Abstract
The transcription factors sinR and abrB are involved in the control of sporulation initiation in Bacillus subtilis. We identified a single homologue to sinR and three highly similar homologues to abrB, designated abrB310, abrB1941, and abrB3647, in Clostridium acetobutylicum ATCC 824. Using reporter vectors, we showed that the promoters of abrB1941 and abrB3647 were not active under the growth conditions tested. The abrB310 promoter was strongly active throughout growth and exhibited a transient elevation of expression at the onset of solventogenesis. Primer extension assays showed that two transcripts of abrB310 and a single, extremely weak transcript for sinR are expressed. Potential −35 and −10 consensus motifs are readily identifiable surrounding the transcription start sites of abrB310 and sinR, with a single putative 0A box present within the promoter of abrB310. In strains of C. acetobutylicum transformed with plasmids to elevate sinR expression or decrease sinR expression, no significant differences in growth or in acid or solvent production were observed compared to the control strains. In C. acetobutylicum strain 824(pAS310), which expressed an antisense RNA construct targeted against abrB310, the acids acetate and butyrate accumulated to approximately twice the normal concentration. This accumulation corresponded to a delay and decrease in acetone and butanol production. It was also found that sporulation in strain 824(pAS310) was delayed but that the morphology of sporulating cells and spores was normal. Based upon these observations, we propose that abrB310 may act as a regulator at the transition between acidogenic and solventogenic growth.
The life cycle of the gram-positive, obligate anaerobe Clostridium acetobutylicum progresses in three phases: acidogenesis, solventogenesis, and sporulation. During acidogenesis, cultures of C. acetobutylicum exhibit exponential growth and produce acetic and butyric acid. After 12 to 24 h of acidogenic growth, the culture undergoes a transition from acidogenesis to solventogenesis, at which point cells enter the stationary growth phase and the acidic metabolites are reassimilated into the solvents acetone and butanol. Sporulation follows solventogenesis, after approximately 48 to 72 h of growth (2, 22, 28).
The genes and enzymes involved in the central acidogenic and solventogenic metabolic pathway have been identified and are well characterized. Less well understood is the control of gene expression at the transition between acidogenic and solventogenic growth, during which the expression of genes involved primarily in acid production is decreased and the expression of genes required for solvent production is greatly elevated, and it is on this transition that we have focused our investigations.
During acidogenesis, the expression of genes coding for phosphotransacetylase (pta), acetate kinase (ack), phosphotransbutyrylase (ptb), and butyrate kinase (buk) is high. As a result, acetate and butyrate accumulate during exponential growth. At the end of exponential growth, the transcriptome of the cell alters, resulting in the transition from acidogenesis to solventogenesis (7, 28).
Four genes located on the pSOL1 megaplasmid are essential to solvent formation (6, 30). adhE, ctfA, and ctfB are arranged in the sol operon and are hence transcribed as a single transcript from the promoter located upstream of adhE. Abutting the 3′ end of the sol operon is adc, which is transcribed from its own promoter in the opposite direction of that of the sol operon (6, 10, 34). At the onset of solventogenesis, the expression of the sol operon and adc increases approximately 10-fold (9). Until relatively recently, little was known about the factors involved in the transcriptional control of these genes.
The stage 0 sporulation protein A (Spo0A) was originally identified in Bacillus subtilis as a key regulator of the onset of sporulation (8). When phosphorylated, Spo0A is activated and binds directly to a conserved DNA promoter motif, designated the 0A box (19, 20). A recent survey of the B. subtilis genome indicated that the expression of 121 genes is directly regulated by activated Spo0A, with 40 of these genes, including spo0A, being upregulated and 81 being downregulated in response to Spo0A binding (29).
Spo0A has been shown to affect the expression of genes at the transition from acidogenic to solventogenic growth in solventogenic clostridia. In strain SKO1 of C. acetobutylicum, in which spo0A has been disrupted, severely decreased levels of the adhE-ctfA-ctfB and adc transcripts were detected, which correlated with extremely low concentrations of acetone and butanol (16, 17). With Clostridium beijerinckii, it was shown that the temporal control of adc and ptb expression was disrupted in the absence of the 0A box motifs (35). A microarray analysis of the C. acetobutylicum transcriptome has identified 123 genes whose expression is significantly altered in strains overexpressing spo0A, compared to the control strain (1). As yet, the Spo0A regulons in B. subtilis and C. acetobutylicum have not been subjected to a detailed comparison.
In our effort to identify putative transcription factors that may interact with Spo0A or may affect the regulation of solventogenesis in C. acetobutylicum, we have adopted a comparative genomics approach using B. subtilis. Spo0A in B. subtilis has been shown to act in concert with several other transcription factors, including AbrB and SinR (14). abrB encodes a 94-amino-acid DNA-binding protein which is expressed during vegetative growth and is positively autoregulated by the binding of AbrB homotetramers to its promoter. The primary function of AbrB is to prevent the premature onset of sporulation by repressing the expression of several sporulation-associated genes (4, 33, 42, 43, 46). As the cell enters the transition phase between growth and sporulation, activated Spo0A binds to two 0A boxes in the abrB promoter independently of AbrB binding, causing repression of abrB expression (41).
For the purposes of this study, AbrB can be described as an inhibitor of sporulation that acts in opposition to Spo0A, but its functions are broader than this description implies. AbrB modulates catabolite repression in circumstances of carbon-source limitation and acts as both a positive and negative regulator of cell competence at various stages in the life cycle (11, 15). AbrB from B. subtilis also controls the expression of anthrax toxin genes, and the gene coding for the antibiotic tyrocidine when expressed in Bacillus anthracis and Bacillus brevis, respectively (37, 38).
In B. subtilis, sinR encodes a 111-amino-acid, helix-turn-helix DNA-binding protein that is transcribed through both vegetative growth and sporulation. In vivo, SinR forms homodimers and directly inhibits the expression of key initiators of sporulation, including spo0A (5, 25). SinR activity is repressed at the onset of sporulation by the binding of its antagonist, SinI, the expression of which is induced at the onset of sporulation (3). The sin operon is arranged such that sinI lies upstream of sinR. Upstream of sinI are two promoters, designated P1 and P2, and a third promoter, P3, is located upstream of sinR but downstream of sinI. P3 is constitutively active and is responsible for the continuous synthesis of SinR. P1 and P2 require functional Spo0A and σH (coded by spo0H) for correct expression; hence, the expression of sinI and the subsequent repression of SinR are under developmental control (3, 12, 13).
We identified homologues to abrB and sinR in C. acetobutylicum. Using both reporter vectors and primer extension techniques, we have characterized the expression of abrB and sinR in C. acetobutylicum. Subsequently, we have examined the effects of abrB and sinR misexpression on solvent production, sporulation, and cell morphology.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used are listed in Table 1.
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Relevant characteristics and description | Reference or source |
|---|---|---|
| Strains | ||
| C. acetobutylicum | ||
| ATCC 824 | Wild type | American Type Culture Collection, Manassas, Va. |
| E. coli | ||
| DH5α | mcrA, ΔmcrBC, recA1 | NEB, Beverly, Mass. |
| DH10β | mcrA, ΔmcrBC, recA1, Strr | NEB |
| TOP10 | mcrA, ΔmcrBC, recA1, Strr | Invitrogen |
| Plasmids | ||
| pDHKM | RSF1030ori, Kmr, Φ3tI | 47 |
| pTrcHisTOPO-TA | Apr, pBR322ori | Invitrogen |
| pTrc310pro | Apr, pBR322ori, abrB310 promoter | This study |
| pTrc1941pro | Apr, pBR322ori, abrB1941 promoter | This study |
| pTrc3647pro | Apr, pBR322ori, abrB3647 promoter | This study |
| pCATP | MLSr, repL, ColE1ori, catP | 38 |
| pCAT310 | MLSr, repL, ColE1ori, catP, abrB310 promoter | This study |
| pCAT1941 | MLSr, repL, ColE1ori, catP, abrB1941 promoter | This study |
| pCAT3647 | MLSr, repL, ColE1ori, catP, abrB3647 promoter | This study |
| pIMP1 | Apr, MLSr, ColE1ori, repL | 26 |
| pM310 | Apr, MLSr, ColE1ori, repL, abrB310 | This study |
| pMsinR | Apr, MLSr, ColElori, repL, sinR | This study |
| pSOS94 | ptb promoter, ctfA-ctfB, adc Apr, MLSr, ColE1ori, repL | 45 |
| pASsos | ptb promoter, Apr, MLSr, ColElori, repL | This study |
| pASsin | ptb promoter, sinRAS, Apr, MLSr, ColElori, repL | This study |
| pAS310 | ptb promoter, abrB310AS, Apr, MLSr, ColElori, repL | This study |
MLSr, macrolide-lincosamide streptogramin B resistant; mcrA, ΔmcrBC, methylcytosine-specific restriction system abolished; recA1, homologous recombination abolished; Strr, streptomycin resistant; RSF1030ori, gram-negative origin of replication; Kmr, kanamycin resistant; Φ3tI, Φ3t methylase; pBR322ori, gram-negative origin of replication; Apr, ampicillin resistant; repL, gram-positive origin of replication; ctfA-ctfB, coenzyme A transferase subunits A and B; adc, acetoacetate decarboxylase; sinRAS, sinR antisense construct; abrB310AS, sinR antisense construct; catP, promoterless CAT open reading frame.
Oligonucleotides.
All oligonucleotides used for PCR, primer extension, antisense vectors construction, or automated DNA sequencing are shown in Table 2.
TABLE 2.
Oligonucleotides used in this studya
| Oligonucleotide name | Sequence |
|---|---|
| 310rev | AGCGGATCCACAAACAATCACCTCTTAAAACAATTATAC |
| 310prom | CCGGAATTCTTTGAATTCCTCCTTAAATTACATA |
| 1941rev | AGCGGATCCACAATAGGGGATGAGAAAAATGCCTAG |
| 1941prom | CCGGAATTCTTTTAATTCCTCCTTTAATCTAATA |
| 3647rev | AGCGGATCCACAAATCATAATAAAGGAAAATACGCGATAC |
| 3647prom | CCGGAATTCTAATAAATACCTCCTAAAGTAACAAC |
| 310for | AGCGGATCCACATAATTTGAATATAATAAATAGACTGCCAAG |
| sinRrev | AGCGGATCCACACAATTTCTTCGCCTCCCTATAC |
| sinRfor | AGCGGATCCACATGTTATCAATCCATTCCATTAACATC |
| 310astop | GATCCCCTAATTCATCTACTCTTCTAACTACACCTGTTGATTTCATTTTGAATTCCTCCTAAAAGTAATTACATTAC |
| 310asbtm | CTAGGTAATGTAATTACTTTTAGGAGGAATTCAAAATGAAATCAACAGGTGTAGTTAGAAGAGTAGATGAATTAGGG |
| sinRastop | GATCCGTCATATAGAATTCGATTCTATCTTTGTTAAACATATAAAAGACCTCCGTAAAAGTAATTACATTAC |
| sinRasbtm | CTAGGTAATGTAATTACTTTTACGGAGGTCTTTTATATGTTTAACAAAGATAGAATCGAATTCTATATGACG |
| 310ext | TTAACTCTATAGGTATTACAATTCTACCTAATTC |
| sinext | AGATAACGTCGACTGACCTATATTAGCTTCTTT |
Underlined regions correspond to a restriction site for the restriction endonucleases BamHI (GGATCC) or EcoRI (GAATTC).
Growth conditions.
Escherichia coli was grown aerobically in Luria-Bertani medium at 37°C (27). For recombinant strains, liquid or agar-solidified medium was appropriately supplemented with ampicillin (100 μg/ml), erythromycin (ERY; 200 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (35 μg/ml). Strains were stored at −80°C in medium supplemented with 50% glycerol.
C. acetobutylicum was grown anaerobically in clostridial growth medium (CGM) at 37°C (18). For recombinant strains, liquid or agar-solidified medium was appropriately supplemented with ERY (40 μg/ml). Strains were stored as horse serum-supplemented lyophilized stocks at room temperature or at −80°C in medium supplemented with 10% glycerol. For the sporulation and morphology assays, strains were grown on agar-solidified clostridial basal medium (CBM) supplemented with ERY (40 μg/ml) at 37°C (32).
DNA isolation, manipulation, and transformation into C. acetobutylicum.
All commercial enzymes used in this study (Taq polymerase [Fisher Scientific, Pittsburgh, Pa.] and restriction endonucleases, calf intestinal phosphatase, T4 DNA ligase, and Klenow fragment of DNA polymerase I [New England Biolabs, Inc., Bethesda, Md.]) were used according to the manufacturers' recommendations.
Plasmids were purified from E. coli by using the QIAprep Miniprep protocols. DNA was purified from agarose gels by using a QIAquick gel extraction kit, and the PCR product or enzymatically manipulated DNA was purified by using a QIAquick PCR purification kit (QIAGEN, Inc., Valencia, Calif.). Plasmids were purified from C. acetobutylicum according to the protocol developed by Harris (16). Automated DNA sequencing was performed by automated DNA sequencing (LoneStar Laboratories Inc., Houston, Tex.).
Prior to transformation into C. acetobutylicum, plasmid was methylated by transformation of the required plasmid into E. coli DH10β harboring vector pDHKM (47). Electrotransformation of C. acetobutylicum was carried out according the protocol developed by Mermelstein et al. (26). All transformed strains were designated 824 followed by the transformed plasmid in parentheses.
Vector construction.
The entire region upstream of abrB310, abrB1941, and abrB3647, spanning from the base immediately preceding the start codon of the open reading frame to the last base prior to the open reading frame of the upstream gene, was amplified by PCR with the following primers: abrB310-310rev and 310prom, abrB1941-1941rev and1941prom, and abrB3647-3647rev and 3647prom. The promoter fragments were cloned into the pTrcHisTOPO-TA vector (Invitrogen, Carlsbad, Calif.). If the fragment was inserted into pTrcHisTOPO-TA in the required orientation, digestion with BamHI yielded the promoter fragment with cohesive ends. The reporter vector pCATP harbors a promoterless copy of the chloramphenicol acetyltransferase (CAT) gene, located downstream of a unique BamHI restriction site (39). Ligation of the promoter fragments into BamHI-digested pCATP formed the promoter reporter plasmids pCAT310, pCAT1941, and pCAT3647. The correct orientation of promoter insertion was confirmed by sequencing plasmids with the appropriate “rev” primer.
Primers 310rev and 310for or sinRrev and sinRfor were used to amplify by PCR the entire abrB310 or sinR open reading frames, including all upstream and downstream intergenic bases from C. acetobutylicum genomic DNA. The ∼0.8-kb abrB310 fragment and the ∼0.7-kb sinR fragment were digested by using BamHI and ligated into BamHI-digested pIMP1 (26) to form vectors pM310 and pMsinR, respectively. Plasmid constructs were confirmed by sequence analysis using both primers used in their construction.
Oligonucleotides 310astop and 310asbtm or sinRastop and sinRasbtm were used to create the abrB310 and sinR antisense constructs in plasmid pSOS94 according to the method previously described (39a, 45). Plasmids were designated pAS310 and pASsin, and the control plasmid pASsos was also used.
Batch fermentations of C. acetobutylicum.
Single colonies of transformed C. acetobutylicum were grown in closed-cap batch fermentations of 100 ml CGM supplemented with the appropriate antibiotic at 37°C in a Forma Scientific anaerobic chamber (Thermo Forma, Marietta, Ohio). To allow for differences in lag time following inoculation, zero hour (T0) was determined when the culture had reached an optical density at 600 nm (OD600) of 0.1. Fermentations were allowed to proceed for 120 h. The acid and solvent content of each sample was analyzed by gas chromatography as previously described (39).
CAT assays.
In strains harboring vectors pCATP, pCAT310, pCAT1941, or pCAT3647, samples of 5 ml were taken at 8, 18, and 24 h after T0 was reached, which corresponded to mid-acidogenic, early-solventogenic, and mid-solventogenic growth phases, respectively. For the 120-h fermentation of the strain harboring pCAT310, samples of 5 ml were taken at the time points indicated in Fig. 1.
FIG. 1.
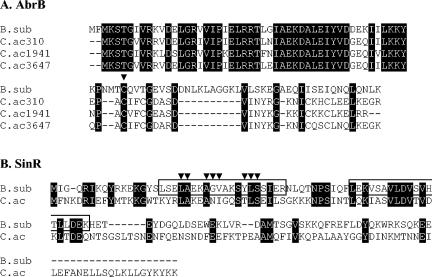
Alignment of the amino acid sequences of AbrB and SinR in B. subtilis and C. acetobutylicum. C.ac310, C.ac1941, and C.ac3647 refer to the three homologues of AbrB identified in C. acetobutylicum as defined by their position in the genome. Alignment was performed using the CLUSTALW tool of the Biology Workbench (San Diego Supercomputer Center). White letters on black background, single, fully conserved residue in all homologues; −, position with no amino acid present (23, 31). (A) Cysteine-56 in the AbrB homologues is indicated by the symbol ▾. (B) The helix-turn-helix DNA-binding domain of sinR in B. subtilis is shown in the first box, with key residues indicated by the symbol ▾ (13). The second boxed region corresponds to the multimerization domain of sinR in B. subtilis (3). B.sub, B. subtilis; C.ac, C. acetobutylicum.
For all samples, the OD600 was recorded, the product formation was assayed, and the sample was prepared and assayed for CAT expression as previously described (39). CAT expression was quantified as units of CAT per milligram of protein in the extract.
Primer extensions.
For mRNA collection, it was determined that the transcripts of both sinR and abrB310 were most abundant in cells in the mid- to late acidogenic phase (as determined by OD600 readings and acid-solvent concentrations). Samples of 10 ml were collected and centrifuged at 6,000 rpm (4,350 × g) at 4°C for 5 min with the JA-20 rotor of a Beckman J2-21 centrifuge. Cell pellets were resuspended in 200 μl of lysis buffer (20 mg of lysozyme/ml of 25% sucrose-0.05 M Tris-HCl-0.05 M EDTA buffer) and incubated at 37°C for 5 min. One milliliter of ice-cold TRIzol solution (Invitrogen) and 0.3 ml of chloroform were added, and the lysis solution was mixed vigorously for 15 seconds and then centrifuged at 16,000 × g at 4°C for 15 min. The aqueous upper layer containing RNA (∼1 ml) was collected, 0.5 ml of ice-cold isopropanol was added, and the solution was centrifuged for a further 10 min as before. The RNA pellet was washed in 1 ml of ice-cold 70% ethanol and air dried prior to resuspension in 22 μl of water.
Primer extension reactions were performed with primers 310ext or sinext by using an AMV reverse transcriptase primer extension system (Promega, Madison, Wis.). To determine the start sites of abrB310 and sinR transcription, vectors pMsinR and pM310 were sequenced by using sinext and 310ext, respectively. Sequencing, based on the Sanger dideoxy method, was performed by using a Thermo Sequenase radiolabeled terminator cycle sequencing kit (USB Corp., Cleveland, Ohio). Sequencing and primer extension products were then subjected to electrophoresis on a 6% acrylamide gel using the Otter sequencing system (Owl Separation Systems, Portsmouth, N.H.). Sequencing gels were run at 45°C at 1,100 V for 3 h. All reactions and sequencing were performed according to the manufacturers' instructions.
Morphology and sporulation assay.
Strains harboring pASsos and pAS310 were grown simultaneously in liquid medium and transferred at 16 to 24 h after T0 to CBM plates supplemented with ERY. At intervals after inoculation (24, 48, 72, and 140 h), cells were picked from the plates by using a sterile toothpick and resuspended in 20 μl of liquid CGM supplemented with 10% glycerol. Samples were frozen at −80°C until all samples had been collected.
Sample preparation and microscopy was carried out as already described (39a).
The following genomes were accessed as part of this study: B. subtilis, NC_000964; C. acetobutylicum, NC_003030; Clostridium perfringens, NC_003366; Bacillus cereus, NC_004722; B. anthracis, NC_003997; Bacillus halodurans, NC_002570; and Bacillus thuringiensis, NC_005957.
RESULTS
Identification of abrB and sinR homologues in C. acetobutylicum.
The initial identification of AbrB and SinR homologues was based on amino acid sequence homology between those proteins from B. subtilis and the predicted open reading frame sequences from C. acetobutylicum (23, 31). Figure 1 illustrates the alignment of the amino acid sequences of AbrB and SinR in B. subtilis and C. acetobutylicum, which was generated by using the CLUSTALW tool of the Biology Workbench (San Diego Supercomputer Center, University of California, San Diego, Calif.).
Three putative homologues of abrB were identified in C. acetobutylicum and were designated abrB310, abrB1941, and abrB3647 based upon their genome position and CAC notation (31). Amino acid sequence homology between the clostridial AbrBs and AbrB from B. subtilis is extremely high. The most homologous regions, exhibiting 72% identity and greater than 90% similarity, are the N-terminal 50 amino acids, which form the DNA-binding domain of AbrB in B. subtilis (33, 46). The C-terminal amino acids, which have been shown in B. subtilis to be necessary for AbrB homotetramerization, exhibit a lesser degree of similarity, although the cysteine residue at position 56, which is critical for correct multimerization, is conserved through all of the AbrB homologues (4, 46). All of the AbrB homologues in C. acetobutylicum as well as the putative AbrB homologue in C. perfringens (40) lack the 10-residue DNLKLAGGKL motif present in AbrB of B. subtilis. A similar 10-residue motif is present in other AbrB homologues identified in B. cereus, B. anthracis, B. halodurans, and Bacillus thuringiensis (21, 36, 44; T. S. Brettin, D. Bruce, J. F. Challacombe, P. Gilna, C. Han, K. Hill, P. Hitchcock, P. Jackson, P. Keim, J. Longmire, S. Lucas, R. Okinaka, P. Richardson, E. Rubin, and H. Tice, submitted for publication). The significance of these differences remains to be determined.
A single homologue of sinR (CAC0549) in C. acetobutylicum was identified, but no obvious homologue to sinI was identified either directly upstream of sinR or elsewhere on the genome (31). Excluding the C-terminal 20 amino acids in the C. acetobutylicum homologue, the sequence is 29% identical and 68% similar to the B. subtilis counterpart. Within the DNA-binding domain, there is strong homology between the essential hydrophobic residues required for the formation of the helix-turn-helix motif (13). Additionally, the multimerization domain sequence is highly homologous, although residues critical for multimerization have not been identified in B. subtilis (3).
CAT assays of abrB homologue promoters.
CAT reporter vector analysis of the promoters of abrB310, abrB1941, and abrB3647 was used to determine whether any of the abrB homologues in C. acetobutylicum were transcriptionally active. In strains 824(pCAT1941) or 824(pCAT3647), CAT activity did not differ significantly from that in the control strain 824(pCATP). In strain 824(pCAT310), CAT activity was elevated during all stages of growth that were assayed. This finding indicated that abrB310 is the active copy of abrB in C. acetobutylicum under the growth conditions tested.
Figure 2 shows CAT activity in strain 824(pCAT310) throughout the course of 120-h culture. During the first 12 to 16 h, CAT activity is detectable at a high basal level of 100 to 150 U of CAT/mg of protein. Between 12 and 24 h, CAT activity rapidly increases from approximately 100 U of CAT/mg of protein at 12 h to a maximum of approximately 285 U of CAT/mg of protein at 24 h. This increase in CAT activity is coincident with the onset of solventogenesis and the transition between the exponential and stationary phases of growth (data not shown in Fig. 2). Between 24 and 72 h, CAT activity declines at an approximate rate of 3 U of CAT/mg of protein per h. From 72 h to 120 h, CAT activity remains relatively constant, between 100 and 150 U of CAT/mg of protein.
FIG. 2.
CAT activity and solvent production in strain 824(pCAT310). ▪, CAT activity; ▵, acetone; and ◊, butanol. Other products and OD600 are omitted for clarity. Data are shown ± 1 standard error; n = 9.
Primer extension analysis of abrB310 and sinR expression.
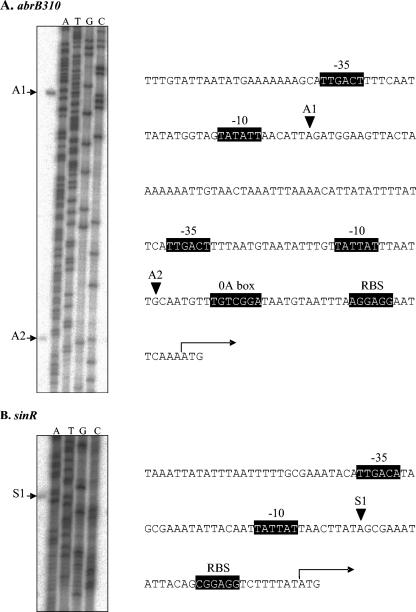
The primer extension products from abrB310 and sinR, sampled at the mid- to late acidogenic phase, are shown beside the appropriate sequencing gels to indicate the transcription start site of each gene in Fig. 3. There are two detectable transcripts of abrB310, with the larger transcript (A1) being approximately 10 times more abundant than the smaller transcript (A2). For sinR, there is a single weak transcript detectable at approximately one-quarter the intensity of the smaller transcript of abrB310.
FIG. 3.
Transcription start sites and promoter region sequence of abrB310 and sinR. The transcripts identified by primer extension for abrB310 (panel A) and sinR (panel B) correspond to the transcription start sites shown on the sequences. The sequencing reaction products (lanes A, T, G, and C) were generated with the same primers as those used for the extension reactions. Also identified within the promoter region sequences are putative −35 and −10 consensus motifs, a potential binding site for Spo0A in abrB310 (0A box) and ribosome-binding sites (RBS). The arrows on both sequences indicate the first codon (ATG) of the open reading frame.
Figure 3 also illustrates the location of the transcription start sites of abrB310 (A1 and A2) and sinR (S1) and putative regulatory sequences within the promoter region sequences of abrB310 and sinR. Upstream of both transcription start sites of abrB310, −10 and −35 consensus sequences are readily identifiable, with a potential 0A box located downstream of the second transcription start site (A2). There are typical −10 and −35 consensus sequences located upstream of the transcription start site of sinR, but there is no obvious 0A box located anywhere within the sinR promoter. Likely ribosome-binding sites are located approximately 10 bases upstream of the start codon of the open reading frames of both abrB310 and sinR.
Phenotypic effects of altered sinR expression.
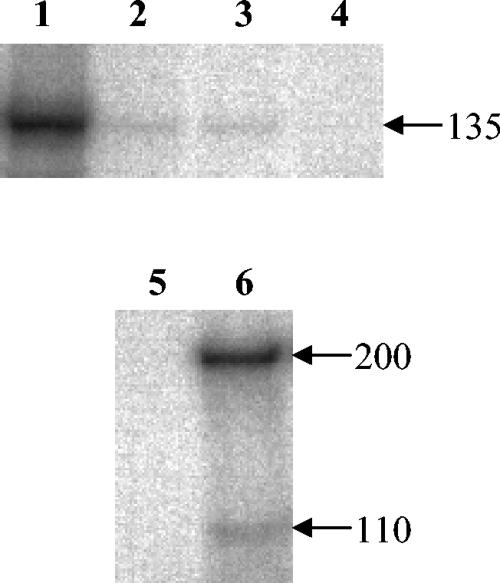
mRNA extracted from the control strains 824(pIMP1) and 824(pASsos), strain 824(pASsin) harboring the sinR antisense construct and the sinR overexpression strain 824(pMsinR) was probed with the sinext primer for sinR transcript. Figure 4 shows that a single weak transcript was detected in the control strains, no transcript could be detected in 824(pASsin), and a significantly elevated level of the sinR transcript was detected in 824(pMsinR). Throughout the 120-h culture, there were no significant differences in growth or acid and solvent production between the control strains and 824(pASsin) or 824(pMsinR) (data not shown).
FIG. 4.
Transcript levels in strains 824(pMsinR), 824(pASsin) and 824(pAS310). sinR transcript was probed with primer sinext in mRNA extracted from the following strains: lane 1, 824(pMsinR); lane 2, 824(pIMP1); lane 3, 824(pASsos); and lane 4, 824(pASsin). abrB310 transcript was probed with primer 310ext in mRNA extracted from the following strains: lane 5, 824(pAS310); and lane 6, 824(pASsos). Arrows indicate the approximate sizes of transcripts (in bases).
Phenotypic, morphological, and sporulation effects of decreased abrB310 expression.
Despite repeated attempts, plasmid pM310 could not be successfully transformed into C. acetobutylicum. Fig. 4 shows that in strain 824(pAS310), both transcripts of abrB310 were absent compared to the control strain 824(pASsos).
Growth and product formation in strains 824(pASsos) and 824(pAS310) are shown in Fig. 5. In both strains, growth is similar, with a peak OD600 of approximately 4.5 after 16 h of exponential growth, followed by a slight decline throughout the remainder of the 120-h fermentation.
FIG. 5.
Growth and product formation in fermentations of strains 824(pASsos) and 824(pAS310). The measured quantities for each profile of strain 824(pASsos) (□) and strain 824(pAS310) (▪) are shown. Data are shown ± 1 standard error. For each data point, n = 4.
Strain 824(pAS310) produces significantly higher levels of acetate and butyrate than the control strain. For both acetate and butyrate, maximum concentrations of approximately twice those in the control strain are observed in fermentations of 824(pAS310) after 16 to 24 h. Acetone and butanol production was slower in 824(pAS310) than that in the control strain, and the maximum concentration of ethanol, acetone, and butanol in 824(pAS310) was less than half of that in the control strain.
After 24 h of culture on CBM, the identification of cells of both 824(pASsos) and 824(pAS310) in all stages of division was possible. After 48 h, the occasional sporulating cell was present in the 824(pASsos) samples, but none was observed in samples of 824(pAS310). After 72 h, approximately 50% of the cells in the 824(pASsos) samples exhibited the typical, swollen morphology associated with sporulation, whereas no sporulating cells could be seen in any of the 824(pAS310) samples. After 140 h, both samples were composed of either sporulating cells or free endospores.
DISCUSSION
Following the completion of the C. acetobutylicum genome sequencing project, gene CAC3647 was identified as the functional copy of abrB in C. acetobutylicum, with CAC310 and CAC1941 designated abrB homologues (31). The basis for making this distinction is not clear, but all of the data presented here show that abrB310 is the functional copy of abrB. It is proposed that the two additional, apparently nonexpressed copies of abrB within the genome of C. acetobutylicum are pseudogenes.
The CAT assay data from the 120-h fermentation of strain 824(pCAT310) show a temporally specific upregulation of expression of abrB310 at the transition from acidogenesis to solventogenesis. However, beyond 24 h after T0, it is difficult to interpret the results due to high CAT concentrations. The CAT protein itself is relatively stable, hence its use in these assays, and it is possible that large amounts of residual CAT are not removed from the system rapidly enough for changes in transcriptional activity to be detected. This problem may be compounded by the cessation of growth and general decrease in cell metabolism during mid- and late solventogenic phases, which may further reduce the rate at which CAT is removed from the cell. It therefore seems likely that changes in the activity of the abrB310 promoter during mid- and late growth phases are masked by high background levels of CAT.
In B. subtilis, primer extensions performed on abrB also revealed two transcription start sites, and other regulatory elements within the abrB promoter have been identified (33). However, the transcription start sites are located only 13 bases apart, with two 0A boxes located downstream of the second site which act to repress transcription in response to spo0A binding at the onset of sporulation (41). Conversely, in C. acetobutylicum, the two transcription start sites of abrB310 are located 89 bases apart, with a single putative 0A box sequence located downstream of the A2 site. No obvious secondary structure was observed with in silico analysis of the 5′end of both transcripts of abrB310.
In stark contrast to the high expression of abrB310, primer extension showed that sinR is expressed weakly in C. acetobutylicum throughout the first 24 h of culture. Nevertheless, it is possible to identify likely promoter elements surrounding the sinR transcription start site, although it does not form part of a sin operon similar to sinR and sinI in B. subtilis. Results indicate that although sinR expression can be significantly modified in C. acetobutylicum, it has no effect on solventogenesis.
Although it is not possible to determine exactly why pM310 would not transform into C. acetobutylicum, it is possible that the overexpression of abrB310 is lethal to the cell. The CAT assay and primer extension studies showed high endogenous expression of abrB310, and the amount of AbrB310 produced from the plasmid pM310, which has an expected copy number of 8 per cell, may have been too great for the cell to survive (24, 26).
Strain 824(pAS310), which expresses no abrB310 transcript, is delayed in both the onset of solventogenesis and the conversion of acetate and butyrate into acetone and butanol. The high levels of acetate and butyrate throughout the fermentation of 824(pAS310) is similar to that observed in other nonsolventogenic mutants, such as M5 and SKO1, where the pathways to convert acids to solvents are not functional and the acids therefore accumulate (16, 30). It was also found that although strain 824(pAS310) was delayed in sporulation, the morphology of both the sporulating cells and the spores was normal.
Our data show that despite high amino acid sequence homology between AbrB310 in C. acetobutylicum and AbrB in B. subtilis, the mode of action of AbrB is subtly different between the species. In B. subtilis, increased levels of activated Spo0A cause abrB expression to be repressed at the transition between vegetative growth and sporulation (41). In C. acetobutylicum, abrB310 expression is elevated at the transition between vegetative acidogenic growth and the stationary phase of solventogenesis. When abrB310 expression is disrupted in strain 824(pAS310), the onset of solventogenesis is delayed, suggesting that AbrB plays a positive role in expediting this transition in growth phases.
In strain 824(pAS310), the onset of sporulation was delayed. Despite this delay, spores appear to develop normally, thus implying that the absence of AbrB310 does not have negative effects on sporulation. It is possible that at the transition to sporulation, AbrB310 may have a similar role to AbrB in B. subtilis in that abrB is no longer transcribed to allow the derepression of sporulation-related genes (29, 41). We are currently continuing our investigations into the relationship between abrB310 expression and the regulation of the transitions between the growth phases in C. acetobutylicum.
Acknowledgments
This work was supported by U.S. Department of Agriculture grant 00-35504-9269, National Science Foundation grants BES-0418289 and BES-0001288, and Robert A. Welch Foundation grants C-1268 (G.N.B.) and C-1372 (F.B.R.).
Footnotes
This paper is dedicated to the memory of Frederick B. Rudolph.
REFERENCES
- 1.Alsaker, K. V., T. R. Spitzer, and E. T. Papoutsakis. 2004. Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress. J. Bacteriol. 186:1959-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Awang, G. M., G. A. Jones, and W. M. Ingledew. 1988. The acetone-butanol-ethanol fermentation. Crit. Rev. Microbiol. 15(Suppl. 1):S33-S67. [DOI] [PubMed] [Google Scholar]
- 3.Bai, U., I. Mandic-Mulec, and I. Smith. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 7:139-148. [DOI] [PubMed] [Google Scholar]
- 4.Benson, L. M., J. L. Vaughn, M. A. Strauch, B. G. Bobay, R. Thompson, S. Naylor, and J. Cavanagh. 2002. Macromolecular assembly of the transition state regulator AbrB in its unbound and complexed states probed by microelectrospray ionization mass spectrometry. Anal. Biochem. 306:222-227. [DOI] [PubMed] [Google Scholar]
- 5.Cervin, M. A., R. J. Lewis, J. A. Brannigan, and G. B. Spiegelman. 1998. The Bacillus subtilis regulator SinR inhibits spoIIG promoter transcription in vitro without displacing RNA polymerase. Nucleic Acids Res. 26:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornillot, E., R. V. Nair, E. T. Papoutsakis, and P. Soucaille. 1997. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J. Bacteriol. 179:5442-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durre, P., M. Bohringer, S. Nakotte, S. Schaffer, K. Thormann, and B. Zickner. 2002. Transcriptional regulation of solventogenesis in Clostridium acetobutylicum. J. Mol. Microbiol. Biotechnol. 4:295-300. [PubMed] [Google Scholar]
- 8.Ferrari, F. A., K. Trach, D. LeCoq, J. Spence, E. Ferrari, and J. A. Hoch. 1985. Characterization of the spo0A locus and its deduced product. Proc. Natl. Acad. Sci. USA 82:2647-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feustel, L., S. Nakotte, and P. Durre. 2004. Characterization and development of two reporter gene systems for Clostridium acetobutylicum. Appl. Environ. Microbiol. 70:798-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer, R. J., J. Helms, and P. Durre. 1993. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J. Bacteriol. 175:6959-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher, S. H., M. A. Strauch, M. R. Atkinson, and L. V. Wray, Jr. 1994. Modulation of Bacillus subtilis catabolite repression by transition state regulatory protein AbrB. J. Bacteriol. 176:1903-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaur, N. K., K. Cabane, and I. Smith. 1988. Structure and expression of the Bacillus subtilis sin operon. J. Bacteriol. 170:1046-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaur, N. K., E. Dubnau, and I. Smith. 1986. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J. Bacteriol. 168:860-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grossman, A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477-508. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, J., M. Roggiani, and D. Dubnau. 1995. The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J. Bacteriol. 177:3601-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, L. M. 2001. Cloning and characterization of the Clostridium acetobutylicum ATCC824 gene encoding the Spo0A transcription regulator and its role in controlling solvent formation and sporulation-specific gene expression. PhD dissertation. Northwestern University, Evanston, Ill.
- 17.Harris, L. M., N. E. Welker, and E. T. Papoutsakis. 2002. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 184:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartmanis, M. G. N., and S. Gatenbeck. 1984. Intermediary metabolism in C. acetobutylicum: levels of enzymes involved in the formation of acetate and butyrate. Appl. Environ. Microbiol. 47:1277-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoch, J. A. 1993. The phosphorelay signal transduction pathway in the initiation of Bacillus subtilis sporulation. J. Cell. Biochem. 51:55-61. [DOI] [PubMed] [Google Scholar]
- 20.Hoch, J. A. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441-465. [DOI] [PubMed] [Google Scholar]
- 21.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T., and D. R. Woods. 1986. Acetone-butanol fermentation revisited. Microbiol. Rev. 50:484-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Lee, S. Y., L. D. Mermelstein, and E. T. Papoutsakis. 1993. Determination of plasmid copy number and stability in Clostridium acetobutylicum ATCC 824. FEMS Microbiol. Lett. 108:319-323. [DOI] [PubMed] [Google Scholar]
- 25.Mandic-Mulec, I., L. Doukhan, and I. Smith. 1995. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J. Bacteriol. 177:4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermelstein, L., N. Welker, G. Bennett, and E. Papoutsakis. 1992. Expression of cloned homologous fermentation genes in Clostridium acetobutylicum ATCC 824. Bio/Technology 10:190-195. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Mitchell, W. J. 1998. Physiology of carbohydrate to solvent conversion by clostridia. Adv. Microb. Physiol. 39:31-130. [DOI] [PubMed] [Google Scholar]
- 29.Molle, V., M. Fujita, S. T. Jensen, P. Eichenberger, J. E. Gonzalez-Pastor, J. S. Liu, and R. Losick. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683-1701. [DOI] [PubMed] [Google Scholar]
- 30.Nair, R. V., and E. T. Papoutsakis. 1994. Expression of plasmid-encoded aad in Clostridium acetobutylicum M5 restores vigorous butanol production. J. Bacteriol. 176:5843-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Brien, R. W., and J. G. Morris. 1971. Oxygen and the growth and metabolism of Clostridium acetobutylicum. J. Gen. Microbiol. 68:307-318. [DOI] [PubMed] [Google Scholar]
- 33.Perego, M., G. B. Spiegelman, and J. A. Hoch. 1988. Structure of the gene for the transition state regulator, AbrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol. Microbiol. 2:689-699. [DOI] [PubMed] [Google Scholar]
- 34.Petersen, D. J., J. W. Cary, J. Vanderleyden, and G. N. Bennett. 1993. Sequence and arrangement of genes encoding enzymes of the acetone-production pathway of Clostridium acetobutylicum ATCC824. Gene 123:93-97. [DOI] [PubMed] [Google Scholar]
- 35.Ravagnani, A., K. C. Jennert, E. Steiner, R. Grunberg, J. R. Jefferies, S. R. Wilkinson, D. I. Young, E. C. Tidswell, D. P. Brown, P. Youngman, J. G. Morris, and M. Young. 2000. Spo0A directly controls the switch from acid to solvent production in solvent-forming clostridia. Mol. Microbiol. 37:1172-1185. [DOI] [PubMed] [Google Scholar]
- 36.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, J. B., M. Gocht, M. A. Marahiel, and P. Zuber. 1989. AbrB, a regulator of gene expression in Bacillus, interacts with the transcription initiation regions of a sporulation gene and an antibiotic biosynthesis gene. Proc. Natl. Acad. Sci. USA 86:8457-8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scotcher, M. C., K. X. Huang, M. L. Harrison, F. B. Rudolph, and G. N. Bennett. 2003. Sequences affecting the regulation of solvent production in Clostridium acetobutylicum. J. Ind. Microbiol. Biotechnol. 30:414-420. [DOI] [PubMed] [Google Scholar]
- 39a.Scotcher, M. C., and G. N. Bennett. 2005. SpoIIE regulates sporulation but does not directly affect solventogenesis in Clostridium acetobutylicum ATCC 824. J. Bacteriol. 187:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strauch, M., V. Webb, G. Spiegelman, and J. A. Hoch. 1990. The SpoOA protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. USA 87:1801-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strauch, M. A., M. Perego, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol. Microbiol. 3:1203-1209. [DOI] [PubMed] [Google Scholar]
- 43.Strauch, M. A., G. B. Spiegelman, M. Perego, W. C. Johnson, D. Burbulys, and J. A. Hoch. 1989. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 8:1615-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterization of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughn, J. L., V. Feher, S. Naylor, M. A. Strauch, and J. Cavanagh. 2000. Novel DNA binding domain and genetic regulation model of Bacillus subtilis transition state regulator abrB. Nat. Struct. Biol. 7:1139-1146. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, Y., L. A. Nurman, A. Chuang, M. Monroe-Augustus, M. Lyristis, M. L. Harrison, F. B. Rudolph, and G. N. Bennett. 2003. Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 69:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]