Abstract
It has been hypothesized that the potential for anaerobic metabolism might be a common feature of bacteria in coastal marine waters (L. Riemann and F. Azam, Appl. Environ. Microbiol. 68: 5554-5562, 2002). Therefore, we investigated whether different phylogenetic groups of heterotrophic picoplankton from the coastal North Sea were able to take up a simple carbon source under anoxic conditions. Oxic and anoxic incubations (4 h) or enrichments (24 h) of seawater with radiolabeled glucose were performed in July and August 2003. Bacteria with incorporated substrate were identified by using a novel protocol in which we combined fluorescence in situ hybridization and microautoradiography of cells on membrane filters. Incorporation of glucose under oxic and anoxic conditions was found in α-Proteobacteria, γ-Proteobacteria, and the Cytophaga-Flavobacterium cluster of the Bacteroidetes at both times, but not in marine Euryarchaeota. In July, the majority of cells belonging to the α-proteobacterial Roseobacter clade showed tracer incorporation both in oxic incubations and in oxic and anoxic enrichments. In August, only a minority of the Roseobacter cells, but most bacteria affiliated with Vibrio spp., were able to incorporate the tracer under either condition. A preference for glucose uptake under anoxic conditions was observed for bacteria related to Alteromonas and the Pseudoalteromonas-Colwellia group. These genera are commonly considered to be strictly aerobic, but facultatively fermentative strains have been described. Our findings suggest that the ability to incorporate substrates anaerobically is widespread in pelagic marine bacteria belonging to different phylogenetic groups. Such bacteria may be abundant in fully aerated coastal marine surface waters.
Is facultatively anaerobic metabolism an exotic or widespread feature of marine bacterioplankton? The majority of heterotrophic microbes in the oceans live in a permanently aerobic environment (47). In the coastal regions, a significant fraction of microbial production is found within hot spots of particulate organic matter (marine snow) that is, e.g., composed of dead or senescent phytoplankton cells (3, 32). Such zones in which there is high turnover potentially represent a niche that might favor anaerobic metabolism (1, 6). In freely suspended or sinking aggregates, the rapid flux of oxygen largely compensates for microbial oxygen consumption, and anoxic microzones within marine snow particles are probably rather short lived (33). However, in shallow habitats, such as the coastal southern North Sea, the sporadic resuspension of detrital material from sediment surfaces (25) might provide a inoculum of bacteria that also thrive in the absence of oxygen.
Currently there is no direct field evidence that supports the hypothesis that facultatively anaerobic metabolism is a common feature of pelagic microbes. Although some bacteria that are readily isolated from the marine water column are capable of fermentative growth (e.g., the γ-proteobacterial genera Moritella and Vibrio [23, 49]), members of these phylogenetic lineages are usually rare in coastal surface waters, as determined by molecular techniques (13). Recently, Riemann and Azam (37) described a significant decrease in [3H]thymidine incorporation into marine bacteria after inhibition of the membrane transfer system responsible for the uptake of N-acetyglucosamine (NAG). These authors argued that this phosphotransferase system (PTS) is a typical feature of facultatively anaerobic bacteria. They speculated that anaerobic metabolism could thus be a widespread feature of bacteria in the marine water column. However, in that study 22 of the 60 marine isolates that incorporated NAG via a PTS were not capable of a facultatively anaerobic metabolism (37). Moreover, the (aerobic) NAG uptake measured in marine water samples was probably not exclusively mediated via the PTS, as indicated by inhibition experiments with other substrates. Thus, it is still not known if pelagic bacteria are capable of substrate incorporation under anoxic conditions in coastal marine waters and, if they are, which bacteria are involved.
Microautoradiography (MAR) is an approach to track the uptake of radiolabeled tracers in single microbial cells (7). It provides a means of studying the facultatively anaerobic metabolic capacities of water column bacteria in situ via short-term incorporation of glucose under experimentally induced anoxic conditions. In order to assign physiological functions to particular bacterioplankton groups, MAR can be combined with single-cell identification by fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes (10, 22, 29, 45). Recently, the FISH technique has been substantially improved for better visualization of small planktonic cells by means of enzymatic signal amplification (catalyzed reported deposition [CARD]) (30). This modified CARD-FISH protocol has been successfully combined with MAR to measure activity in open-ocean microbes (45). However, in current protocols, evaluation of MAR-FISH-stained bacterioplankton samples is still very time-consuming. Moreover, approximately 50% of the cells are lost during preparation (9), which potentially reduces the accuracy and reproducibility of the MAR technique. Therefore, there is a need for a simplified MAR-FISH approach that profits from the superior signal intensities of CARD-FISH staining and at the same time does not cause cell loss during the MAR procedure.
We tested the hypothesis that incorporation of glucose under anoxic conditions may be a widespread ability of bacteria that live in oxic coastal marine surface waters. This was done by performing short-term incubation and enrichment experiments at two times for samples from the German Bight of the North Sea. FISH with group-specific probes was used to assign aerobic and anaerobic glucose incorporation to individual bacterial populations. For this purpose, a modified protocol that combined MAR and CARD-FISH without the critical cell transfer step was developed and combined with a rapid evaluation system in which semiautomated image analysis was used.
MATERIALS AND METHODS
Sample acquisition, incubation, and fixation.
Surface water samples (depth, 1 m) were collected in July and August 2003 at the Helgoland Roads sampling station (54°11′N, 7°54′E; water depth, 8 m), which is 50 km offshore in the German Bay of the North Sea. The sampling dates represented the onset and decline of a phytoplankton bloom that was mainly composed of Thalassiosira spp. and Ceratium spp. The median surface water temperatures were 18°C in July and 19°C in August, and the median salinities were 34.9 and 32.3 ppt, respectively (Mursys Umweltreport [www.bsh.de]). Incubations with radioactively labeled glucose under oxic and anoxic conditions were performed within 2 h after sample collection. Triplicate 10-ml subsamples were incubated for every treatment type. Oxic incubations were performed in sterile 70-ml serum vials with cotton plugs. Anoxic incubation conditions were established in the same type of serum vials by flushing the vials with nitrogen (purity, 4.0; Air Liquide, Stelle, Germany). The nitrogen flushing times required to produce anoxic conditions in the 10-ml subsamples in the serum vials were verified by Winkler titration and by using an oxygen microsensor (8, 36). The procedure used for anoxic incubations was as follows. First, the empty serum vials were preflushed for 2 min with nitrogen. Next, seawater was added without interrupting the flushing, and the water was bubbled with nitrogen for 15 min. Subsequently d-[6-3H]glucose (specific activity, 1,29 TBq/mmol; Amersham) was added to a final concentration of 10 nM. Before the vials were closed with air-tight butyl rubber stoppers, the headspace in the bottles was flushed for 5 min. The bubbling procedure caused a small but significant increase in the pH, from pH 8.05 ± 0.01 to pH 8.28 ± 0.02, likely because of the removal of dissolved CO2.
The preparations were incubated for either 4 or 24 h in the dark at the ambient water temperature (19 to 21°C). For each treatment one additional control sample was fixed at the beginning of the incubation, prior to the addition of radiolabeled substrate. Subsequently, freshly prepared buffered paraformaldehyde fixative (pH 7.0) was added to the samples to a final concentration of 1%. After fixation the samples for MAR were filtered through polycarbonate filters (type GTTP; pore size, 0.2 μm; diameter, 25 mm; Millipore, Eschborn, Germany). The filters were rinsed twice with sterile phosphate-buffered saline and stored at −20°C until they were analyzed.
Population analysis by FISH.
The percentages of different microbial taxa were determined by FISH with horseradish peroxidase-labeled oligonucleotide probes and catalyzed reporter deposition (30). The following probes were used to characterize the microbial community in the original water samples and after incubation with radiolabeled glucose: EUB I-III (most Bacteria) (11), EURY806 (marine Euryarchaeota) (45), ALF968 (most α-Proteobacteria) (16), GAM42a (most γ-Proteobacteria), CF319a (many groups belonging to the Cytophaga-Flavobacterium cluster of the Bacteroidetes) (2), ROS537 (members of the Roseobacter-Sulfitobacter-Silicibacter clade [referred to as the Roseobacter clade below]), NOR5-730 (NOR5 subcluster of the γ-proteobacterial OM60 clade) (13), SAR86-1245 (members of the γ-proteobacterial SAR86 clade), ALT1413 (Alteromonas spp.), PSA184 (Pseudoalteromonas spp. and Colwellia spp.) (12), and GV822 (Vibrio spp.) (15). The EUB antisense probe NON338 (2) was used as a negative control. All probes were purchased from Biomers.net (Ulm, Germany). Hybridization of polycarbonate filter sections and signal amplification with ALEXA488 (Molecular Probes, Eugene, Oreg.)-custom-labeled tyramides were performed as previously described (30). Specific hybridization conditions were established by addition of formamide to the hybridization buffers. Since the hybridizations were carried out at a lower temperature (35°C), the formamide concentrations in the hybridization buffers were increased by 20% compared to the concentrations reported previously for the directly fluorescently labeled probes (e.g., 55% instead of 35% for probe GAM42a). Counterstaining of CARD-FISH preparations with 4,6-diamidino-2-phenylindole (DAPI) (1 μg ml−1) and mounting on microscopic slides were carried out as described previously (30). DAPI- and FISH-stained cells were counted by automated image analysis (31).
Development of a protocol for CARD-FISH and MAR on membrane filters.
Since all previously described protocols for MAR-FISH of marine bacteria (10, 29, 45) include steps that may cause high cell losses (9), we developed a strategy to combine CARD-FISH and MAR staining of microbes on membrane filters without prior transfer of cells to glass slides (Table 1). For this purpose, we first tested different types of membrane filters (polycarbonate, cellulose nitrate, aluminum oxide). Polycarbonate membrane filters were most appropriate, but they were not rigid enough for the MAR procedure. Therefore, filter sections (one-quarter of a 25-mm-diameter filter) were glued onto glass slides prior to processing. Several glues were tested to determine whether they could resist the handling during the MAR-FISH procedure and cause no increase in the fluorescent background. The optimal glue for fixing the membrane filters onto the slides was a two-component epoxy adhesive (UHU plus sofortfest; UHU GmbH, Bühl, Germany).
TABLE 1.
Overview of protocol for MAR and CARD-FISH of marine bacterioplankton on membrane filters
| Stage | Description |
|---|---|
| Incubation and preparation of filters | Add radiolabeled substrates at the desired concentration; end incubation by fixation with freshly prepared buffered paraformaldehyde fixative; filter onto polycarbonate filters. |
| CARD-FISH (can alternatively be performed after MAR) | See reference 30. |
| Microautoradiographic procedure | Glue the filter pieces onto glass slides; coat the slides with molten emulsion (diluted 1:1 with 0.2% agarose); let coated slides dry on top of an ice-cold metal surface; expose slides inside cardboard boxes at 4°C; develop photographic emulsion as described by the manufacturer. |
| DAPI counterstaining | Stain with a 1-μg/ml DAPI solution for 10 min in a refrigerator; rinse for 1 min with distilled water, followed by 1 min with 80% ethanol; air dry inside the refrigerator; mount in a 4:1 mixture of Citifluor and Vectashield. |
| Evaluation | Image capture for DAPI-, FISH-, and MAR-positive cells; offline image evaluation by semiautomated image analysis. |
We subsequently determined whether there were differences between performing the CARD-FISH staining before the MAR procedure and performing the CARD-FISH staining after the MAR procedure. No difficulties were encountered when we performed CARD-FISH of filter sections before the MAR procedure. In contrast, performing MAR and then CARD-FISH resulted in disruption of the photographic emulsion. To avoid this, the photographic emulsion was diluted with agarose instead of gelatin. Different types of agarose with different gel strengths and melting and gelling points were tested. The best results were achieved with Seakem LE agarose (gel strength, 1%; >1,200 g/cm2; gelling temperature at a concentration of 1.5%, 36 ± 1.5°C; Biozym, Oldenburg, Germany). In the final protocol 1 part of agarose was added to 1 part of molten photographic emulsion at a final agarose concentration of 0.1%. This allowed CARD-FISH to be carried out also after the MAR procedure.
Sample preparation for CARD-FISH and MAR.
For evaluation of our samples, CARD-FISH was usually performed before the MAR analysis, because simultaneous hybridization of numerous filter sections was easier before they were glued onto microscopic slides. The standard CARD-FISH procedure was used (30), including prior embedding of filter sections in agarose and permeabilization with lysozyme. After the FISH-stained cells were counted, the filter pieces were glued onto slides, and MAR was performed within 24 h after hybridization. For the MAR procedure we essentially used the protocol described for cells that were transferred to slides (44). The photochemicals employed were purchased from Eastman Kodak (Rochester, N.Y,) and included autoradiography emulsion (NTB-2), developer (Dektol), and fixer. Different MAR exposure times were tested to obtain the maximum number of cells with silver grains while minimizing the number of false-positive cells, as judged from MAR of the prefixed controls. The optimal exposure times were 8 h for the samples obtained in July and 18 to 24 h for the samples obtained in August. For development of the exposed slides we used the instructions of the manufacturer (2 min of development, 10 s of rinsing with distilled water, and 5 min in fixer, followed by 5 min of washing with distilled water).
Evaluation by image analysis.
Evaluations of MAR-FISH preparations were carried out by using an Axioplan II imaging fluorescence microscope (Carl Zeiss, Jena, Germany) equipped with a motorized stage, a z-axis drive, a fluorescent filter wheel, and a digital camera (Orca I; Hamamatsu, Herrsching, Germany) linked to a personal computer. The KS400 image analysis software (Carl Zeiss Vision, Hallbergmoos, Germany) was used to develop a semiautomated image acquisition and evaluation strategy. First, the operator focused on cells with UV excitation at a magnification of ×63. An image pair was acquired with UV and blue excitation. Next, a stack of five bright-field images was acquired automatically in a region that was 1 to 5 μm above the filtered cells, in order to adequately record MAR grains in an emulsion whose thickness was potentially variable. The bright-field images were combined to obtain a single image, and grains from different layers were detected by selecting the minimum brightness of each pixel from the image stack. For each sample, image triplets from at least 10 microscopic fields were acquired and evaluated.
Detection of DAPI- and FISH-double-stained objects in the respective images was performed as previously described (31). The MAR grains in the combined bright-field image were detected by using the following strategy. First, the image was inverted, and the average gray value was subtracted for background correction. Next, the contrast was rescaled to cover 255 grey levels, and the image was binarized with a fixed threshold (gray value, 150). Objects below a custom threshold were removed, and the binary image was further processed by two rounds of sequential object dilation and erosion by 1 pixel (morphological closing). Binary images from each image triplet were combined for automated colocalization of DAPI-stained and hybridized cells and of MAR grains. The binary images were depicted on the screen as a three-color overlay which could be interactively combined with the original images for manual elimination of artifacts before the object-counting procedure.
RESULTS
Quality of the novel MAR-FISH protocol.
The modified MAR and CARD-FISH staining and quantification protocol allowed substantially more rapid processing of preparations than approaches that require transfer of cells to microscopic slides. It specifically eliminated the accidental loss of preparations due to unsuccessful cell transfer, which is a common problem of the MAR technique. Altogether, 99 individual MAR-FISH preparations were evaluated. Since the bacterial cells were embedded in agarose prior to treatment, no significant cell loss was caused by the procedure. On average, 350 FISH-stained cells were counted per sample. The average coefficient of variation of the fraction of cells with visible tracer uptake in triplicate incubations was 0.2 (i.e., 20% of the mean value). The number of MAR-positive cells counted and the variance of triplicate incubations were used to explore the relationship between evaluation effort and experimental precision (Fig. 1). The observed coefficients of variation were usually less than 0.2 if on average more than 50 MAR-positive cells were counted for each replicate (Fig. 1). Negative controls yielded similar percentages of false-positive MAR-active cells irrespective of the sampling time, incubation time, or incubation conditions (<2% of all FISH-stained cells).
FIG. 1.
Relationship between the mean number of counted cells with visible tracer uptake (MAR+) and the coefficient of variation (CV) of the fraction of these cells (percentage of all FISH-stained cells) in triplicate incubations.
Microbial community composition.
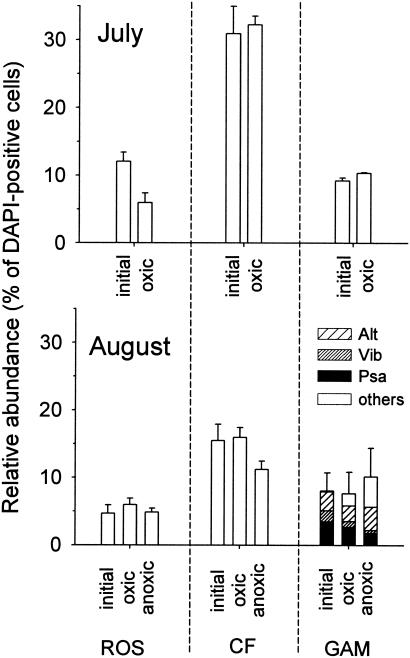
In July, 87% of all DAPI-stained objects could be detected by FISH, but only 60% could be detected in August. Euryarchaeota accounted for 19 and 12% of the total counts in July and August, respectively. Bacteria that hybridized with probes ALF968, GAM42a, and CF319a accounted for around 90% of the cells targeted by the general bacteria probe EUB I-III at both times. Most α-Proteobacteria (as detected with probe ALF968) could be assigned to the Roseobacter clade in both July and August (data not shown). Therefore, in our subsequent evaluations we focused on this group rather than on α-Proteobacteria in general. In July, Alteromonas spp., Pseudoalteromonas spp., and Vibrio spp. were rare in the water column, whereas these groups accounted for the majority of γ-Proteobacteria in the August samples (Fig. 2). Members of the Cytophaga-Flavobacterium cluster of the Bacteroidetes were substantially more abundant in July than in August (Fig. 2).
FIG. 2.
Relative amounts of members of the Roseobacter clade (ROS), the Cytophaga-Flavobacterium cluster (CF), and the γ-Proteobacteria (GAM) in oxic and anoxic incubations (4 h) of coastal North Sea surface water. Alt, Alteromonas; Psa, Pseudoalteromonas-Colwellia; Vib, Vibrio; others, other γ-Proteobacteria. The term initial refers to a sample prior to incubation.
Short-term (4-h) incorporation of radiolabeled glucose.
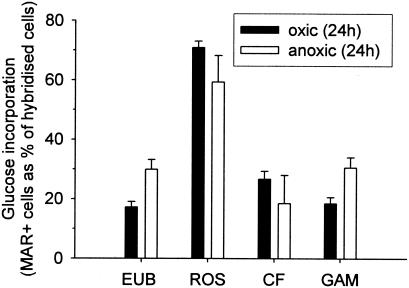
In July, short-term glucose uptake experiments were carried out only under oxic conditions, whereas in August, both oxic and anoxic incubations were performed. Under oxic conditions, aerobic glucose incorporation was found in 29% ± 2% and 12% ± 5% of the bacteria in July and August, respectively (Fig. 3). Euryarchaeota did not show glucose incorporation under either oxic or anoxic conditions (data not shown). In July 84% ± 4% of Roseobacter spp. cells took up the tracer under oxic conditions, but in August the percentage was only 11% ± 1% (Fig. 3). The frequency of MAR-positive γ-Proteobacteria and members of the Cytophaga-Flavobacterium cluster ranged from <10% to approximately 20%, and there were no clear differences between the two times.
FIG. 3.
Fractions of cells with visible tracer uptake (MAR+) affiliated with the Bacteria (EUB), the Roseobacter clade (ROS), the Cytophaga-Flavobacterium cluster (CF), and the γ-Proteobacteria (GAM) in oxic and anoxic incubations (4 h).
In August, the fractions of bacteria and of Roseobacter spp. that took up glucose were similar in oxic and anoxic incubations (Fig. 3). For the γ-Proteobacteria the tracer uptake was significantly higher under anoxic conditions than under oxic conditions. In contrast, fewer members of the Cytophaga-Flavobacterium cluster were found to incorporate radiolabeled glucose after anoxic incubation (Fig. 3).
Enrichment under oxic and anoxic conditions.
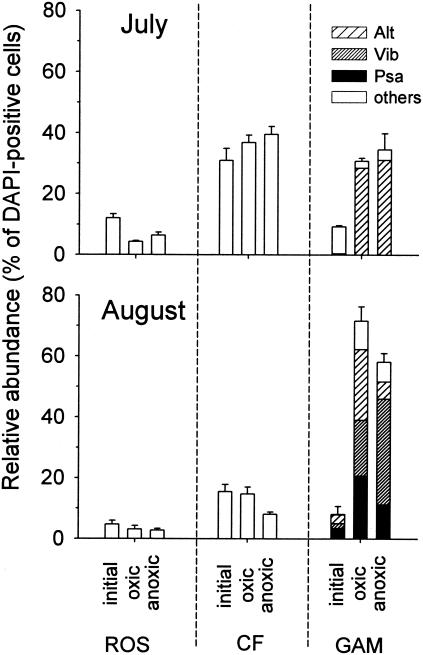
During 24 h of incubation under oxic conditions, the total cell concentration increased from 0.6 × 106 ± 0.2 × 106 to 1.3 × 106 ± 0.3 × 106 cells ml−1 in July and from 1.2 × 106 ± 0.2 × 106 to 2.9 × 106 ± 0.6 × 106 cells ml−1 in August. Under anoxic conditions, the increase was less pronounced; in July the concentration increased to 1.0 × 106 ± 0.2 × 106 cells ml−1, and in August the concentration increased to 1.7 × 106 ± 0.3 × 106 cells ml−1. Under both incubation conditions, mainly γ-Proteobacteria were enriched (Fig. 4). In July, Alteromonas spp. constituted >90% of the newly grown γ-Proteobacteria, whereas in August the numbers of Pseudoalteromonas spp. and Vibrio spp. cells also significantly increased. In August the relative contribution of Alteromonas spp. to the γ-Proteobacteria was substantially smaller under anoxic conditions, whereas bacteria affiliated with Vibrio spp. were more numerous under these conditions (Fig. 4).
FIG. 4.
Relative amounts of members of the Roseobacter clade (ROS), the Cytophaga-Flavobacterium cluster (CF), and the γ-Proteobacteria (GAM) in oxic and anoxic enrichments (24 h). Alt, Alteromonas; Psa, Pseudoalteromonas-Colwellia; Vib, Vibrio; others, other γ-Proteobacteria.
Glucose incorporation during the 24-h enrichments.
Tracer uptake into microbial cells over a 24-h period was only studied with the July samples. The frequency of MAR-positive bacteria was approximately one-third higher under anoxic incubation conditions than under oxic incubation conditions, which mirrored the higher numbers of glucose-incorporating γ-Proteobacteria in the former treatments (Fig. 5). Roseobacter spp. showed the highest fraction of MAR-positive cells irrespective of the incubation conditions (under oxic conditions, 71% ± 2%; under anoxic conditions, 59% ± 9% [mean ± standard deviation]). Approximately 20% of the cells belonging to the Cytophaga-Flavobacterium cluster were capable of taking up glucose under anoxic enrichment conditions. In August, after 24 h of incubation a substantial fraction (>50%) of active cells was found in small aggregates that were composed of different γ-Proteobacteria. While the numbers of cells belonging to these bacterial groups could still be determined by FISH (Fig. 4), it was not possible to assign the MAR signals from such aggregated cells to individual populations.
FIG. 5.
Fractions of cells with visible tracer uptake (MAR+) affiliated with the Bacteria (EUB), the Roseobacter clade (ROS), the Cytophaga-Flavobacterium cluster (CF), and the γ-Proteobacteria (GAM) in oxic and anoxic enrichments (24 h) in July 2003.
Glucose incorporation for specific genera of the γ-Proteobacteria.
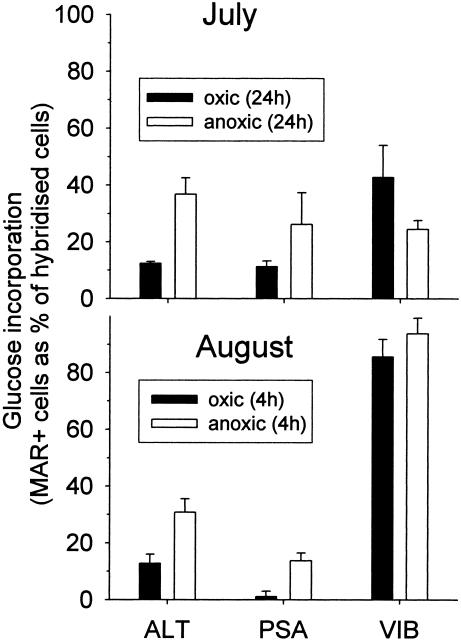
Tracer uptake was specifically investigated for the three genera of γ-Proteobacteria that were found to be enriched over a 24-h period (Fig. 4). At both sampling times, significantly larger fractions of MAR-positive Alteromonas spp. and Pseudoalteromonas spp. cells were present after anoxic incubation for 24 h (July) and 4 h (August), respectively (Fig. 6). In July, nearly twice as many Vibrio spp. took up glucose under oxic conditions as under anoxic conditions. In contrast, little difference between oxic glucose uptake and anoxic glucose uptake by Vibrio spp. was observed in August. At that sampling time, approximately 80 to 95% of all Vibrio spp. cells were MAR positive. No visible glucose incorporation could be detected in cells belonging to other γ-proteobacterial groups (related to the NOR5 and SAR86 clades) under any conditions or at any time.
FIG. 6.
Fractions of cells with visible tracer uptake (MAR+) affiliated with the γ-proteobacterial genera Alteromonas (ALT), Pseudoalteromonas and Colwellia (PSA), and Vibrio (VIB) in oxic and anoxic incubations (4 h) and enrichments (24 h).
DISCUSSION
Method development.
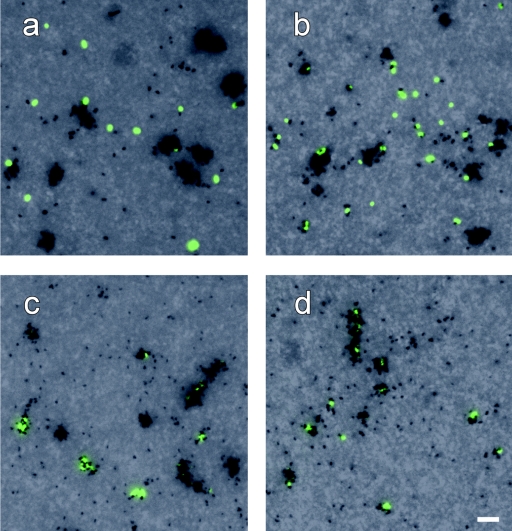
We successfully combined CARD-FISH and MAR to detect glucose uptake in coastal bacterioplankton populations under oxic and anoxic conditions. A satisfactory level of reproducibility was obtained for counts of active cells from triplicate incubations (Fig. 1). We addressed one of the most problematic issues of current MAR approaches by directly performing FISH and MAR with microbes concentrated on membrane filters without prior transfer of cells to glass slides. This strategy was originally introduced by Meyer-Reil (26), but it was later dismissed. Tabor and Neihof (44) stated that such an approach would result in background fluorescence that was too high to perform total cell counting by acridine orange staining and that the filter pores would interfere with the recognition of bacterial cells and silver grains. In our MAR preparations the background fluorescence with blue excitation was minimal and did not interfere with the CARD-FISH staining (Fig. 7). The filter pores did not present an obstacle to the visualization of MAR grains by bright-field illumination (Fig. 7) if (i) the aperture and field stops were sufficiently opened and (ii) the condenser was slightly misaligned from Köhler illumination to decrease contrast. Such empirical optimization of bright-field illumination is comparable to adjusting optimal Köhler illumination.
FIG. 7.
Photomicrographs of hybridized bacteria from 24-h enrichments (July) with different levels of tracer uptake after MAR and CARD-FISH. (a and b) Probe ALT1413 (Alteromonas spp.) under oxic (a) and anoxic (b) conditions. (c and d) Probe ROS537 (members of the Roseobacter clade) under oxic (c) and anoxic (d) conditions.
In this context we also compared MAR preparations on normal glass slides and on Cyto-Clear slides (GE Osmonics, Minnetonka, Minn.). These slides make polycarbonate filters nearly optically transparent with bright-field illumination (34). No substantial enhancement in the quality of the microscopic images was observed when Cyto-Clear slides were used. However, the quality of DAPI staining was indeed lower after MAR than after CARD-FISH alone. This was not a critical problem, since the fractions of hybridized cells could be readily determined before the actual MAR procedure was performed. Other DNA stains, such as SYBR Green I (28), might result in reduced background values for counts of total MAR-active cells.
In addition, our method also allowed us to perform the MAR procedure before the actual FISH staining (Table 1). While this is not important if substrates are labeled with 14C or 3H, it may be a great advantage for preserving the activity signal when isotopes with relatively short half-lives are used (e.g., in studies of the incorporation of dimethylsulfoniopropionate labeled with 35S) (52).
Anoxic glucose incorporation as an indication of facultatively anaerobic metabolism.
Our results suggest that a substantial fraction of microbes in coastal North Sea surface waters are capable of substrate uptake in the absence of oxygen. During 4-h incubations in August, equal fractions of bacterial cells incorporated glucose under anoxic and oxic conditions (Fig. 3). Thus, it seems likely that the slight increase in the ambient pH due to the oxygen removal procedure did not negatively affect microbial viability.
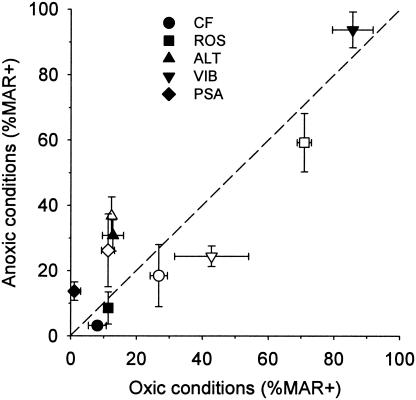
The ability to incorporate glucose in the absence of oxygen was present in bacterioplankton populations belonging to different phylogenetic lineages (Fig. 3 and 6). At our level of analysis, strictly oxic uptake was not observed in any of the groups studied, and some bacterial lineages preferentially incorporated the tracer under anoxic conditions (Fig. 8). Glucose uptake is an active transport mechanism, and there must be a simultaneous process to provide the required energy (e.g., fermentation). In addition, some planktonic bacteria might be able to use electron acceptors other than oxygen for respiration. Isolates related to the Alteromonadaceae, Vibrionaceae, and Roseobacter spp. are capable of denitrification (14, 23, 41). However, the median monthly nitrate concentrations at the Helgoland Roads sampling station in July and August 2003 were <5 μmol liter−1 (MURSYS-report [www.bsh.de]). These concentrations were probably too low to explain the growth of these groups during the 24-h enrichments (Fig. 2 and 4).
FIG. 8.
Comparison of glucose incorporation under oxic and anoxic conditions. Open symbols, July, 4-h incubations; solid symbols, August, 24-h enrichments. ROS, Roseobacter clade; CF, Cytophaga-Flavobacterium cluster; ALT, Alteromonas; VIB, Vibrio; PSA, Pseudoalteromonas-Colwellia; MAR+, fraction of cells with visible tracer uptake.
Facultatively anaerobic marine γ-proteobacteria.
The observed enrichment and high levels of tracer incorporation in Vibrio spp. under anoxic conditions (Fig. 6) support the interpretation that our experimental setup tested for facultatively anaerobic metabolism. Marine Vibrio spp. are known for their ability to ferment sugars in the absence of oxygen (23). Surprisingly, a higher fraction of cells that hybridized with probes ALT1412 and PSA184 exhibited glucose incorporation under anoxic conditions than under oxic conditions (Fig. 6). The genus Alteromonas was originally defined on the basis of a collection of predominantly aerobic gram-negative isolates (5), and the described isolates belonging to the genera Pseudoalteromonas and Colwellia are also known as obligately aerobic bacteria (38, 54). However, recently Riemann and Azam (37) reported a potential for anaerobic metabolism in several strains that were affiliated with the Alteromonadaceae, including a close relative of Alteromonas macleodii. 16S rRNA sequence types related to Pseudoalteromonas spp. have been obtained from the anoxic zone of the Cariaco Basin (24) and from the suboxic layers overlying the sulfide-rich deeper waters of the Black Sea (51). This suggests that the well-studied, strictly aerobic isolates from this family might not be ecophysiologically representative of Alteromonas and Pseudoalteromonas spp. in the marine water column.
Glucose uptake by members of the Roseobacter clade.
In July, a high fraction of microbes affiliated with the Roseobacter clade showed incorporation of radiolabeled glucose. They accounted for <10% of the total picoplankton but >25% of all bacteria that took up detectable amounts of tracer within 4 h during aerobic incubation. In contrast, only a minor fraction of cells belonging to the Roseobacter clade was able to incorporate glucose in August, although the total amounts of such bacteria were similar on the two sampling dates (Fig. 2). This suggests that the activities of pelagic microbes are substantially more dynamic than their population sizes and that a small number of active cells can be responsible for a significant fraction of the turnover of particular substrates. The Roseobacter clade as defined by the probe ROS537 harbors a number of subgroups with strikingly different physiological properties (41, 43, 53). Thus, the observed changes in activity might also reflect a succession of species between the sampling dates.
Bacteria belonging to the Roseobacter clade are widely distributed in the temperate oceans (17, 40), and some microbes related to the Roseobacter group are readily enriched and isolated from marine waters (13, 18). Planktonic members of this group are known to be major consumers of the algal osmolyte dimethylsulfoniopropionate, as observed, e.g., during a coccolithophore bloom in the North Sea (55). Substantial fractions of 16S rRNA gene sequence types and isolates from nonaxenic cultures of marine algae are affiliated with this lineage (35, 39). This close association of bacteria belonging to the Roseobacter lineage with primary producers agrees with the seasonally high numbers of tracer-incorporating cells (Fig. 3). In marine waters glucose originates mainly from the degradation of phytoplankton-derived exudates (20).
A large fraction of cells related to the Roseobacter group incorporated the tracer under anoxic conditions during the July 24-h enrichments (Fig. 5 and 7). The potential for anoxic glucose uptake may shed new light on the ecology of members of the Roseobacter clade in coastal surface waters. Although this clade comprises bacteria with very different physiological capacities (e.g., heteroorganotrophs, anoxygenic phototrophs, sulfite oxidizers, and denitrifiers) (41, 43, 53), no facultatively fermentative strains have been described. Recently, representatives of a new genus belonging to this group have been isolated from the German Bight (53), but these strains are also strictly aerobic heterotrophs.
Ecological role of facultatively anaerobic water column bacteria.
The apparently widespread ability to take up glucose under anoxic conditions (Fig. 8) raises the question of why this is so common in bacteria from fully aerated marine surface waters. We support the argument of Riemann and Azam that this feature likely is an adaptation to growth in nutrient-rich microenvironments in which oxygen is periodically depleted (37). Zooplankton fecal pellets seasonally constitute the most common type of particulate organic matter in the coastal North Sea (50), and carbon from chitinous particles may account for a substantial fraction of bacterial production in coastal marine habitats (21). In contrast to entirely alga-derived marine snow, such compact particles might represent a habitat in which oxygen fluxes are sufficiently low to allow temporary anoxia (1), although this is disputed by other authors (42). Vibrio spp. are commonly found in fecal pellets of copepods (19), and some strains produce specific proteins to adhere to chitin (27). Isolates related to Alteromonas and Pseudoalteromonas readily colonize and degrade chitinous particles (4, 46), and a large fraction of the cells of members of the Cytophaga-Flavobacterium cluster from a coastal pelagic assemblage were able to incorporate chitin degradation products (10).
Even if marine snow aggregates do not become permanently anoxic while they are freely suspended, their sedimentation times are probably rather short. The average depth of the German Bight is only 20 m, and suspended particles and fecal pellets may sink through the water column at speeds of 100 m or more per day (48). Thus, bacteria associated with aggregated senescent algae (e.g., bacteria belonging to the Roseobacter lineage) might experience temporary anoxia while they are on the sediment surface. Subsequent reintroduction of such bacteria into the water column could be induced by periodic vertical mixing and resuspension of particulate organic matter (25). Altogether, the observed anaerobic uptake of glucose by different pelagic bacteria (Fig. 8) suggests that there might be a substantial overlap among the microbial assemblages that inhabit the water column, organic particles, and the sediment surfaces in shallow coastal systems.
Acknowledgments
We thank Josef Franzoi and Armin Gieseke for help with oxygen measurements, Friedrich Widdel and Jens Harder for advice on anoxic incubations, Heidi Pirkner and Annelie Pernthaler for introduction to MAR and CARD-FISH, and Citlali Guerra for help during sample processing. Gunnar Gerdts is gratefully acknowledged for hosting us in Helgoland. We also thank Rudi Amann for continued support and helpful comments on the manuscript.
This work was supported by the European Union (grant EVK3-2001-00194 BASICS) and by the Max Planck Society.
REFERENCES
- 1.Alldredge, A. L., and Y. Cohen. 1987. Can microscale chemical patches persist in the sea—microelectrode study of marine snow, fecal pellets. Science 235:689-691. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 4.Baty, A. M., C. C. Eastburn, Z. Diwu, S. Techkarnjanaruk, A. E. Goodman, and G. G. Geesey. 2000. Differentiation of chitinase-active and non-chitinase-active subpopulations of a marine bacterium during chitin degradation. Appl. Environ. Microbiol. 66:3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumann, L., P. Baumann, M. Mandel, and R. D. Allen. 1972. Taxonomy of aerobic marine eubacteria. J. Bacteriol. 110:402-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianchi, M., D. Marty, J. L. Teyssie, and S. W. Fowler. 1992. Strictly aerobic and anaerobic bacteria associated with sinking particulate matter and zooplankton fecal pellets. Mar. Ecol. Prog. Ser. 88:55-60. [Google Scholar]
- 7.Brock, T. D. 1967. Bacterial growth rate in the sea—analysis by thymidine autoradiography. Science 155:81-83. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, J. H. 1965. The Chesapeake Bay Institute technique for the Winkler dissolved oxygen method. Limnol. Oceanogr. 10:141-143. [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 10.Cottrell, M. T., and D. L. Kirchman. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 12.Eilers, H., J. Pernthaler, F. O. Glöckner, and R. Amann. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eilers, H., J. Pernthaler, J. Peplies, F. O. Glöckner, G. Gerdts, and R. Amann. 2001. Isolation of novel pelagic bacteria from the German Bight and their seasonal contribution to surface picoplankton. Appl. Environ. Microbiol. 67:5134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gauthier, G., M. Gauthier, and R. Christen. 1995. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit ribosomal RNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of 12 new species combinations. Int. J. Syst. Bacteriol. 45:755-761. [DOI] [PubMed] [Google Scholar]
- 15.Giuliano, L., E. De Domenico, M. G. Höfle, and M. M. Yakimov. 1999. Identification of culturable oligotrophic bacteria within naturally occurring bacterioplankton communities of the Ligurian Sea by 16S rRNA sequencing and probing. Microb. Ecol. 37:77-85. [DOI] [PubMed] [Google Scholar]
- 16.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez, J. M., and M. A. Moran. 1997. Numerical dominance of a group of marine bacteria in the alpha-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez, J. M., W. B. Whitman, R. E. Hodson, and M. A. Moran. 1996. Identifying numerically abundant culturable bacteria from complex communities: an example from a lignin enrichment culture. Appl. Environ. Microbiol. 62:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, B., and G. Bech. 1996. Bacteria associated with a marine planktonic copepod in culture. 1. Bacterial genera in seawater, body surface, intestines and fecal pellets and succession during fecal pellet degradation. J. Plankton Res. 18:257-273. [Google Scholar]
- 20.Ittekkot, V., U. Brockmann, W. Michaelis, and E. T. Degens. 1981. Dissolved free and combined carbohydrates during a phytoplankton bloom in the northern North Sea. Mar. Ecol. Prog. Ser. 4:299-305. [Google Scholar]
- 21.Kirchman, D. L., and J. White. 1999. Hydrolysis and mineralization of chitin in the Delaware Estuary. Aquat. Microb. Ecol. 18:187-196. [Google Scholar]
- 22.Lee, N., P. Nielsen, K. Andreasen, S. Juretschko, J. Nielsen, K. Schleifer, and M. Wagner. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macian, M. C., W. Ludwig, K. H. Schleifer, M. J. Pujalte, and E. Garay. 2001. Vibrio agarivorans sp. nov., a novel agarolytic marine bacterium. Int. J. Syst. Evol. Bacteriol. 51:2031-2036. [DOI] [PubMed] [Google Scholar]
- 24.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCandliss, R. R., S. E. Jones, M. Hearn, R. Latter, and C. F. Jago. 2002. Dynamics of suspended particles in coastal waters (southern North Sea) during a spring bloom. J. Sea Res. 47:285-302. [Google Scholar]
- 26.Meyer-Reil, L. A. 1978. Autoradiography and epifluorescence microscopy combined for determination of number and spectrum of actively metabolizing bacteria in natural waters. Appl. Environ. Microbiol. 36:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montgomery, M. T., and D. L. Kirchman. 1993. Role of chitin-binding proteins in the specific attachment of the marine bacterium Vibrio harveyi to chitin. Appl. Environ. Microbiol. 59:373-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 29.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler, J., A. Pernthaler, and R. Amann. 2003. Automated enumeration of groups of marine picoplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ploug, H., H. P. Grossart, F. Azam, and B. B. Jorgensen. 1999. Photosynthesis, respiration, and carbon turnover in sinking marine snow from surface waters of Southern California Bight: implications for the carbon cycle in the ocean. Mar. Ecol. Prog. Ser. 179:1-11. [Google Scholar]
- 33.Ploug, H., M. Kühl, B. Buchholz Cleven, and B. B. Jörgensen. 1997. Anoxic aggregates: an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 13:285-294. [Google Scholar]
- 34.Posch, T., J. Pernthaler, A. Alfreider, and R. Psenner. 1997. Cell-specific respiratory activity of aquatic bacteria studied with the tetrazolium reduction method, cyto-clear slides, and image analysis. Appl. Environ. Microbiol. 63:867-873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prokic, I., F. Brümmer, T. Brigge, G. H. D., G. Gerdts, C. Schütt, M. Elbrächter, and M. E. G. Müller. 1998. Bacteria of the genus Roseobacter associated with the toxic dinoflagellate Prorocentrum lima. Protist 149:347-357. [DOI] [PubMed] [Google Scholar]
- 36.Revsbech, N. P. 1989. An oxygen microsensor with a guard cathode. Limnol. Oceanogr. 34:474-478. [Google Scholar]
- 37.Riemann, L., and F. Azam. 2002. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol. 68:5554-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawabe, T., H. Makino, M. Tatsumi, K. Nakano, K. Tajima, M. M. Iqbal, I. Yumoto, Y. Ezura, and R. Christen. 1998. Pseudoalteromonas bacteriolytica sp. nov., a marine bacterium that is the causative agent of red spot disease of Laminaria japonica. Int. J. Syst. Bacteriol. 48:769-774. [DOI] [PubMed] [Google Scholar]
- 39.Schäfer, H., B. Abbas, H. Witte, and G. Muyzer. 2002. Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42:25-35. [DOI] [PubMed] [Google Scholar]
- 40.Selje, N., M. Simon, and T. Brinkhoff. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445-448. [DOI] [PubMed] [Google Scholar]
- 41.Shiba, T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol. 14:140-145. [Google Scholar]
- 42.Simon, M., H. P. Grossart, B. Schweitzer, and H. Ploug. 2002. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 28:175-211. [Google Scholar]
- 43.Sorokin, D. Y. 1995. Sulfitobacter pontiacus gen. nov., sp. nov.—a new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Microbiology 64:295-305. [Google Scholar]
- 44.Tabor, P. S., and R. A. Neihof. 1982. Improved micro-auto-radiographic method to determine individual microorganisms active in substrate uptake in natural waters. Appl. Environ. Microbiol. 44:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teira, E., T. Reinthaler, A. Pernthaler, J. Pernthaler, and G. J. Herndl. 2004. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl. Environ. Microbiol. 70:4411-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsujibo, H., H. Orikoshi, N. Baba, M. Miyahara, K. Miyamoto, M. Yasuda, and Y. Inamori. 2002. Identification and characterization of the gene cluster involved in chitin degradation in a marine bacterium, Alteromonas sp. strain O-7. Appl. Environ. Microbiol. 68:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turley, C. M., and P. J. Mackie. 1994. Biogeochemical significance of attached and free-living bacteria and the flux of particles in the NE Atlantic Ocean. Mar. Ecol. Prog. Ser. 115:191-203. [Google Scholar]
- 48.Turner, J. T. 2002. Zooplankton fecal pellets, marine snow and sinking phytoplankton blooms. Aquat. Microb. Ecol. 27:57-102. [Google Scholar]
- 49.Urakawa, H., K. Kita-Tsukamoto, S. E. Steven, K. Ohwada, and R. R. Colwell. 1998. A proposal to transfer Vibrio marinus (Russell 1891) to a new genus, Moritella gen. nov., as Moritella marina comb. nov. FEMS Microbiol. Lett. 165:373-378. [DOI] [PubMed] [Google Scholar]
- 50.Urban-Rich, J., E. Nordby, I. J. Andreassen, and P. Wassmann. 1999. Contribution by mesozooplankton fecal pellets to the carbon flux on Nordvestbanken, north Norwegian shelf in 1994. Sarsia 84:253-264. [Google Scholar]
- 51.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vila, M., R. Simo, R. P. Kiene, J. Pinhassi, J. A. Gonzalez, M. A. Moran, and C. Pedros-Alio. 2004. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wagner-Döbler, I., H. Rheims, A. Felske, R. Pukall, and B. J. Tindall. 2003. Jannaschia helgolandensis gen. nov., sp nov., a novel abundant member of the marine Roseobacter clade from the North Sea. Int. J. Syst. Evol. Bacteriol. 53:731-738. [DOI] [PubMed] [Google Scholar]
- 54.Yumoto, I., K. Kawasaki, H. Iwata, H. Matsuyama, and H. Okuyama. 1998. Assignment of Vibrio sp. strain ABE-1 to Colwellia maris sp. nov., a new psychrophilic bacterium. Int. J. Syst. Bacteriol. 48:1357-1362. [DOI] [PubMed] [Google Scholar]
- 55.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. A. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulfoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]