Abstract
Filamentous bacteria with a conspicuous morphology were found in the majority of the bacterioplankton samples from a variety of freshwater habitats that were studied. These heterotrophic filaments typically account for <1 to 11% of the total number of bacteria. The biovolume of this morphotype can exceed 40% of the biovolume for all bacteria. Surprisingly, we found hardly any data on these morphologically conspicuous filaments in the literature. Mixed cultures containing these filamentous bacteria were established by cultivation and isolation experiments with samples from different freshwater lakes. Nearly full-length 16S rRNA gene sequences were obtained from several mixed cultures and environmental samples from habitats in Europe, Africa, China, Australia, and New Zealand. Phylogenetic analysis of the sequences showed that three groups form a single monophyletic cluster, the SOL cluster, in the family Saprospiraceae. We developed a set of six nested probes for fluorescence in situ hybridization. Of the six probes, one probe was specific for Haliscomenobacter hydrossis, three probes were specific for the three subclusters (each probe was specific for one subcluster), one probe was specific for the entire SOL cluster, and another probe targeted almost the entire Saprospiraceae family. Specific hybridization of environmental samples and enrichments showed that the members of the three subclusters exhibited the same filamentous morphology. So far, using the subcluster-specific probes, we have not been able to detect any bacteria with a differing morphology. We conclude that the SOL cluster bacteria are an integral part of bacterioplankton in many freshwater habitats. They potentially account for a large fraction of the total bacterial biomass but have been underrepresented in molecular diversity studies so far.
In the last 9 years, several studies provided insights into the phylogenetic composition of freshwater bacterioplankton communities (5, 6, 7, 8, 11, 13, 21, 23, 30, 31, 35, 37). Most of these studies investigated a single habitat or a few habitats, and only a few studies investigated a larger number of ecosystems (8, 37). Despite the limited number of comprehensive studies, a rough idea surfaced as to which groups of bacteria are typically present in freshwater bacterioplankton (37).
This study focused on a group of planktonic bacteria defined by their morphological characteristics. The conspicuous filamentous morphotype was observed in samples from many freshwater habitats, but we did not find detailed reports on the presence of these bacteria in freshwater in the literature, and not much is known about the phylogenetic affiliation of these striking filamentous bacteria. Haliscomenobacter hydrossis (32), a member of the Saprospiraceae family (which is found exclusively in wastewater treatment plants [34]), is known to exhibit a similar morphology.
The rationale of this work was to reveal the phylogenetic affiliation and ecological significance of this distinct group of filamentous bacteria. Therefore, molecular tools and culture approaches for the identification and cultivation of these prominent members of the bacterioplankton were developed. Due to the similarity in morphology, the initial steps of the investigation presented here were directed against H. hydrossis. A large number of freshwater habitats were systematically investigated for the presence of the target bacteria. The systems investigated represent a broad range of habitats; habitats differing in size, geographic location, limnological status, etc., were studied. Substantial numbers of the conspicuous filamentous bacteria were detected in the majority of habitats investigated.
(This research was conducted by M. Schauer in partial fulfillment of the requirements for a Ph.D. degree of the University of Salzburg, Salzburg, Austria, 2005.)
MATERIALS AND METHODS
Study sites and sampling.
We investigated samples from the pelagic zones of freshwater habitats in Europe, East Africa, China, Australia, and New Zealand. Samples from freshwater lakes and ponds were taken from a depth of 1 to 2 m with a 10-liter Schindler sampler and kept in 5-liter plastic bottles for less than 2 h until processed in the laboratory. Subsamples for total bacterial counts were fixed with formaldehyde (final concentration, 2%). For fluorescence in situ hybridization (FISH), samples of water (10 to 30 ml) were fixed with paraformaldehyde (final concentration, 2%) for 2 h at room temperature, passed through 0.2-μm-pore-size polycarbonate filters (45 mm long; Millipore), dried, and frozen until further processing. To collect microbial biomass, samples of water (between 150 and 500 ml) were passed through 0.2-μm-pore-size polycarbonate filters (45 mm long; Millipore). The filters were frozen at −70°C until the extraction of DNA (biomass filters).
Bacterial abundance and biovolume.
Formaldehyde-fixed samples were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (final concentration, 6 μg ml−1) and enumerated by epifluorescence microscopy (Zeiss Axioplan; Zeiss, Jena, Germany). Between 300 and 700 DAPI-stained bacterial cells were counted at a magnification of ×1,250. High-resolution images (752 by 548 pixels; 8-bit dynamic range) were saved with an analogue monochrome charge-coupled device camera (Hitachi Denshi, Ltd., Tokyo, Japan) and processed with the image analysis system LUCIA G (Lucia 4.51; Laboratory Imaging, Ltd., Prague, Czech Republic). To determine bacterial cell volumes, the images of 50 filaments and 100 other bacteria per sample were analyzed with LUCIA G. Bacterial cell volumes were calculated by using the formula of Andersson et al. (3). Because of a lack of suitable conversion factors, we did not calculate bacterial biomass.
Cultivation and isolation experiments.
In order to isolate and cultivate bacteria of the SOL cluster, a variety of cultivation and isolation experiments were performed. Filaments of the SOL morphotype were generally cultivated and isolated in sterile glassware at 15°C with continuous illumination without agitation. For base media, a complex organic NSY medium (3 g of organic substrate liter−1) (for details, see reference 9), an H. hydrossis-specific medium (3.3 g of organic substrate liter−1; DSM 134), and an inorganic IBM medium (10) enriched with different amounts of complex organic substrates (NSY medium) and sterile lake water (15) were tested. Briefly, sterile lake water was prepared by filtering the water through presterilized 0.22-μm-pore-size filtration towers (Steritop; Millipore) directly into sterile 1-liter Schott bottles. These bottles were heated in a microwave to a temperature slightly under the boiling point (heating 1 liter of lake water for 7 min 30 s at 1,000 W) to avoid precipitation of inorganic substances. Microwave treatment was performed twice with subsequent cooling phases to suppress bacterial growth effectively. Media were supplemented with vitamin B12 and thiamine (final concentration, 10 μg liter−1). Growth of the target bacteria was also tested with the following organic substrates added at a concentration of 100 mg liter−1 to an inorganic IBM medium: sodium acetate, deoxycholic acid, pyruvic acid, n-caproic acid, caprylic acid, capric acid, d-glucose, d-galactose, potassium citrate, glycolic acid, glutamic acid, and Tween 80. The resistance of H. hydrossis strain DSM1100 antibiotics was tested in DSM 134 medium. Antibiotics were added at concentrations of 50 mg liter−1 (for polymyxin thiosulfate, gentamicin sulfate, and neomycin) and 100 mg liter−1 (for nalidixic acid, streptomycin sulfate, and ampicillin). Mixed cultures obtained from environmental samples by the methods mentioned above were diluted or subcultured in new flasks every 4 to 8 weeks. Substrates were added at low concentrations (100 to 200 mg liter−1) in weekly to biweekly intervals. Bacterial growth was monitored by counting samples stained with DAPI with an epifluorescence microscope (Zeiss Axioplan).
DNA extraction.
Nucleic acids were extracted either by a standard protocol (including incubation with lysozyme and proteinase K and subsequent extraction with phenol-chloroform-isoamyl alcohol [25:24:1, pH 8] and chloroform-isoamyl alcohol [24:1]) or alternatively with a much faster beadbeater extraction routine (FastDNA kit and FastPrep instrument FP120; QBioGene) following the manufacturer's recommendations. Qualitative comparison of extracts by PCR and denaturing gradient gel electrophoresis (DGGE) did not yield significant differences.
Primer design and evaluation.
Reverse primers HAL1R and HAL3R targeting the 16S-23S internal transcribed spacer (ITS) were designed after sequencing the internal transcribed spacers (ITSs) of H. hydrossis, Saprospira grandis, Flexibacter elegans, Flexibacter sancti, Cytophaga hutchinsonii, Chitinophaga pinensis, and Flectobacillus major with primers 1406F (19) and L189R (36). The newly developed, more-specific primers 26F, HAL-1473R, LD2-1473R, GKS-1473R, and SAP-1490R (primer sequences and specificities shown in Table 1), were designed on the basis of the first set of SOL cluster bacterial 16S rRNA gene sequences. Optimization of PCRs for newly designed primers was performed in a Mastercycler gradient (Eppendorf).
TABLE 1.
Primers used in this study
| Primer | Sequence (5′→3′) | Length (nt)a | GC content (%) | Tm (°C)b | Specificity | Reference |
|---|---|---|---|---|---|---|
| 1406F | TGY ACA CAC CGC CCG T | 16 | 65.6 | 55.6 | Bacteria | 19 |
| L189R | TAC TGA GAT CYT TMA RTT C | 19 | 34.2 | 49.1 | Bacteria | 36 |
| 27F | AGA GTT TGA TCM TGG CTC AG | 20 | 47.5 | 56.3 | Bacteria | 20 |
| 530FC | TCC GTG CCA GCA GCC GCG G | 19 | 78.9 | 67.5 | Bacteria | 20c |
| Hal1R | CTC TAA AAG GAG GTA TTC | 18 | 38.9 | 49.1 | Saprospiraceae | This study |
| Hal3R | AGC TAT GAG ACT TTG ACT | 18 | 38.9 | 49.1 | Saprospiraceae | This study |
| 26F | AGG ATG AAC GCT AGC GGG | 18 | 61.1 | 58.2 | Bacteria | This study |
| HAL-1473R | CTT AGC CCC AGT TAC TGG TTT T | 22 | 45.5 | 58.4 | HAL subcluster | This study |
| LD2-1473R | CTT AGC CCC AGT CAC TAG TTT T | 22 | 45.5 | 58.4 | LD2 subcluster | This study |
| GKS-1473R | CTT AGC CCC AGT CAT TGG TTT T | 22 | 45.5 | 58.4 | GKS2-217 subcluster | This study |
| SAP-1490R | CGG CTA CCT TGT TAC GAC TTA G | 22 | 50 | 60.3 | Bacteria | 20c |
nt, nucleotides.
Tm, melting temperature [Tm = 69.3 + 41 × [(nG + nC)/s] − (650/s), where nG is the number of G’s in the sequence, nC is the number of C’s in the sequence, and s is the length of the sequence].
Primers were modified to amplify preferentially target bacterial sequences.
PCR and sequencing.
Bacterial 16S rRNA genes were amplified in a Primus 96plus thermal cycler (MWG Biotech, Ebersberg, Germany). The reaction mixtures (50 μl) contained 100 μM concentrations of each of the deoxynucleotide triphosphates, 0.3 μM concentrations of each of the primers, 3 mM MgCl2, 1× PCR buffer, and 1.25 U of Taq DNA polymerase (QIAGEN). The universal bacterial primer 27F (20) and the two reverse primers (HAL1R and HAL3R) were used for the amplification of nearly full-length 16S rRNA gene sequences. The PCR program follows: (i) an initial denaturation step (2 min at 94°C); (ii) 30 cycles, with 1 cycle consisting of denaturation (1 min at 94°C), annealing (1 min at 56°C), and extension (2 min at 72°C); and (iii) a final extension step (5 min at 72°C). Specific amplification of nearly full-length 16S rRNA gene sequences of subclusters HAL, GKS2-217, and LD2 in the Saprospiraceae family was performed with primers 26F and HAL-1473R, GKS-1473R, and LD2-1473R, respectively. The PCR program follows: (i) an initial denaturation (2 min at 94°C); (ii) 30 cycles, with 1 cycle consisting of denaturation (1 min at 94°C), annealing (1 min at 62°C), and extension (2 min at 72°C); and (iii) a final extension step (5 min at 72°C). For a template, 1-μl samples of the total microbial DNA extracts or small pieces (∼5 mm2) of biomass filters were used. Filter PCR was performed by the protocol of Kirchman et al. (17). Comparison of bacterial DNA amplified from PCRs with extracted template DNA and reactions with small pieces of 0.2-μm-pore-size polycarbonate filters from the same sample by DGGE (according to the protocol described in reference 24) did not yield any qualitative differences in the microbial fingerprints obtained. PCR products were sequenced by MWG Biotech. Primer 530FC (modified from the primer in reference 20) was used to sequence the middle section of the 16S rRNA gene. For details on the primers used in this survey, refer to Table 1. Two sequences (MS-Falk1-L and MS-oKlaff1-L) were retrieved from two clone libraries. For the construction of clone libraries, nearly full-length bacterial 16S rRNA gene sequences of a subset of the bacterial community were amplified by PCR using primers 26F and SAP-1490R (modified from the primers in reference 20). Cloning was performed with the QIAGEN PCR Cloningplus kit following the manufacturer's recommendations. Plasmid DNA was extracted using the Nucleospin plasmid kit (Macherey-Nagel, Düren, Germany).
Phylogenetic analysis.
The BLAST program (2) (http://www.ncbi.nlm.nih.gov/BLAST/) was used to search for related sequences with high similarity values. A comparative analysis of all newly obtained sequences and relatives found in the databases was done with the ARB software package (http://www.arb-home.de). Sequences imported into the ARB database were aligned with the Fast Aligner tool of ARB and checked manually for misaligned positions. Phylogenetic trees were constructed for stretches of 1,357 homologous nucleotides. Evolutionary distances were corrected using the algorithm of Jukes and Cantor (14). Partial sequences shorter than 1,357 nucleotides were added to the existing neighbor-joining tree by the ARB parsimony tool without changing the tree topology. Bootstrap values were calculated using the Mega V2.1 program (18) (http://www.megasoftware.net). The tree topologies of neighbor-joining trees were validated using maximum-parsimony and maximum-likelihood algorithms as alternative tree construction methods. Sequence similarities were calculated as uncorrected base identities between sequences on the basis of sequence alignments.
Probe design and FISH.
A nested set of six oligonucleotide probes with different specificities for our target bacteria was designed using the ARB software package. The stringency of the hybridization conditions for the probes was adjusted by a series of solutions of formamide (0 to 60% formamide concentration). The conditions were adjusted with filters from several habitats for all newly designed probes. Signal intensities were evaluated by eye. Probes labeled with the indocarbocyanine dye Cy3 were purchased from Interactiva (Ulm, Germany). FISH was performed on polycarbonate filter sections by the protocol of Alfreider et al. (1). Counting bacteria on FISH filter sections was performed by epifluorescence microscopy (Zeiss Axioplan) at a magnification of ×1,250.
Nucleotide sequence accession numbers.
The nearly full-length 16S rRNA gene sequences obtained were deposited in the EMBL nucleotide sequence database under the accession numbers AJ784892 and AJ786318 to AJ7863342.
RESULTS
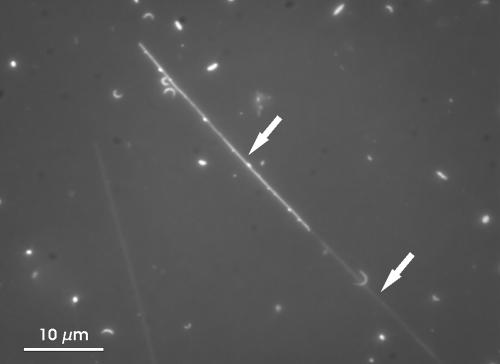
Water samples from the pelagic zones of 102 different freshwater habitats located on three continents and in three different climatic zones were investigated. In 78 of the 102 habitats (76%), bacteria with a conspicuous morphology were found. These filamentous bacteria exhibit a very distinct morphology and are easily distinguished from the rest of the heterotrophic bacterioplankton. The large cells resemble thin, unbowed sticklike filaments. While the cell diameter is quite stable (between 0.25 and 0.35 μm), the length of the filaments is highly variable (8 to >100 μm). A sheath covering the filaments was sometimes visible in DAPI-stained preparations (Fig. 1). In some of the samples, these filaments featured small yellowish particles, which seemed to stick to the surfaces of the cells. Altogether, the described morphotype reminded us remarkably of an Austrian brand of pretzels, so we called this distinct morphology the SOL morphotype.
FIG. 1.
Microscopic image of a DAPI-stained bacterioplankton sample from Lake Victoria (31 May 2001) presenting a filament of the SOL morphotype. These filaments always possess blunt ends, and flagella were never observed. These characteristics together with the unbowed rigid habit of the filaments allows for a reliable discrimination of members of the SOL morphotype against other filamentous bacteria by standard microscopic techniques. The upper arrow marks the filament with yellowish granule. The lower arrow marks a sheath covering the filament. A similar sheath is also known to cover the wild-type cells of H. hydrossis. Magnification, ×1,250.
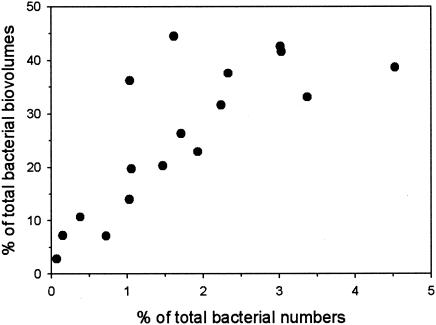
Bacteria with the conspicuous SOL morphotype were found in plankton samples from a broad range of habitats. Freshwater lakes and rivers inhabited by bacteria of the SOL morphotype were found in the following varied locations: large tropical lakes (e.g., Lake Victoria and Lake Tanganjika in East Africa), subtropical shallow eutrophic lakes and rivers (e.g., Lake Taihu, Changjiang river [Yangtze] in the People's Republic of China), large shallow lowland lakes (e.g., Lake Loosdrecht, The Netherlands), large oligotrophic prealpine lakes (e.g., Lake Attersee, Austria), and small deep ultraoligotrophic high mountain lakes (e.g., Lake Oberer Klaffersee and Lake Kapuzinersee in Austria). SOL morphotype bacteria were not detected in acidic habitats (pH 4.4 to 5.9; seven habitats) or in habitats with short water retention times (4 to 10 days; three habitats) or in nonpermanent freshwater bodies (small, shallow water bodies; two habitats). The other freshwater ecosystems (12 habitats) where it was not possible to detect these filamentous bacteria did not fit in any of those generalized types of habitats. Bacteria of the SOL morphotype were not detected in five acidified Bohemian forest lakes studied, which are known to be frequently dominated by filamentous bacteria (33). The absolute numbers of target bacteria ranged from several hundreds to 200,000 cells ml−1. The relative contribution of the SOL morphotype to the total number of bacteria ranged from 0.1 to 10.8%, with a typical value of around 1% of the total bacterioplankton. Biovolume estimations for the SOL morphotype in several habitats ranged from 2.8 to 44.5% of the total bacterial biovolume (Fig. 2).
FIG. 2.
Contribution of SOL morphotype bacteria to the number and biovolume of all bacteria in 17 samples from 16 different freshwater habitats. Selected samples represent a broad range of ecologically different habitat types.
Cultivation and isolation experiments.
In order to cultivate and isolate bacteria of the SOL morphotype, we attempted several different strategies. (i) Dilution cultures were established in the wells of cell culture plates and in sterile glass flasks. Sterile lake water, NSY medium (3 g of organic substrate liter−1), and Haliscomenobacter medium (DSM 134; 3.3 g of organic substrate liter−1) were used as the base media. The plates and flasks were incubated for several days to weeks and screened by epifluorescence microscopy. No pure isolates could be obtained from these dilution experiments. (ii) Live filaments were sorted with a MoFlo cell sorter (Cytomation, Fort Collins, Colo.) with the assistance of Stefan Andreatta. For details on the cytometer settings, see reference 4. SOL morphotype filaments were sorted with a low-salt sheath fluid in cell culture plates containing sterile filtered lake water or inorganic IBM medium enriched with small amounts of organic substrates (25 or 50 μl of NSY medium to 2 ml of IBM medium). One, 10, 100, and 1,000 cells were sorted into the different media in replicate wells. No growth of filaments was observed for any of the different treatments after incubation of the plates for 2 weeks at 15°C. (iii) In another experiment, a series of environmental samples and enrichment cultures were plated on agar plates containing NSY medium (3 g liter−1), and Haliscomenobacter medium (DSM 134; 3.3 g liter−1). After 1 week of incubation at 15°C, colonies were screened by epifluorescence microscopy. No colony formation of filamentous bacteria was detected.
While the attempt to establish pure cultures failed, it was possible to enrich and cultivate (long-term) the mixed cultures. The enrichment of SOL morphotype bacteria was accomplished partly by reducing the diversity and total number of bacteria in samples and partly by increasing the number of target bacteria through modified growing conditions. The most successful treatments were the addition of glutamic acid (concentration of 100 mg liter−1) as an organic substrate to filtered (0.2-μm-pore-size filters) and microwave-treated lake water and inhibition of other bacteria by treatment with nalidixic acid (final concentration, 100 mg liter−1). Cultivation of bacteria of the SOL morphotype normally resulted in a substantial increase of the target bacteria (up to 42% of the total number of bacteria). With this type of treatment, we were able to obtain several mixed cultures of members of the HAL and LD2 subclusters (details on phylogenetic analysis given below) of the SOL cluster bacteria. Mixed cultures were subcultured every 4 to 8 weeks. Cultures were maintained for several weeks to more than 1 year. For more details on the samples used for the enrichment experiments, see Table 2.
TABLE 2.
Environmental samples used for cultivation and enrichment experiments and details on the sequences retrieved
| Geographic origin of sample | Total no. of bacteria (106 ml−1)a | No. of SOL morphotype bacteria (104 ml−1)a | Sequence | Affiliation (subcluster) | Accession no. | Length (nt)b |
|---|---|---|---|---|---|---|
| Lake Egelsee | 1.59 | 0.19 | MS-Egel1-L | LD2 | AJ786318 | 940 |
| Lake Egelsee | 1.59 | 0.19 | MS-Egel1-H | HAL | AJ786324 | 765 |
| Jaegerteich pond | 5.94 | 7.13 | MS-Jaeger1-L | LD2 | AJ786319 | 1,135 |
| Lake Wolfgangsee | 1.30 | 1.11 | MS-Wolf1-H | HAL | AJ786322 | 1,414 |
| Lake Wolfgangsee | NDc | 2.00 | MS-Wolf2-H | HAL | AJ786323 | 1,478 |
On the sampling day.
nt, nucleotides.
ND, not determined.
Primer design and specific direct sequencing.
The first specific primers developed, primers HAL1R and HAL3R (Table 1), were derived from 16S-23S ITS sequences of H. hydrossis and several related bacterial strains available in culture collections. The primer binding positions are at the very beginning of the ITS region and the first tRNA gene located in the ITS. These primers were designed with a broader phylogenetic specificity to recover sequences of H. hydrossis and close relatives in the family Saprospiraceae. These reverse primers were combined with the universal bacterial primer, primer 27F. First, nearly full-length sequences of the target bacteria allowed the development of more-specific primers 26F, SAP-1490R, HAL-1473R, LD2-1473R, and GKS-1473R. Primer 26F was designed to match all of the available sequences from members of the Saprospiraceae family but is not strictly specific for this group. Primer SAP-1490R was derived from universal primer 1490R (20) to preferentially fit known Saprospiraceae sequences but also matches a broad spectrum of other bacterial 16S rRNA gene sequences. The combination of the 26F primer with subcluster-specific primers HAL-, LD2-, and GKS-1473R allowed a subcluster-specific amplification (details on phylogenetic analysis given below) of 16S rRNA gene sequences from a variety of environmental samples (Table 3) and mixed cultures. The sequences consisted of 765 to 1,478 nucleotides and contained 0 to 7 ambiguous positions (average, 1.17). Resequencing H. hydrossis strain DSM1100 with primers 27F and HAL1R yielded a slightly longer 16S rRNA gene sequence (1,509 nucleotides) with fewer ambiguous positions than a previously published sequence of the same strain (accession no. M58790).
TABLE 3.
Details on habitats from which sequences were retrieved by direct sequencing without enriching SOL cluster bacteria
| Sampling site | Trophic status | Country | Climatic zone | Total no. of bacteria (106 ml−1) | No. of SOL morphotype bacteria (104 ml−1) | SOL morphotype bacteria (% of total) | % of bacterial biovolume | Sequence | Affiliation (subcluster) | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| Lake Irrsee | Oligomesotrophic | Austria | Temperate | 1.05 | 1.09 | 1.04 | 36.2 | MS-Irr1-H | HAL | AJ786320 |
| Lake Irrsee | Oligomesotrophic | Austria | Temperate | 1.05 | 1.09 | 1.04 | 36.2 | MS-Irr2-H | HAL | AJ786329 |
| Lake Mondsee | Oligomesotrophic | Austria | Temperate | 2.66 | 0.39 | 0.15 | 7.2 | MS-Mond1-H | HAL | AJ786321 |
| Lake Ahomsee | Oligotrophic | Austria | Temperate | 0.62 | 1.39 | 2.24 | 31.6 | MS-Ahom1-G | GKS2-217 | AJ786325 |
| Lake Kapuzinersee | Oligotrophic | Austria | Temperate | 0.3 | 0.91 | 3.03 | 41.6 | MS-Kapu1-G | GKS2-217 | AJ786326 |
| Lake Oberer Klaffersee | Oligotrophic | Austria | Temperate | 0.55 | 1.66 | 3.02 | 42.6 | MS-oKlaff1-G | GKS2-217 | AJ786327 |
| Lake unterer Sonntagkarsee | Oligotrophic | Austria | Temperate | 0.18 | 0.59 | 3.28 | 33.1 | MS-uSonn1-G | GKS2-217 | AJ786328 |
| Lake Wolfgangsee | Oligotrophic | Austria | Temperate | 1.12 | 2.61 | 2.33 | 37.6 | MS-Wolf1-L | LD2 | AJ786330 |
| Lake Toplitzsee | Oligotrophic | Austria | Temperate | 1.39 | 1.43 | 1.03 | 14.0 | MS-Topli1-L | LD2 | AJ786332 |
| Lake Schwarzensee | Oligotrophic | Austria | Temperate | 1.05 | 0.4 | 0.38 | 10.7 | MS-Schwa1-L | LD2 | AJ786334 |
| Lake Mondsee | Oligomesotrophic | Austria | Temperate | 2.19 | 1.57 | 0.72 | 7.1 | MS-Mond1-L | LD2 | AJ786337 |
| Lake Irrsee | Oligomesotrophic | Austria | Temperate | 1.87 | 1.98 | 1.06 | 19.7 | MS-Irr1-L | LD2 | AJ786338 |
| Lake Attersee | Oligotrophic | Austria | Temperate | 1.12 | 5.07 | 4.53 | 38.7 | MS-Atter1-L | LD2 | AJ786340 |
| Lake Falkertsee | Oligotrophic | Austria | Temperate | 0.96 | 1.85 | 1.93 | 22.9 | MS-Falk1-L | LD2 | AJ786341 |
| Lake Krottensee | Oligotrophic | Austria | Temperate | 1.13 | 1.66 | 1.47 | 20.3 | MS-Krott1-L | LD2 | AJ786342 |
| Lake Loosdrecht | Eutrophic | The Netherlands | Temperate | 3.13 | 5.07 | 1.62 | 44.5 | MS-Loos1-L | LD2 | AJ786339 |
| Pond in Dunedin | NDa | New Zealand | Temperate | ND | ND | ND | ND | MS-NZ1-L | LD2 | AJ786335 |
| Pond in Sydney | ND | Australia | Temperate | ND | ND | ND | ND | MS-Aus1-L | LD2 | AJ786336 |
| Lake Taihu | Hypertrophic | Chinab | Subtropical | ND | ND | ND | ND | MS-Taihu1-L | LD2 | AJ786333 |
| Lake Tanganjika | Oligotrophic | Zambia | Tropical | 1.75 | 0.12 | 0.07 | 2.8 | MS-Tang1-L | LD2 | AJ786331 |
ND, not determined.
People's Republic of China.
Comparative analysis of 16S rRNA gene sequences.
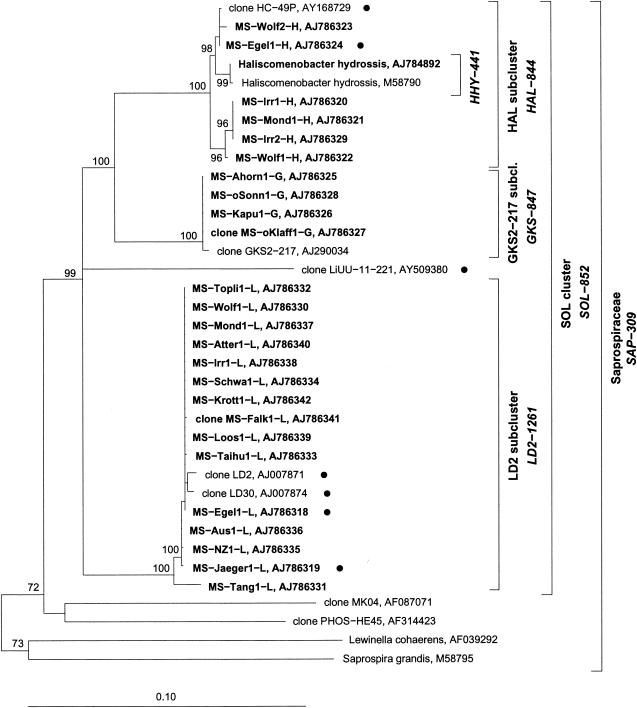
The alignment and phylogenetic analysis of the 25 16S rRNA gene sequences obtained and previously published sequences yielded three phylogenetic groups. Only the first group contains a described species, namely, H. hydrossis; therefore, it was named the HAL subcluster. The second group of sequences joined the LD2 cluster introduced by Zwart et al. (37). The rest of the retrieved sequences formed a phylogenetic group with a sequence obtained from Lake Gossenköllesee. We named this new group GKS2-217 according to the clone name introduced by Glöckner et al. (8). These three groups form a monophyletic cluster (the SOL cluster) affiliated with the family Saprospiraceae (Bacteroidetes phylum). Aside from the sequence of the only described species (H. hydrossis), all other previously published sequences that were found to be affiliated with the SOL cluster were retrieved only by culture-independent methods from freshwater habitats. Some shorter sequences that were also affiliated with the SOL cluster (Table 4) were omitted in the calculation of the phylogenetic tree (presented in Fig. 3), due to their limited length.
TABLE 4.
Sequences affiliated with the SOL cluster but not presented in the tree shown in Fig. 3 due to insufficient sequence lengtha
| Sequence name | Accession no. | Length (nt)b | Affiliation (subcluster) | Sample type | Reference |
|---|---|---|---|---|---|
| Clone BCM3S-7B | AY102912 | 719 | HAL | Subtropical freshwater marsh | Unpublished |
| Clone csbio160175 | AY187353 | 942 | HAL | Drinking-water biofilm | 28 |
| Clone Urk0-01 | AJ416169 | 709 | LD2 | The Netherlands, Lake IJssel | 37 |
| Clone Med0-02 | AJ416157 | 699 | LD2 | The Netherlands, Lake IJssel | 37 |
| Clone DC5-0-5 | AY145578 | 506 | LD2 | Germany, Weser estuary | Unpublished |
| Clone CR98-5-53 | AF428783 | 600 | LD2 | China, Changjiang River (Yangtze) | 29 |
| Clone CR98-35-61 | AF428943 | 600 | LD2 | China, Changjiang River (Yangtze) | 29 |
| Clone CR98-5-24 | AF428754 | 600 | LD2 | China, Changjiang River (Yangtze) | 29 |
| Clone CRP99-04 | AF428658 | 600 | LD2 | China, Lake Poyang | 29 |
| Clone LiUU-5-268 | AY509298 | 763 | LD2 | Sweden, freshwater bacterioplankton | Unpublished |
| Clone LiUU-9-278 | AY509321 | 800 | LD2 | Sweden, freshwater bacterioplankton | Unpublished |
| Clone LiUU-9-266 | AY509369 | 798 | LD2 | Sweden, freshwater bacterioplankton | Unpublished |
| Clone LiUU-3-375 | AY509275 | 800 | LD2 | Sweden, freshwater bacterioplankton | Unpublished |
The affiliation of the sequences with the SOL cluster was proven by detailed phylogenetic analysis.
nt, nucleotides.
FIG. 3.
Phylogenetic tree showing the affiliation of new bacteria of the Saprospiraceae family (in bold type) from mixed cultures and environmental samples. The neighbor-joining tree was calculated with almost full-length 16S rRNA gene sequences. Seven shorter sequences (765 to 1,165 nucleotides) (indicated by a small solid circle) were added by the ARB parsimony method without changing the tree topology. H. hydrossis sequence AJ784892 is the resequenced sequence. Bootstrap values (1,000 iterations) of >50% are given. Brackets indicate the specificity of FISH probes (probe names in italic type). Probe SAP-309 targets all SOL cluster bacteria and >80% of the remaining sequences within the Saprospiraceae family.
Trees constructed by alternative methods (maximum-parsimony and maximum-likelihood algorithms) validated the topology of the neighbor-joining tree shown in Fig. 3. The high bootstrap values for the SOL cluster and the HAL, LD2, and GKS2-217 subcluster levels support the topology shown in Fig. 3. The short sequence of the clone LiUU-11-221 was added to the tree by way of the ARB parsimony method. The exact position of this sequence inside the SOL cluster could not be determined. To validate the position of this clone in the SOL cluster, we truncated the sequences of our data set in order to include some shorter sequences in the tree-building process. This additional tree building (data not shown) supported the position of this clone in the SOL cluster with high bootstrap values for the SOL cluster in any case. The minimal sequence similarity values inside the subclusters are 98.7% for the HAL subcluster, 97.6% for the LD2 subcluster, and 99.7% for the GKS2-217 subcluster. The maximum similarities between subclusters are 92.7% for the HAL and LD2 subclusters, 95.5% for the HAL and GKS2-217 subclusters, and 92.1% for the LD2 and GKS2-217 subclusters. The overall minimum similarity value inside the SOL cluster is 88% (Table 5).
TABLE 5.
Similarity values within and between subclusters HAL, LD2, and GKS2-217
| Subcluster | 16S rRNA gene sequence similarity valuea
|
|||||
|---|---|---|---|---|---|---|
| HAL
|
LD2
|
GKS2-217
|
||||
| Min | Max | Min | Max | Min | Max | |
| HAL | 98.7 | 100 | ||||
| LD2 | 90.1 | 92.7 | 97.6 | 100 | ||
| GKS2-217 | 93.8 | 95.5 | 89.9 | 92.1 | 99.7 | 100 |
| LiUU-11-221b | 88 | 91.5 | 88.7 | 91.5 | 89.5 | 89.7 |
Minimum (Min) and maximum (Max) similarity values within and between subclusters.
Representative of a potential fourth subcluster inside the SOL cluster.
FISH.
We used nested FISH probes to detect phylogenetic groups of various levels in lake water samples (Table 6). The new probe HHY-441 is specific for the H. hydrossis species. All other sequences inside the broader HAL subcluster contain one to two mismatches at the probe binding position. We used this probe in order to determine whether we could find filaments of H. hydrossis in lake water samples. Hybridization of samples from natural freshwater habitats did not yield any positive signals with this very specific probe. This indicates the absence of H. hydrossis as found in wastewater treatment plants (34) in all of the natural freshwater ecosystems tested. Nevertheless, we were able to detect bacteria of the HAL subcluster in a broad range of habitats with the subcluster-specific probe HAL-844.
TABLE 6.
Oligonucleotide probes used in this study
| Probe | Sequence (5′→3′) | GC content (%) | Tm (°C)a | Length (nt)b | Specificity | Formamide concn (%) | No. of unspecific matchesc |
|---|---|---|---|---|---|---|---|
| SAP-309 | TCT CAG TAC CCG TGT GGG | 61.1 | 48.3 | 18 | Saprospiraceae | 25 | 8 |
| SOL-852 | ACG CTT TCG CTT GGA CAC | 55.6 | 50.9 | 18 | Soletticluster | 30 | 20 |
| HAL-844 | CGC TTG GAC ACT CAC TCC | 61.1 | 48.1 | 18 | HAL subcluster | 25 | 1 |
| GKS-847 | TTC GCT TGG ACA CAC AAT C | 47.4 | 48.9 | 19 | GKS2-217 subcluster | 25 | 1 |
| LD2-1261 | GGC TCC GCT TCA CAG CTT | 61.1 | 52.7 | 18 | LD2 subcluster | 30 | 3 |
| HHY-441 | CCA GAT TTC TTC CCA AGC | 50 | 46.8 | 18 | Haliscomenobacter hydrossis | 20 | 17 |
Tm, melting temperature (according to the manufacturer).
nt, nucleotides.
Number of known sequences matching (<1 weighted mismatch) the probes but not affiliated to the respective target groups.
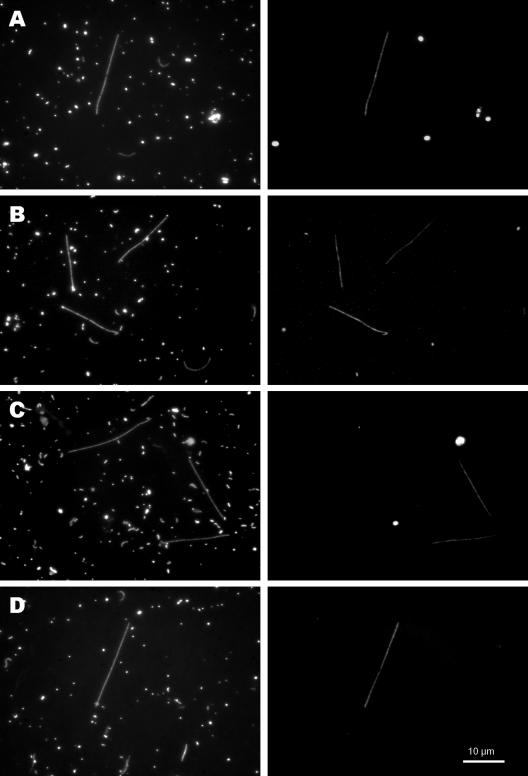
Probe CF-319a (22), which was designed for the broad Cytophaga-Flavobacteria-Bacteroides group shows one mismatch with all available SOL cluster 16S rRNA sequences. This results in weak to undetectable signals of SOL cluster filaments under stringent hybridization conditions. We designed a probe for the family Saprospiraceae (probe SAP-309), which targets all of the members of the SOL cluster as well as more than 80% of all other available sequences from members of the Saprospiraceae family. This probe detected all filaments of the SOL morphotype in each of our samples. The SOL-852 probe was designed to cover all SOL cluster sequences in our database, yet it also perfectly matched the sequence of the clone PHOS-HE19 (accession no. AF314428), which does not belong to the SOL cluster. Additionally, some members of the Bacteroidetes show only weak mismatches with this probe. This reduced specificity of the probe results in signals from bacteria with different morphologies (rods and cocci) in some environmental samples. However, we did not observe FISH signals of these rods and cocci with the SAP-309 probe (which covers all SOL cluster sequences), so we can exclude the possibility that members of these morphotypes belong to the SOL cluster. Probe SOL-852 detected all SOL filaments in any of the environmental samples investigated so far. The subcluster-specific probes HAL-844, LD2-1261, and GKS-847 (Fig. 4) did not yield any signals from cells other than the SOL morphotype.
FIG. 4.
Pairs of microscopic images of the same microscopic field with UV excitation (DAPI stained; left column) and green excitation (hybridized with Cy3-labeled oligonucleotide probes; right column). Probes SAP-309 (A), SOL-852 (B), HAL-844 (C), and LD2-1261 (D) were used. All coccoid signals on the right-hand images arise from phototrophic organisms (autofluorescent signals). All photomicrographs were taken at a magnification of ×1,250. The scale bar is valid for all microscopic images.
DISCUSSION
Between several hundred to more than 100,000 individuals of the remarkable SOL morphotype per ml were found in most of the bacterioplankton samples that were analyzed. This morphotype typically accounted for around 1% of the total bacteria. The use of standard procedures for direct counts of microbial abundance (16) should lead to the detection of some of these striking organisms in almost every sample from a freshwater habitat inhabited by SOL bacteria. Given the observed abundance of SOL cluster bacteria and the fact that these filamentous bacteria produce quite bright and conspicuous signals when they were stained by standard procedures (26), one would think that these conspicuous filamentous bacteria would be mentioned in a number of articles in journals with a microbial and limnological focus. Surprisingly, the reverse is true. There are few mentions of these filamentous forms, and if they are mentioned or depicted, it looks rather accidental. Schmaljohann et al. (27) detected long straight filaments with diameters of 0.4 μm and lengths of up to 30 μm in Lake Kinneret in Israel. Alfreider and coworkers (1) pictured some of these filamentous members of bacterioplankton hybridized with eubacterial probe EUB338 on a figure presenting FISH data on Lake Gossenköllesee, a mountain lake high in the Austrian Alps. Hofer et al. (12) worked on the same lake and demonstrated that filamentous viruses infect bacteria of the SOL morphotype. Recently, Pernthaler et al. (25) also demonstrated that members of the LD2 subcluster derived from Lake Schöhsee in Germany possess the conspicuous SOL morphology.
We demonstrate here that this distinct morphotype consists of a genetically quite diverse (Table 5) yet monophyletic group. In view of the high frequency of occurrence of members of the SOL cluster in a broad range of different freshwater habitats, as well as their remarkable contribution to the total bacterial biomass, this group of bacteria was clearly underrepresented in bacterial diversity studies performed by culture-independent methods. Apart from the sequence of the only described species (H. hydrossis), there was only a limited number of 16S rRNA gene sequences affiliated with the SOL cluster available up to now. The sequence of clone HC-49P (accession no. AY168729) was obtained from an arsenite-oxidizing biofilm (unpublished data). The short sequences of clones BCM3S-7B (accession no. AY102912) and csbio160175 (accession no. AY187353) were retrieved from a subtropical freshwater marsh (unpublished data) and a drinking water biofilm (28), respectively. The GKS2-217 clone comes from a clone library of the high mountain Lake Gossenköllesee (8). Zwart and coworkers (38) retrieved some sequences from Lake Loosdrecht (clone LD2 [accession no. AJ007871] and clone LD30 [accession no. AJ007874]) and Lake IJssel (clone Urk0-01 [accession no. AJ416169] and clone Med0-02 [accession no. AJ416157]) and designated these four sequences as the typical freshwater bacterial cluster LD2 (37). Sequencing of a DGGE band retrieved from a dilution culture of the Weser estuary in Germany yielded the short sequence of clone DC5-0-5 (unpublished data) (accession no. AY145578). Sekiguchi et al. (29) obtained short sequences from the Changjiang (Yangtze) river and Lake Poyang in the People's Republic of China (accession no. AF428658, AF428754, AF428783, and AF428943). Five sequences 763 to 1,038 nucleotides long were obtained from different lakes in Sweden by Eiler et al. (unpublished data) (accession no. AY509298, AY509321, AY509369, AY509275, and AY509380). The monophyletic SOL cluster was found by phylogenetic analysis of the 25 16S rRNA sequences retrieved from a variety of different habitats in this study together with some of the sequences already known. The three subclusters HAL, LD2, and GKS2-217 form this monophyletic SOL cluster. The hybridization of a broad spectrum of bacterioplankton samples with subcluster-specific probes proved that members of all three subclusters have the same conspicuous morphology.
However, in some environmental samples, a small fraction of typical SOL morphotype bacteria did not hybridize with any of the three subcluster-specific probes. This finding together with the phylogenetic position of the LiUU-11-221 clone in the neighbor-joining tree shown in Fig. 3 gives rise to the question of whether there are more than three subclusters belonging to the SOL cluster. Taking into account the high genetic diversity of this group and the scarce data on the bacterioplankton community of freshwater habitats in major parts of the world, one would not be surprised by the appearance of additional phylogenetic subclusters of bacteria also possessing the SOL morphology. Nevertheless, with probes SAP-309 and SOL-852, we were able to securely identify all SOL morphotype bacteria in all environmental samples investigated so far.
Members of the SOL cluster were found in a wide spectrum of different freshwater habitats. Samples were taken on three continents over temperate, subtropical, and tropical zones. The limnological characterization of the habitats ranged from ultraoligotrophic high mountain lakes (e.g., Lake Zwerfenbergsee, Austria) to hypertrophic parts of large shallow lakes (e.g., Mailing bay, Lake Taihu, People's Republic of China). Bodies of water where we were able to find SOL cluster bacteria ranged from sea level (e.g., Lake Loosdrecht, The Netherlands) to up to 2,309 m above sea level (e.g., Lake Oberer Klaffersee, Austria). The surface area of the habitats ranged from around 1 ha (e.g., Lake Egelsee, Austria) to 68,870 km2 (Lake Victoria, East Africa). SOL cluster filaments were also found in small artificial ponds used for aquaculture (e.g., Jägerteich pond, Austria). Considering the large number of different habitats where it was possible to identify these conspicuous filaments, we conclude that these SOL cluster bacteria form an integral part of the bacterioplankton in diverse freshwater habitats. This group potentially makes up a significant portion of the total number of bacteria. These facts, together with the filamentous morphology, lead to a significant share of the total bacterial biomass of this group in many freshwater habitats. However, we still do not know details about the ecology of this intriguing group of bacterioplankton. Nevertheless, this study established a phylogenetic basis for these monophyletic SOL cluster bacteria, with at least three subclusters with the same conspicuous morphology. On this basis, we will try to elucidate more details of the ecology of this interesting group of bacteria.
Acknowledgments
This work was supported in part by the Austrian Science Fund (project P15655).
We thank Eva Schober, Rainer Kurmayer, Quinglong Wu, Johann Knoll, Gabriel Zwart, Paul Bodelier, Eva Lindström, Silke Langenheder, Günther Stadler, Doris Hummer, Robert Sinyinza, Jaroslav Vrba, and all of the IPGL participants for providing samples. Peter Stadler and Matthias Pöckl were of great help during sampling and provided technical assistance, as did Johanna Schmid. Stefan Andreatta helped us with the sorting of filaments in Innsbruck, Austria. We are grateful to Hannes Höllerer for building some of these small technical gadgets that make research much easier. Christian Kamenik was an excellent guide to some high mountain lakes in the Austrian Alps.
REFERENCES
- 1.Alfreider, A., J. Pernthaler, R. Amann, B. Sattler, F.-O. Glöckner, A. Ville, and R. Psenner. 1996. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake using in situ hybridization. Appl. Environ. Microbiol. 62:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, A., U. Larsson, and A. Hagström. 1986. Size-selective grazing by a microflagellate on pelagic bacteria. Mar. Ecol. Prog. Ser. 33:51-57. [Google Scholar]
- 4.Andreatta, S. 2001. Cytometry of aquatic bacteria: analyses at the community and subgroup level. Ph.D. thesis. University of Innsbruck, Innsbruck, Austria.
- 5.Bahr, M., J. E. Hobbie, and M. L. Sogin. 1996. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat. Microb. Ecol. 11:271-277. [Google Scholar]
- 6.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crump, B. C., G. W. Kling, M. Bahr, and J. E. Hobbie. 2003. Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Appl. Environ. Microbiol. 69:2253-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glöckner, F. O., E. Zeichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn, M. W., H. Lünsdorf, Q. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, M. W., P. Stadler, Q. L. Wu, and M. Pöckl. 2004. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57:379-390. [DOI] [PubMed] [Google Scholar]
- 11.Hiorns, W. D., E. A. Methé, S. A. Nierzwickibauer, and J. P. Zehr. 1997. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl. Environ. Microbiol. 63:2957-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hofer, J. S., and R. Sommaruga. 2001. Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat. Microb. Ecol. 26:1-11. [Google Scholar]
- 13.Höfle, M. G., H. Haas, and K. Dominik. 1999. Seasonal dynamics of bacterioplankton community structure in a eutrophic lake as determined by 5S rRNA analysis. Appl. Environ. Microbiol. 65:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y. [Google Scholar]
- 15.Keller, M. D., W. K. Bellows, and R. R. L. Guillard. 1988. Microwave treatment for sterilization of phytoplankton culture media. J. Exp. Mar. Biol. Ecol. 117:279-283. [Google Scholar]
- 16.Kirchman, D. L. 1993. Statistical analysis of direct counts of microbial abundance, p. 117-119. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, London, United Kingdom.
- 17.Kirchman, D. L., L. Yu, B. M. Fuchs, and R. Amann. 2001. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26:13-22. [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobson, and M. Nei. 2001. Mega2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. L. Sogin, and N. R. Pace. 1985. Rapid determination of 16S RNA sequences for phylogenetic analysis. Proc. Natl. Acad. Sci. USA 82:6955-6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing. Nucleic acid techniques, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Bacterial systematics. John Wiley and Sons, New York, N.Y.
- 21.Lindström, E. S. 2000. Bacterioplankton community composition in five lakes differing in trophic status and humic content. Microb. Ecol. 40:104-113. [DOI] [PubMed] [Google Scholar]
- 22.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 23.Methé, B. A., W. D. Hiorns, and J. P. Zehr. 1998. Contrasts between marine and freshwater bacterial community composition—analyses of communities in Lake George and six other Adirondack lakes. Limnol. Oceanogr. 43:368-374. [Google Scholar]
- 24.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. C. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 25.Pernthaler, J., E. Zöllner, F. Warnecke, and K. Jürgens. 2004. Bloom of filamentous bacteria in a mesotrophic lake: identity and potential controlling mechanism. Appl. Environ. Microbiol. 70:6272-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 27.Schmaljohann, R., U. Pollingher, and T. Bermann. 1987. Natural populations of bacteria in Lake Kinneret: observations with scanning electron and epifluorescence microscopy. Microb. Ecol. 13:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Schmeisser, C., C. Stockigt, C. Raasch, J. Wingender, K. N. Timmis, D. F. Wenderoth, H. C. Flemming, H. Liesegang, R. A. Schmitz, K. E. Jaeger, and W. R. Streit. 2003. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 69:7298-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semenova, E. A., and K. D. Kuznedelov. 1998. A study of the biodiversity of Baikal picoplankton by comparative analysis of 16S rRNA gene 5′-terminal regions. Mol. Biol. 32:754-760. [PubMed] [Google Scholar]
- 31.Urbach, E., K. L. Vergin, L. Young, A. Morse, G. L. Larson, and S. J. Giovannoni. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557-572. [Google Scholar]
- 32.van Veen, W. L., D. van der Kooij, E. C. Geuze, and A. W. van der Vlies. 1973. Investigations of the sheathed bacterium Haliscomenobacter hydrossis gen. n., sp. n., isolated from activated sludge. Antonie Leeuwenhoek 39:207-216. [DOI] [PubMed] [Google Scholar]
- 33.Vrba, J., J. Nedoma, L. Kohout, J. Kopáček, L. Nedbalová, P. Ráčková, and K. Šimek. 2003. Massive occurrence of heterotrophic filaments in acidified lakes: seasonal dynamics and composition. FEMS Microbiol. Ecol. 46:281-294. [DOI] [PubMed] [Google Scholar]
- 34.Wagner, M., R. Amann, P. Kämpfer, B. Assmus, A. Hartmann, P. Hutzler, N. Springer, and K. H. Schleifer. 1994. Identification and in situ detection of Gram-negative filamentous bacteria in activated sludge. Syst. Appl. Microbiol. 17:405-417. [Google Scholar]
- 35.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, Z., and W. W. Mohn. 2001. Bacterial diversity and community structure in an aerated lagoon revealed by ribosomal intergenic spacer analyses and 16S ribosomal DNA sequencing. Appl. Environ. Microbiol. 67:1565-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwart, G. J. M., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 38.Zwart, G. J. M., W. D. Hiorns, B. A. Methé, M. P. van Agterveld, R. Huismans, S. C. Nold, J. P. Zehr, and H. J. Laanbroek. 1998. Nearly identical 16S rRNA sequences recovered from lakes in North America and Europe indicate the existence of clades of globally distributed freshwater bacteria. Syst. Appl. Microbiol. 21:546-556. [DOI] [PubMed] [Google Scholar]