Abstract
Background
Clear-cell renal cell carcinoma (ccRCC) is the most common and aggressive form of renal cancer and a paradigm of inter- and intratumor heterogeneity. We carried out an exploratory digital spatial profiling of the tumor interior and periphery of two ccRCC tumor specimens and mapped spatially the molecular and cellular composition of their tumor microenvironment and ecosystem.
Materials and methods
Digital spatial profiling of the whole transcriptome of 19 regions of interest (ROIs) was carried out from two selected highly immunogenic stage pT3a/grade 3 (G3) and stage pT3a/grade 4 (G4) ccRCC. A total of 9-10 ROIs were selected from distinct areas from each tumor, including tumor interior and tumor periphery, and differences in gene expression were analyzed by RNA sequencing, pathway enrichment analysis, and cell deconvolution.
Results
The distinct areas from the two locally advanced tumors displayed unique gene expression spatial patterns defining distinct biological pathways. Dimensional reduction analysis showed that the G3 ccRCC, compared to the G4 ccRCC, correlated with more variability between regions from the tumor interior and tumor periphery. Cell deconvolution analysis illustrated higher abundance of immune cells, including macrophages, myeloid dendritic cells, and CD4 T cells, and lower abundance of regulatory T cells in the tumor periphery compared to the tumor interior.
Conclusions
Transcriptome spatial profiling revealed high inter- and intratumor heterogeneity in the analyzed tumors and provided information with potential clinical utility. This included the finding of less intratumor heterogeneity and more tumor-infiltrated T cells in the ccRCC tumor specimen with a higher grade.
Key words: digital spatial profiling, whole transcriptome, clear-cell renal cell carcinoma
Highlights
-
•
Whole transcriptome analysis reveals distinct spatial landscapes in ccRCC.
-
•
Transcriptome spatial profiling revealed high inter- and intratumor heterogeneity in ccRCC.
-
•
Spatial transcriptomics and cell type deconvolution showed differential distribution of immune cells in ccRCC tumor interior and periphery.
Introduction
Clear-cell renal cell carcinoma (ccRCC) is a common aggressive renal cancer subtype in both male and female patients, with an increasing incidence in the Western world.1 The varied spectrum of ccRCC morphological features, immunohistochemical patterns, and genomic profiles reveals the high level of intertumor heterogeneity occurring in this neoplasm.2 Also, ccRCC is a paradigmatic example of intratumor heterogeneity, where different eco-evolutionary models (linear, branching, and punctuated) have been identified.3 Geographical, histological, and molecular diversification may be high in some cases and this fact makes tumor sampling a crucial issue in unveiling tumor complexity and possible targetable clones.4
Classical cytological (clear, eosinophilic, syncytial, and rhabdoid cells) and histological (sarcomatoid phenotype and tumor necrosis) features characterize high-grade [grade 4 (G4)] tumors correlating with aggressive behavior and shorter survivals.2 Besides, recent studies have shown a correlation between environmental features and specific mutations with prognostic implications making the microscopic analysis a reliable surrogate of genomic malfunctions with therapeutic impact.3
ccRCC shows a high degree of molecular and cell population intratumor heterogeneity. Systematic studies of ccRCC evolutionary features have revealed that ccRCC can be grouped into at least seven evolutionary subtypes that correlate with clinical outcomes.5, 6, 7 Furthermore, specific patterns of proliferation and necrosis could explain clonal expansion and emergence of parallel evolution and micro-diversity in tumors.8 Different clinical outcomes of ccRCC patients are observed in association with specific gene expression and cell subsets’ intratumor heterogeneity, including populations of tumor-infiltrating lymphocytes.9, 10, 11 An inverse correlation between intratumor heterogeneity, both at histological and molecular levels, and tumor aggressiveness has been recently proposed in ccRCC, with the tumor interior displaying a more aggressive and metastasizing subclonal growth phenotype.4,12 In addition, an inverse correlation between ccRCC intratumor heterogeneity and therapy response has also been reported.13 Thus, the clinical significance of ccRCC intratumor heterogeneity is starting to be understood, and diagnostic tools that consider ccRCC intratumor heterogeneity are expected to be implemented in the near future.
Current state-of-the-art methodologies to study intratumor heterogeneity have incorporated the precise spatial analysis at molecular level of individual tumor specimens,14,15 which is of special relevance in solid tumors such as locally advanced renal cancer. ccRCC is a paradigm of a highly immunogenic and heterogeneous cancer. We have analyzed the molecular and cellular composition of the interior and the periphery from two ccRCC stage pT3a specimens, one G4 and one G3. Our results illustrate correlations between spatial intratumor heterogeneity and tumor malignancy, and provide insights into the diagnostic potential of spatial digital analysis from tumor biopsies in this malignancy.
Materials and methods
Tumor samples
Formalin-fixed paraffin-embedded (FFPE) sections were analyzed in parallel by hematoxylin-eosin (H/E). Whole tissue sections were selected for the analysis by an experienced uropathologist (JIL), who identified tumor areas with well-preserved tissue representative of the whole tissue sections from FFPE tissue blocks. H/E staining was carried out on consecutive tissue sections to verify the presence of tumor content. Ethical approval was obtained (CEIC number PI2022085). Patients had a total nephrectomy from which the pathologist selected the samples taking into account their exact position within the tumor. Pathologist sampling includes topographic information about where the samples come from: center means interior, >1 cm away from the tumor capsule, and periphery means <1 cm from the capsule.
Preparation for digital spatial profiling
Briefly, 5 μm FFPE sections from two patient tissues were profiled using the NanoString (Seattle, Washington, USA) digital spatial profiler, GeoMx®, for whole transcriptome analysis (WTA with 18 676 total targets). Slides were baked at 60°C for 1-2 h, deparaffinized, rehydrated and subjected to antigen retrieval in 1x Tris-EDTA (pH 9) at 100°C for 15 min, digested in 1 μg/ml proteinase K (ThermoFisher Scientific (Waltham, Massachusetts, USA), AM2548) for 15 min, postfixed in 10% neutral-buffered formalin for 10 min at room temperature, and hybridized to UV-cleavable barcode-conjugated RNA in situ hybridization probe set in Buffer R (NanoString) overnight at 37°C. After the overnight hybridization, the slides were washed to remove off-target and excess probes and counterstained with morphology markers (1 : 25 SYTO13 for DNA in 488 blue, ThermoFisher Scientific #57575; 1 : 40 PanCK in 532 green, NanoString commercially available lot #042608302; 1 : 400 CD68 in 594 yellow, Santa Cruz Biotechnology (Dallas, Texas, USA) mouse monoclonal antibody sc-20060, clone KP1, lot #B1422; and 1 : 300 CD3 in 647 red, OriGene (Rockville, Maryland, USA) mouse monoclonal antibody UM500048CF, clone UMAB54, lot #F005) in blocking buffer W (NanoString) for 2 h at room temperature.
The stained slides were scanned using the GeoMx® instrument to produce a digital fluorescent image of the tissues. Next, individual geometric regions of interest (ROIs) covering abundant CD68+ areas of inflammation were identified within distinct areas from each tumor, including the tumor periphery and interior. Representative images of ROIs with morphology markers of tumor interior and tumor periphery are included in Supplementary Figure S1, available at https://doi.org/10.1016/j.iotech.2023.100690.
Sequencing libraries and PCR reactions were carried out according to the manufacturer’s instructions. PCR reactions were pooled and purified twice using AMPure XP beads (A63881, Beckman Coulter, Brea, California, USA). Each ROI was uniquely indexed and then pooled for sequencing on the NovaSeq 2000 platform (Illumina, San Diego, California, USA). No ROI failed standard quality control (QC) methods, which exclude areas with <50% sequencing saturation and those with genes above the limit of quantitation (LOQ) in 5% of ROIs (Supplementary Figure S2, available at https://doi.org/10.1016/j.iotech.2023.100690). All ROIs were then moved on through Q3 normalization by a standard NanoString pipeline. Q3 (third quartile of all selected targets) normalization was used for all targets that are above the LOQ. Q3 normalization uses the top 25% of expressed genes to normalize across ROIs/segments (Supplementary Figure S3, available at https://doi.org/10.1016/j.iotech.2023.100690).
The count matrices after QC were analyzed in R software (version 4.0.1) for further differential expression analysis and data visualization.
Statistics
P values of the NanoString data were calculated by false discovery rate-corrected empirical Bayes moderated T statistics using R package Limma, hypergeometric distribution using R package ClusterProfiler, or permutation test using R package ClusterProfiler. Cell deconvolution analysis was carried out using the SpatialDecon v1.0.0 package (https://github.com/Nanostring-Biostats/SpatialDecon/). P < 0.05 was considered as significant.
Results
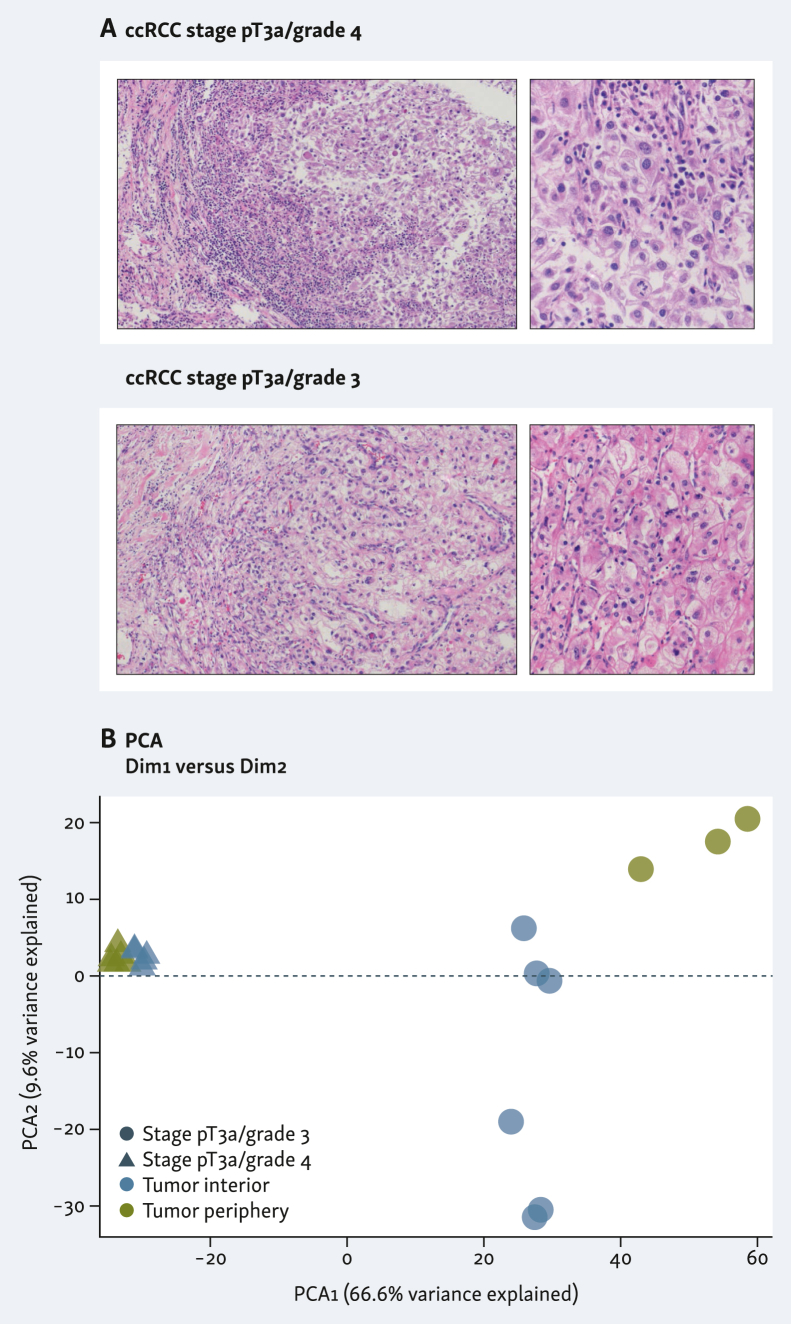
FFPE tissue specimens from two patients with locally advanced ccRCC treated with radical nephrectomy alone were evaluated. Both were large-size stage pT3a lesions with perinephric fat invasion. Hematoxylin–eosin staining showed that both tumors were high grade, G3 and G4 [World Health Organization/International Society of Urological Pathology (WHO/ISUP) grading system], respectively. G3 ccRCC displayed enlarged nuclei with prominent nucleoli and large eosinophilic cytoplasm. Tumor cells were arranged in solid nests with heavy inflammatory infiltrates. G4 ccRCC showed enlarged and odd-shaped nuclei with prominent, sometimes multiple, nucleoli and large clear cytoplasm. Occasionally, syncytial-type cells were identified. Tumor cells were arranged in large solid lobes with prominent inflammatory infiltrates (Figure 1A). Both patients were followed up for 2 years without tumor recurrence or progression. Spatial digital profile analysis across 19 ROIs by whole transcriptome sequencing analysis was carried out in each tumor specimen.
Figure 1.
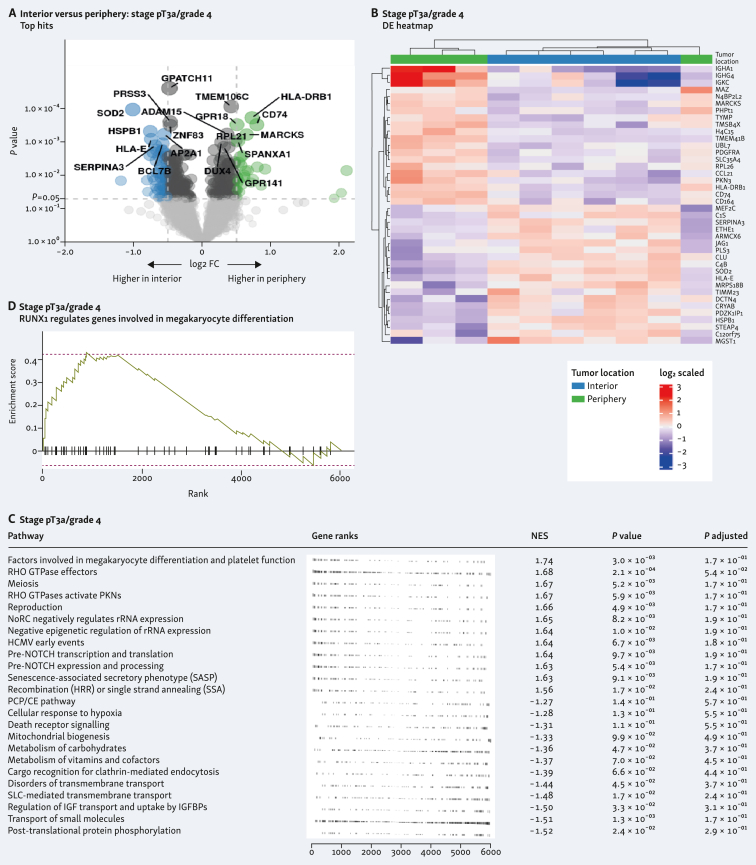
Histological samples used in this study, and sample similarity. (A) Hematoxylin–eosin staining of clear-cell renal cell carcinoma (ccRCC) tumor tissue sections from two patients. Left images ×100 magnification, and right panels ×400 magnification. (B) Sample similarity by principal component analysis (PCA) displaying tumor interior and tumor periphery in blue and green, respectively, and the two sample types in circles (stage pT3a/grade 3) or triangles (stage pT3a/grade 4).
Principal component analysis of all selected regions showed a more dispersed distribution of similarity of the G3 tumor interior versus periphery when compared with G4 tumor interior versus periphery, indicating higher intratumor heterogeneity in the G3 specimen (Figure 1B).
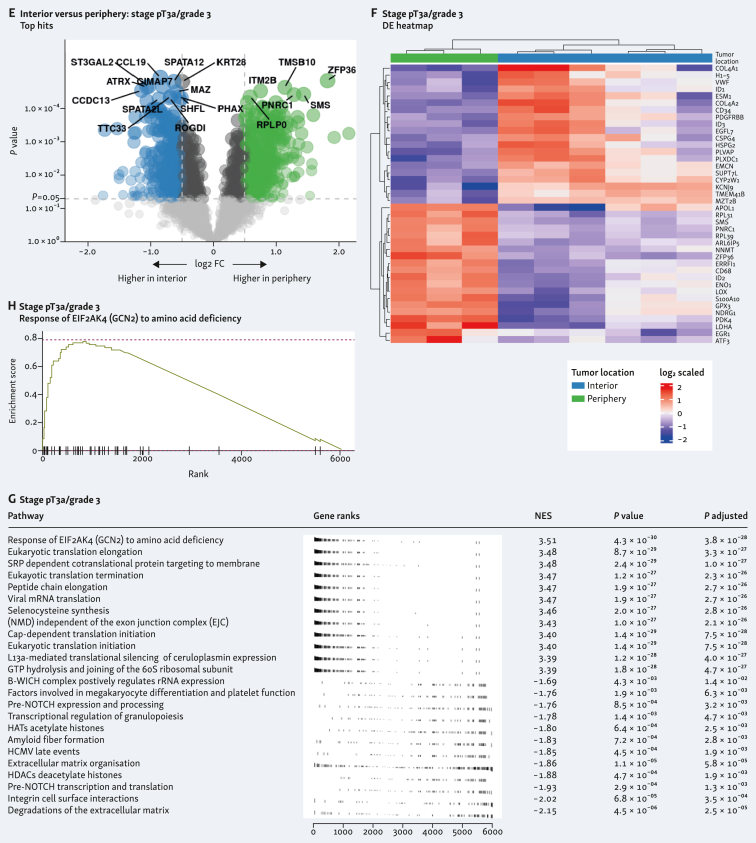
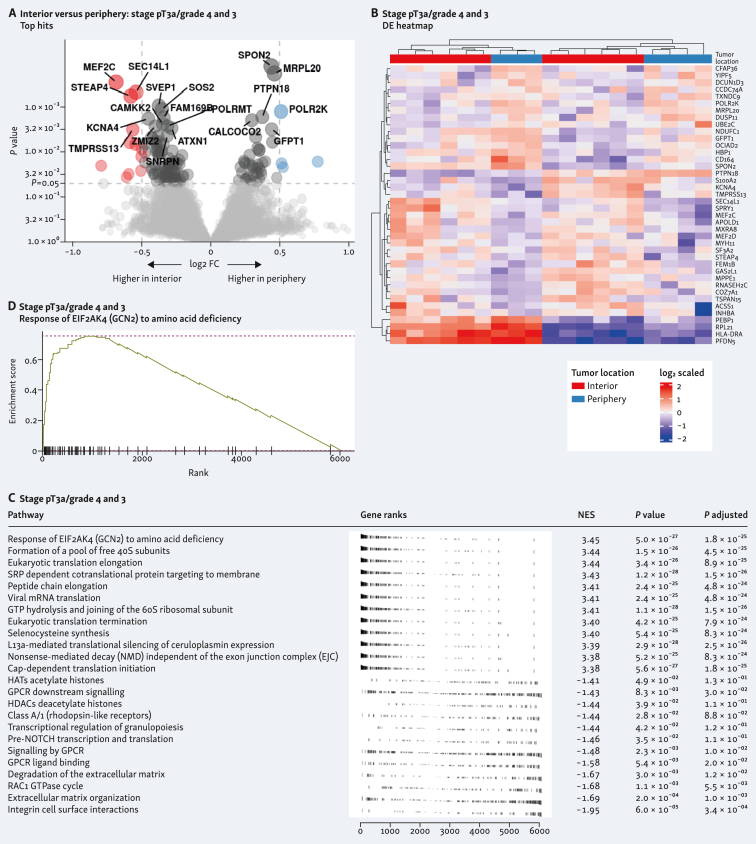
Differentially expressed genes in the tumor interior and periphery in both samples is shown in Figure 2A and E. Individual heatmap of the differentially expressed genes from each tumor interior and periphery showed clustering according to the different spatial location (Figure 2B and F). Pathway analysis showed different routes involved in both tumors (Figure 2C and G). In the G4 tumor, this included the most significant run-related transcription factor 1 (RUNX1) pathway, which regulates genes involved in megakaryocyte differentiation (Figure 2D), and in the G3 tumor, this included response of EIF2AK4 (GCN2) to amino acid deficiency pathway (Figure 2H). A volcano plot of the differentially expressed genes from the combined analysis of both tumor samples is shown in Figure 3A. Heatmap of the combined analysis of the differentially expressed genes from both tumors clustered the ROIs according to both tumor sample and spatial distribution (Figure 3B). Pathway enrichment analysis from the combined specimens revealed differentially expressed genes involved in the response of EIF2AK4 to amino acid deficiency, as observed with the individual G3 tumor (Figure 3C and D).
Figure 2.
Differentially expressed genes and pathway analysis within clear-cell renal cell carcinoma (ccRCC) tumor types comparing tumor interior and tumor periphery. Differential expression of genes displayed in volcano plots of interior versus periphery is shown in A (stage pT3a/grade 4) and E (stage pT3a/grade 3). Heatmaps corresponding with interior versus periphery are shown in B (stage pT3a/grade 4) and F (stage pT3a/grade 3). Gene set enrichment analysis (GSEA) for each tumor type is shown in C (stage pT3a/grade 4) and G (stage pT3a/grade 3) comparing tumor interior versus periphery and top differentially expressed pathway shown in D (stage pT3a/grade 4) and H (stage pT3a/grade 3). CE, convergent extension; FC, fold change; HATs, histone acetyltrasnsferases; HCMV, human cytomegalovirus; HDACs, histone deacetylaces; mRNA, messenger RNA; NoRC, nucleolar remodeling complex; PCP, planar cell polarity; PKN, protein kinase N; rRNA, ribosomal RNA; SLC, solute carrier.
Figure 3.
Differentially expressed genes and pathway analysis in combined tumor type analysis comparing tumor interior and tumor periphery. (A) Differential gene expression shown in volcano plots of interior versus periphery in stage pT3a/grade 4 and stage pT3a/grade 3. (B) Heatmap of differential expression analysis as in A. (C) Gene set enrichment analysis (GSEA) in stage pT3a/grade 4 and stage pT3a/grade 3, comparing tumor interior versus periphery. (D) Top differentially expressed pathway is shown. GPCR, G protein-coupled receptor; HATs, histone acetyltransferases; HDACs, histone deacetylases; mRNA, messenger RNA; NMD, nonsense-mediated decay.
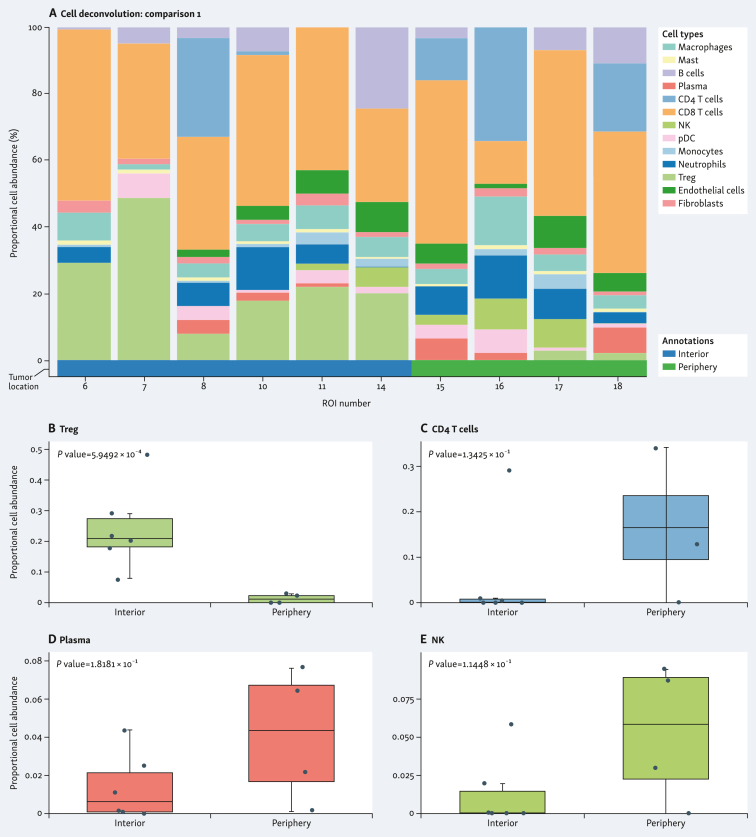
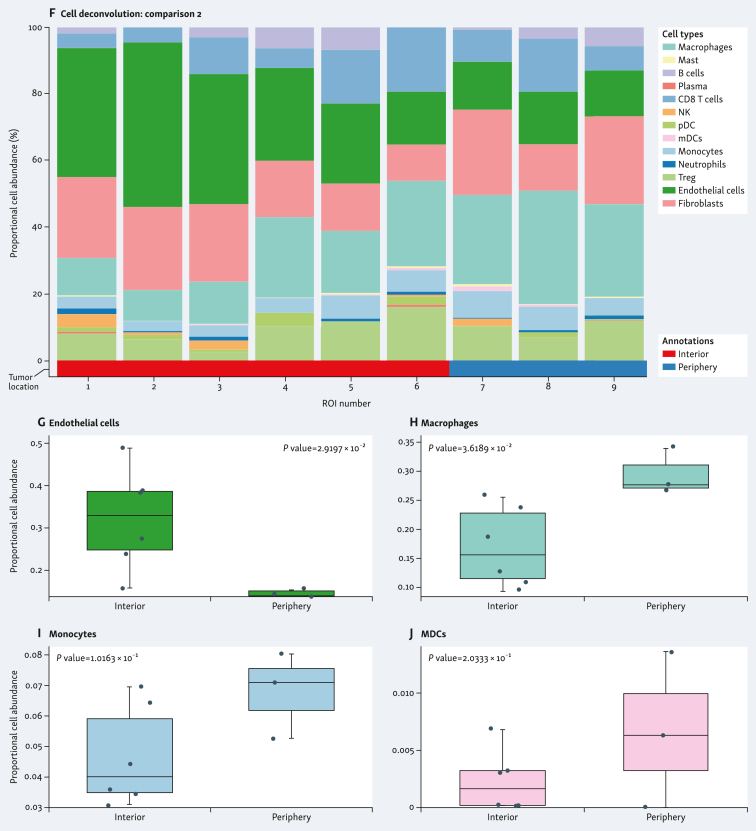
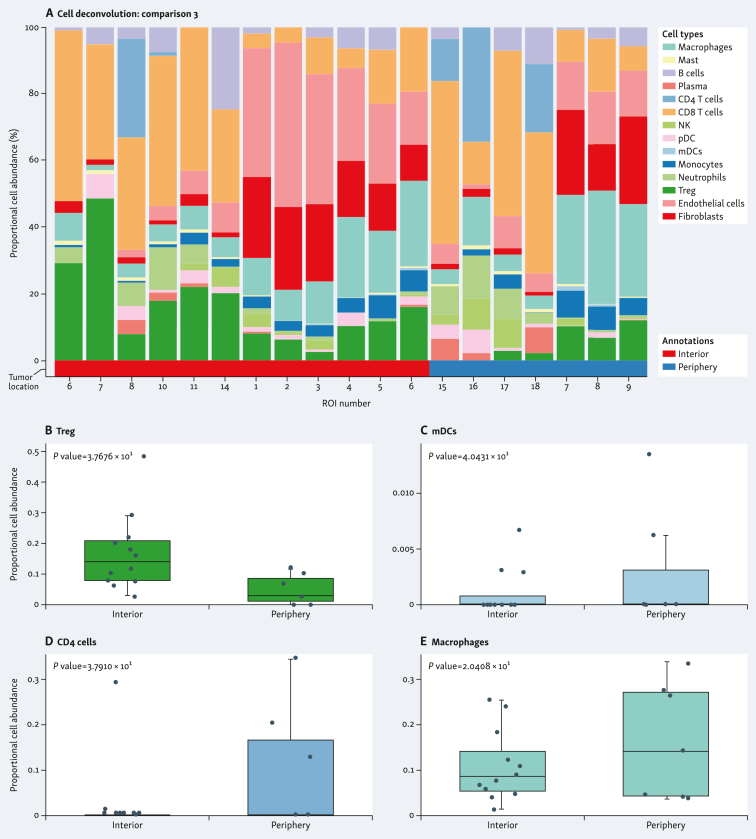
The spatial distribution of cell subpopulations showed differential patterns of cell type abundance. This included immune cell abundance, which was observed in both samples. When comparing the individual tumors, the G4 tumor was enriched in CD8+ T cells, whereas the G3 tumor was enriched in endothelial cells and fibroblasts (Figure 4A and F). G4 tumor showed higher abundance of regulatory T cells (Treg) (Figure 4B), and lower abundance of CD4+ T cells (Figure 4C), plasma cells (Figure 4D), and natural killer (NK) cells (Figure 4E) in the tumor interior versus the tumor periphery. G3 tumor showed higher abundance of endothelial cells (Figure 4G) and lower abundance of macrophages (Figure 4H), monocytes (Figure 4I), and myeloid dendritic cells (MDCs) (Figure 4J) in the tumor interior versus the tumor periphery. Cell deconvolution analysis when combining the two tumors is shown in Figure 5A. The tumor interior showed higher abundance of Treg than the tumor periphery (Figure 5B). On the contrary, the tumor periphery showed higher abundance of MDCs (Figure 5C), CD4+ T cells (Figure 5D), and macrophages (Figure 5E).
Figure 4.
Spatial cell type deconvolution in sample specimens. Cell deconvolution within tumor types (A-E, stage pT3a/grade 4; F-J, stage pT3a/grade 3) comparing tumor interior [blue in stage pT3a/grade 4 (A); red in stage pT3a/grade 3 (F)] and tumor periphery [green in stage pT3a/grade 4 (A); blue in stage pT3a/grade 3 (F)]. Proportional cell abundance is shown in each graph. (B-E and G-J) Most differentially expressed cell types in each tumor type are shown. MDCs, myeloid dendritic cells; NK, natural killer; ROI, region of interest; Treg, regulatory T cells.
Figure 5.
Spatial cell type deconvolution in combined sample specimens. Cell deconvolution within tumor types [A-E, stage pT3a/grade 4 and stage pT3a/grade 3 comparing tumor interior (red) and tumor periphery (blue)]. Proportional cell abundance is shown in each graph. (B-E) Most differentially expressed cell types in each spatial location are shown.mDCs, myeloid dendritic cells; NK, natural killer; pDC, plasmacytoid dendritic cell; ROI, region of interest; Treg, regulatory T cells.
Discussion
Non-metastatic locally advanced renal cancer is a neoplasm in which the prediction of prognosis is complex. The rationale to optimally guide the selection of treatments and accurately predict the oncological outcome is based on routine parameters, including pathological staging and grade evaluation. The most widely used grading system for ccRCC has been the nuclear grading system described by Fuhrman et al. and by WHO/ISUP that concurrently evaluates nuclear size and shape and nucleolar prominence.16,17 However, discordance and grading imprecision may occur among these three parameters, and the predictive performance of prospectively applied Fuhrman/WHO/ISUP grade in non-metastatic ccRCC is currently under revision.18
Additionally, studies reporting inconsistent oncological outcomes have demonstrated the heterogeneity of the definition and raised a controversy regarding whether the classification of stage pT3a ccRCC is appropriate.19 Validated models available in clinical practice and machine learning approaches are being evaluated to predict disease recurrence and metastatic progression following surgical resection of ccRCC, but there is still a need for improvement from the integration of novel biological biomarkers.20,21 In this sense, a comprehensive analysis of tumor immune infiltration and the tumor microenvironment can facilitate development of biomarkers in ccRCC.22,23
Single-cell transcriptomes have been used to evaluate intratumor heterogeneity and microenvironment, thus revealing gene sets and specific macrophage and T-cell clusters with potential to be used as prognostic markers in ccRCC.24, 25, 26 Not only cancer cells, but also tumor-infiltrating cells, participate in the process of drug resistance and response to tyrosine-kinase inhibitors and anti-programmed cell death protein 1 (PD-1) antibody treatment.9,27 There is evidence to consider that low intratumor heterogeneity is associated with better response and clinical benefit of anti-PD-1 immunotherapy.7,13,28
Our findings using spatial whole transcriptome profiling of the intratumoral and peritumoral tissue of ccRCC further support the notion expressed recently in a histological-based study4 that low intratumor heterogeneity is inversely correlated with aggressiveness. The significance of immune topographies evaluating intratumoral and peritumoral regions separately in ccRCC remains unclear. However, distinct immuno-spatial profiles in tumor center and periphery are likely associated with clinicopathological differences.29 Whole transcriptome profiling using digital spatial profiling in user-defined ROIs is a promising tool to better evaluate intratumor heterogeneity in renal malignancies and its clinical meaning.
With our spatial transcriptome approach, we identified the RUNX1 pathway as one of the pathways most affected in the tumor interior compared to the periphery of G4 renal cancer sample. RUNX is a transcription factor family of embryonic development master regulators composed of three mammalian transcription factors. Known as RUNX genes, RUNX1, RUNX2, and RUNX3, they play a pivotal role in many cellular growth and development processes.30 In renal cancer, RUNX protein expression has been found to correlate with clinical outcome and metastasis,31, 32, 33 and RUNX1 is considered a driver of renal cancer with potential as a predictive marker.31,34,35 RUNX1 is predominantly expressed in hematopoietic cells,36 which parallels our cell deconvolution analysis with an increase in CD4+ T and NK cells in the tumor periphery.
Despite promising new findings, our study has some limitations. Firstly, we have included in the study a small number of patients, despite extensive multisampling used. Due to the small number of patients, the results cannot be generalized but are hypothesis generating. Secondly, we could not analyze the prognostic implication of our findings as we analyzed primary tumor samples in patients without metastatic progression. The spatial intratumor heterogeneity pattern revealed will require further validation in other cohorts of ccRCC, especially to evaluate the clinicopathological implications suggested.
Acknowledgments
Funding
This study has been supported by Instituto de Salud Carlos III through the projects PI22/00386 and Miguel Servet Research Grant [grant number CP20/00008] (co-funded by European Union); and Stiftelsen til Fremme av Forskning Innen Nyresykdommer/The Norwegian Foundation for the Promotion of Research in Kidney Diseases. We thank the European Association for Cancer Research (EACR), and NanoString in collaboration with Illumina co-sponsor for the European Whole Transcriptome Atlas grant.
Disclosure
The authors have declared no conflicts of interest.
Supplementary data
References
- 1.Padala S.A., Barsouk A., Thandra K.C., et al. Epidemiology of renal cell carcinoma. World J Oncol. 2020;11:79–87. doi: 10.14740/wjon1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trpkov K., Hes O., Williamson S.R., et al. New developments in existing WHO entities and evolving molecular concepts: the Genitourinary Pathology Society (GUPS) update on renal neoplasia. Mod Pathol. 2021;34:1392–1424. doi: 10.1038/s41379-021-00779-w. [DOI] [PubMed] [Google Scholar]
- 3.Brugarolas J., Rajaram S., Christie A., Kapur P. The evolution of angiogenic and inflamed tumors: the renal cancer paradigm. Cancer Cell. 2020;38:771–773. doi: 10.1016/j.ccell.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manini C., Lopez-Fernandez E., Lawrie C.H., Laruelle A., Angulo J.C., Lopez J.I. Clear cell renal cell carcinomas with aggressive behavior display low intratumor heterogeneity at the histological level. Curr Urol Rep. 2022;23:93–97. doi: 10.1007/s11934-022-01095-9. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell T.J., Turajlic S., Rowan A., et al. Timing the landmark events in the evolution of clear cell renal cell cancer: TRACERx Renal. Cell. 2018;173:611–623.e17. doi: 10.1016/j.cell.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turajlic S., Xu H., Litchfield K., et al. Tracking cancer evolution reveals constrained routes to metastases: TRACERx Renal. Cell. 2018;173:581–594.e12. doi: 10.1016/j.cell.2018.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turajlic S., Xu H., Litchfield K., et al. Deterministic evolutionary trajectories influence primary tumor growth: TRACERx Renal. Cell. 2018;173:595–610.e11. doi: 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu X., Zhao Y., Lopez J.I., et al. Spatial patterns of tumour growth impact clonal diversification in a computational model and the TRACERx Renal study. Nat Ecol Evol. 2022;6:88–102. doi: 10.1038/s41559-021-01586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J., Chen Z., Bao L., et al. Single-cell transcriptome analysis reveals intratumoral heterogeneity in ccRCC, which results in different clinical outcomes. Mol Ther. 2020;28:1658–1672. doi: 10.1016/j.ymthe.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senbabaoglu Y., Gejman R.S., Winer A.G., et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. doi: 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghatalia P., Gordetsky J., Kuo F., et al. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J Immunother Cancer. 2019;7:139. doi: 10.1186/s40425-019-0621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao Y., Fu X., Lopez J.I., et al. Selection of metastasis competent subclones in the tumour interior. Nat Ecol Evol. 2021;5:1033–1045. doi: 10.1038/s41559-021-01456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran X., Xiao J., Zhang Y., et al. Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma. Ther Adv Med Oncol. 2020;12 doi: 10.1177/1758835920977117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhoundova D., Rubin M.A. Clinical application of advanced multi-omics tumor profiling: shaping precision oncology of the future. Cancer Cell. 2022;40:920–938. doi: 10.1016/j.ccell.2022.08.011. [DOI] [PubMed] [Google Scholar]
- 15.Li Q., Zhang X., Ke R. Spatial transcriptomics for tumor heterogeneity analysis. Front Genet. 2022;13 doi: 10.3389/fgene.2022.906158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuhrman S.A., Lasky L.C., Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 17.Delahunt B., McKenney J.K., Lohse C.M., et al. A novel grading system for clear cell renal cell carcinoma incorporating tumor necrosis. Am J Surg Pathol. 2013;37:311–322. doi: 10.1097/PAS.0b013e318270f71c. [DOI] [PubMed] [Google Scholar]
- 18.Galtung K.F., Lauritzen P.M., Baco E., Berg R.E., Naas A.M., Rud E. Predictive performance of prospectively applied ISUP and Fuhrman grade in nonmetastatic renal cell carcinoma. Anticancer Res. 2022;42:2967–2975. doi: 10.21873/anticanres.15780. [DOI] [PubMed] [Google Scholar]
- 19.Guo P., Wang Y., Han Y., et al. Development and validation of a nomogram to predict postoperative cancer-specific survival of patients with nonmetastatic T3a renal cell carcinoma. Urol Oncol. 2021;39:835.e19–835.e27. doi: 10.1016/j.urolonc.2021.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Khene Z.E., Bigot P., Doumerc N., et al. Application of machine learning models to predict recurrence after surgical resection of nonmetastatic renal cell carcinoma. Eur Urol Oncol. 2023;6:323–330. doi: 10.1016/j.euo.2022.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H., Yang S., Xie T., et al. Risk factors, prognostic factors, and nomograms for bone metastasis in patients with newly diagnosed clear cell renal cell carcinoma: a large population-based study. Front Surg. 2022;9 doi: 10.3389/fsurg.2022.877653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu K., Wu Y., Chi H., et al. SLC22A8: an indicator for tumor immune microenvironment and prognosis of ccRCC from a comprehensive analysis of bioinformatics. Medicine (Baltimore) 2022;101 doi: 10.1097/MD.0000000000030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Braun D.A., Street K., Burke K.P., et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell. 2021;39:632–648.e8. doi: 10.1016/j.ccell.2021.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu K., Gao R., Wu H., Wang Z., Han G. Single-cell analysis reveals metastatic cell heterogeneity in clear cell renal cell carcinoma. J Cell Mol Med. 2021;25:4260–4274. doi: 10.1111/jcmm.16479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M., Zhai W., Miao J., et al. Single cell analysis reveals intra-tumour heterogeneity, microenvironment and potential diagnosis markers for clear cell renal cell carcinoma. Clin Transl Med. 2022;12:e713. doi: 10.1002/ctm2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jikuya R., Murakami K., Nishiyama A., et al. Single-cell transcriptomes underscore genetically distinct tumor characteristics and microenvironment for hereditary kidney cancers. iScience. 2022;25 doi: 10.1016/j.isci.2022.104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bi K., He M.X., Bakouny Z., et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell. 2021;39:649–661.e5. doi: 10.1016/j.ccell.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris L.G., Riaz N., Desrichard A., et al. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7:10051–10063. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruck O., Lee M.H., Turkki R., et al. Spatial immunoprofiling of the intratumoral and peritumoral tissue of renal cell carcinoma patients. Mod Pathol. 2021;34:2229–2241. doi: 10.1038/s41379-021-00864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mevel R., Draper J.E., Lie A.L.M., Kouskoff V., Lacaud G. RUNX transcription factors: orchestrators of development. Development. 2019;146:dev148296. doi: 10.1242/dev.148296. [DOI] [PubMed] [Google Scholar]
- 31.Rooney N., Mason S.M., McDonald L., et al. RUNX1 is a driver of renal cell carcinoma correlating with clinical outcome. Cancer Res. 2020;80:2325–2339. doi: 10.1158/0008-5472.CAN-19-3870. [DOI] [PubMed] [Google Scholar]
- 32.Gao K., Zhang F., Chen K., et al. Expression patterns and prognostic value of RUNX genes in kidney cancer. Sci Rep. 2021;11 doi: 10.1038/s41598-021-94294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen F., Liu X., Cheng Q., Zhu S., Bai J., Zheng J. RUNX3 regulates renal cell carcinoma metastasis via targeting miR-6780a-5p/E-cadherin/EMT signaling axis. Oncotarget. 2017;8:101042–101056. doi: 10.18632/oncotarget.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Y., Sun S., Man X., Kong C. Increased expression of RUNX1 in clear cell renal cell carcinoma predicts poor prognosis. PeerJ. 2019;7 doi: 10.7717/peerj.7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Z., Yu H., Ding Y., et al. RNA sequencing reveals upregulation of RUNX1-RUNX1T1 gene signatures in clear cell renal cell carcinoma. Biomed Res Int. 2014;2014 doi: 10.1155/2014/450621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yzaguirre A.D., de Bruijn M.F., Speck N.A. The role of Runx1 in embryonic blood cell formation. Adv Exp Med Biol. 2017;962:47–64. doi: 10.1007/978-981-10-3233-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.