Abstract
Comparative analysis of bacterial diversity in freshwater sediment collected from a shallow eutrophic lake was performed by using 16S rRNA gene clone library and improved cultivation-based techniques. Our study demonstrated that the use of gellan gum as a gelling reagent instead of agar was more effective at increasing culturability, cultivating a diverse array of novel microbes, and reducing the gaps of the results between molecular and cultivation-based analyses.
Prokaryotes are among the most important contributors to the transformation of complex organic compounds and minerals in freshwater sediments (22, 34). Therefore, investigating microbial structure and function in freshwater sediments is of great importance for gaining a better general understanding of aquatic ecosystems.
Although efforts have been made to reveal the microbial ecosystems in freshwater sediments on the basis of traditional cultivation methods (21, 45), it is now widely recognized that only 0.001 to 15% (0.25% in sediments) of the total cell counts in environmental samples can be cultured (3). Hence, the traditional cultivation methods cannot be directly applied to whole microbial diversity analyses. However, cultivation-based study remains important, since the ecological role of prokaryotes in natural environments can be estimated only when they are successfully cultivated and characterized.
The culture-independent molecular approaches based on small-subunit rRNA have also been used for studies of microbial ecology in freshwater sediments (1, 31, 38, 39, 45). However, information on microbial compositions in freshwater sediments is limited, since previous studies have mainly focused on some particular functional groups such as nitrifiers, denitrifiers, sulfate reducers, methanogens, and methanotrophs (8, 16, 29, 38, 45, 49). There are several studies on overall bacterial communities in freshwater sediments (33, 45, 50), but all of them are based solely on molecular analyses.
In this study, we performed comparative analyses of the bacterial diversity of freshwater sediment of a shallow eutrophic lake on the basis of 16S rRNA gene analysis and modified cultivation techniques that improved the culturability of hitherto unknown species present in the ecosystem. In particular, we demonstrated the effectiveness of using gellan gum as a gelling reagent in a culture-dependent analysis of bacterial diversity in freshwater sediment.
Sampling site and sediment samples.
Sediment samples were collected with an Eckman grabber from Lake Kasumigaura (36°08.07′N, 140°20.62′E), an important reservoir of water and a typical eutrophic freshwater lake in Japan, on 30 November 2002. Lake Kasumigaura is the second largest lake (lake area, 219.9 km2; catchment area, 2,135 km2) in Japan, and it is sufficiently shallow (maximum depth, 7 m; average, 4 m) that the biological processes in the sediment exert a significant effect on the aquatic environment. The temperature, pH, concentration of dissolved oxygen, and turbidity of the water column at the sampling site were 7°C, 8.1, 7.2 mg/liter, and 65 mg/liter, respectively. After the sampling, the sediment samples (depth, 0 to 10 cm) were cooled immediately on ice and stored at −20°C for molecular analysis. The analytical data of the sediment were as follows: water content, 68.1%; pH 6.4; ignition loss, 18.1%; total carbon, 5.04%; total nitrogen, 0.58%; particle classification, silty clay (7% sand, 52% silt, 41% clay).
Phylogenetic analysis of bacterial diversity based on a 16S rRNA gene clone library.
Total nucleic acids were extracted and purified from the sediment as described by Purdy et al. (37) and Koizumi et al. (27). The PCR amplification of 16S rRNA genes (1,175 bp, Escherichia coli positions 338 to 1513) from the purified genomic DNA was carried out with bacterial universal primers: mixtures of EUB338, EUB338I, EUB338II, and EUB338III designated by Daims et al. (9) for recovering almost all known bacterial lineages, and 1492R (28). The PCR conditions were as follows: initial denaturation at 95°C for 9 min, followed by 18 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 2 min. To minimize the PCR bias (25, 46), the number of PCR cycles was decreased to 18 (43, 48). The purified rRNA gene fragments were cloned with a pT7 Blue T-vector kit (Novagen). The clonal DNAs were amplified from randomly selected recombinants by direct PCR with M13 primers (M4, 5′-GTTTTCCCAGTCACGAC-3′; RV, 5′-CAGGAAACAGCTATGAC-3′), purified with a MicroSpin S-400 HR column, and then used as templates for sequencing. Sequencing was performed with primer 907R (5′-CCGYCAATTCMTTTRAGTTT-3′), a DTCS-Quick Start kit (Beckman), and a CEQ-2000 automated sequence analyzer (Beckman).
The sequences of all bacterial 16S rRNA gene clones with a range of about 500 to 600 bases were determined. All sequences were compared with those in the GenBank database (www.ncbi.nlm.nih.gov/BLAST) by using the BLAST program (2). To detect and omit chimeric DNAs, the CHECK-CHIMERA program (32) of the Ribosomal Database Project was used. Phylogenetic analysis was performed with the ARB program package (http://www.arb-home.de/). All clonal sequences and the reference sequences from the GenBank database were imported into a database of the ARB program. After automatic and manual sequence alignment, phylogenetic trees were constructed by the neighbor-joining method (41) and bootstrap analyses for 1,000 replicates were performed.
A total of 112 clones from the 16S rRNA gene library were analyzed in order to estimate the bacterial diversity in the freshwater sediment (Table 1). The sequence analysis grouped the clones into 86 distinct types (i.e., sequences with greater than 97% similarity were treated as identical). The coverage value derived from the equation described by Giovannoni (15) was 23.2%. On the basis of the phylogenetic analysis, the clonal sequences were affiliated with at least 10 classes of the domain Bacteria. The most dominant group of our clone library was allocated to the phylum Proteobacteria (47% of the total number of clones), and the dominant organisms within this class were in the delta, beta, and gamma classes (23.2, 12.5, and 9.8%, respectively). The second most dominant group of the clone library, represented by 15 clones (13.4%), was classified into the phylum Nitrospira. The other groups of the library were determined to be, in order of abundance, in the phyla Acidobacteria, Chloroflexi, Bacteroidetes, Chlorobi, Planctomycetes, Actinobacteria, Verrucomicrobia, and Cyanobacteria. Some clones affiliated with the candidate phyla WS3 and OP8, which were represented solely by environmental clones (11, 19), were also observed.
TABLE 1.
Phylogenetic affiliations of 16S rRNA gene clones belonging to the domain Bacteria
| Phylum | No. of clones | % of total | No. of typesa |
|---|---|---|---|
| Proteobacteria | |||
| Alpha class | 2 | 1.8 | 2 |
| Beta class | 14 | 12.5 | 12 |
| Gamma class | 11 | 9.8 | 7 |
| Delta class | 26 | 23.2 | 20 |
| Nitrospira | 15 | 13.4 | 6 |
| Acidobacteria | 9 | 8.0 | 8 |
| Bacteroidetes | 7 | 6.3 | 6 |
| Chlorobi | 5 | 4.5 | 4 |
| Actinobacteria | 2 | 1.8 | 2 |
| Cyanobacteria | 1 | 0.9 | 1 |
| Verrucomicrobia | 1 | 0.9 | 1 |
| Planctomycetes | 3 | 2.7 | 3 |
| Chloroflexi | 8 | 7.1 | 8 |
| OP8 | 1 | 0.9 | 1 |
| WS3 | 2 | 1.8 | 1 |
| Unidentified | 4 | 3.6 | 4 |
| Total | 112 | 100 | 86 |
An identical type was defined as a group of sequences with >97% similarity.
Sequences allocated to the delta and beta groups of the phylum Proteobacteria (delta and beta Proteobacteria, respectively) have also frequently been retrieved from freshwater sediments in previous studies (33, 45, 50). In particular, delta Proteobacteria has been indicated as the representative bacterial lineage in benthic environments, since this group was more frequently recovered from sediments than from water columns, in which alpha, beta, and gamma Proteobacteria; Bacteroidetes; and Actinobacteria were observed as the dominant groups (18, 30, 45). This finding may be due to the oxidation-reduction potential gradient between the water and sediment environments. In fact, our study, as well as previous reports (29, 39, 45), frequently detected clones moderately related to the strict anaerobes, such as sulfate reducers (the genera Desulfococcus, Desulfomonile, and Desulfonema) and syntrophic bacteria (e.g., members of the genus Syntrophus), within delta Proteobacteria (supplemental Fig. A2 [all of the supplemental material cited in this report is available online at http://staff.aist.go.jp/tamaki-hideyuki/]).
There was no clone whose 16S rRNA gene sequence was 100% identical to those of known bacterial species. Only three clones had more than 97% sequence similarity to known bacterial species. One such clone, KTS75, was closely related to Pelobacter propionicus within delta Proteobacteria (supplemental Fig. A2) (45). The remaining two clones, KTS29 and KTS25, were affiliated with Skeletonema pseudocostatum within Cyanobacteria and Novosphingomonas subarcticum within alpha Proteobacteria. In addition, there were only three clones (KTS104, KTS6, and KTS9) having more than 95% sequence similarity to the previously described species (supplemental Fig. A1 and A3). Apart from the above-mentioned six clones, all of the remaining clones showed less than 95% 16S rRNA gene sequence similarity to any other identified species. The majority of the remaining clones were related to previously described uncultured environmental clone clusters, i.e., RBF8 within beta Proteobacteria (4), subclass 6 within Acidobacteria, and subclass 1 within Chloroflexi (19). Some other clones formed clusters (designated KTS II, KTS III, and KTS IV) with some previously known environmental sequences: KTS II within gamma Proteobacteria, KTS III within Nitrospira, and KTS IV within Bacteroidetes (supplemental Fig. A1 and A3). Moreover, some of the remaining clone sequences were not closely related to any of the previously published environmental sequences and thus formed independent unique clusters, designated KTS I and KTS V, which are affiliated with delta Proteobacteria and Chlorobi, respectively (supplemental Fig. A2 and A3[b]).
These results indicate that the bacterial community in the freshwater sediment of Lake Kasumigaura is remarkably diverse and is primarily composed of unknown bacterial species. Therefore, cultivation-based study was necessary to reveal the function of the unknown bacteria, as determined by 16S rRNA gene clone analysis.
Direct enumeration of microbial cells in sediment samples.
Direct counting of visible cells in sediment samples was performed under an epifluorescence microscope with ethidium bromide staining, which allowed a significant reduction in the autofluorescence background derived from sediment particles. The number of microbial cells was calculated to be 8.37 × 109/g (dry weight) of sediment (standard deviation [SD] = 3.12 × 108 cells per g).
Cultivation of sediment microbes with agar- and gellan gum-based media.
In this study, to capture a diverse array of microorganisms, we focused on three factors: constituents of the nutrient media (PE03 and DR2A), pH conditions (pHs 5.5, 6.0, and 7.0), and gelling reagents (agar and gellan gum). Moderately low-nutrient media DR2A and PE03 were used for cultivation. The composition of DR2A was as follows (per liter): 0.05 g each of yeast extract, peptone, acid hydrolysate of casein, dextrose, and soluble starch; 0.03 g each of dipotassium phosphate and sodium pyruvate; and 0.0024 g of magnesium sulfate. The composition of PE03 was as follows (per liter): 0.05 g each of sodium glutamate, sodium succinate, sodium acetate, yeast extract, Casamino Acids, sodium thiosulfate, and ammonium sulfate; 5 ml of basal salts solution (17); and 0.2 ml of vitamin solution (17). The pH values of these media were adjusted with 10 mM potassium phosphate buffer to 5.5, 6.0, and 7.0. As gelling reagents, gellan gum (Wako, Tokyo, Japan) and agar (Noble agar; Difco) were used for solidification of the media at final concentrations of 1.0 and 1.5%, respectively. The media were designated PE03-7G, PE03-6G, and PE03-55G for PE03 media solidified with gellan gum (pHs 7.0, 6.0, and 5.5, respectively). PE03 agar media (pH 7.0, 6.0, and 5.5) were designated PE03-7A, PE03-6A, and PE03-55A, respectively. In the same manner, the DR2A media were referred to as DR2A-7G, DR2A-6G, DR2A-55G, DR2A-7A, DR2A-6A, and DR2A-55A, respectively.
Sediment samples (10 g) were suspended in sterile water and diluted in 10-fold steps. A series of medium plates (12 different types, as described above) was inoculated with 100-μl aliquots from different dilutions and incubated at 20°C for 11 weeks in the dark under aerobic and anaerobic conditions. Anaerobic cultivation was performed with an AnaeroPack system (Mitsubishi Gas Chemical, Tokyo, Japan).
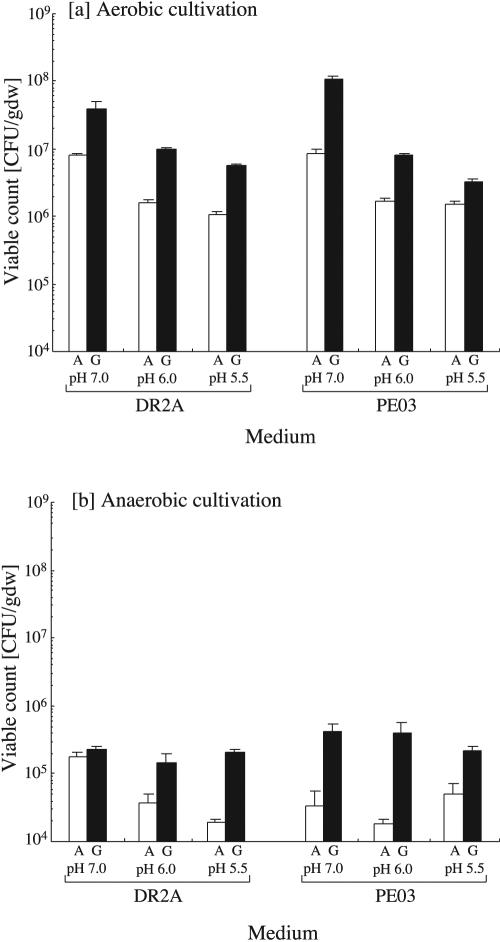
The number of CFU on all types of media continuously increased for more than 8 weeks. The viable counts shown in Fig. 1 were determined after 11 weeks of cultivation. Under aerobic conditions, viable counts from all of the types of gellan gum-based media used were 2.2 to 12.6 times greater than those from all of the agar media used (Fig. 1a). As regards the pH conditions, the viable counts tended to decrease as the pH decreased. The composition of the media had little influence on the viable counts. The maximum viable count was obtained on PE03-7G (gellan gum) and reached 1.10 × 108 CFU/g (dry weight) of sediment (SD = 1.14 × 107 CFU/g, 1.3% of the mean microscopically determined total cell count). This value was 12.6 times higher than the viable count, 8.68 × 106 CFU/g (SD = 1.23 × 106 CFU/g, 0.1% of the total cell count), as determined by using the same medium solidified with agar, PE03-7A. Likewise, under anaerobic conditions, the viable counts from all of the gellan gum media were also 1.3 to 21.3 times greater than those from all of the agar media (Fig. 1b). The effects of pH and the composition of the media on the viable counts were not remarkable. In general, the viable counts observed under anaerobic conditions were lower than those obtained under aerobic conditions by 2 to 3 orders of magnitude. Although the maximum viable count under anaerobic conditions was found on PE03-7G, the value was only 4.15 × 105 CFU/g (SD = 1.33 × 105 CFU/g).
FIG. 1.
Effects of medium composition, pH conditions, and gelling reagents on viable counts after 11 weeks of incubation under aerobic (a) and anaerobic (b) conditions. White and black bars represent CFU counts from agar-based (A) and gellan gum-based (G) media, respectively. The vertical bars indicate 1 SD from the mean.
In the previous studies of soil ecosystems, Sait et al. (40) reported only slight differences between the viable counts from gellan gum-based and agar-based media. Janssen et al. (20) demonstrated that the CFU counts obtained from gellan gum media were higher than those obtained from agar media. However, the differences in CFU counts between the two types of media in our study were much higher than those observed in their studies and were statistically significant (P < 0.05) on the basis of Student's t test except for the result obtained at pH 5.5 under anaerobic conditions. The use of gellan gum as a gelling reagent can be more effective than that of agar in improving the culturability of sediment microbes.
Phylogenetic distribution of cultivated sediment microbes.
Identification of the microbes grown on the media was performed on the basis of the 16S rRNA gene sequences. Thirty colonies were randomly selected from all of the medium types with gellan gum, and their 16S rRNA gene sequences were determined. 16S rRNA genes were directly PCR amplified from single colonies grown on plates with bacterial universal primers 8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (28). The 16S rRNA gene sequences determined were compared with those in the GenBank database by using the BLAST program (2). Furthermore, to investigate the effect of gelling reagents on the spectrum of culturable microbes, 30 colonies were also selected from the agar-based medium PE03-7A, which showed the highest CFU count among the agar-based media. The phylogenetic distributions of agar-cultured microbes were compared to those retrieved from the gellan gum-based medium PE03-7G, which gave the highest CFU count among the gellan gum-based media.
The cultivated microbes under aerobic conditions were affiliated with seven major bacterial lineages: alpha, beta, gamma, and delta Proteobacteria; Firmicutes; Actinobacteria; Bacteroidetes; and Planctomycetes. Growth of microbes affiliated with four to six major lineages was observed on all types of media. Although the differences in cultivable bacteria between those cultured on PE03 and DR2A media were not remarkable, the shifts to acidic pH (pH 5.5 and 6.0) led to the preferential growth of microbes classified into alpha Proteobacteria and Firmicutes (supplemental Fig. A4).
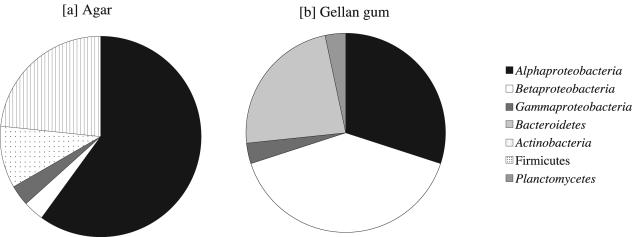
The differences between gelling reagents exerted a significant influence on the diversity of cultivable microbes in the freshwater sediment (Fig. 2). The randomly picked colonies from gellan gum medium PE03-7G were affiliated with alpha, beta, and gamma Proteobacteria (30.0, 40.0, and 3.3% of the total number of colonies selected from this medium, respectively), Bacteroidetes (23.3%), and Planctomycetes (3.3%), whereas those from agar medium PE03-7A belonged to alpha, beta, and gamma Proteobacteria (60.0, 3.3 and 3.3%), Firmicutes (10.0%), and Actinobacteria (23.3%). Thus, on agar plates, the microbes classified into alpha Proteobacteria, Firmicutes, and Actinobacteria accounted for 93.3% of the total isolates from PE03-7A. Although there was no statistical evidence because of the small sample size, such remarkable differences in microbial composition between agar- and gellan gum-based cultivations suggest that a gelling reagent has a great influence on capturing a diverse array of sediment microbes.
FIG. 2.
Effects of gelling reagents (agar [a] and gellan gum [b]) on the phylogenetic distribution of microbes cultivated from freshwater sediment with PE03 medium (pH 7.0) over various bacterial lineages on the basis of 16S rRNA gene analysis.
Under anaerobic conditions, 28 or 29 of 30 colonies grown on each medium type were affiliated with the family Clostridiaceae within the phylum Firmicutes (data not shown).
Effects of the improved cultivation method on culturing of novel microbes.
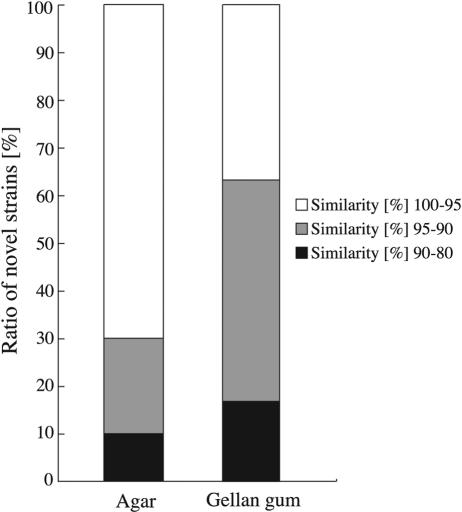
For each medium, the percentage of novel bacterial strains among selected microbes was determined. We considered those isolates whose 16S rRNA gene sequences were less than 95% similar to those of any known bacterial species to be novel microbes, at least at the species level. The percentage of novel bacterial strains obtained under anaerobic conditions was low, accounting for less than 13.3% of the total number of examined microbes cultivated on each type of medium (data not shown). Under aerobic conditions, the greatest number of strains that could be considered novel was retrieved from PE03-7G (gellan gum), and the percentage of novel microbes reached 63.3% of the total number of isolates examined on this medium (Fig. 3). These novel isolates consisted of 13 different phylotypes (i.e., strains with greater than 97% seqence similarity were treated as the same type), and they were classified as alpha and beta Proteobacteria, Bacteroidetes, and Planctomycetes (Table 2). On the other hand, the percentage of novel microbes found on PE03-7A (agar) was 30% of the total number of colonies selected from the medium (Fig. 3). These novel strains consisted of only three different phylotypes, and they were affiliated with only two groups: alpha Proteobacteria and Firmicutes (Table 3). These results clearly indicate the effectiveness of using gellan gum as a gelling reagent instead of agar for the cultivation of novel microbes from freshwater sediment.
FIG. 3.
Effects of gelling reagents in PE03 medium (pH 7.0) on the cultivation of novel microbes from freshwater sediment under aerobic conditions. The similarity percentages shown are the 16S rRNA gene sequence similarities between the cultivated microbes and their closest relatives in the GenBank database.
TABLE 2.
Phylogenetic affiliations of microbes grown on PE03-7G (gellan gum) medium on the basis of 16S rRNA gene sequences by using the BLAST program in the GenBank database
| Taxonomic group and strain | Closest species | Accession no. | Similarity (%) | Length (bp) |
|---|---|---|---|---|
| Alpha Proteobacteria | ||||
| PE70G13 | Hyphomicrobium zavarzinii | Y14306 | 88 | 219 |
| PE70G23 | Magnetospirillum gryphiswaldense | Y10109 | 91 | 521 |
| PE70G8 | Roseomonas gilardii | AY150045 | 94 | 558 |
| PE70G9 | Roseomonas gilardii | AY150045 | 94 | 558 |
| PE70G30 | Roseomonas gilardii | AY150045 | 94 | 558 |
| PE70G3 | Methylocystis parvus | Y18945 | 96 | 560 |
| PE70G16 | Novosphingobium subarcticum | AY167828 | 98 | 593 |
| PE70G25 | Novosphingobium subarcticum | AY167828 | 99 | 591 |
| PE70G24 | Agrobacterium sanguineum | AB062105 | 99 | 589 |
| Beta Proteobacteria | ||||
| PE70G26 | Gallionella ferruginea | L07897 | 89 | 615 |
| PE70G6 | Burkholderia glathei | AY154379 | 90 | 561 |
| PE70G19 | Propionivibrio pelophilus | AF016690 | 91 | 553 |
| PE70G15 | Paucimonas lemoignei | X92555 | 91 | 526 |
| PE70G17 | Paucimonas lemoignei | X92555 | 92 | 561 |
| PE70G5 | Herbaspirillum seropedicae | AJ238361 | 92 | 553 |
| PE70G20 | Caenibacterium thermophilum | AJ512945 | 96 | 626 |
| PE70G28 | Caenibacterium thermophilum | AJ512945 | 96 | 626 |
| PE70G14 | Leptothrix cholodnii | X97070 | 96 | 560 |
| PE70G7 | Leptothrix cholodnii | X97070 | 97 | 561 |
| PE70G22 | Leptothrix cholodnii | X97070 | 97 | 612 |
| PE70G2 | Thiobacillus denitrificans | AJ243144 | 97 | 560 |
| Gamma Proteobacteria PE70G27 | Thermomonas haemolytica | AJ300185 | 97 | 617 |
| Bacteroidetes | ||||
| PE70G4 | Flexibacter aggregans | AB078038 | 87 | 560 |
| PE70G11 | Flexibacter sancti | AB078067 | 90 | 542 |
| PE70G10 | Flexibacter sancti | AB078067 | 91 | 561 |
| PE70G12 | Flexibacter sancti | AB078067 | 91 | 560 |
| PE70G29 | Flexibacter sancti | AB078067 | 91 | 561 |
| PE70G18 | Hymenobacter actinosclerus | Y17356 | 93 | 610 |
| PE70G21 | Hymenobacter actinosclerus | Y17356 | 93 | 622 |
| Planctomycetes PE70G1 | Planctomyces limnophilus | X62911 | 93 | 483 |
TABLE 3.
Phylogenetic affiliations of microbes grown on PE03-7A (agar) medium on the basis of 16S rRNA gene sequences by using the BLAST program in the GenBank database
| Taxonomic group and strain | Closest species | Accession no. | Similarity (%) | Length (bp) |
|---|---|---|---|---|
| Alpha Proteobacteria | ||||
| PE70A15 | Rhodoplanes elegans | D25311 | 89 | 624 |
| PE70A22 | Rhodoplanes elegans | D25311 | 89 | 624 |
| PE70A19 | Paracraurococcus ruber | D85827 | 93 | 641 |
| PE70A1 | Roseomonas gilardii | AY150045 | 94 | 609 |
| PE70A5 | Roseomonas gilardii | AY150045 | 94 | 610 |
| PE70A11 | Roseomonas gilardii | AY150045 | 94 | 602 |
| PE70A20 | Roseomonas gilardii | AY150045 | 94 | 632 |
| PE70A6 | Hyphomicrobium zavarzinii | Y14306 | 94 | 348 |
| PE70A10 | Methylosinus sporium | Y18946 | 95 | 603 |
| PE70A24 | Methylosinus sporium | Y18946 | 96 | 620 |
| PE70A3 | Methylosinus sporium | Y18946 | 97 | 634 |
| PE70A17 | Rhodobacter capsulatus | D16427 | 97 | 608 |
| PE70A18 | Microvirga subterranea | AY078053 | 97 | 617 |
| PE70A25 | Bradyrhizobium elkanii | U35000 | 99 | 630 |
| PE70A29 | Bradyrhizobium elkanii | U35000 | 99 | 601 |
| PE70A23 | Bradyrhizobium elkanii | U35000 | 100 | 623 |
| PE70A30 | Bradyrhizobium elkanii | U35000 | 100 | 635 |
| PE70A9 | Methylobacterium radiotolerans | D32227 | 100 | 603 |
| Beta Proteobacteria PE70A28 | Roseateles depolymerans | AB003624 | 96 | 612 |
| Gamma Proteobacteria PE70A8 | Pseudomonas putida | D85999 | 99 | 626 |
| Firmicutes | ||||
| PE70A13 | Fusibacter paucivorans | AF050099 | 88 | 552 |
| PE70A16 | Paenibacillus hongkongensis | AF433165 | 95 | 595 |
| PE70A14 | Bacillus cereus | AE017013 | 99 | 506 |
| Actinobacteria | ||||
| PE70A2 | Mycobacterium manitobense | AY082001 | 98 | 616 |
| PE70A7 | Mycobacterium manitobense | AY082001 | 98 | 616 |
| PE70A26 | Rhodococcus equi | M29574 | 98 | 643 |
| PE70A4 | Arthrobacter oxydans | AJ243423 | 99 | 615 |
| PE70A12 | Mycobacterium brumae | AF480576 | 99 | 616 |
| PE70A27 | Janibacter limosus | Y08539 | 99 | 641 |
| PE70A21 | Nocardia veterana | AF430059 | 100 | 638 |
Of the novel strains grown on PE03-7G medium, the most abundant strains (seven) were affiliated with Bacteroidetes (Table 2). Of these seven strains, five (PE70G4, PE70G10, PE70G11, PE70G12, and PE70G29) were related to the genus Flexibacter, although they showed considerably lower similarities (87 to 91%) to any of the known bacterial species. The remaining two strains, PE70G18 and PE70G21, were related to Hymenobacter actinosclerus, with 93% 16S rRNA gene similarity. The second most abundant novel strains (i.e., six strains) retrieved from PE03-7G medium were affiliated with beta Proteobacteria. We found that three strains (PE70G5, PE70G15, and PE70G17) were distantly related to the family Oxalobacteraceae; one strain (PE70G5) was related to members of the genus Herbaspirillum, with 92% identity; and two strains (PE70G15 and PE70G17) were related to Paucimonas lemoignei, with 92% similarity. The growth of three strains (PE70G6, PE70G19, and PE70G26) that were affiliated with uncultured cluster RBF8 (4), representing a novel order within beta Proteobacteria, was also observed on PE03-7G. We also obtained a novel strain (PE70G1) belonging to the genus Planctomycetes. This strain was distantly related to Planctomyces limnophilus, with a 16S rRNA gene similarity of 93%. A novel strain affiliated with this class was also grown on DR2A-7G medium (data not shown). Although the species closest to this strain was Gemmata obscuriglobus, the similarity was very low (88%). We are currently performing continuous cultivation of these novel strains to further investigate their physiological features, which will facilitate a better understanding of their functional role in freshwater sediments.
Comparison of bacterial diversity between the 16S rRNA gene clone library- and culture-based methods.
Modifications of either pH conditions or gelling reagents had a significant influence on the examination of viable counts and the spectrum of microbes cultivated from freshwater sediment. The shifts to acidic pH conditions (pHs 5.5 and 6.0) reduced the viable counts and led to the preferential growth of bacteria belonging to alpha Proteobacteria, Actinobacteria, and Firmicutes, which were found to be insignificant groups by 16S rRNA gene clone analysis (supplemental Fig. A4). Under slightly acidic pH conditions, Janssen et al. (20) and Sait et al. (40) successfully cultivated globally distributed unknown bacteria affiliated with Proteobacteria, Actinobacteria, Acidobacteria, and Verrucomicrobia from soil samples. However, in this study, modification of the pH conditions appeared to be ineffective for increasing culturability and thereby at isolating novel bacterial strains from the freshwater sediment.
The preferential growth of bacteria belonging to alpha Proteobacteria, Actinobacteria, and Firmicutes, which were minor groups in the 16S rRNA gene clone library, was observed on the agar medium (Fig. 2). In contrast, the use of gellan gum medium led to the growth of a number of microorganisms belonging to beta Proteobacteria and Bacteroidetes (Fig. 2), which were more abundant in the clone library. In addition, 11 of 30 colonies selected from the gellan gum medium showed more than 95% sequence similarity to the 16S rRNA gene clones, while only 1 of 30 colonies from the agar medium showed such a sequence similarity.
The significant discrepancy between bacterial community compositions determined by culture-dependent and -independent analyses has been revealed by many previous studies of a wide variety of natural ecosystems (10, 12-14, 24, 26, 36, 42, 44, 47). Our study clearly demonstrated that the use of gellan gum instead of agar medium would enable closure of the gaps between the two approaches. However, differences between the phylogenetic distributions of microbes shown by molecular and cultivation-based studies still remain and several explanations for these remaining differences appear plausible. First, a well-recognized primary reason for such differences is the bias associated with the use of molecular methods (12, 13, 24, 26, 36, 44, 47). Second, the anaerobic culture techniques used in this study were not suitable to cover anaerobes, which are thought to be quite abundant in freshwater sediment. Third, a percentage of the unknown bacteria in freshwater sediment may indeed be extremely difficult to cultivate in general. Recently, a number of researchers succeeded at increasing culturability and they subsequently cultivated hitherto uncultured bacteria by modifying the traditional cultivation techniques as follows: use of plant polysaccharides as an energy source (7), addition of signal molecules related to cell-cell communication (5, 6, 35), and effective use of background bacteria as aids to growth (23, 51). Our study strongly suggested that the gelling reagent used is an important factor in the successful cultivation of unknown bacteria. Certainly, the use of gellan gum alone is not sufficient for a culture-based study. However, the combination of the use of gellan gum and modifications of previously designed methods might lead to the cultivation of yet-to-be-isolated organisms and might also increase the opportunity to study bacterial diversity in a wide variety of ecosystems by culture-dependent analyses.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of both the clone library and culture collection have been submitted to public databases under accession numbers AB127608 to AB127929.
Supplementary Material
Acknowledgments
We are grateful to Yoshikazu Koizumi at the Department of Biological Sciences, Tokyo Metropolitan University, for instruction regarding nucleic acid extraction. We thank Akiko Sunaga at the National Institute of Advanced Industrial Science and Technology for help with the cultivation procedure. We also acknowledge Wataru Kitagawa, Yasuhiro Tanaka, and Naoya Shinzato at the Advanced Industrial Science and Technology for reviewing the manuscript.
Footnotes
Supplemental material for this article can be found at http://aem.asm.org.
REFERENCES
- 1.Altmann, D., P. Stief, R. Amann, D. De Beer, and A. Schramm. 2003. In situ distribution and activity of nitrifying bacteria in freshwater sediment. Environ. Microbiol. 5:798-803. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brummer, I. H., A. Felske, and I. Wagner-Dobler. 2003. Diversity and seasonal variability of beta-proteobacteria in biofilms of polluted rivers: analysis by temperature gradient gel electrophoresis and cloning. Appl. Environ. Microbiol. 69:4463-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bussmann, I., B. Philipp, and B. Schink. 2001. Factors influencing the cultivability of lake water bacteria. J. Microbiol. Methods 47:41-50. [DOI] [PubMed] [Google Scholar]
- 7.Chin, K. J., D. Hahn, U. Hengstmann, W. Liesack, and P. H. Janssen. 1999. Characterization and identification of numerically abundant culturable bacteria from the anoxic bulk soil of rice paddy microcosms. Appl. Environ. Microbiol. 65:5042-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costello, A. M., A. J. Auman, J. L. Macalady, K. M. Scow, and M. E. Lidstrom. 2002. Estimation of methanotroph abundance in a freshwater lake sediment. Environ. Microbiol. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 9.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 10.Dees, P. M., and W. C. Ghiorse. 2001. Microbial diversity in hot synthetic compost as revealed by PCR-amplified rRNA sequences from cultivated isolates and extracted DNA. FEMS Microbiol. Ecol. 35:207-216. [DOI] [PubMed] [Google Scholar]
- 11.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis, R. J., P. Morgan, A. J. Weightman, and J. C. Fry. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl. Environ. Microbiol. 69:3223-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felske, A., A. Wolterink, R. van Lis, W. M. de Vos, and A. D. Akkermans. 1999. Searching for predominant soil bacteria: 16S rDNA cloning versus strain cultivation. FEMS Microbiol. Ecol. 30:137-145. [DOI] [PubMed] [Google Scholar]
- 15.Giovannoni, S. J. 1995. Microbial diversity in oceanic systems: rRNA approaches to the study of unculturable microbes, p. 217-248. In I. Joint (ed.), Molecular ecology of aquatic microbes. Springer-Verlag KG, Berlin, Germany.
- 16.Gregory, L. G., P. L. Bond, D. J. Richardson, and S. Spiro. 2003. Characterization of a nitrate-respiring bacterial community using the nitrate reductase gene (narG) as a functional marker. Microbiology 149:229-237. [DOI] [PubMed] [Google Scholar]
- 17.Hanada, S., A. Hiraishi, K. Shimada, and K. Matsuura. 1995. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int. J. Syst. Bacteriol. 45:676-681. [DOI] [PubMed] [Google Scholar]
- 18.Hiorns, W. D., R. C. Hastings, I. M. Head, A. J. McCarthy, J. R. Saunders, R. W. Pickup, and G. H. Hall. 1995. Amplification of 16S ribosomal RNA genes of autotrophic ammonia-oxidizing bacteria demonstrates the ubiquity of nitrosospiras in the environment. Microbiology 141:2793-2800. [DOI] [PubMed] [Google Scholar]
- 19.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssen, P. H., P. S. Yates, B. E. Grinton, P. M. Taylor, and M. Sait. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones, J. G., and B. M. Simon. 1981. Differences in microbial decomposition processes in profundal and littoral lake sediments, with particular references to the nitrogen cycle. J. Gen. Microbiol. 123:297-312. [Google Scholar]
- 22.Jurgens, G., F. Glockner, R. Amann, A. Saano, L. Montonen, M. Likolammi, and U. Munster. 2000. Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiol. Ecol. 34:45-56. [DOI] [PubMed] [Google Scholar]
- 23.Kaeberlein, T., K. Lewis, and S. S. Epstein. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 296:1127-1129. [DOI] [PubMed] [Google Scholar]
- 24.Kaiser, O., A. Puhler, and W. Selbitschka. 2001. Phylogenetic analysis of microbial diversity in the rhizoplane of oilseed rape (Brassica napus cv. westar) employing cultivation-dependent and cultivation-independent approaches. Microb. Ecol. 42:136-149. [DOI] [PubMed] [Google Scholar]
- 25.Kanagawa, T. 2003. Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96:317-323. [DOI] [PubMed] [Google Scholar]
- 26.Kisand, V., and J. Wikner. 2003. Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl. Environ. Microbiol. 69:3607-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koizumi, Y., J. J. Kelly, T. Nakagawa, H. Urakawa, S. El-Fantroussi, S. Al-Muzaini, M. Fukui, Y. Urushigawa, and D. A. Stahl. 2002. Parallel characterization of anaerobic toluene- and ethylbenzene-degrading microbial consortia by PCR-denaturing gradient gel electrophoresis, RNA-DNA membrane hybridization, and DNA microarray technology. Appl. Environ. Microbiol. 68:3215-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 29.Li, J. H., K. J. Purdy, S. Takii, and H. Hayashi. 1999. Seasonal changes in ribosomal RNA of sulfate-reducing bacteria and sulfate reducing activity in a freshwater lake sediment. FEMS Microbiol. Ecol. 28:31-39. [Google Scholar]
- 30.Lindstrom, E. S., and E. Leskinen. 2002. Do neighboring lakes share common taxa of bacterioplankton? Comparison of 16S rDNA fingerprints and sequences from three geographic regions. Microb. Ecol. 44:1-9. [DOI] [PubMed] [Google Scholar]
- 31.MacGregor, B. J., D. P. Moser, B. J. Baker, E. W. Alm, M. Maurer, K. H. Nealson, and D. A. Stahl. 2001. Seasonal and spatial variability in Lake Michigan sediment small-subunit rRNA concentrations. Appl. Environ. Microbiol. 67:3908-3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miskin, I. P., P. Farrimond, and M. Head. 1999. Identification of novel bacterial lineages as active members of microbial populations in a freshwater sediment using a rapid RNA extraction procedure and RT-PCR. Microbiology 145:1977-1987. [DOI] [PubMed] [Google Scholar]
- 34.Nealson, K. H. 1997. Sediment bacteria: who's there, what are they doing, and what's new? Annu. Rev. Earth Planetary Sci. 25:403-434. [DOI] [PubMed] [Google Scholar]
- 35.Olson, J. B., C. C. Lord, and P. J. McCarthy. 2000. Improved recoverability of microbial colonies from marine sponge samples. Microb. Ecol. 40:139-147. [DOI] [PubMed] [Google Scholar]
- 36.Pearce, D. A., C. J. van der Gast, B. Lawley, and J. C. Ellis-Evans. 2003. Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol. Ecol. 45:59-70. [DOI] [PubMed] [Google Scholar]
- 37.Purdy, K. J., T. M. Embley, S. Takii, and D. B. Nedwell. 1996. Rapid extraction of DNA and rRNA from sediments using a novel hydroxyapatite spin column method. Appl. Environ. Microbiol. 62:3905-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purdy, K. J., D. B. Nedwell, and T. M. Embley. 2003. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69:3181-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purdy, K. J., D. B. Nedwell, T. M. Embley, and S. Takii. 1997. Use of 16S rRNA-targeted oligonucleotide probes to investigate the occurrence and selection of sulfate-reducing bacteria in response to nutrient addition to sediment slurry microcosms from a Japanese estuary. FEMS Microbiol. Ecol. 24:221-234. [Google Scholar]
- 40.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 41.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 42.Sekiguchi, H., H. Koshikawa, M. Hiroki, S. Murakami, K. Xu, M. Watanabe, T. Nakahara, M. Zhu, and H. Uchiyama. 2002. Bacterial distribution and phylogenetic diversity in the Changjiang estuary before the construction of the Three Gorges dam. Microb. Ecol. 43:82-91. [DOI] [PubMed] [Google Scholar]
- 43.Sekiguchi, Y., Y. Kamagata, K. Syutsubo, A. Ohashi, H. Harada, and K. Nakamura. 1998. Phylogenetic diversity of mesophilic and thermophilic granular sludges determined by 16S rRNA gene analysis. Microbiology 144:2655-2665. [DOI] [PubMed] [Google Scholar]
- 44.Smit, E., P. Leeflang, S. Gommans, J. van den Broek, S. van Mil, and K. Wernars. 2001. Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spring, S., R. Schlze, J. Overmann, and K. H. Schleifer. 2000. Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: molecular and cultivation studies. FEMS Microbiol. Rev. 24:573-590. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki, M. T., M. S. Rappe, Z. W. Haimberger, H. Winfield, N. Adair, J. Strobel, and S. J. Giovannoni. 1997. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl. Environ. Microbiol. 63:983-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urakawa, H., K. Kita-Tsukamoto, and K. Ohwada. 1999. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology 145:3305-3315. [DOI] [PubMed] [Google Scholar]
- 49.Whitby, C. B., J. R. Saunders, R. W. Pickup, and A. J. McCarthy. 2001. A comparison of ammonia-oxidiser populations in eutrophic and oligotrophic basins of a large freshwater lake. Antonie Leeuwenhoek 79:179-188. [DOI] [PubMed] [Google Scholar]
- 50.Wise, M. G., J. V. McArthur, and L. J. Shimkets. 1997. Bacterial diversity of a Carolina Bay as determined by 16S rRNA gene analysis: confirmation of novel taxa. Appl. Environ. Microbiol. 63:1505-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.