.
- 54.Slattery, M., I. Rajbhandari, and K. Wesson. 2001. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb. Ecol. 41:90-96. [DOI] [PubMed] [Google Scholar]
- 55.Tait, K., and I. W. Sutherland. 2002. Antagonistic interactions among bacteriocin-producing enteric bacteria in dual species biofilms. J. Appl. Microbiol. 93:345-352. [DOI] [PubMed] [Google Scholar]
- 56.van Loosdrecht, M. C., J. J. Heijnen, H. Eberl, J. Kreft, and C. Picioreanu. 2002. Mathematical modeling of biofilm structures. Antonie Leeuwenhoek 81:245-256. [DOI] [PubMed] [Google Scholar]
- 57.Wahl, M. 1989. Marine epibosis. 1. Fouling and antifouling: some basic aspects. Mar. Ecol. Prog. Ser. 58:175-189. [Google Scholar]
- 58.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan, L., K. G. Boyd, D. R. Adams, and J. G. Burgess. 2003. Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl. Environ. Microbiol. 69:3719-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]