Abstract
The natural product rapamycin, produced during fermentation by Streptomyces hygroscopicus, is known for its potent antifungal, immunosuppressive, and anticancer activities. During rapamycin biosynthesis, the amino acid l-pipecolate is incorporated into the rapamycin molecule. We investigated the use of precursor-directed biosynthesis to create new rapamycin analogs by substitution of unusual l-pipecolate analogs in place of the normal amino acid. Our results suggest that the l-pipecolate analog (±)-nipecotic acid inhibits the biosynthesis of l-pipecolate, thereby limiting the availability of this molecule for rapamycin biosynthesis. We used (±)-nipecotic acid in our precursor-directed biosynthesis studies to reduce l-pipecolate availability and thereby enhance the incorporation of other pipecolate analogs into the rapamycin molecule. We describe here the use of this method for production of two new sulfur-containing rapamycin analogs, 20-thiarapamycin and 15-deoxo-19-sulfoxylrapamycin, and report measurement of their binding to FKBP12.
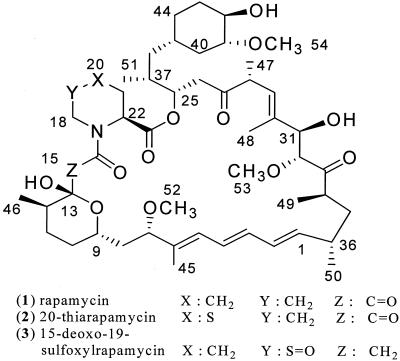
Rapamycin (Fig. 1) is a macrocyclic polyketide produced during fermentation by the bacterium Streptomyces hygroscopicus ATCC 29253, which was isolated from a soil sample from Easter Island (24, 27). This compound was originally discovered as an antifungal agent and has since been studied for its potent immunosuppressive and anticancer properties. Rapamycin is currently manufactured and marketed by Wyeth as Rapamune for the prevention of organ graft rejection in kidney transplant patients and is also used as a coating for vascular stents to prevent restenosis of coronary arteries following angioplasty (13). The rapamycin derivative CCI-779 is currently in development for treatment of a variety of cancers (8, 28).
FIG. 1.
Structures of rapamycin, 20-thiarapamycin, and 15-deoxo-19-sulfoxylrapamycin.
Novel analogs of rapamycin have been generated in attempts to create analogs with improvements over the natural product and to study structure-activity relationships. Such analogs have been created by using synthetic methods (2, 16), semisynthetic methods (21), and precursor feeding (9, 10, 12, 18), and there have been substantial efforts to achieve this goal by manipulation of the biosynthetic gene cluster (4, 6, 7, 9). In our efforts to generate novel rapamycin analogs, we have evaluated feeding S. hygroscopicus with alternative rapamycin precursor molecules (precursor-directed biosynthesis).
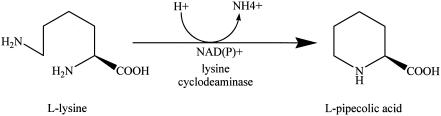
The rapamycin macrolide is synthesized via a mixed polyketide synthase-nonribosomal peptide synthetase enzymatic complex (15). During rapamycin biosynthesis, the enzyme lysine cyclodeaminase, encoded by the gene rapL, catalyzes the cyclization and conversion of the amino acid l-lysine to l-pipecolate (Fig. 2) (9, 19). l-Pipecolate is then incorporated into the molecule just prior to final closure of the macrocyclic ring (6, 19). A pipecolate-incorporating enzyme, encoded by the rapP gene, is believed to be responsible for the catalysis of l-pipecolate incorporation, as well as the ring closure (11, 15, 17). It is already known that the amino acid l-proline and some of its analogs can be substituted for l-pipecolate at this site, suggesting that the pipecolate-incorporating enzyme can accept some structural changes in the amino acid substrate (9, 10, 17). We exploited this relaxed substrate specificity to generate novel rapamycin analogs by incorporating unusual analogs of l-pipecolate into the rapamycin molecule.
FIG. 2.
Biosynthesis of l-pipecolic acid. l-Pipecolic acid is derived from l-lysine via a lysine cyclodeaminase-catalyzed reaction (9).
MATERIALS AND METHODS
Strain.
S. hygroscopicus ATCC 29253 was obtained from the Lederle Culture Collection, Wyeth Research, Pearl River, N.Y.
Culture conditions.
S. hygroscopicus was routinely cultivated at 28°C on an agar medium containing 0.5% dextrose, 0.25% yeast extract (Difco), 1% soluble starch (Difco), 0.25% NZ-Amine A (Quest), 0.05% CaCO3, and 1.5% agar. The seed culture medium and production medium used were the media described previously by Sehgal et al. (23). Seed cultures were initiated by inoculating a loopful of culture from agar into 10 ml of seed inoculum broth. Seed cultures were incubated at 28°C with shaking at 200 rpm with a 2-in. throw for 3 days. For production of rapamycin and 21-nor-rapamycin (prolyl-rapamycin), 0.1 ml of seed culture broth was inoculated into a 50-ml Erlenmeyer shake flask containing 5 ml of production medium. For the directed biosynthesis of rapamycin analogs, the production medium was made without lysine or pipecolic acid and was supplemented with 5% (±)-nipecotic acid (Sigma). The unusual pipecolate analogs 1,4-thiazane-(3S)-carboxylic acid and 1,3-thiazane-(4S)-carboxylic acid were added to this modified production medium at a final concentration of 0.2%. Fermentation was performed as described above, and cultures were harvested after 6 days.
Analogs of l-pipecolate.
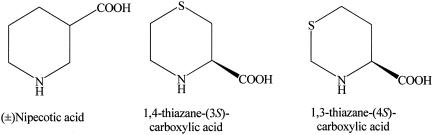
(±)-Nipecotic acid (Fig. 3) was obtained from Sigma-Aldrich. The sulfur-containing analogs 1,4-thiazane-(3S)-carboxylic acid and 1,3-thiazane-(4S)-carboxylic acid (Fig. 3) were synthesized by previously described methods (1, 14).
FIG. 3.
Structures of pipecolate analogs used in this study, including (±)-nipecotic acid, 1,4-thiazane-(3S)-carboxylic acid, and 1,3-thiazane-(4S)-carboxylic acid.
Detection of rapamycin analogs.
Fermentation broth (5 ml) was centrifuged at 3,000 rpm (1,090 × g) for 20 min, and the supernatant was discarded. The pellet was extracted by shaking in 5 ml of methanol, followed by centrifugation at 3,000 rpm (1,090 × g). The solvent was collected and evaporated, and the residue was reconstituted in 0.5 ml of methanol. Samples were analyzed by high-performance liquid chromatography (HPLC), and the production of new rapamycin analogs was monitored by comparison of the peak retention times and UV spectra to those of authentic rapamycin standards. The incorporation of pipecolate analogs into the rapamycin compounds was also revealed by liquid chromatography-mass spectrometry (MS) analysis of fermentation extracts (see below). The molecular weights of new peaks (the retention times differed from those of native compounds) were compared with the predicted molecular weights of rapamycin derivatives containing the pipecolate analog. Peaks corresponding to new rapamycin analogs were purified and assayed for bioactivity.
HPLC analysis of rapamycin analogs.
Reverse-phase HPLC was performed by using a Hewlett-Packard model HP1100 liquid chromatograph with photodiode array detection. Extracts were resolved by reverse-phase chromatography by using a YMC ODS-A HPLC column (4.6 by 150 mm; 5 μm; 120 Å) with mobile phases of 0.025% formic acid in water (solvent A) and 0.025% formic acid in acetonitrile (solvent B). For elution, we used a linear gradient from 5% solvent B to 95% solvent B in 25 min, with holding at 95% acetonitrile for 10 min and a flow rate of 0.8 ml/min.
Liquid chromatography-MS analysis of rapamycin analogs.
The molecular weights of new rapamycin species were determined by using a Hewlett-Packard model HP1100 HPLC system coupled with a ThermoFinnigan LCQ-Classic ion trap mass spectrometer equipped with an electrospray ionization source. Extracts were resolved by reverse-phase HPLC as described above. A split was used to divert 25% of the flow into the mass spectrometer. UV detection was performed by using a photodiode array with a scan range of 200 to 600 nm. The MS electrospray analysis was performed in the positive mode with a scan range of 200 to ∼2,000 m/z.
Purification of rapamycin analogs and structure elucidation.
The purification and structural identification of 20-thiarapamycin and 15-deoxo-19-sulfoxylrapamycin have been described previously (5).
FKBP12-binding assay.
His6-tagged human FKBP12 protein was cloned, expressed, and purified by using previously described methods (3, 25). FKBP12 binding was monitored by an assay performed with His6-tagged human FKBP12 protein, the FKBP12 ligand FK506 (3H-labeled FK506; catalog no. NET1095; Perkin-Elmer-NEN), and nickel-coated scintillation plates (nickel chelate FlashPlate; catalog no. SMP107; Perkin-Elmer-NEN). In this assay we measured specific binding of 3H-labeled FK506 to FKBP12 by using a scintillation counter to detect the binding complex captured on a nickel chelate FlashPlate through the His6 tag of FKBP12. Binding of test compounds to FKBP12 was identified by detecting the inhibition of 3H-labeled FK506-FKBP12 complex formation. The binding screening procedure was performed in a 96-well plate as follows. To each well of the 96-well FlashPlate we added 50 μl of binding buffer (50 mM HEPES [pH 7.4], 0.1% Tween 20, 0.25 mg of sodium azide per ml) containing 5 nM (final concentration) His6-FKBP12. The test compound or control vehicle was then added. Rapamycin (10 nM) was also tested as a positive control for comparison. The plates were incubated for 10 min at room temperature with gentle shaking, after which 50 μl of binding buffer containing 2 nM (final concentration) 3H-labeled FK506 was added. The plates were sealed with plastic film. Binding reactions were allowed to proceed for 1.5 h at room temperature with gentle shaking. The assay results were read with a microbeta plate reader (Perkin-Elmer-NEN). The specific binding value was defined as the value (in scintillation counts) after subtraction of the nonspecific background value (an average value obtained from reaction mixtures not containing the FKBP12 receptor). Data obtained from the plate reader were expressed as percentages of binding relative to the control (untreated) wells on the same assay plate. Binding data are expressed below as percentages of inhibition of 3H-labeled FK506-FKBP12 complex formation relative to untreated control wells. The positive control compound rapamycin at a concentration of 10 nM typically gave a value of 10 to 30% of control binding.
RESULTS
Effect of nipecotic acid.
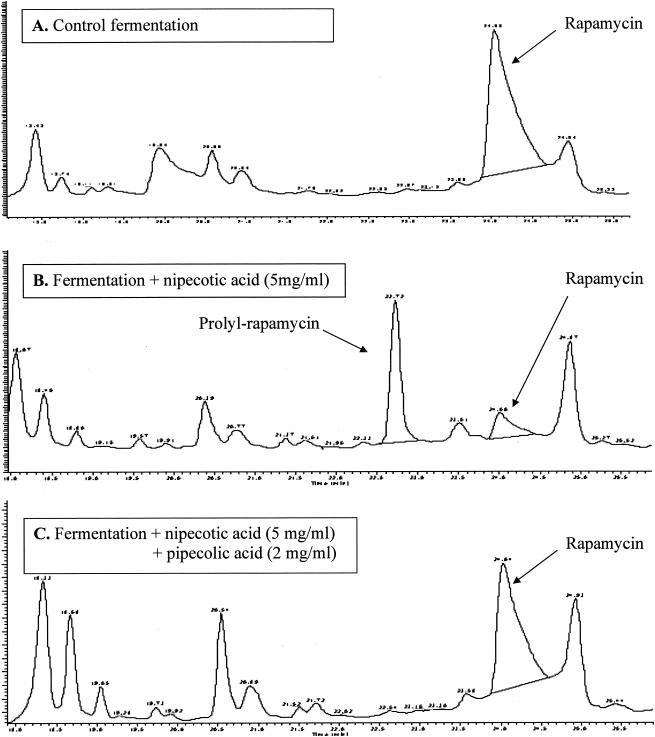
HPLC profiles revealed a trend in fermentations that indicated that racemic (±)-nipecotic acid had an effect on rapamycin production (Fig. 4). Individual isomers of nipecotic acid [(+)-nipecotic acid or (−)-nipecotic acid] were not tested. Under standard production conditions, the major product of fermentation was rapamycin, and prolyl-rapamycin was a minor product. The ratio of rapamycin to prolyl rapamycin produced under these conditions was approximately 16:1. In the presence of (±)-nipecotic acid, rapamycin production was reduced approximately fourfold, while the prolyl-rapamycin titer increased approximately 12-fold. Under these modified conditions, the ratio of rapamycin to prolyl-rapamycin produced was approximately 1:3. Addition of l-pipecolic acid under these fermentation conditions restored rapamycin production and reduced prolyl-rapamycin titers to the levels seen under standard production conditions. These results suggested that (±)-nipecotic acid inhibited the formation and subsequent availability of pipecolate, thereby allowing greater incorporation of l-proline into the rapamycin molecule.
FIG. 4.
Effect of nipecotic acid. (A) Under normal conditions for rapamycin fermentation, rapamycin was the primary product. (B) With the addition of (±)-nipecotic acid to the fermentation, prolyl-rapamycin became the major product. (C) Adding l-pipecolic acid to the production medium restored rapamycin production. The retention times were as follows: prolyl-rapamycin, 22.9 min; rapamycin, 24.1 min.
Feeding of unusual pipecolate analogs in the presence of (±)-nipecotic acid.
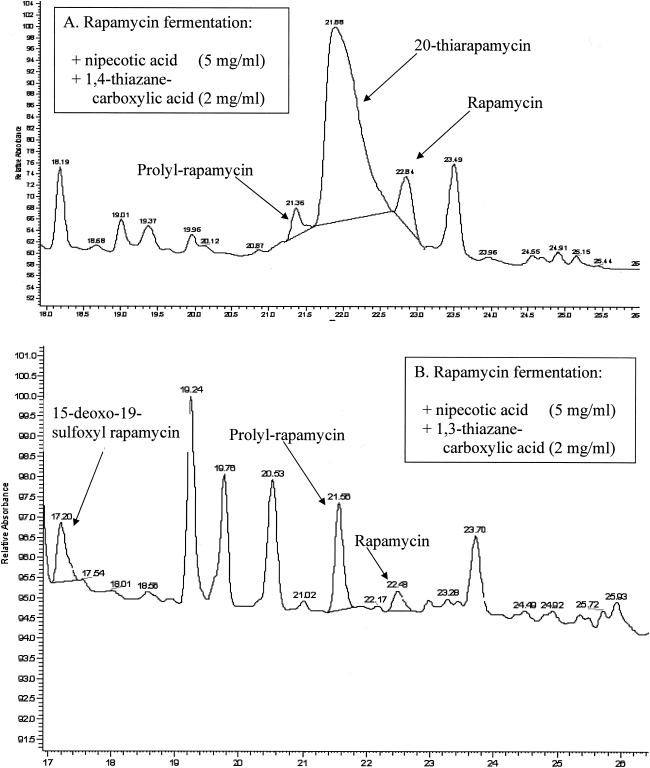
HPLC-MS analysis of fermentation extracts revealed new peaks in fermentations that had been supplemented with pipecolate analogs (Fig. 5). These new peaks did not correspond to known rapamycins with respect to retention time, but they were identified as rapamycin-related compounds by their characteristic UV spectra. Mass spectral data indicated the presence of novel rapamycin analogs (5). Purification and subsequent nuclear magnetic resonance analysis of these molecules permitted elucidation of the novel rapamycin structures (5).
FIG. 5.
Detection of 20-thiarapamycin (A) and 15-deoxo-19-sulfoxylrapamycin (B) by HPLC-UV analysis. (A) Retention times: prolyl-rapamycin, 21.4 min; 20-thiarapamycin, 21.9 min; rapamycin, 22.8 min. (B) Retention times: 15-deoxo-19-sulfoxylrapamycin, 17.2 min; prolyl-rapamycin, 21.6 min; rapamycin, 22.5 min.
20-Thiarapamycin (Fig. 1) was isolated from fermentations supplemented with 0.2% 1,4-thiazane-(3S)-carboxylic acid and 0.5% (±)-nipecotic acid. The yield of 20-thiarapamycin in these fermentations was approximately 100 mg/liter. 15-Deoxo-19-sulfoxylrapamycin (Fig. 1) was isolated from fermentations supplemented with 0.2% 1,3-thiazane-(4S)-carboxylic acid and 0.5% (±)-nipecotic acid. The yield of 15-deoxo-19-sulfoxylrapamycin in these fermentations was approximately 10 mg/liter.
FKBP12 binding.
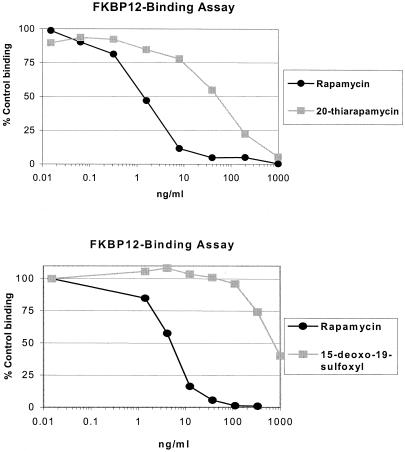
20-Thiarapamycin and 15-deoxo-19-sulfoxylrapamycin were tested in an FKBP12 binding assay (Fig. 6). Binding of analogs was expressed as percent inhibition of 3H-labeled FK506-FKBP12 (control) complex formation. In each experiment, rapamycin was also tested for comparison of binding affinity. 20-Thiarapamycin had a 50% inhibitory concentration (IC50) of 53.6 nM (50 ng/ml) in this assay, while rapamycin had an IC50 of 1.6 nM (1.5 ng/ml). In a separate experiment, 15-deoxo-19-sulfoxylrapamycin had an IC50 of 800 nM (747 ng/ml), while rapamycin had an IC50 of 4.9 nM (4.5 ng/ml).
FIG. 6.
FKBP12 binding of novel rapamycin analogs, expressed as percent inhibition of 3H-labeled FK506-FKBP12 (control) complex formation. (A) Binding of 20-thiarapamycin to FKBP12 was reduced 33-fold compared to binding of rapamycin. The IC50 of the rapamycin control was 1.6 nM, and the IC50 of 20-thiarapamycin was 53.6 nM. (B) Binding of 15-deoxo-19-sulfoxylrapamycin to FKBP12 was reduced 166-fold compared to binding of rapamycin. The IC50 of the rapamycin control was 4.9 nM, and the IC50 of 15-deoxo-19-sulfoxylrapamycin was 800 nM.
DISCUSSION
Because the enzyme responsible for the incorporation of l-pipecolate into the rapamycin molecule has a relaxed substrate specificity, other amino acids, including l-proline and certain proline analogs, can be incorporated in place of l-pipecolate (9, 10). Under standard fermentation conditions in media containing l-proline, S. hygroscopicus ATCC 29253 produces a small amount of prolyl-rapamycin in addition to the major product, rapamycin (9, 10). Our aim was to generate new analogs of rapamycin by feeding unusual analogs of l-pipecolate to the rapamycin fermentation, so that the analogs could be incorporated into the molecule at the l-pipecolate site.
During rapamycin biosynthesis, the conversion of l-lysine to l-pipecolate is catalyzed by the enzyme lysine cyclodeaminase. The gene encoding this enzyme, rapL, is in the rapamycin biosynthetic gene cluster (15, 20). It has been demonstrated previously that elimination of lysine cyclodeaminase activity, either by rapL gene knockout (9) or by inhibition of the enzyme itself with certain analogs of l-proline (10), results in a reduction in the rapamycin titer and an increase in the prolyl-rapamycin titer. It has also been shown that this disruption of pipecolate biosynthesis can be used to enhance the incorporation of certain l-proline analogs into the rapamycin molecule (9, 10).
It was observed during our studies that the addition of (±)-nipecotic acid to the fermentation medium reduced the rapamycin yield, while it increased the prolyl-rapamycin titer. We also observed that the addition of l-pipecolic acid to these fermentations reversed this effect. These results suggested that (±)-nipecotic acid somehow limited the availability of the preferred substrate, l-pipecolic acid, thereby allowing increased incorporation of l-proline into the rapamycin molecule. As a result, all further studies involving the feeding of unusual l-pipecolate analogs were performed with (±)-nipecotic acid present in the fermentation in order to enhance incorporation of the desired analog.
We observed incorporation of two different sulfur-containing analogs of l-pipecolate into the rapamycin molecule. The analogs 1,4-thiazane-(3S)-carboxylic acid and 1,3-thiazane-(4S)-carboxylic acid were both successfully incorporated in fermentations containing 0.5% (±)-nipecotic acid, yielding the new rapamycin analogs 20-thiarapamycin and 15-deoxo-19-sulfoxylrapamycin, respectively. The biosynthetic differences in the incorporation of these two structurally similar analogs are particularly interesting. 1,4-Thiazane-(3S)-carboxylic acid was incorporated directly into the rapamycin molecule in place of l-pipecolate. There was no apparent modification to the analog, and the downstream tailoring enzymes of the biosynthetic pathway were not disturbed. As a result, the structure of the product, 20-thiarapamycin, matched the predicted structure and was identical to the structure of rapamycin except for the presence of a sulfur atom on the pipecolate/thiazane ring. 1,3-Thiazane-(4S)-carboxylic acid was incorporated at a 10-fold-lower yield than 1,4-thiazane-(3S)-carboxylic acid, and the structure of the product, 15-deoxo-19-sulfoxylrapamycin, differed significantly from the predicted structure. The thiazane sulfur of 15-deoxo-19-sulfoxylrapamycin is oxidized to a sulfoxide, presumably after incorporation of 1,3-thiazane-(4S)-carboxylic acid. Also, the final product lacks the C-15 carbonyl that is present in both rapamycin and 20-thiarapamycin, which indicates that the usual activity of the downstream cytochrome P-450 monooxygenase is somehow disrupted. Perhaps, because of the modified structure of this compound, this cytochrome P-450 now acts on the thiazane sulfur instead of the C-15 carbon, but this hypothesis has not been confirmed experimentally.
It is known that the pipecolate-containing region of the rapamycin molecule is involved in the binding of rapamycin to FKBP12 (5, 26). This binding is required for further binding of rapamycin to the kinase mTOR, which is responsible for many of the biological activities of rapamycin (22, 28). We have previously reported (5) that both of these molecules are weaker FKBP12 binders than rapamycin. In our FKBP12-binding assay, the IC50 of 20-thiarapamycin was approximately 33-fold higher than that of rapamycin, indicating a correspondingly weaker binding affinity. 15-Deoxo-19-sulfoxylrapamycin was an even weaker binder of FKBP12, with an IC50 that was approximately 166-fold higher than that of rapamycin. It is known that the C-15 carbonyl of rapamycin plays an important role in FKBP12 binding (26). The lack of this carbonyl in 15-deoxo-19-sulfoxylrapamycin may account for the great reduction in binding affinity. In brief, one can surmise that it is difficult to modify this domain of the rapamycin molecule without negatively affecting its FKBP12 interaction.
REFERENCES
- 1.Carson, J. F., and F. F. Wong. 1964. The synthesis of l-1,4-thiazane-3-carboxylic acid 1-oxide. J. Org. Chem. 29:2203-2205. [Google Scholar]
- 2.Caufield, C. E. 1995. Structure-activity relationships involving modifications to the macrolides FK-506 and rapamycin. Curr. Pharm. Des. 1:145-160. [Google Scholar]
- 3.Choi, J., J. Chen, S. L. Schreiber, and J. Clardy. 1996. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273:239-242. [DOI] [PubMed] [Google Scholar]
- 4.Chung, L., L. Liu, S. Patel, J. R. Carney, and C. D. Reeves. 2001. Deletion of rapQONML from the rapamycin gene cluster of Streptomyces hygroscopicus gives production of the 16-O-desmethyl-27-desmethoxy analog. J. Antibiot. 54:250-256. [DOI] [PubMed] [Google Scholar]
- 5.Graziani, E. I., F. V. Ritacco, M. Y. Summers, T. M. Zabriskie, K. Yu, V. S. Bernan, M. Greenstein, and G. T. Carter. 2003. Novel sulfur-containing rapamycin analogs prepared by precursor-directed biosynthesis. Org. Lett. 5:2385-2388. [DOI] [PubMed] [Google Scholar]
- 6.Gregory, M. A., S. Gaisser, R. E. Lill, H. Hong, R. M. Sheridan, B. Wilkinson, H. Petkovic, A. J. Weston, I. Carletti, H.-L. Lee, J. Staunton, and P. F. Leadlay. 2004. Natural product biosynthesis: isolation and characteristics of pre-rapamycin, the first macrocyclic intermediate in the biosynthesis of the immunosuppressant rapamycin by S. hygroscopicus Angew. Chem. 43:2551-2553. [DOI] [PubMed] [Google Scholar]
- 7.Gregory, M. A., S. Gaisser, H. Petkovic, and S. Moss. January2004. Genetic engineering of Streptomyces hygroscopicus and other microbes for production of polyketides and other natural products. World patent 2004007709.
- 8.Hidalgo, M., and E. K. Rowinsky. 2000. The rapamycin-sensitive signal transduction pathway as a target for cancer therapy. Oncogene 19:6680-6686. [DOI] [PubMed] [Google Scholar]
- 9.Khaw, L. E., G. A. Böhm, S. Metcalfe, J. Staunton, and P. F. Leadlay. 1998. Mutational biosynthesis of novel rapamycins by a strain of Streptomyces hygroscopicus NRRL 5491 disrupted in rapL, encoding a putative lysine cyclodeaminase. J. Bacteriol. 180:809-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kojima, I., and A. L. Demain. 1998. Preferential production of rapamycin versus prolylrapamycin by Streptomyces hygroscopicus J. Ind. Microbiol. Biotechnol. 20:309-316. [Google Scholar]
- 11.König, A., T. Schwecke, I. Molnár, G. A. Böhm, P. A. S. Lowden, J. Staunton, and P. F. Leadlay. 1997. The pipecolate-incorporating enzyme for the biosynthesis of the immunosuppressant rapamycin. Nucleotide sequence analysis, disruption and heterologous expression of rapP from Streptomyces hygroscopicus. Eur. J. Biochem. 247:526-534. [DOI] [PubMed] [Google Scholar]
- 12.Lowden, P. A. S., G. A. Böhm, S. Metcalfe, J. Staunton, and P. F. Leadlay. 2004. New rapamycin derivatives by precursor-directed biosynthesis. ChemBioChem 5:535-538. [DOI] [PubMed] [Google Scholar]
- 13.Marx, S. O., and A. R. Marks. 2001. The development of rapamycin and its application to stent restenosis. Circulation 104:852-855. [DOI] [PubMed] [Google Scholar]
- 14.Miyazaki, H., A. Ohta, N. Kawakatsu, Y. Waki, Y. Gogun, T. Shiraiwa, and H. Kurokawa. 1993. Preparations of optically active homocysteine and homocystine by asymmetric transformation of (RS)-1,3-thiazane-4-carboxylic acid. Bull. Chem. Soc. Jpn. 66:536-540. [Google Scholar]
- 15.Molnár, I., J. F. Aparicio, S. F. Haydock, L. E. Khaw, T. Schwecke, A. König, J. Staunton, and P. F. Leadlay. 1996. Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of genes flanking the polyketide synthase. Gene 169:1-7. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, F. C., S. J. Stachel, C. P. Eng, and S. N. Sehgal. 1999. Manipulation of the C(22)-C(27) region of rapamycin: stability issues and biological implications. Bioorg. Med. Chem. Lett. 9:295-300. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, J. B., M. J. Hsu, K. M. Byrne, and L. Kaplan. 1991. Biosynthesis of the immunosuppressant immunomycin: the enzymology of pipecolate incorporation. Biochemistry 30:5789-5796. [DOI] [PubMed] [Google Scholar]
- 18.Nishida, H., T. Sakakibara, F. Aoki, T. Saito, K. Ichikawa, T. Inagaki, Y. Kojima, Y. Yamauchi, L. H. Huang, M. A. Guadliana, et al. 1995. Generation of novel rapamycin structures by microbial manipulations. J. Antibiot. 48:657-666. [DOI] [PubMed] [Google Scholar]
- 19.Paiva, N. L., A. L. Demain, and M. F. Roberts. 1993. The immediate precursor of the nitrogen-containing ring of rapamycin is free pipecolic acid. Enzyme Microb. Technol. 15:581-585. [Google Scholar]
- 20.Schwecke, T., J. F. Aparicio, I. Molnár, A. König, L. E. Khaw, S. F. Haydock, M. Oliynyk, P. Caffrey, J. Cortés, J. B. Lester, et al. 1995. The biosynthetic gene cluster for the polyketide immunosuppressant rapamycin. Proc. Natl. Acad. Sci. USA 92:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sedrani, R., L. H. Jones, A.-M. Jutzi-Eme, W. Schuler, and S. Cottens. 1999. Cleavage of the cyclohexyl-subunit of rapamycin results in loss of immunosuppressive activity. Bioorg. Med. Chem. Lett. 9:459-462. [DOI] [PubMed] [Google Scholar]
- 22.Sehgal, S. N. 1998. Rapamune (RAPA, rapamycin, sirolimus): mechanism of action immunosuppressive effect results from blockade of signal transduction and inhibition of cell cycle progression. Clin. Biochem. 31:335-340. [DOI] [PubMed] [Google Scholar]
- 23.Sehgal, S. N., H. Baker, C. P. Eng, K. Singh, and C. Vézina. 1983. Demethoxyrapamycin (AY-24,668), a new antifungal antibiotic. J. Antibiot. 36:351-354. [DOI] [PubMed] [Google Scholar]
- 24.Sehgal, S. N., H. Baker, and C. Vézina. 1975. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation, and characterization. J. Antibiot. 28:727-732. [DOI] [PubMed] [Google Scholar]
- 25.Van Duyne, G. D., R. F. Standaert, P. A. Karplus, S. L. Schreiber, and J. Clardy. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229:105-124. [DOI] [PubMed] [Google Scholar]
- 26.Van Duyne, G. D., R. F. Standaert, S. L. Schreiber, and J. Clardy. 1991. Atomic structure of the rapamycin human immunophilin FKBP-12 complex. J. Am. Chem. Soc. 113:7433-7434. [Google Scholar]
- 27.Vézina, C., A. Kudelski, and S. N. Sehgal. 1975. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 28:721-726. [DOI] [PubMed] [Google Scholar]
- 28.Yu, K., L. Toral-Barza, C. Discafani, W. G. Zhang, J. Skotnicki, P. Frost, and J. J. Gibbons. 2001. mTOR, a novel target in breast cancer: the effect of CCI-779, an mTOR inhibitor, in preclinical models of breast cancer. Endocr. Relat. Cancer 8:249-258. [DOI] [PubMed] [Google Scholar]