Abstract
Bacillus thuringiensis crystal proteins of the Cry34 and Cry35 classes function as binary toxins showing activity on the western corn rootworm, Diabrotica virgifera virgifera LeConte. We surveyed 6,499 B. thuringiensis isolates by hybridization for sequences related to cry35A genes, identifying 78 strains. Proteins of the appropriate molecular mass (ca. 44 kDa) for Cry35 were observed in 42 of the strains. Full-length, or nearly full-length, sequences of 34 cry34 genes and 16 cry35 genes were also obtained from cloning, PCR analysis, and DNA sequencing. These included representatives of all known Cry34A, Cry34B, Cry35A, and Cry35B classes, as well as a novel Cry34A/Cry35A-like pair. Bioassay analysis indicated that cry35-hybridizing strains not producing a ca. 14-kDa protein, indicative of Cry34, were not active on corn rootworms, and that the previously identified Cry34A/Cry35A pairs were more active than the Cry34B/Cry35B pairs. The cry35-hybridizing B. thuringiensis strains were found in locales and materials typical for other B. thuringiensis strains. Comparison of the sequences with the geographic origins of the strains showed that identical, or nearly identical, sequences were found in strains from both Australasia and the Americas. Sequence similarity searches revealed that Cry34 proteins are similar to predicted proteins in Photorhabdus luminescens and Dictyostelium discoidium, and that Cry35Ab1 contains a segment similar to beta-trefoil domains that may be a binding motif. The binary Cry34/Cry35 B. thuringiensis crystal proteins thus appear closely related to each other, are environmentally ubiquitous, and share sequence similarities consistent with activity through membrane disruption in target organisms.
Bacillus thuringiensis is a spore-forming bacterium that produces proteinaceous crystals during sporulation which are typically insecticidal, although crystals with activities on other invertebrates and crystals having no detected biological activity have also been reported (10, 19). Several B. thuringiensis insecticidal crystal proteins (ICPs) have been developed and commercialized as sprayable biopesticides and transgenic plant-incorporated protectants for agricultural applications. Because of this commercial value, numerous B. thuringiensis strain collections have been generated as sources of ICPs with improved or novel activities (29). Various isolates of this bacterium have been obtained worldwide by many investigators from a large number of habitats, including insectaries, many soil types, the phyloplane, grain products and grain processing facilities, aquatic habitats, and animal feces (13, 20, 25, 31).
The strains in any particular collection may be characterized in a number of ways, such as growth physiology, flagellar serotyping, profiling plasmid arrays or proteins, the use of monoclonal antibodies, and hybridization or PCR amplification based on sequences of known crystal protein genes (28, 29). There is only a poor correlation between any one of these characterization methods and the insecticidal activity of a particular strain for a number of reasons, including the presence of multiple genes per strain, variable gene families in a given serotype, differing expression levels of the genes present, and relative solubility in the insect midgut (11, 28). In surveys of several B. thuringiensis collections for a number of crystal protein genes, 40 to 50% of the strains either have lepidopteran activity or contain cry1 genes (6, 25, 28, 29), which is perhaps a slightly higher level of representation than that of the approximately 40 cry1 genes among the more than 130 distinct crystal protein sequences known (8). Studies of the frequency of non-cry1 genes, or of strains having no hybridization with any of the tested probes (suggesting novel sequences), have not provided consistent results due to greater variability and lower frequency of such strains and genes in those studies (28).
The ca. 14-kDa Cry34A and ca. 44-kDa Cry35A proteins are insecticidal proteins from B. thuringiensis that function together in the intoxication and control of the western corn rootworm (WCR), Diabrotica virgifera virgifera LeConte (14, 18). Because both proteins are required for effective mortality of the insects, they have been referred to as binary insecticidal proteins. Soluble preparations of Cry34 and Cry35 proteins may be obtained using 40% NaBr or mildly acidic buffers, because the alkaline and reducing conditions that solubilize many other crystal proteins were ineffective (27; unpublished data). Synthetic genes encoding one binary pair, Cry34Ab1 and Cry35Ab1, have been engineered for expression in corn plants, and they provide high levels of protection to corn roots under typical growing conditions (26).
As an extension of our work on Cry34 and Cry35 proteins, we screened B. thuringiensis collections for strains having related sequences in the hopes of finding more active proteins or proteins sufficiently different in sequence that they might provide resistance management options due to novel molecular interactions in the insect. An additional aspect of this work was that we would also obtain information about the diversity and origin of sequences related to Cry34 and Cry35 and the frequency of B. thuringiensis strains having such genes in our collections.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
The B. thuringiensis strains screened in this study were from the Dow AgroSciences strain collection. They included large and representative proportions of the collection described by Feitelson et al. (16), originating from a multicontinent geographic area, and two other collections, obtained independently, that were predominantly of Asian origin. A number of the Cry34- and Cry35-containing B. thuringiensis strains identified in this study as well as several recombinant B. thuringiensis strains expressing cloned cry34 and cry35 genes have been deposited with the Northern Regional Research Laboratory (NRRL), as noted in Tables 1 and 2.
TABLE 1.
Wild-type B. thuringiensis strains used in this study
| Isolate | NRRL reference | Cry34 accession no. | Cry34 designation | Cry35 accession no. | Cry34 designation | Reference or source |
|---|---|---|---|---|---|---|
| PS80JJ1 | B-18679 | AAG50341 | Cry34Aa1 | AAG50342 | Cry35Aa1 | 14 |
| PS149B1 | B-21553 | AAG41671 | Cry34Ab1 | AAG41672 | Cry35Ab1 | 14 |
| PS167H2 | B-21554 | AAG50118 | Cry34Ac1 | AAG50117 | Cry35Ac1 | 14 |
| PS69Q | B-30175 | AY536899 | Cry34Aa3 | AY536895 | Cry35Aa3 | This study |
| PS185GG | B-30175 | AY536897 | Cry34Aa4 | AY536892 | Cry35Aa4 | This study |
| KR1369 | B-30200 | AY536896 | Cry34Ac3 | AY536891 | Cry35Ab3 | This study |
| PS187G1 | B-30185 | AY552563 | AY552583 | This study | ||
| PS187Y2 | B-30187 | AY552565 | This study | |||
| PS201G | B-30188 | AY552566 | This study | |||
| PS201HH2 | B-30190 | AY536898 | Cry34Ba3 | AY536893 | Cry35Ba3 | This study |
| PS242K10 | B-30195 | AY552575 | AY552581 | This study | ||
| KB54A1-6 | B-30197 | AY552577 | This study | |||
| KR589 | B-30198 | AY552579 | AY552580 | This study | ||
| PS185L12 | B-30179 | AY552559 | This study | |||
| PS185W3 | B-30180 | AY552560 | This study | |||
| PS187L14 | B-30186 | AY552564 | AY552582 | This study | ||
| PS186FF | B-30182 | AY552561 | This study | |||
| PS131W2 | B-30176 | AY552554 | AY552587 | This study | ||
| PS158T3 | B-30177 | AY552556 | AY552586 | This study | ||
| PS158X10 | B-30178 | AY552557 | This study | |||
| PS185FF | B-30182 | AY552558 | AY552585 | This study | ||
| PS187F3 | B-30184 | AY552561 | AY552584 | This study | ||
| PS201L3 | B-30189 | AY536900 | Cry34Ba2 | AY536894 | Cry35Ba2 | This study |
| PS204C3 | B-30191 | AY552569 | This study | |||
| PS204G4 | B-18685 | AY552570 | This study | |||
| PS204I11 | B-30192 | AY552571 | This study | |||
| PS204J7 | B-30193 | AY552572 | This study | |||
| PS236B6 | B-30194 | AY552574 | This study | |||
| PS246P42 | B-30196 | AY552576 | This study | |||
| KR1209 | B-30199 | AY552578 | This study | |||
| PS137A | AY552555 | |||||
| PS201V2 | AY552567 | |||||
| PS203J1 | AY552568 | |||||
| PS207C3 | AY552573 |
TABLE 2.
Recombinant B. thuringiensis strains used in this study
| Isolate | Host (plasmid) | Binary toxin genes | Reference or source |
|---|---|---|---|
| Cry-B | Acrystalliferous cloning host | None | 32 |
| MR529 | Cry-B(pHT370) | None | This study |
| MR539 | Cry-B(pMYC2369) | None | This study |
| MR543 | Cry-B(pMYC2426) | cry34Aa1/cry35Aa1 | 14 |
| MR544 | Cry-B(pMYC2427) | cry34Ab1/cry35Ab1 | 26 |
| MR546 | Cry-B(pMYC2429) | cry34Ac1/cry35Ac1 | 14 |
| MR561 | Cry-B(pMYC2476) | cry34Ba2/cry35Ba2 | This study |
| MR562 | Cry-B(pMYC2477) | cry34Ba3/cry35Ba3 | This study |
Shake flask cultures of B. thuringiensis were grown to sporulation and lysis at 30°C for 72 h in NYS/CAA medium, pH 7.2, containing (per liter) 1.25 g of nutrient broth, 1.25 g of Bacto tryptone, 2 g of Casamino Acids, 0.5 g of yeast extract, and 10 ml of B. thuringiensis salt solution, which consisted of (per liter) 40.7 g of MgCl2 · 6H2O, 20 g of CaCl2 · 2H2O, 2 g of MnCl2 · 4H2O, 0.04 g of FeSO4 · 7H2O, 0.04 g of ZnSO4 · H2O, and 0.04 g of (NH4)2SO4, all in 14 mM HCl. Biomass containing sporulated cells and crystalline protein inclusions was harvested from the cultures by centrifugation at 10,000 × g for 20 min. Recombinant B. thuringiensis strains were prepared similarly in NYS/CAA medium supplemented with erythromycin (10 μg/ml).
ICP preparation and quantitation.
For wild-type B. thuringiensis strains, whole-culture pellets were washed twice with the original culture volume of distilled water and collected by centrifugation as described above. The washed pellet was resuspended to 1/10th its original culture volume in extraction buffer (100 mM sodium citrate, pH 3.8), homogenized, and incubated for 2.5 h on a rocker platform at 4°C. Cell debris was removed by centrifugation at 26,000 × g for 30 min, and the supernatant containing soluble proteins was retained. For bioassays, the supernatant was dialyzed against 20 mM sodium citrate buffer, pH 5.5, to equilibrate the acidity of the sample with the insect diet. The dialyzed protein concentration was determined according to Bradford (5) using bovine serum albumin (BSA) as a standard. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (14) on 4 to 12% gradient Bis-Tris NuPAGE gels (Invitrogen) at 200 V for 50 min. Proteins were visualized by staining with Gelcode Blue Stain G-250 (Pierce). For protein quantitation, laser-scanning gel densitometry was performed with a Molecular Dynamics Personal Densitometer SI with BSA as a standard.
For recombinant B. thuringiensis clones, whole-culture pellets were washed and resuspended in water. Proteins were visualized, and the concentration of binary ICPs was determined by densitometry as described above. Preparations were adjusted to 1 mg of binary ICP/ml and serially diluted for quantitative bioassays.
Bioassays.
Methods for testing insecticidal activity of binary ICPs against western corn rootworm neonates in surface-applied bioassays were previously described (14).
Genomic DNA hybridization.
Total genomic DNA from each B. thuringiensis isolate was prepared using the QIAGEN DNEasy kit. DNA in 96-well plates was denatured prior to blotting by diluting 10 μl of DNA solution and 10 μl of 4 M NaOH in 80 μl of sterile distilled water. Samples were incubated at 70°C for 1 h, after which 100 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (recipes for all DNA hybridization solutions are found in reference 21) was added to each well. Total genomic DNA from the Cry34Ab1/Cry35Ab1 source strain PS149B1 was included with each set of 94 samples as a positive hybridization control for cry35Ab1, and CryB total genomic DNA was included with each set of 94 samples as a negative hybridization control. Each complete set of 96 samples was applied to Magnacharge nylon membranes using two 48-well slot blot manifolds (Hoefer Scientific), followed by two washes with 10× SSC. Membranes were baked at 80°C for 1 h and kept dry until used. Membranes were prehybridized and hybridized in standard formamide solution, which contained 50% formamide, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 2% SDS, and 100 μg of single-stranded DNA/ml, at 42°C. Membranes were probed with an approximately 1.3-kbp DNA fragment that included the entire cry35Ab1 coding sequence amplified by PCR from genomic DNA of PS149B1. The DNA probe was amplified by PCR, using as the forward primer 5′-ATGYTWGATACWAATAAAGTWTATGAAAT-3′ and as the reverse primer 5′-GTAGAAGCAGAACAAGAAGGTATT-3′, corresponding to nucleotide coordinates 784 to 812 and 2106 to 2129, respectively, of GenBank accession no. AYO11120. The probe was radioactively labeled using the Prime-it II kit (Stratagene) and [32P]dCTP, purified on Sephadex columns, denatured at 94°C, and added to fresh hybridization solution. Membranes were washed under two conditions: 2× SSC-0.1% SDS at 42°C (low stringency) and 0.2× SSC-0.1% SDS at 65°C (moderate to high stringency). Strains containing genes with homology to the cry35Ab1 probe were identified by autoradiography.
PCR and DNA sequencing.
Degenerate oligonucleotides for PCR were designed to amplify polynucleotides corresponding to open reading frame positions 1 to 341 of cry34Ab1 and 1 to 1103 of cry35Ab1 from B. thuringiensis strains identified by hybridization. The oligonucleotides are homologous to conserved sequence blocks identified by alignment of previously described cry34A and cry35A genes (14, 26). Forward primers for both genes were designed to begin at the ATG initiation codon. Reverse primers were designed from sequences as close to the 3′ end of each respective gene as possible. The sequence of the cry34 forward primer was 5′-ATG TCA GCW CGY GAA GTW CAY ATT G-3′, while the sequence of the cry34 reverse primer was 5′-GTY TGA ATH GTA TAH GTH ACA TG-3′. The sequence of the cry35 forward primer was 5′-ATG TTA GAT ACW AAT AAA RTW TAT G-3′, and the sequence of the cry35 reverse primer was 5′-GTW ATT TCT TCW ACT TCT TCA TAH GAA G-3′. PCR products were fractionated on 1% agarose gels and purified from the gel matrix using the QiaexII kit (QIAGEN). Purified DNA fragments were cloned using the TOPO TA kit (Invitrogen). Cloned PCR-derived fragments were then sequenced using Applied Biosystems automated sequencing systems and associated software.
Gene cloning and coexpression.
The cry34 and cry35 genes from strains PS149B1, PS167H2, and PS80JJ1 were obtained previously (14), and the related genes from PS69Q, PS185GG, and PS201L3 were isolated using the same methods. The related genes from PS187G1, PS201HH2, and KR1369 were obtained using cosmid libraries constructed in the SuperCos1 vector (Stratagene), having inserts prepared by partial digestion with NdeII. XL1-Blue MR cells (Stratagene) were transfected with the packaged cosmids to obtain clones resistant to carbenicillin and kanamycin and were screened by hybridization with a cry35-specific probe generated from each strain produced as described above. Standard methods were used to subclone cry34/cry35 operons into pHT370 (2) for expression of the Cry34 and Cry35 proteins in B. thuringiensis, as previously described (14). Acrystalliferous strain Cry-B was transformed with the pHT370-derived expression plasmids by electroporation as described for coexpression of binary ICPs from native B. thuringiensis promoters (14).
Sequence comparisons.
Sequence similarity searches were performed using the PSI-BLAST (1) facility available at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/BLAST/) during January 2004. Searches were performed against the nonredundant databases using default parameters. Sequences of the Cry34 and Cry35 proteins were aligned using ClustalW, and unweighted paired groupwise mean averaging (UPGMA) dendrograms were generated as described previously (7). For this analysis, the less than full-length sequences resulting from cloned PCR fragments were trimmed to remove the common, conserved primer sequences, because the sequence hybridizing with the primer may not represent the amplified gene and amplification of certain combinations of redundant bases in the primers may have resulted in nonnative sequence. For Cry34 proteins, this trimming results in sequences about 75 to 81% of full length (missing the 8 N-terminally and 9 C-terminally located conserved residues and a 6- to 15-residue variable C-terminus segment). The trimmed Cry35 sequences (missing the 9 N-terminally and 7 C-terminally located conserved residues and a 15- to 17-residue variable C-terminus segment) are about 90% of full length.
Nucleotide sequence accession numbers.
Nucleotide sequence accession numbers are listed in Table 1.
RESULTS
Distribution of binary insecticidal crystal genes and proteins in B. thuringiensis strain collections.
Out of 6,499 B. thuringiensis strains from our collections, slot blot hybridization analysis of B. thuringiensis total genomic DNA using a full-length cry35Ab1 gene probe was used to identify 78 strains that contained sequences putatively related to cry35Ab1. The cry35Ab1 probe had previously been shown to hybridize to related cry35 genes but not to other known classes of cry genes or to genomic B. thuringiensis DNA that were included as controls (data not shown).
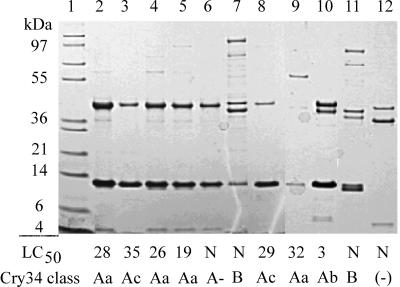
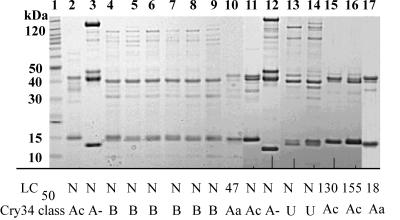
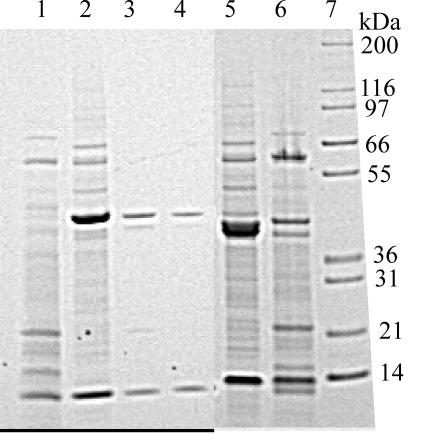
These hybridizing strains were subsequently examined for the presence of proteins similar in size and solubility to Cry34 and Cry35 (14). Proteins were extracted from sporulated culture pellets by solubilization in mildly acidic buffer (pH 3.8) and analyzed by SDS-PAGE for the presence of ca. 14- and 44-kDa proteins. Under the culture conditions used in this study, 42 of the 78 hybridizing strains produced proteins of 40 to 44 kDa. Of these, 29 strains produced both 14- and 44-kDa proteins (Fig. 1, lanes 2 to 11, Fig. 2, lanes 2 to 17, and Fig. 3, lanes 2 and 9 to 12), while 11 strains produced proteins of 40 to 44 kDa in mass but lacked a copurifying 14-kDa protein (Fig. 1, lane 12; Fig. 3, lanes 3 to 8 and 13 to 16). Data for two additional strains are not shown. Further, several strains produced additional proteins, mostly greater than 70 kDa in mass, which were not examined further. Some prominent examples of these are found in lanes 7, 9, and 11 of Fig. 1, lanes 3 and 12 to 14 of Fig. 2, and lanes 9, 10, and 12 of Fig. 3. Strains producing Cry34B seem to consistently produce such proteins. The presence of additional crystal proteins has also been visualized by standard SDS-PAGE extraction and analysis of Cry34- and Cry35-producing strains by us (14, 27, and unpublished data) and by Baum et al. (3).
FIG. 1.
Presence of binary ICPs in B. thuringiensis isolates identified by cry35 nucleic acid hybridization. Lanes: 1, molecular mass markers; 2, PS69Q; 3, PS167H2; 4, PS185FF; 5, PS185GG; 6, PS187G1; 7, PS201L3; 8, PS242K10; 9, PS80JJ1; 10, PS149B1; 11, PS204G4; and 12, KB65A15-8. The 50% lethal concentration (LC50) (in micrograms per squared centimeters) or N (indicating a dose response could not be calculated) is indicated below each lane. A− denotes sequences that would be a new tertiary rank, if complete. A (−) indicates the absence of a 14-kDa protein.
FIG. 2.
Presence of binary ICPs in B. thuringiensis isolates identified by cry35 nucleic acid hybridization. Lanes: 1, molecular mass markers; 2, PS158T3; 3, PS187G1; 4, PS201G; 5, PS201HH2; 6, PS204C3; 7, PS204J7; 8, PS204I11; 9, PS236B6; 10, PS246P42; 11, PS158X10; 12, PS187F3; 13, PS201H2; 14, PS203G2; 15, KR589; 16, KR1209; and 17, PS131W2. The 50% lethal concentration (LC50) (in micrograms per squared centimeters) or N (indicating a dose response could not be calculated) is indicated below each lane. U indicates an undetermined cry34 allele sequence. A− denotes sequences that would be a new tertiary rank, if complete.
FIG. 3.
Presence of binary ICPs in B. thuringiensis isolates identified by cry35 nucleic acid hybridization. Lanes: 1, molecular mass markers; 2, PS149B1; 3, PS223L2; 4, KB10H-5; 5, KB59A54-4; 6, KB59A54-5; 7, KB59A58-4; 8, KB65A14-1; 9, PS201V2; 10, PS207C3; 11, KR1369; 12, PS137A; 13, PS147U2; 14, KB65A15-2; 15, KB65A15-3; and 16, KB65A15-7. The 50% lethal concentration (LC50) (in micrograms per squared centimeters) or N (indicating a dose response could not be calculated) is indicated below each lane. A (−) indicates the absence of a 14-kDa protein.
Protein preparations containing both 14- and 44-kDa protein species demonstrated various degrees of activity on WCR neonates, as summarized in Fig. 1 to 3. Preparations from strains that contained Cry34A-type ICPs as determined by DNA sequencing (see below) appeared generally more active toward WCR neonates than preparations from strains containing Cry34B-type ICPs. Further, no WCR activity was observed for those protein preparations containing a 40- to 44-kDa protein species but were lacking a 14-kDa protein (Fig. 1, lane 12; Fig. 3, lanes 3 to 8 and 13 to 16).
Operon cloning, expression, and insecticidal activity of Cry34B/Cry35B ICPs.
Because the Cry34B/Cry35B subfamily is the most divergent among the binary ICPs described to date, we chose to examine representatives from this phylogenic group in more detail. Genes encoding two examples of Cry34B/Cry35B ICPs were cloned from strains PS201L3 and PS201HH2 and completely sequenced. Comparison of full-length holotype binary ICPs revealed that the deduced Cry34 protein sequences clustered with about 50% amino acid sequence identity, while the Cry35 proteins clustered with about 60% amino acid sequence identity (see Fig. 5 and 6).
FIG. 5.
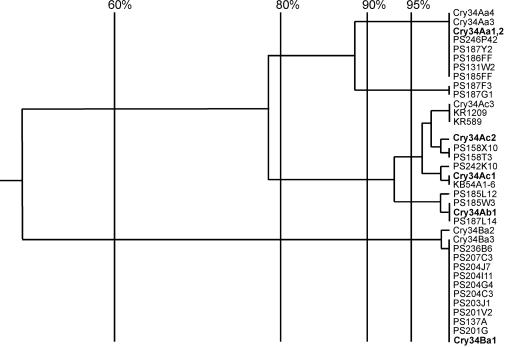
Dendrogram showing the relatedness of Cry34 sequences. Vertical bars indicate the percent sequence identity on the dendrogram. Sequences with nomenclature assignments are full length (previously published assignments are in boldface); PCR-amplified subsequences are indicated by strain (see Materials and Methods for details).
FIG. 6.
Dendrogram showing the relatedness of Cry35 sequences. Vertical bars indicate the percent sequence identity on the dendrogram. Sequences with nomenclature assignments are full length (previously published assignments are in boldface); PCR-amplified subsequences are indicated by strain (see Materials and Methods for details).
To assess bioactivity of Cry34B/Cry35B ICPs, the PS201L3 and PS201HH2 operons encoding these proteins were subcloned into the shuttle vector pHT370 and transformed into acrystalliferous B. thuringiensis strain Cry-B to enable ICP expression from the native promoters. Recombinant B. thuringiensis strains MR561 and MR562, encoding Cry34Ba2/Cry35Ba2 and Cry34Ba3/Cry35Ba3, respectively, both produced crystalline inclusions comprised of ca. 14- and 44-kDa proteins when grown to sporulation (Fig. 4). Unfractionated protein preparations from recombinant strains expressing representative Cry34A/Cry35A and Cry34B/Cry35B binary ICPs were tested for activity against WCR neonates in surface-applied bioassays. As noted for the B. thuringiensis strains expressing Cry34B/Cry35B, the cloned, expressed Cry34B/Cry35B binary ICPs were much less active than Cry34A/Cry35A binary ICPs in comparative assays (Table 3).
FIG. 4.
SDS-PAGE analysis of coexpressed recombinant binary ICPs. Lanes: 1, MR529 negative control; 2, Cry34Aa1/Cry35Aa1; 3, Cry34Ac1/Cry35Ac1; 4,Cry34Ab1/Cry35Ab1; 5, Cry34Ba2/Cry35Ba2; 6, Cry34Ba3/Cry35Ba3; and 7, molecular mass markers.
TABLE 3.
Activity of coexpressed recombinant binary ICPs
| Bacterial strain | Binary ICP | LC50 (μg/cm2) | % Mortalitya (highest dosage, in μg/cm2) |
|---|---|---|---|
| MR543 | Cry34Aa1/Cry35Aa1 | 34 (21-130)b | |
| MR544 | Cry34Ab1/Cry35Ab1 | 3 (2-5) | |
| MR546 | Cry34Ac1/Cry35Ac1 | 7 (3-44) | |
| MR561 | Cry34a2B/Cry35Ba2 | NDRc | 55 (110) |
| MR562 | Cry34Ba3/Cry35Ba3 | NDR | 44 (170) |
| MR529 (negative control) | None | NDR |
The effects of treatments for which the 50% lethal concentration (LC50) could not be calculated are expressed in percent mortality at the top dose.
95% Confidence interval values are in parentheses.
NDR, dose response could not be calculated.
Sequence diversity of binary ICP genes.
We obtained sequence information from a number of the cry34- and cry35-hybridizing strains to assess the sequence diversity and geographic distribution of the various members of this binary toxin family. To determine the relatedness of binary ICP sequences among the hybridizing strains, nearly full-length genes corresponding to nucleotides 1 to 341 of cry34Ab1 and 1 to 1103 of cry35Ab1 were amplified by PCR. Sequences of 34 cry34 genes were obtained from 29 of the 42 expressing strains (including the two expressing strains not shown in Fig. 1 to 3) and 5 nonexpressing strains. Note that the 11 expressing strains lacking a 14-kDa protein did not yield PCR products with either the cry34 or cry35 oligonucleotide primers used in this study, and two strains producing Cry34 proteins were not sequenced. We sequenced 16 cry35 genes from strains having cry34 sequences, including one nonexpressing strain. The nearly full-length PCR-generated sequences (identified by strain designations in Fig. 5 and 6) were N- and C-terminally truncated to remove sequences corresponding to the primers and were compared to the full-length sequences of several of the genes also obtained by standard genomic cloning. Analysis of the deduced polypeptide sequences was performed essentially as described previously (7) such that percent identity (or difference) is calculated based on the shorter sequence, allowing reasonable comparison of sequences of differing lengths. The analysis includes the sequences reported by Ellis et al. (14) and Rupar et al. (30), which have previous nomenclature assignments. The results revealed that the 34 Cry34 and 16 Cry35 sequences analyzed here form distinct families, as shown in Fig. 5 and 6. For the Cry34 (Fig. 5) sequences, nearly all classes of proteins have multiple identical sequences obtained from distinct bacterial isolates; for example, 8 instances for Cry34Aa and 11 instances for Cry34Ba. A similar pattern holds for Cry35 (Fig. 6), although in this case there are fewer identical sequences. For sequence novelty, aside from the more divergent Cry34B and Cry35B sequences, the truncated PCR-derived sequences of PS187G1 and PS187F3 are Cry34A-like and Cry35A-like sequences that would qualify for a new tertiary rank (7), being 89% identical to Cry34Aa and 87% identical to Cry35Aa. By contrast to the striking uniformity of the Cry34Aa and Cry35Ba sequences, there is more sequence variation of the Cry34Ab, Cry34Ac, Cry35Ab, and Cry35Ac sequences from roughly 93% or greater identity. Additionally, Cry34Ab1 and Cry35Ac1 appear to be the more divergent members of these groups, so that in most cases cry34Ac-type genes are in operons with cry35Ab-type genes.
Geographic distribution of cry34 and cry35.
The hybridization screening reported here encompassed three B. thuringiensis collections obtained by different individuals from samples with differing geographic distributions. The rates of recovery of Cry35-hybridizing strains in the three collections were 0.5, 1.4, and 1.9%, for an overall rate of 1.2%. The strains containing the cry34 and cry35 genes were isolated from several continents (North and South America, Australia, and Asia). Table 4 summarizes the general distribution of strains containing the Cry34 sequences shown in Fig. 5. For Cry34Aa and Cry34Ba, the same sequence is found in B. thuringiensis strains isolated in multiple locations on essentially opposite sides of the world. A similar pattern may hold for the Cry34Ab and Cry34Ac sequences; however, there are fewer identical sequences.
TABLE 4.
Geographic distribution of different classes of Cry34 protein
| Protein class | Sequence similarity (%) | Continent(s) | No. of geographically distinct areasa |
|---|---|---|---|
| Cry34Aa | 100 | Australia and Asia | 2 |
| North America | 2b | ||
| Cry34Ab/c | >93 | Australia and Asia | 4 |
| North America | 1 | ||
| South America | 1 | ||
| Cry34Ba | >98 | Australia and Asia | 1 |
| North America | 2 |
Separate large geographic regions of at least several hundred miles or those separated by bodies of water.
Includes information from Rupar et al. (30).
The materials from which the strains containing cry34 and/or cry35 were isolated included dust, soil, leaf litter, nematodes (15), and insects. In most cases the materials were from an agricultural setting either inside or outside of structures. In two cases, however, the habitats were not associated with human activity. Therefore, cry34 and cry35 genes appear to have a broad habitat distribution in nature and are found in materials and environments that are typical for B. thuringiensis (31).
Flagellar serotyping was attempted, essentially by the method of de Barjac (9), on 15 of the cry34- and cry35-containing strains. Of these, 10 were not testable because they were not motile. Of the motile strains, three were serotype 4 (sotto or kenyae), one was subtyped to serovar 4a,4b (sotto), and one was serotype 5 (galleriae or canadensis). Except for one serotype 4 strain for which no sequence information is available, the motile strains were all Cry34Aa-expressing strains.
Sequence relatedness to other proteins.
As noted previously, the Cry35 proteins are similar in sequence to the mosquitocidal B. sphaericus BinA and BinB proteins (14). Also included in this family is the Cry36 protein, which is a 58-kDa polypeptide homologous to Cry35 proteins over C-terminally located ca. 280 residues, and it is as closely related to the Bin proteins as it is to the Cry35 proteins (8, 10). A conserved domain search (22, 23) using Cry35Ab1 as a query also revealed that the N-terminal 146 residues of this protein contain two repeats of the tripartite beta-trefoil carbohydrate-binding domain (17), including two signature QxW motifs. Figure 7 shows the location and sequences of the homology of Cry35Ab1 to the beta-trefoil consensus.
FIG. 7.
Conserved domain search for Cry35Ab1. (A) The Cry35Ab1 protein sequence is indicated by a horizontal line, with blackened ovals indicating the position of two partial alignments of the tripartate beta-trefoil domain similarity (Conserved Domain Database [CDD] entry cd00161) and a white oval indicating a conserved domain based on the B. sphaericus 42-kDa mosquitocidal toxin (CDD entry pfam05431). (B) Multiple-sequence alignment showing two possible alignments of the cd00161 beta-trefoil consensus sequence against the Cry35Ab1 sequence. The QxW motifs of the beta-trefoil sequence are underlined below the consensus line. Lines with arrows denote the internal repeat sequences in the beta-trefoil domain. Consensus similarity groups are the following: 1-D,N; 3-S,T; 4-K,R; 6-L,I,V,M.
Using Cry34Ab1 as a query sequence, the BLAST program (1) returns significant similarity values for other Cry34 proteins and additional non-B. thuringiensis proteins (Fig. 8). A hypothetical protein from Dictyostelium discoidium (GenBank accession no. AAO52140.1) is 1,107 residues in length and contains a conserved domain for the endonuclease/exonuclease/phosphatase family, which includes magnesium-dependent endonucleases and phosphatases involved in intracellular signaling from about residue 550 to 750. The Cry34 homology is in two blocks: residues 336 to 452 of AAO52140 align to residues 3 to 118 of Cry34Aa1 at 27% identity, and residues 15 to 76 of AAO52140 align to residues 2 to 63 of Cry34Aa1 at 31% identity. The overall E value is ∼10−7. Two additional proteins related to Cry34 are from Photorhabdus luminescens (12) and may be detected using iterated PSI-BLAST (1). Both exhibit E values below the 0.005 significance cutoff. Plu1537 (GenBank accession no. NP_928828) is a 136-residue polypeptide that is 27% identical to Cry34Ab1. Plu2230 (GenBank accession no. NP_929487) is a 465-residue polypeptide that has a segment homologous to Cry34 near the N terminus and a more C-terminal segment again containing a phosphatase-like module.
FIG. 8.
Sequence homologies to Cry34. (A) Non-Bacillus sequences in GenBank showing homology to Cry34Ab1 in PSI-BLAST searches. Source organisms and protein accession numbers are indicated. The protein sequence is indicated by a horizontal line, with blackened ovals indicating the position of Cry34 similarity; significant gaps are indicated by segmentation, and white ovals indicating a phosphatase conserved domain (CDD entries COG5555, KOG1378, and KOG2679) for NP_929487 and a endonuclease/exonuclease/phosphatase domain (CDD entry pfam03372) for AAO52140. Truncated ovals indicate incomplete alignment of the conserved domain to the respective sequence. (B) Multiple-sequence alignment of unique Cry34 sequences and homologues. Black highlighting indicates conserved identity or similarity across all sequences, and dark and light gray shading indicates lesser degrees of conservation among the sequences. Consensus similarity groups are the following: 3-S,T and 6-L,I,V,M. The two different alignments with the D. discoidium sequence (accession no. AAO52140) are indicated by the start positions chosen for this alignment based on the PSI-BLAST alignment.
DISCUSSION
Identification and characteristics of strains containing cry34 and cry35 genes.
One objective of this study was to identify additional B. thuringiensis strains containing cry34 and cry35 genes in a broad survey of our B. thuringiensis collection. The use of a hybridization-based method allowed this to be done quickly and inexpensively. We anticipated that this method would allow the identification of genes having 45 to 50% sequence identity with the initial genes and without the strict sequence dependence of 3′ priming sites required for PCR-based analysis (28). The fact that the cry34B and cry35B genes were identified, having 50 to 60% sequence identity, and that some additional hybridizing strains clearly produced 44-kDa proteins but failed in PCR amplification with the primers described here, indicates that the general expectations of the screening strategy were met.
Of the strains identified by hybridization, less than half produced proteins of the expected size and extraction properties. It may be that somewhat different growth or extraction conditions would have allowed the identification of more strains producing proteins of the correct size; however, the presence of genes is known to not necessarily correlate with activity (28), and there are numerous reasons why Cry34/Cry35 proteins were not identified from some hybridizing strains. It is notable that we have cry34 sequence information on five of the strains that did not produce readily identifiable 14-kDa proteins, reinforcing the greater frequency for detecting genes than expressed proteins. A number of cry35-hybridizing strains also produced 44-kDa proteins without the presence of a 14-kDa protein, and they failed in amplification attempts with either cry34-specific or cry35-specific PCR primers. Because these strains failed to give a clear dose response in our corn rootworm assays, they were not pursued further; however, they may contain yet more distant relatives of the cry35 gene family. Compared to the previously described Cry34A/Cry35A binary toxins (14), corn rootworm bioassays also indicated that the Cry34B/Cry35B combination was much less active. This conclusion differs from that of Baum et al. (3), who found roughly similar activity for the Cry34A/Cry35A and Cry34B/Cry35B toxins. However, there were several differences in the assays, including weight reduction versus mortality-based scoring of the assays and the use of purified crystals versus either soluble or whole-culture materials, that may account for the different conclusions.
A number of the cry35-hybridizing strains producing 14- and/or 44-kDa proteins also produced proteins of other sizes. The identity of these proteins was not determined; however, we noted previously (14) that Cry34/Cry35Aa-producing strain PS80JJ1 also produces the Cry14Aa protein. A comparison of the hybridization screening results with a survey of the same collection for lepidopteran-active cry9-like genes showed that six strains were common to the two surveys. Further genotyping of these six strains by restriction analysis of gene-class-specific PCR products revealed the presence of some other common Bacillus insecticidal protein genes (cry1, cry2, cry9, and vip3; data not shown). While none of the identified strains was among the strains producing the 14- and/or 44-kDa proteins shown in Fig. 1 to 3, cry34/cry35Ab-like sequences were obtained from one of them: strain PS187L14 (Fig. 5 and 6). Thus, it is possible to find B. thuringiensis strains that contain genes for several well-known lepidopteran ICPs and corn rootworm binary ICPs.
It has been noted previously that a number of B. thuringiensis crystal protein genes having identical sequences have been found to have a worldwide distribution (7). In the present study, multiple, identical sequences were found for the Cry34 proteins, with Cry34Aa and Cry34Ba having numerous representatives. A similar trend seems likely for the smaller number of Cry35 sequences presented, although the proportion of identical sequences seems lower than that for Cry34, which may partly be due to the use of PCR in some cases and the larger in vivo mutational target of the cry35 genes. As indicated in Table 4, the three major groupings of Cry34 sequences each contained representatives from strains obtained from widely separated geographies. The discovery of strains containing cry34 and cry35 at several locations in Australasia is in agreement with the conclusion of Martin and Travers (25) that this area is a rich source of B. thuringiensis strains and reinforces the notion made earlier (24) that the broad environmental distribution of B. thuringiensis strains was not related to specific insect targets, because western corn rootworm is presently found only in North America and places in eastern Europe, where it was recently introduced. Taken together with the relatively typical materials from which the strains reported here were obtained, this indicates that B. thuringiensis strains containing genes related to cry34 and cry35 are not unusual in terms of geographic distribution or habitat.
Frequency of strains containing genes related to cry34 and cry35.
The overall rate of recovery of cry35-hybridizing strains was 1.2%, ranging from 0.5 to 1.9% for the three collections screened. The overall rate for strains from which Cry34 sequences were obtained was 0.52%, which is a more conservative indicator of the frequency of strains containing cry34 (and cry35) in the collections. This compares with a roughly 40 to 50% frequency for cry1 genes or common lepidopteran-active strains (6, 25, 28, 29).
As a point of comparison, PCR screening for a rarer yet familiar gene, cry3A, has also been reported in the literature. This gene and its corresponding beetle active protein have been registered in both sprayable B. thuringiensis and CellCap-killed recombinant insecticides as well as in transgenic potatoes. Ben-Dov et al. (4) found no cry3-containing strains, by PCR screening, among 215 environmental isolates (<0.5%). A rate of 2.5% was reported for strains having the flat, square crystal morphology typical of Cry3Aa-containing strains of the 3,500 isolates in the International Entomopathogenic Bacillus Centre Collection at Institute Pasteur (19). Bravo et al. (6) found 6 cry3A-containing strains in a collection of 496 isolates (1.2%); however, they also screened for cry3B and cry3C, genes yielding an overall cry3 rate of 5.6%. The latter figure would represent sequence diversity closer to the diversity of Cry34 and Cry35 sequences reported here.
Relatedness of Cry34 and Cry35 to other proteins.
Sequence comparisons (8, 14) with other known B. thuringiensis insecticidal crystal proteins failed to reveal homology between Cry34 or Cry35 proteins with other previously described Cry, Cyt, or Vip insecticidal proteins. However, database searches revealed that Cry35 proteins are homologous, at 26 to 29% sequence identity, respectively, to the 42- and 51-kDa crystalline mosquitocidal proteins from B. sphaericus strain 2362 and share distinct blocks of conserved sequence homology with the B. sphaericus proteins (14). An extension of this search to conserved domain alignments (22, 23) using Cry35Ab1 as a query revealed segments of similarity to two different domains. Residues ∼60 to ∼147 are similar to the beta-trefoil, or QxW, domain typified by the binding subunit of ricin. This common carbohydrate-binding domain was formed by an apparent gene triplication, and the aligning segment of Cry35Ab1 represents two of the three triplicated elements (both the first or second two of the three elements align; Fig. 7A and B). Residues 146 through 348 are similar to a conserved domain P42, derived from the previously noted B. sphaericus 42- and 51-kDa proteins, other Cry35 proteins, and Cry36, a protein with some coleopteran activity that appears to be more related to the B. sphaericus proteins than to Cry35 (8, 30). Because of the close juxtaposition of the beta-trefoil and P42 conserved domain alignments, it seems more likely that a third beta-trefoil repeat would occur in the first 60 residues, despite the low sequence similarity, than in the P42 segment following residue 147.
Upon iterated PSI-BLAST searches of GenBank using Cry34 proteins as queries, some non-Bacillus proteins are also found with significant sequence similarity. One of the proteins is similar in size to the Cry34 proteins, but it is from the gram-negative insect pathogen P. luminescens (12). A second open reading frame in this organism encodes a protein of 465 residues having a segment of sequence similarity to the Cry34 proteins at its predicted N terminus and similarity to certain phosphatases near its predicted C terminus. A more surprising result comes from the eukaryotic organism Dictyostelium discoidium, where an 1,107-residue hypothetical protein encodes essentially complete and partial homologues of Cry34 and also a segment resembling magnesium-dependent endonucleases and phosphatases involved in intracellular signaling. The discovery of a homologue of a B. thuringiensis insect toxin in a gram-negative insect pathogen is somewhat unexpected, given the extensive study of other types of toxins in P. luminescens. However, the discovery of a homologue in D. discoidium and the presence of Cry34-like modules in putative proteins that also contain endonuclease and/or phosphatase modules is also intriguing. Because the only described role for Cry34-like polypeptides is as an insect toxin acting together with an unrelated protein, Cry35, it is interesting that Cry34-like modules are found covalently linked to additional functional domains. Whether the 136-amino-acid P. luminescens putative protein functions as an insect toxin will require experimental determination. However, the possible role of the Cry34-like modules in the longer P. luminescens and D. discoidium putative proteins is less clear. Because the B. thuringiensis binary toxins must interact with, and probably disrupt, membranes, it seems likely that the Cry34-like modules would perform some membrane anchoring or translocation function for the endonuclease and/or phosphatase modules in those proteins, whether they function as toxins, in cellular regulation, or in some other function.
Acknowledgments
We thank Penny Hunst for her support and encouragement of this work as well as Marc Farrow and Rosa Reynolds for their assistance with the insect bioassays.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, and Z. Zhang. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 3.Baum, J. A., C. R. Chu, M. Rupar, G. R. Brown, W. P. Donovan, J. E. Huesing, O. Ilagan, T. M. Malvar, M. Pleau, M. Walters, and T. Vaughn. 2004. Binary toxins from Bacillus thuringiensis active against the western corn rootworm, Diabrotica virgifera virgifera LeConte. Appl. Environ. Microbiol. 70:4889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Dov, E., A. Zaritsky, E. Dahan, Z. Barak, R. Sinai, R. Manasherob, A. Khamraev, E. Troitskaya, A. Dubitsky, N. Berezina, and Y. Margalith. 1997. Extended screening by PCR for seven cry-group genes from field-collected strains of Bacillus thuringiensis. Appl. Environ. Microbiol. 63:4883-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Bravo, A., S. Sarabia, L. Lopez, H. Ontivero, C. Abarca, A. Ortiz, M. Ortiz, L. Lina, F. J. Villalobos, G. Pena, M. E. Nunez-Valdez, M. Soberon, and R. Quintero. 1998. Characterization of cry genes in a Mexican Bacillus thuringiensis strain collection. Appl. Environ. Microbiol. 64:4965-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crickmore, N., D. R. Zeigler, J. Feitelson, E. Schnepf, J. Van Rie, D. Lereclus, J. Baum, and D. H. Dean. 1998. Revision of the nomenclature for the Bacillus thuringiensis pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crickmore, N., D. R. Zeigler, E. Schnepf, J. van Rie, D. Lereclus, J. Baum, A. Bravo, and D. H. Dean. 2003. Bacillus thuringiensis toxin nomenclature. [Online.] http://www.biols.susx.ac.uk/Home/Neil_Crickmore/Bt/index.html. Accessed 13 December 2003.
- 9.de Barjac, H. 1981. Identification of H-serotypes of Bacillus thuringiensis, p. 35-43. In H. D. Burges (ed.), Microbial control of pests and plant diseases. Academic Press, Inc., London, United Kingdom.
- 10.De Maagd, R. A., A. Bravo, C. Berry, N. Crickmore, and H. E. Schnepf. 2003. Structure, diversity, and evolution of protein toxins from spore-forming entomopathogenic bacteria. Annu. Rev. Genet. 37:409-433. [DOI] [PubMed] [Google Scholar]
- 11.Du, C., P. A. W. Martin, and K. W. Nickerson. 1994. Comparison of disulfide contents and solubility at alkaline pH of insecticidal and noninsecticidal Bacillus thuringiensis protein crystals. Appl. Environ. Microbiol. 60:3847-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 13.Ejiofor, A. O., and T. Johnson. 2002. Physiological and molecular detection of crystalliferous Bacillus thuringiensis strains from habitats in the south central United States. J. Ind. Microbiol. Biotechnol. 28:284-290. [DOI] [PubMed] [Google Scholar]
- 14.Ellis, R. T., B. A. Stockhoff, L. Stamp, H. E. Schnepf, G. E. Schwab, M. Knuth, J. Russell, G. A. Cardineau, and K. E. Narva. 2002. Novel Bacillus thuringiensis binary insecticidal crystal proteins active on western corn rootworm, Diabrotica virgifera virgifera LeConte. Appl. Environ. Microbiol. 68:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feitelson, J. S. December1999. Means for discovering microbes. U.S. patent 5,997,269.
- 16.Feitelson, J., J. Payne, and L. Kim. 1992. Bacillus thuringiensis: insects and beyond. Bio/Technology. 10:271-275. [Google Scholar]
- 17.Hazes, B. 1996. The (QxW)3 domain: a flexible lectin scaffold. Protein Sci. 5:1490-1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman, R. A., P. N. Scherer, D. L. Young, C. A. Mihaliak, T. Meade, A. T. Woodsworth, B. A. Stockhoff, and K. E. Narva. 2002. Binary insecticidal crystal protein from Bacillus thuringiensis, strain PS149B1: effects of individual protein components and mixtures in laboratory bioassays. J. Econ. Entomol. 95:635-639. [DOI] [PubMed] [Google Scholar]
- 19.Lecadet, M. M., E. Frachon, V. C. Dumanoir, H. Ripouteau, S. Hamon, P. Laurent, and I. J. Thiery. 1999. Updating the H-antigen classification of Bacillus thuringiensis. Appl. Microbiol. 86:660-672. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. H., N. Shisa, N. Wasano, A. Ohgushi, and M. Ohba. 2003. Characterization of flagellar antigens and insecticidal activities of Bacillus thuringiensis populations in animal feces. Curr. Microbiol. 46:287-290. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchler-Bauer, A., A. R. Panchenko, B. A. Shoemaker, P. A. Thiessen, L. Y. Geer, and S. H. Bryant. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 30:281-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, P. A. W. 1994. An iconoclastic view of Bacillus thuringiensis ecology. Am. Entomol. 40:85-90. [Google Scholar]
- 25.Martin, P. A. W., and R. S. Travers. 1989. Worldwide abundance and distribution of Bacillus thuringiensis isolates. Appl. Environ. Microbiol. 55:2437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moellenbeck, D. J., M. L. Peters, J. W. Bing, J. R. Rouse, L. S. Higgins, L. Sims, T. Nevshemal, L. Marshall, R. T. Ellis, P. G. Bystrak, B. A. Lang, J. L. Stewart, K. Kouba, V. Sondag, V. Gustafson, K. Nour, D. Xu, J. Swenson, J. Zhang, T. Czapla, G. Schwab, S. Jayne, B. A. Stockhoff, K. Narva, H. E. Schnepf, S. J. Stelman, C. Poutre, M. Koziel, and N. Duck. 2001. Insecticidal proteins from Bacillus thuringiensis protect corn from corn rootworms. Nat. Biotechnol. 19:668-672. [DOI] [PubMed] [Google Scholar]
- 27.Narva, K. E., H. E. Schnepf, M. Knuth, M. R. Pollard, G. A. Cardineau, G. E. Schwab, T. E. Michaels, S. F. Lee, P. Burmeister, and J. Dojillo. April2002. Pesticidal proteins. U.S. patent 6,372,480.
- 28.Porcar, M., and V. Juarez-Perez. 2003. PCR-based identification of Bacillus thuringiensis pesticidal crystal genes. FEMS Microbiol. Rev. 26:419-432. [DOI] [PubMed] [Google Scholar]
- 29.Prieto-Samsonov, D. L., R. I. Vazquez-Padron, C. Ayra-Pardo, J. Gonzalez-Cabrera, and G. A. de la Riva. 1997. Bacillus thuringiensis: from biodiversity to biotechnology. J. Ind. Microbiol. Biotechnol. 19:202-219. [DOI] [PubMed] [Google Scholar]
- 30.Rupar, M. J., W. P. Donovan, C.-R. Chu, E. Pease, A. C. Slaney, T. M. Malvar, and J. A. Baum. April2003. Coleopteran-toxic polypeptide compositions and insect-resistant transgenic plants. U.S. patent 6,555,655.
- 31.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stahly, D. P., D. W. Dingman, L. A. Bulla, Jr., and A. I. Aronson. 1978. Possible origin and function of the parasporal crystal in Bacillus thuringiensis. Biochem. Biophys. Res. Commun. 84:581-588. [DOI] [PubMed] [Google Scholar]