Abstract
Agrobacterium tumefaciens was used to transform Aspergillus fumigatus by either random or site-directed integration of transforming DNA (T-DNA). Random mutagenesis via Agrobacterium tumefaciens-mediated transformation (ATMT) was accomplished with T-DNA containing a hygromycin resistance cassette. Cocultivation of A. fumigatus conidia and Agrobacterium (1:10 ratio) for 48 h at 24°C resulted in high frequencies of transformation (>100 transformants/107 conidia). The majority of transformants harbored a randomly integrated single copy of T-DNA and were mitotically stable. We chose alb1, a polyketide synthase gene, as the target gene for homologous integration because of the clear phenotype difference between the white colonies of Δalb1 mutant strains and the bluish-green colonies of wild-type strains. ATMT with a T-DNA-containing alb1 disruption construct resulted in 66% albino transformants. Southern analysis revealed that 19 of the 20 randomly chosen albino transformants (95%) were disrupted by homologous recombination. These results suggest that ATMT is an efficient tool for transformation, random insertional mutagenesis, and gene disruption in A. fumigatus.
Aspergillus fumigatus, an airborne fungal pathogen, is the major cause of allergic bronchopulmonary aspergillosis, aspergilloma, and invasive aspergillosis. Of particular importance is the invasive aspergillosis that starts with inhalation of conidia and progresses to life-threatening infection in immunocompromised patients (11). Due to the rising prevalence of cancer, organ transplantation, and other causes of immunosuppression, the number of patients at risk of invasive aspergillosis is on the rise. Despite aggressive antifungal therapy, the overall mortality of invasive aspergillosis remains high, and new strategies to prevent and treat this disease are urgently needed (7). Several factors such as polyketide synthase, which is involved in melanin synthesis (12, 17), and a Ras-related protein, RhbA, presumably involved in nutrient sensing (15), have been reported to be associated with A. fumigatus virulence. Further identification of the genes necessary for virulence will enable us to address the key pathobiological questions for this fungus and provide a foundation for better strategies to manage aspergillosis.
As genomic sequencing of A. fumigatus is in its final stages, identification of potential virulence genes can be more rapidly assessed. Investigation of the functions of such genes can be accomplished via mutational analysis either by reverse or forward genetic approaches. Transformation of fungi with DNA that does not possess homology with the fungal genome results in random integration into the fungal genome and can cause gene disruption as an insertional mutagen. A decisive advantage of insertional mutagenesis over chemical or radiation mutagenesis is that the mutated genes are tagged by the transforming DNA (T-DNA), which can be used to identify the disrupted genes. Although recombinational analysis by classical genetics is not possible in A. fumigatus, the corresponding wild-type genes can be disrupted by homologous integration of a knockout construct to confirm its function.
To disrupt a gene by homologous integration in A. fumigatus, transformation via the spheroplast method has been most widely used; this is a laborious and time-consuming method, and the homologous recombination frequency can be relatively low (3). Electroporation and biolistic methods were also used to transform A. fumigatus; these methods, however, resulted in a high frequency of multiple integrations and a low frequency of homologous recombination (3). As an alternative method, Agrobacterium tumefaciens-mediated transformation (ATMT) has been used to transform several Aspergillus species such as Aspergillus niger, Aspergillus awamori, and Aspergillus giganteus (5, 10, 13). de Groot and colleagues used A. tumefaciens to deliver T-DNA containing a hygromycin resistance gene into conidia as well as spheroplasts of A. awamori (5). The transformation frequency via ATMT was significantly higher in comparison to frequency with the traditional method, regardless of whether conidia or spheroplasts were used for transformation. Most of the transformants obtained by ATMT contained a randomly integrated single T-DNA copy. Homologous integration via ATMT was also accomplished in A. awamori. The frequency of homologous recombination with T-DNA containing the pyrG gene of A. awamori was higher compared to results with conventional transformation methods (10). A comparison of ATMT with biolistic, electroporation, and spheroplast methods showed that ATMT is superior to the other methods for the transformation of A. giganteus. The transformation frequency was enhanced, and all the transformants contained a randomly integrated single copy of T-DNA (13). In contrast, ATMT was described as being less efficient than conventional transformation methods for A. niger (5). Here we report transformation of A. fumigatus mediated by Agrobacterium tumefaciens as a tool for insertional mutagenesis as well as for gene disruption.
MATERIALS AND METHODS
Strains and media.
A clinical strain of A. fumigatus, B-5233, and transformants obtained by ATMT were maintained on Aspergillus minimal medium (17). Agrobacterium tumefaciens strain EHA105 (a gift from Seogchan Kang, Pennsylvania State University, University Park, Pa.) was grown either on Luria-Bertani broth supplemented with 50 μg of kanamycin per ml or induction medium (4) supplemented with 0.2 mM acetosyringone (IMAS). Transformants were selected on Aspergillus minimal medium (17) supplemented with 200 μg of hygromycin per ml and 200 μg of cefotaxime (SM) per ml.
Plasmid construction.
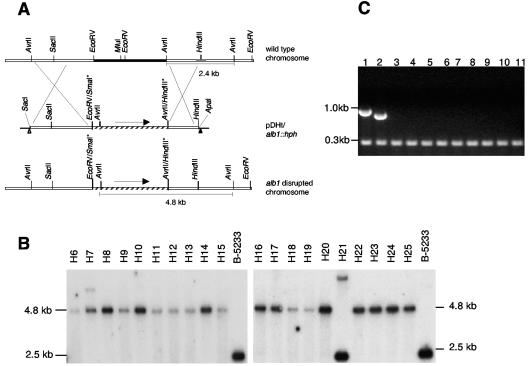
The plasmid used for ectopic mutagenesis, pDHt/hph, was constructed by insertion of a 2.7-kb HindIII/SacI fragment from pAN7 containing the hygromycin resistance gene (hph) (16) into the HindIII/SacI-restricted pDHt/SK plasmid. pDHt/SK is a gift from Seogchan Kang (Pennsylvania State University) and is derived from the plasmid pDHt (14) by inserting a 0.8-kb HpaI/StuI fragment from pGreenII between the two PvuII sites in pDHt. Gene disruption by homologous recombination was performed with the plasmid pDHt/alb1::hph. This plasmid was constructed by inserting a 7.2-kb SacI/ApaI fragment from pRGD12 (17), which contained the hph gene flanked by alb1 sequence, into the SacI/ApaI-restricted plasmid pDHt/SK (Fig. 2A). Agrobacterium tumefaciens strain EHA105 was transformed by electroporation (6) with pDHt/hph or pDHt/alb1::hph plasmids. Strains of Agrobacterium harboring pDHt/hph and pDHt/alb1::hph were named EHA105A and EHA105B, respectively.
FIG. 2.
The alb1 gene disruption via ATMT. (A) Schematic representation of the alb1 native gene, the disruption construct, and the disrupted locus. The deleted fragment of alb1 (filled black line), the fragment used as the probe for Southern analysis (gray line), and the hygromycin resistance cassette (hatched line) are indicated. An asterisk indicates the destroyed restriction sites. Triangles represent left- and right-border repeats in the T-DNA. (B) Southern hybridization of 20 randomly chosen albino transformants. AvrII-restricted DNA was probed with the alb1 wild-type gene. (C) RT-PCR of alb1 transcript in albino mutants. Lane 1, B-5233 genomic DNA; lane 2, RNA derived from B-5233; lanes 3 to 11, RNA derived from transformants H6 through H14. The 232-bp fragment is the PCR product of β-tubulin.
ATMT transformation.
Strain EHA105A or EHA105B was grown on Luria-Bertani broth supplemented with 50 μg of kanamycin per ml on a rotatory shaker (200 rpm) at 28°C for 12 h. One milliliter of the Agrobacterium culture was inoculated into 9.0 ml of IMAS and incubated on a shaker (200 rpm) at 28°C for 6 h or until an optical density at 660 nm of 0.8 was reached. Conidia of B-5233 and Agrobacterium were cocultivated at a ratio of 1:10 (conidia to bacteria), unless otherwise specified. A total of 100 μl of the Agrobacterium culture (108 CFU) was mixed with 100 μl of B-5233 conidia (107 conidia) and spread onto a filter placed on an IMAS agar plate. The filters used were nitrocellulose (Protran; Schleicher and Schuell, Keene, N.H.), cellulose (Whatman no. 1; Maidstone, United Kingdom) and nylon (Hybond-N; Amersham Biosciences, Little Chalfont, United Kingdom). Plates containing the filters were incubated at 24°C in the dark for 16, 26, 40, or 48 h. Initial incubations for 16 and 26 h were followed by further incubation at 28°C for 24 h. To select ATMT transformants, filters containing the transformants were transferred to SM agar plates and incubated at 37°C for 3 days.
DNA analysis.
Conidia were inoculated into yeast nitrogen base, (Difco, Sparks, Md.), supplemented with 0.5% glucose, and grown on a shaker (200 rpm) at 37°C for 24 h. DNA extractions from hyphae and Southern hybridizations were carried out as previously described (17). SpeI-restricted DNA from transformants with random insertions was probed with the 32P-labeled pAN7 plasmid. In the case of alb1-targeted disruption, AvrII-restricted DNA from albino transformants was probed with an alb1 fragment amplified from the template pks33 (14) by using the primers ALB9 (GATCTGGAATCGTCCGTGTT) and ALB10 (C CCTGGAGAAGAATCGAGGT). For reverse transcription PCR (RT-PCR), total RNA was isolated from albino conidia as described (17). RT-PCR with a One-Step RT-PCR kit (QIAGEN, Valencia, Calif.) was performed according to the manufacturer's protocol. Specific primers used were INT3.1 (ATGGAGTGGCTCTTACGCCAT) and INT4.1 (ATCTTTCGAGCCAGCGCTTG). Since the amplicon contains two introns in the genomic DNA, the wild-type DNA template (positive control) produced a fragment 120 bp larger than the one observed with the total RNA. Tubulin primers were added to the reaction mixture as a control for the amount and quality of RNA. Tubulin primer sequences were as follows: for β-tubulin2F, AAAGGTCTCATCTGCGTGCT; for β-tubulin2R, GGCGGAGAGCTGTGACTATC.
RESULTS AND DISCUSSION
Agrobacterium tumefaciens, a plant-pathogenic bacteria, has the ability to transfer a fragment of DNA (T-DNA) from its tumor-inducing plasmid to a recipient's genome. The tumor-inducing plasmid also contains a set of genes termed vir that are essential for the T-DNA transfer. Induction of the vir genes by certain chemical signals, such as acetosyringone, initiates the process of transfer and integration of the T-DNA into the recipient's genome (9). Based on previous reports of Agrobacterium-mediated transformation in certain species of Aspergillus (5, 10, 13), we developed an ATMT system in A. fumigatus by using hygromycin resistance (hph gene) as the selectable marker. Initially, we inserted the hph gene into T-DNA and aimed for its random integration into the A. fumigatus genome.
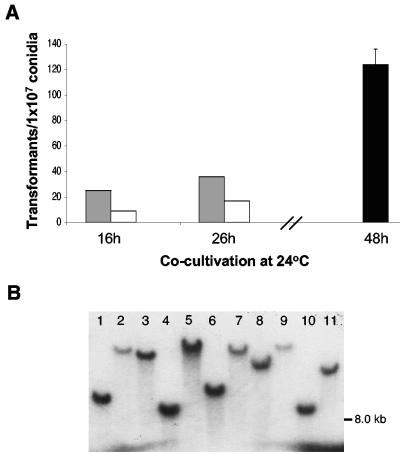
Contact between recipient and Agrobacterium is an essential prerequisite for the T-DNA transfer. A solid substrate allows the recipient and Agrobacterium to grow in close proximity so that the T-DNA can be transferred. Three different filters (nitrocellulose, cellulose, and nylon) were tested as substrates for cocultivation of A. fumigatus and Agrobacterium tumefaciens. The best results were obtained with nylon and cellulose filters (Fig. 1A). Although nitrocellulose filters are most commonly used as the substrates for cocultivation of Agrobacterium and filamentous fungi for ATMT (5), our results with A. fumigatus indicate that nitrocellulose is not as efficient as nylon or cellulose filters (data not shown). The reason for this difference is unclear. It is possible that the chemical properties of these membranes may affect the distribution of the Agrobacterium cells and conidia or inhibit their interaction, thereby resulting in differences in transformation efficiency. Initially, conidia and Agrobacterium were cocultivated at 28°C for 72 h. The number of transformants was low under these conditions, averaging 10 transformants per 107 conidia. The cocultivation temperature was then lowered to 24°C for the first 16 or 26 h, followed by incubation at 28°C for 24 h. The results showed that extending the incubation time at 24°C led to higher numbers of transformants (Fig. 1A). When the cocultivation time at 24°C was extended to 48 h without further incubation at 28°C, over 100 transformants per 107 conidia were obtained. This was approximately a fourfold increase compared to the number of transformants from shorter incubations at 24°C (Fig. 1A). Although 28°C is optimal for growth of A. tumefaciens, this temperature was not appropriate for T-DNA transfer. It has been proposed that the T-DNA transfer machinery is greatly affected by temperature. For example, the expression of some vir genes of Agrobacterium that are necessary for interaction between the bacteria and the recipient is impaired at temperatures above 26°C, thus affecting the transformation efficiency (2, 8).
FIG. 1.
A. fumigatus transformation frequency by ATMT method. (A) Agrobacterium and conidia were cocultivated on either a nylon (gray bars) or cellulose (white bars) filter at 24°C for the indicated time period, followed by further incubation at 28°C for 24 h. Cocultivation on nylon (black bar) was at 24°C for 48 h without subsequent incubation at 28°C. (B) Southern analysis. DNAs of A. fumigatus transformants were digested with SpeI and hybridized with pAN7 plasmid DNA as a probe containing the hph gene.
To analyze the number of T-DNA copies integrated in the genome, 15 transformants were randomly selected. Southern blot analysis revealed that most of the transformants harbored a single copy of T-DNA integrated randomly in the genome (Fig. 1B). Similar results have been reported in A. giganteus (13) and A. awamori (10), where a majority of transformants contained a single copy of T-DNA in the genome. The efficiency of generating mutants by ATMT in A. fumigatus was demonstrated in further experiments where we obtained six albino conidial mutants in about 9,000 transformants (data not shown). According to our data, the best conditions for random insertion of T-DNA via ATMT in A. fumigatus involve the cocultivation of conidia and Agrobacterium on nylon filters at 24°C for 48 h, followed by transfer of the filter to SM agar for further incubation at 37°C for 3 days.
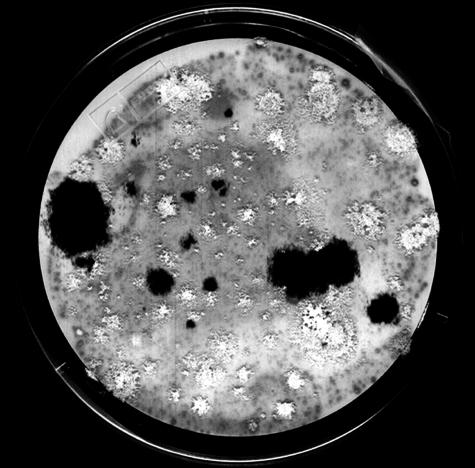
Our second aim was to assess whether a T-DNA-containing A. fumigatus gene can be integrated at a specific site via homologous recombination. The gene chosen as the target for homologous integration was alb1, a polyketide synthase gene. This gene was chosen because the colonies of Δalb1 mutants are readily distinguishable from the wild-type colonies (17). When alb1 is disrupted, the mutant fails to synthesize the bluish-green conidial pigment and produces albino colonies. Disruption of the alb1 gene was carried out by using a plasmid construct containing a hygromycin resistance cassette flanked by alb1 sequence (17) (Fig. 2A). Initially A. tumefaciens strain EHA105B was cocultivated with conidia of the wild-type strain B-5233 at 24°C for 48 h. However, the number of transformants was too numerous, and the filters were completely covered with transformants (data not shown). Since previous experiments had shown that cocultivation time influences transformation efficiency, we reduced the time to 40 h in order to obtain a manageable number of transformants. With a 40-h cocultivation time, the number of transformants was significantly lower, averaging 60 colonies per 107 conidia (Fig. 3). Among the transformants, 66% produced albino conidia, while the other 34% produced bluish-green conidia. Further experiments with a 40-h incubation time with a conidia-to-bacteria ratio of either 1:100 or 1:1,000 resulted in a greater reduction in transformation frequency. The average numbers of transformants obtained with the 1:100 and 1:1,000 ratios were 15 and 4, respectively. A reduction in the transformation frequency resulting from an increase in the number of Agrobacterium cells relative to conidia has also been described in A. giganteus ATMT; a fivefold increase of input Agrobacterium above the optimum levels resulted in a 16-fold reduction in transformation efficiency (13).
FIG. 3.
Gene disruption of alb1 via ATMT. The cocultivation plate shows transformants generated by targeted mutagenesis. Wild-type colonies (black) presumably contain ectopically integrated T-DNA. Albino colonies suggest the disruption of the alb1 gene. Adobe Photoshop version 3.0 was used for the image.
Twenty randomly selected albino mutants were analyzed by Southern hybridization. Upon hybridization with a 560-bp wild-type alb1 fragment, the strain B-5233 yielded a 2.4-kb band corresponding to the wild-type alb1 gene (Fig. 2A), while the albino transformants resulting from a double crossover at the homologous site yielded a 4.8-kb band (Fig. 2A). Homologous recombination between the disruption construct and the alb1 gene occurred in 19 transformants (95%) (Fig. 2B). One of the transformants, H7, contained an extra band in addition to the expected deletion pattern, suggesting that H7 has an additional copy of the disruption construct, one in the alb1 locus and the other at an ectopic site. Transformant H21 revealed two bands, the wild-type fragment and a fragment of approximately 7 kb (Fig. 2B). The presence of a wild-type-sized band suggested that the albino phenotype was not the result of a double crossover. Further analysis indicated that the albino phenotype in this transformant was due to integration of a T-DNA copy at the alb1 locus by single crossover (data not shown). Gene disruption was confirmed by a loss of the alb1 transcript. RT-PCR from nine randomly selected transformants revealed that all strains lacked the alb1 transcript (Fig. 2C), confirming the gene disruption. High frequencies of gene disruption via ATMT have been reported in various other fungi. For example, ATMT carried out with a disruption construct of the β-1,6-glucanase gene (VFGlu1) in Verticillium fungicola resulted in a targeted gene deletion in 75% of the transformants (1). In Trichoderma atroviride, approximately 60% of ATMT transformants showed targeted disruptions in the tmk1 as well as the tga3 genes (18).
The gene alb1 has been previously disrupted in the wild-type B-5233 by the spheroplast transformation method (17). The disruption construct contained a cassette similar to the one used in the present study, with the same alb1 flanking regions as well as the selectable marker. Spheroplast transformation yielded 30% of the transformants with albino phenotype (17). Among the albino transformants, only 25% contained a single copy of the disruption construct integrated via double crossover; for the rest, either multiple copies of the disruption construct were present, or the disruption resulted from a single crossover event (unpublished data). Because the precise number of viable spores as well as the input of transforming DNA cannot be standardized, direct comparisons of homologous integration frequencies between spheroplast transformation and ATMT are not possible. As a general outcome, however, the yield of albino transformants by the ATMT method was twofold higher relative to wild-type transformants when compared with results from the spheroplast transformation method. Furthermore, the frequency of homologous recombination via a double crossover without additional ectopic integrations of the disruption construct is higher among albino mutants obtained by the ATMT than by the spheroplast method. In general, the frequency of homologous integration in A. fumigatus (B-5233) by spheroplast transformation is between 10 to 30%, depending on the genes targeted, and multiple integrations can range between 40 to 60% (unpublished observations). In conclusion, the ATMT method not only simplifies the transformation procedure but also offers an efficient tool for random as well as targeted integration in A. fumigatus.
Acknowledgments
We thank Seogchan Kang at the Pennsylvania State University for his supply of A. tumefaciens strains and the plasmid pDHt/SK. We also thank A. Varma for reading the manuscript.
REFERENCES
- 1.Amey, R. C., P. R. Mills, A. Bailey, and G. D. Foster. 2003. Investigating the role of a Verticillium fungicola beta-1,6-glucanase during infection of Agaricus bisporus using targeted gene disruption. Fungal Genet. Biol. 39:264-275. [DOI] [PubMed] [Google Scholar]
- 2.Baron, C., N. Domke, M. Beinhofer, and S. Hapfelmeier. 2001. Elevated temperature differentially affects virulence, VirB protein accumulation, and T-pilus formation in different Agrobacterium tumefaciens and Agrobacterium vitis strains. J. Bacteriol. 183:6852-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brakhage, A. A., and K. Langfelder. 2002. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 56:433-455. [DOI] [PubMed] [Google Scholar]
- 4.Bundock, P., A. den Dulk-Ras, A. Beijersbergen, and P. J. Hooykaas. 1995. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14:3206-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Groot, M. J., P. Bundock, P. J. Hooykaas, and A. G. Beijersbergen. 1998. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16:839-842. [DOI] [PubMed] [Google Scholar]
- 6.den Dulk-Ras, A., and P. J. J. Hooykaas. 1995. Electroporation of Agrobacterium tumefaciens. Methods Mol. Biol. 55:63-72. [DOI] [PubMed] [Google Scholar]
- 7.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-805. [DOI] [PubMed] [Google Scholar]
- 8.Fullner, K. J., and E. W. Nester. 1996. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J. Bacteriol. 178:1498-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67:16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gouka, R. J., C. Gerk, P. J. Hooykaas, P. Bundock, W. Musters, C. T. Verrips, and M. J. de Groot. 1999. Transformation of Aspergillus awamori by Agrobacterium tumefaciens-mediated homologous recombination. Nat. Biotechnol. 17:598-601. [DOI] [PubMed] [Google Scholar]
- 11.Kwon-Chung, K. J., and J. E. Bennett. 1992. Medical mycology. Lea and Febiger, Philadelphia, Pa.
- 12.Langfelder, K., B. Jahn, H. Gehringer, A. Schmidt, G. Wanner, and A. A. Brakhage. 1998. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187:79-89. [DOI] [PubMed] [Google Scholar]
- 13.Meyer, V., D. Mueller, T. Strowig, and U. Stahl. 2003. Comparison of different transformation methods for Aspergillus giganteus. Curr. Genet. 43:371-377. [DOI] [PubMed] [Google Scholar]
- 14.Mullins, E. D., X. Chen, P. Romaine, R. Raina, D. M. Geiser, and S. Kang. 2001. Agrobacterium-mediated transformation of Fusarium oxysporum: an efficient tool for insertional mutagenesis and gene transfer. Phytopathology 91:173-180. [DOI] [PubMed] [Google Scholar]
- 15.Panepinto, J. C., B. G. Oliver, J. R. Fortwendel, D. L. Smith, D. S. Askew, and J. C. Rhodes. 2003. Deletion of the Aspergillus fumigatus gene encoding the Ras-related protein RhbA reduces virulence in a model of invasive pulmonary aspergillosis. Infect. Immun. 71:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56:117-124. [DOI] [PubMed] [Google Scholar]
- 17.Tsai, H. F., Y. C. Chang, R. G. Washburn, M. H. Wheeler, and K. J. Kwon-Chung. 1998. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeilinger, S. 2004. Gene disruption in Trichoderma atroviride via Agrobacterium-mediated transformation. Curr. Genet. 45:54-60. [DOI] [PubMed] [Google Scholar]