Abstract
Sleep is a deeply conserved, yet fundamentally mysterious behavioral state. Knowledge of the circuitry of sleep–wake regulation is an essential step in understanding the physiology of the sleeping brain. Recent efforts in Drosophila have revealed new populations which impact sleep, as well as provided unprecedented mechanistic and electrophysiological detail of established sleep-regulating neurons. Multiple, distributed centers of sleep–wake circuitry exist in the fly, including the mushroom bodies, central complex and the circadian clock cells. Intriguingly, certain populations have been implicated in specific roles in homeostatic rebound sleep, occurring after sleep loss. In short, our knowledge of fly sleep circuitry advances towards a greater view of brain-wide connectivity and integration of the signals and correlates of the state of sleep.
Introduction
The use of Drosophila as a model for sleep research has grown for more than 15 years [1,2] to become a vital component of our efforts to understand the molecular and cellular mechanisms and functions of this enigmatic behavioral state. The ease and power of genetic manipulation in this model has enabled targeted examination of neurotransmitter/neuromodulator systems [3–5] and neuropeptides [6] for a role in sleep–wake states, and led the way in forward-genetic screens to identify novel genes that regulate sleep [7–9].
The focus of this review will be on the circuitry of sleep–wake states, which has also been comprehensively studied in Drosophila. Advances in our understanding of fly brain anatomy, coupled with technology to manipulate activity of specific neurons, have facilitated the identification of neural populations required for daily baseline sleep and arousal as well as those that function in sleep homeostasis. The latter is typically assayed by depriving flies of sleep and monitoring the increased sleep (rebound) that follows. Interestingly, as discussed below, baseline and homeostatic sleep may be controlled by distinct neurons.
Sleep and wake-promoting circuits in the mushroom bodies
Located in the protocerebrum of the Drosophila brain, the mushroom body (MB) is a structure which has classically been established as an essential associative center for olfactory learning and memory, although more recently, this network has begun to be appreciated for much broader roles in behavior [10,11]. The mushroom bodies are comprised of Kenyon cells (KCs) whose axons project together to form the three MB lobes – α/β, α′/β′, and γ – which can be further divided into 2–3 types, depending on projection pattern within each lobe. KCs synapse with the mushroom body output neurons (MBONs), whose dendrites cover the lobes as distinct ‘compartments’ [10].
Early circuitry studies identified both sleep and wake-promoting populations in the mushroom body [12,13]. Manipulation of signaling or neural activity by the Gal4-UAS system in multiple MB lobes revealed that sleep effects arise, at least in part, from α/β populations. Examination of cellular mechanisms implicates Go signaling as important for sleep regulation through the MBs [14,15]. Limiting expression of a constitutively active Go to adulthood with a specific MB driver decreased total sleep, while blocking Go signaling via pertussis toxin led to increased day sleep and nighttime fragmentation, attributable to a cholinergic population [15]. Interestingly, pertussis toxin produced loss of sleep with other MB drivers, supporting varied effects of multiple sleep-regulating MB neurons [15].
In these, as in other circuit examinations, promiscuity of driver expression pattern is a constant caveat. As example, the 201y-Gal4 driver exhibits broad MB expression and promotes sleep when driving PKA [12]. In addition, conditional depolarization of 201Y neurons by activation of the heat-sensitive TrpA1 channel [16] increases total sleep, while silencing with Shibire, a temperature-sensitive dominant negative dynamin [17], decreases sleep [18]. Nevertheless, Gal80 (an inhibitor of Gal4) experiments show that these effects come from populations external to the mushroom body [18].
More recent studies of the MB have taken advantage of improved tools for the dissection of Drosophila circuitry, namely the ‘split-Gal4’ system [19], which considerably refines expression by segregating Gal4 domains to two enhancers whose expression pattern must overlap. Using such drivers to target the KCs, TrpA1 activation revealed multiple split-Gal4 combinations localized to α′/β′ KCs to be wake-promoting [20••]. The γmain KCs were also wake-promoting, while γdorsal KCs promoted sleep. Interestingly, no effects were shown with α/β lobe split-Gal4s. While comparative strength of classic and split-Gal4 drivers may be questioned, previous works implicating α/β [12,13,15] concluded so based on overlapping driver expression, which often also contained γ lobe expression.
Output from the mushroom bodies
Regarding the postsynaptic targets of KCs, the GABAergic MBONs which synapse with γ3 and β′1 KCs (MBON-γ3β′1 and MBON-γ3) were found to promote sleep upon TrpA1-mediated activation, as did the cholinergic MBON-calyx and MBON-γ2α′1. Wake-promoting MBONs (γ4 > γ1γ2, β′2mp, γ5β′2α, β′2mp_bilateral) were also discovered, with all being glutamatergic [11,20••]. Curiously, although MBONs are convergent outputs of KC signaling, neither the wake-promoting MBON-γ5β′2α/β′2mp/β′2mp_bilateral nor the sleep-promoting MBON-γ2α′1 produced significant baseline sleep alteration when conditionally silenced with Shibire [20••], suggesting that they, or at least sleep-promoting MBONs, have a greater role in homeostatic sleep (see below).
Since sleep-promoting (MBON-γ2α′1) and wake-promoting (MBON-γ5β′2α/β′2mp/β′2mp_bilateral) output neurons receive input from both sleep and wake-promoting KCs, the question of how these signals are integrated arises [10,20••]. The sleep-loss produced by α′/β′ KC activation could be abrogated by simultaneously silencing MBON-γ5β′2α/β′2mp/β′2mp_bilateral, but not MBON-γ2α′1. This behavior correlated with a stronger Ca2+ response to local excitation of α′/β′ KCs in MBON-γ5β′2α/β′2mp/β′2mp_bilateral than in MBON-γ2α′1 [20••]. On the other hand, using a genetically encoded voltage indicator, greater spontaneous activity was seen in certain sleep-promoting populations (MBON-γ2α′1 and γdorsal KCs) following sleep-deprivation, with diminished activity in wake-promoting populations (α′/β′ KCs and MBON-γ5β′2α/β′2mp/β′2mp_bilateral). Silencing sleep-promoting MBON-γ2α′1 with Shibire following sleep deprivation, also limited the resultant rebound sleep [20••]. Together these findings indicate that sleep and wake signals can be traced through KCs to MBONs, and certain MBONs are important for rebound.
Inputs to the mushroom bodies
The MB lobes are known to receive numerous modulatory inputs, such as from dorsal paired medial neurons (DPM) and dopaminergic neurons (DANs). DANs innervate distinct sections which largely cover the MB lobes, and localize with the separate MBON dendrites, allowing the MB lobes to be portioned into ‘compartments’ defined by MBON and DAN arborization [10,11]. TrpA1 activation revealed a number of DANs to be wake-promoting, while none were sleep-promoting [21]. DANs projecting to the γ5, γ4 and β′2 compartments all increased wake, matching the arborization patterns of wake-promoting MBONs. At the same time, stimulating certain MB lobe compartments through DANs increased wake while activation of the respective MBONs did not [21]. Additionally, a sub-population of the dopaminergic PAM neurons, which project to the MBs, is necessary for the wake-promoting effect of caffeine [22], likely through Dop1R1 receptors [23].
A prominent input to the MBs are the DPMs—which project to all lobes, and are important for memory [24•,25]. Lui et al. [25] demonstrated that inhibiting synaptic release from DPMs with tetanus toxin (TNT) leads to shorter, more numerous sleep bouts. This phenotype was similar to that of mutants lacking the peptide amnesiac, although DPMs are not the sole source of amnesiac, and also package other neurotransmitters [24•]. Consistent with a sleep-regulating function of DPMs, TrpA1 activation increased total sleep, and conditional Kir2.1 inhibition lead to a decrease in sleep at night [24•]. DPM input is inhibitory to the MB and RNAi knockdown of either the vesicular GABA transporter (VGAT) or tryptophan hydroxylase (serotonin synthesis) caused a loss of nighttime sleep—suggesting GABA and serotonin signaling. The relevant serotonin receptor is likely d5-HT1a, previously implicated in sleep consolidation through its action in the MBs [26].
In short, the mushroom body has a complex role in sleep–wake regulation (Figure 1). Kenyon cells of all three major lobes have been implicated, although initial use of broader drivers may necessitate validation of α/β involvement, with more recent findings focusing on the α′/β′ and γ divisions. MBONs integrate diverse KC innervations to drive either sleep or wake. Modulatory inputs from DANs appear to solely be wake-promoting, while inhibitory DPM input is sleep-promoting.
Figure 1. The mushroom body exhibits complicated influence on sleep and wake.
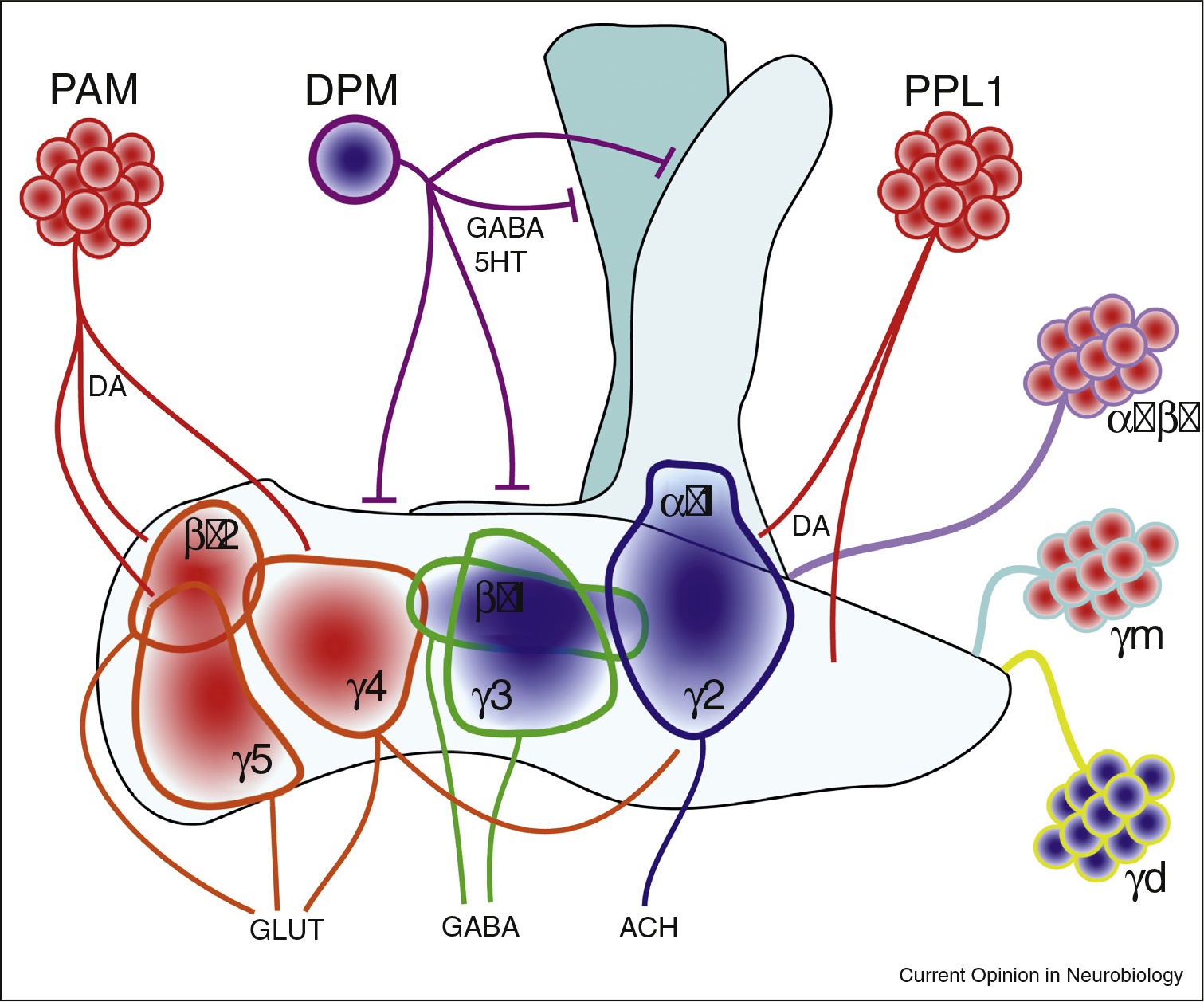
Mushroom body α′/β′ and γm KCs are wake-promoting, while γd promote sleep. All of these KCs synapse at MBONs within the MB lobes, and the signal is integrated by glutamatergic wake-promoting MBONs (γ5,β′2,γ4) or sleep-promoting GABAergic (γ3,β′1) or cholinergic (α′ γ2) MBONS. DANs project to MBONs, and are wake-promoting through dopamine. DPMs are sleep promoting, and signal through GABA and serotonin. DPMs project to all lobes and are thought to be inhibitory. Blue fills indicate a sleep-promoting population. Red fills indicate a wake-promoting population. DA = dopamine. 5HT = serotonin. GLUT = glutamate. ACH = acetylcholine. Mushroom body and DAN depiction based on representations in Aso et?al. [11] and Sitaraman et?al. [21].
Components of the central complex drive sleep
Fan-shaped body
The central complex in Drosophila refers to a group of central brain neuropils, including the fan-shaped and ellipsoid body which, collectively, are implicated in processes such as sensory integration and locomotion, memory, and most recently, sleep [27]. Neuronal activation revealed ExFl2 cells of the dorsal fan-shaped body (dFSB) to be profoundly sleep-promoting [28]. Acute activation by TrpA1 demonstrated that sleep could be driven in defined windows [28,29], with flies showing increased arousal threshold over the course of activation [28], while constitutively silencing this population resulted in sleep loss [30].
Numerous studies have implicated dopamine in the promotion of wake and arousal in Drosophila [23,29–31]. DANs promote wake through the MBs, but dopaminergic input to the dFSB has also been established anatomically [29,30] as well as functionally through cAMP imaging and electrophysiology [29,32••]. Using spatially-restricted DAN-Gal4s to drive TrpA1, Liu et al. [30] reported that dFSB-projecting DANs of the PPL1 cluster are wake-promoting. Further, silencing through conditional Kir2.1 expression produced an opposing increase in sleep [30], and multiple DANs exhibited greater vesicular release during the day (vs night) and following sleep deprivation. Alternatively, employing stochastic targeting through MARCM [33], Ueno et al. [29] expressed TrpA1 in individual DANs to show that dFSB-projecting PPM3 cluster neurons promote wake. Regulation of the DAN-dFSB circuit also accounts for heightened sleep of newly eclosed flies [2,34•], which display diminished DAN activity and concurrently elevated dFSB activty [34•].
Consistently, these groups identified Dop1R1 (also referred to as DA1 or DopR) as mediating the effects of dopamine in the dFSB [29,30]. Dop1R1 expression restricted to the dFSB supports the low-sleep phenotype of a fumin (dopamine transporter) mutant [29], as well as increased wake in response to L-dopa feeding [30]. However, others found that dFSB knockdown of Dop1R2, but not Dop1R1 or other dopamine receptors, led to an increase in total sleep [32••]. Ueno et al. [29] also did not see an effect of dFSB Dop1R1 knockdown in otherwise wild-type background flies. It is possible that both receptors function in the dFSB, with Dop1R1 being sufficient but not necessary.
In agreement with dopamine operating through a Gs-coupled D1-like receptor, expression of a constitutively active Gαs in the dFSB produces a decrease in sleep [29]. Further, the dFSB responds directly to dopamine application by increasing cAMP, which is eliminated with concurrent Dop1R1 knockdown [29]. Driving constitutively active PKA also results in a loss of sleep [30], suggesting that dopamine-dependent inhibition of the dFSB operates through cAMP-dependent signaling. In contrast, while Pimentel et al. [32••] show that dopamine application indeed hyperpolarizes dFSB neurons through potassium conductance, this effect could be eliminated by pertussis toxin, arguing for a Gi/o-coupled mechanism downstream of Dop1R2 [32••]. As noted above, both mechanisms are possible.
Regardless of the relevant receptor(s), sustained dopaminergic tone, as would be expected during times of wake, is thought to switch dFSB neurons from an excitable, sleep-promoting ‘ON’ state to an electrically quiet ‘OFF’ state [32••]. In the ON state, potassium conductance is dominated by A-type current from Shaker and Shab channels, while the OFF state is marked by a non-A type current based in Sandman. Appropriately, knockdown of Shaker or Shab in the dFSB produces a loss of sleep, while decreasing Sandman leads to greater sleep amount. Impressively, the membrane properties of dFSB neurons correlate with homeostatic sleep pressure, as the excitability of these neurons was heightened in sleep-deprived flies and normalized with recovery sleep [31]. This sensitivity depends on the Rho-GTPase-activating protein crossveinless-c, and dFSB knockdown of crossveinless-c impairs the rebound sleep following deprivation.
In sum, the dorsal fan-shaped body and its dopaminergic inputs (Figure 2), represent one of the best-characterized centers of Drosophila sleep–wake circuitry. Although the exact identity of the cells providing dopaminergic input as well as the receptor(s) through which dopamine acts on this population may still be debatable, it is clear that dopamine promotes wake in the fly, in part, through inhibition of the dFSB.
Figure 2. The central complex promotes sleep: dorsal fan-shaped body and ellipsoid body.
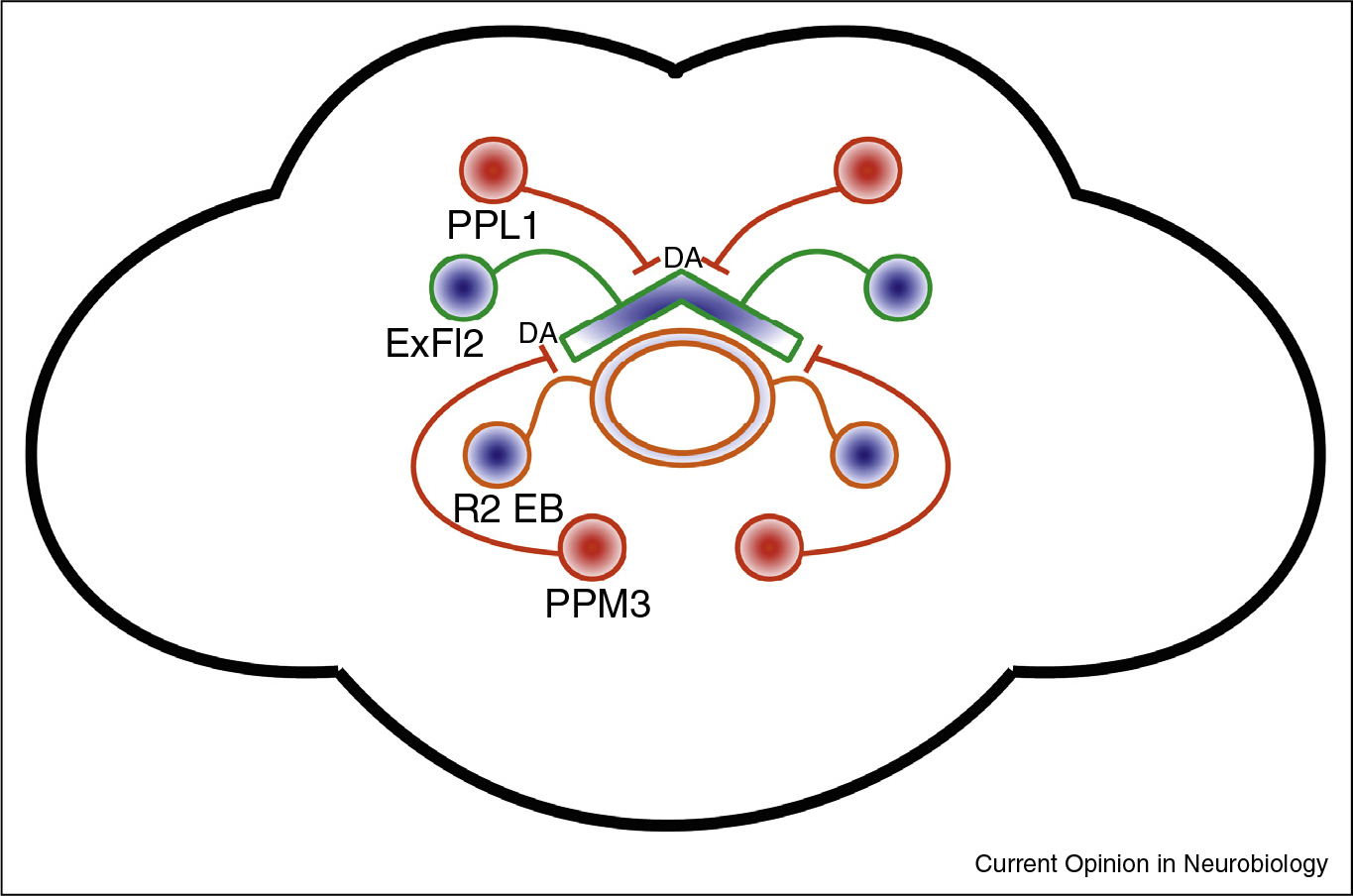
ExFl2 neurons of the dFSB and R2 neurons of the ellipsoid body are sleep-promoting. dFSB neurons are inhibited by dopaminergic neurons of the PPL1 and PPM3 cluster. The sleep-promoting effect of R2 depends on dFSB activity; the connectivity is unknown, but not monosynaptic. Blue fills indicate a sleep-promoting population. Red fills indicate a wake-promoting population. DA = dopamine.
Ellipsoid body
While the ellipsoid body (EB) was not originally found to impact baseline sleep [28], subsequent work identified R2 EB neurons as mediators of homeostatic sleep pressure [35••]. R2 activation produces lasting sleep increases, while TNT silencing during sleep deprivation significantly abrogates rebound sleep. As in the dFSB, excitability of R2 neurons builds across the day, and is further heightened by sleep deprivation. Additionally, sleep need correlates with heightened Ca2+ levels in R2 neurons, and surprisingly, greater synaptic density as measured by the active zone protein, Brp. This plasticity is likely dependent on NMDA-receptors, as dNR1 or NR2 knockdown in R2 neurons reduced rebound sleep following deprivation. Silencing dFSB neurons prevents the sleep increase produced by R2 activation, although the circuit is likely not monosynaptic [35••].
Circadian clock cells
The clock neurons, which possess the molecular components of the circadian clock, are a diffuse network of groups, comprised of the PDF-positive small and large ventral lateral neurons (sLNVs and lLNVs), the dorsal neurons (DN1s, DN2s, and DN3s) and the dorsal lateral neurons (LNds) (for detailed review see Refs. [36,37]).
Constitutive hyperpolarization of PDF-positive cells increases total sleep, while activating the same population results in sleep loss [38,39]. A much subtler loss of sleep, limited to the early night, was seen when excitation of the LNvs was limited to adulthood by TrpA1 [38]. In support, mutants lacking PDF [38], the PDF receptor, or PDF neurons altogether [39], exhibit greater sleep.
Since both lLNVs and sLNVs express PDF, multiple groups have attempted to differentiate the effects of each population on sleep [38,40,41]. These studies have been limited by a lack of drivers for lNVs alone, necessitating different strategies, which may account for differences in results. Multiple groups have employed the c929-Gal4 driver, which targets a large span of peptidergic neurons, including the lLNVs but not the sLNVs [38,40,41]. Consistently, activating c929-Gal4 produced decreases in nighttime sleep [38,40], although non-clock cells also contribute to the phenotype. A separate strategy ablated Pdf-G4-expressing sLNvs with poly-Q Huntingtin protein while activating the lLNvs with NaChBac, and found that lLNv hyperexcitability is sufficient to decrease nighttime sleep [41]. Knocking down PDFR in the PDF-Gal4 population is argued to mainly impact sLNVs, as the lLNVs are not as responsive to the neuropeptide, and doing so produces an increase in day and night sleep [38], but another group used an sLNV-specific driver (R6-Gal4) and found no effect on sleep with NaChBac activation [40]. The very widely expressed, sleep-promoting short neuropeptide F (sNPF) may have a weak role in sleep-promotion through sLNVs [42]. In short, multiple findings demonstrate that lLNvs are wake-promoting cells, while the role of sLNvs is not well defined.
GABAergic input to the LNvs is important for sleep as knock down of the GABAA receptor, Resistant to dieldrin (Rdl) by PDF-Gal4 decreased total sleep [38]. Given that Rdl expression was largely detected in lLNvs, these findings suggest that GABA may promote sleep by inhibiting lLNvs, which otherwise promote wake, at least in part, though PDF signaling (Figure 3) [39]. Interestingly, WIDE AWAKE acts in lLNVs to control Rdl levels as a function of the circadian clock, thereby dampening arousal-promoting effects of these cells at dusk [43].
Figure 3. The circadian clock network influences both sleep and wake.
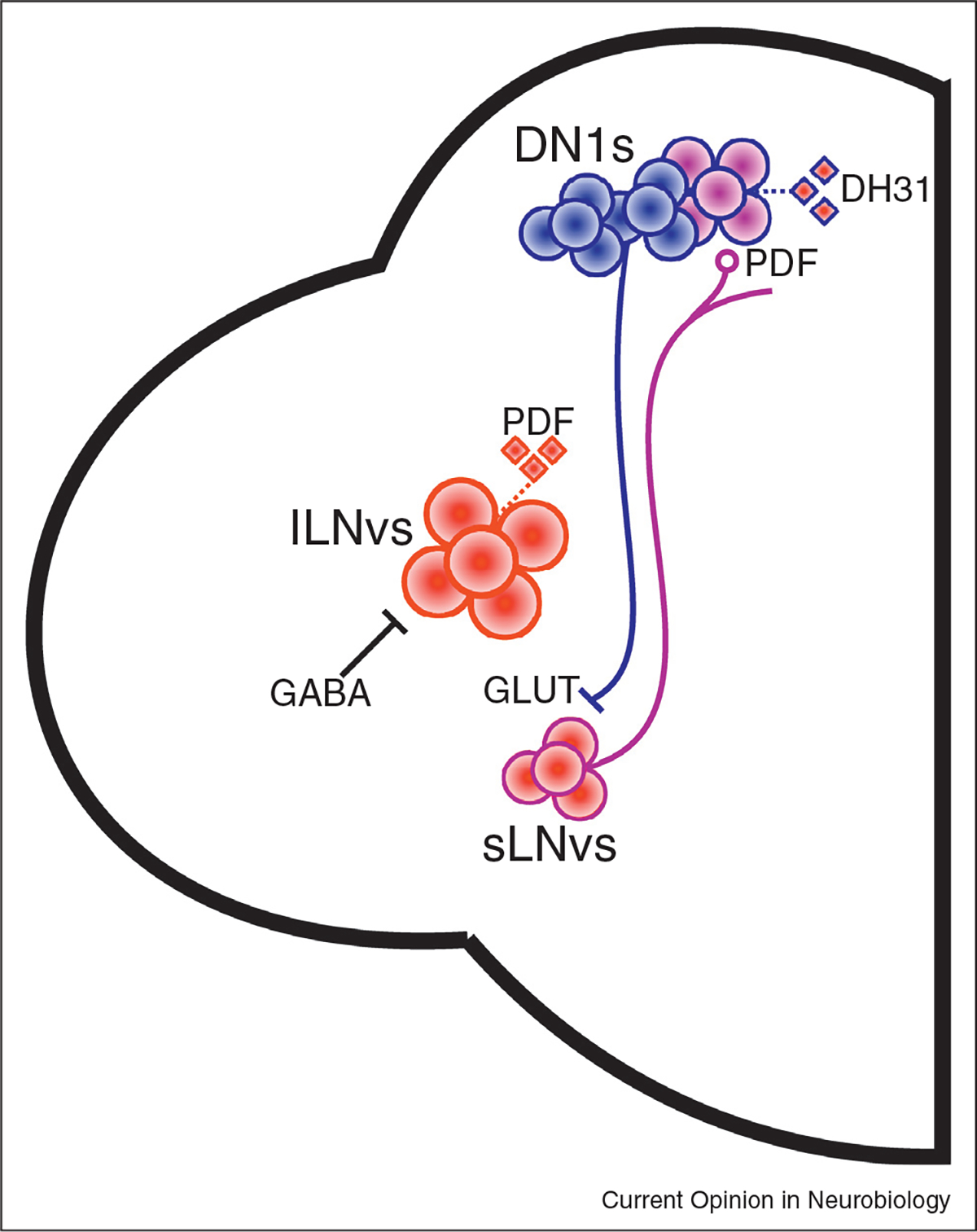
The LNVs have been found to promote wake, at least in part through release of wake-promoting PDF. lLNvs are inhibited by GABA, although the source is unknown. DN1s are both wake-promoting through DH31+ neurons, as well as sleep-promoting through inhibition of sLNvs by glutamate. Whether these populations are distinct is unknown. Blue fills indicate a sleep-promoting population. Red fills indicate a wake-promoting population. Purple fills indicate populations which may promote sleep and wake. Diamonds are neuropeptides, with red fill indicating wake-promotion. GLUT = glutamate.
More recently, the DN1 neurons, which connect anatomically [44•,45] and functionally [44•] to sLNVs and LNds, were shown to modulate sleep–wake patterns. Kunst et al. [45] showed that the fly ortholog of calcitonin gene-related peptide, DH31, acts in a small sub-set of DN1s to promote wakefulness. Expressing DH31 in this population in a DH31 mutant background produced a modest increase in wake in the hours before dawn, a phenotype mimicked by tethered-PDF expression. However, this DN1 sub-population is not the sole mediator of PDF influence on sleep. Using the same tethered-PDF construct, others have found that the PLP sub-type of allatostatin-A (AstA) positive cells increases sleep in response to PDF, as well as TrpA1 activation [46]. Consistently, AstA dendrites are found in close association to PDF-positive terminals and exhibit a cAMP response to PDF. While TrpA1 activation of DN1s produced an even greater sleep loss than DH31 expression [45], another group used the same DN1 driver for optogenetic manipulation and instead found an increase in daytime sleep with excitation, and a loss of nighttime sleep with inhibition [44•]. This discrepancy may come from time-of-day-specific effects of DN1s, in particular reflecting their interaction with light [44•,47]. DN1s inhibit multiple clock populations through glutamate, suggesting a mechanism for the behavioral effects [44•]. Together, these results show a DN1 contribution to wake, through DH31 signaling, and sleep by inhibition of the clock neuron network (Figure 3).
The pars intercerebralis and other sleep–wake regulating populations
One other region of well-described sleep–wake circuitry lies in the pars intercerebralis (PI), a diverse, neuropeptidergic population which has been compared to the mammalian hypothalamus [48].
The insulin-producing cells (IPCs) in the PI are wake-promoting [49]. This population is depolarized by octopamine, a wake-promoting neuromodulator, which produces an increase in cAMP, and affects the Slowpoke potassium current. Thereby, the model posits that octopamine-mediated excitation of the IPCs reduces sleep and this is likely mediated by the OAMB receptor, as OAMB rescue by IPC-G4 in a receptor null background restored sleep loss due to octopamine feeding. The ASM subset of octopaminergic neurons is sufficient to promote wake and is located close to the PI [49] (Figure 4).
Figure 4. The PI is a diverse neurosecretory center which can promote sleep and wake.
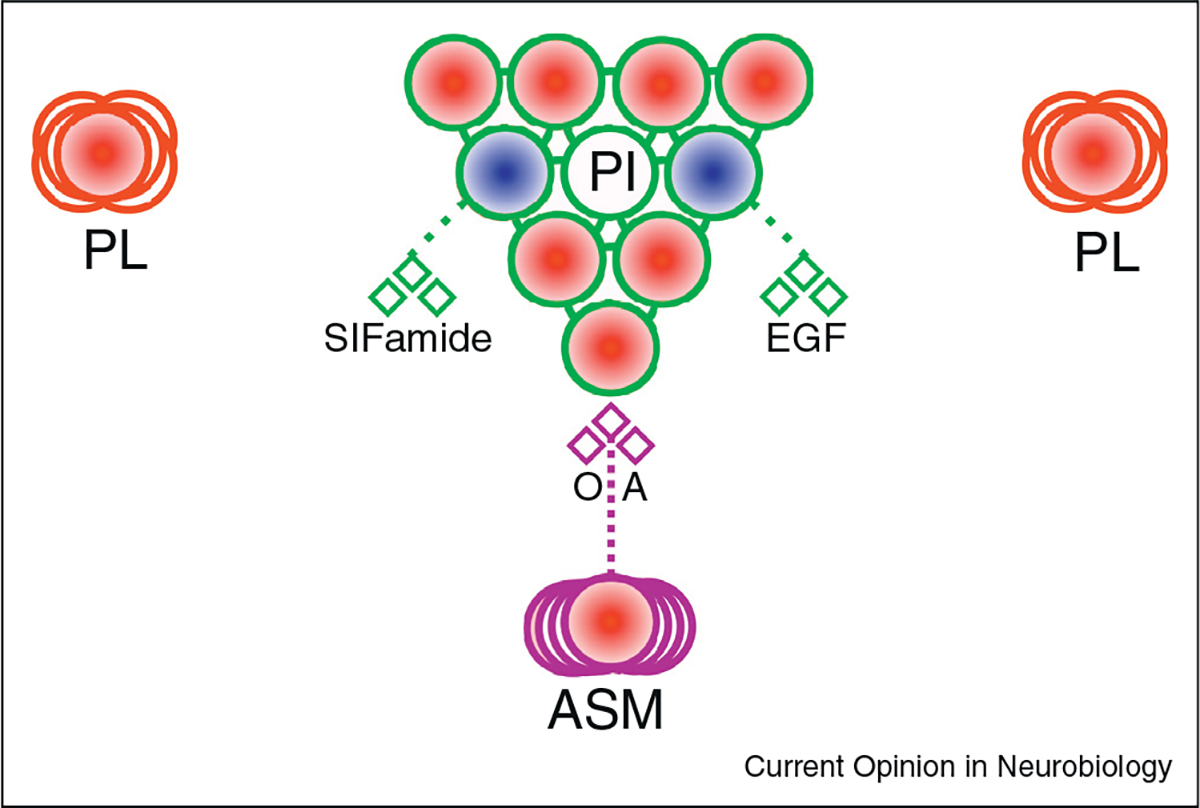
IPCs of the PI are wake-promoting and are depolarized by the wake-promoting neuromodulator, octopamine. Other neurons within or adjacent to the PI are sleep promoting, through EGF and/or SIFamide signaling. A PL population is wake promoting, although not linked to currently known circuitry. Blue fills indicate a sleep-promoting population. Red fills indicate a wake-promoting population. Diamonds are neurotransmitters/peptides. OA = octopamine.
Additionally, the PI may impact sleep through epidermal growth factor (EGF) [48] and SIFamide signaling [50]. Using several drivers which overlap in the PI, Foltenyi et al. [48] demonstrated that conditional expression of the secreted EGFR ligand, s-Spitz, increased total sleep, while knockdown of its processing enzyme, Rho, reduced day and night sleep. SIFamide is expressed in four PI cells [51] and its knockdown resulted in a loss of sleep [50]. In sum, the PI contains wake-promoting IPCs and sleep-promoting neuropeptidergic cells (Figure 4).
Still other sleep–wake regulating populations have been identified that, to date, lack placement in a particular circuit (Figure 4). A split-Gal4 population common to ppk-Gal4 and 24C10-Gal4 projects to the thoracic and gnathal ganglia, promotes wake and elicits a homeostatic sleep increase following its activation. However, these neurons do not produce loss of sleep when silenced and are not involved in baseline sleep [52•]. Interestingly, the ppk-Gal4 population has also been found to be involved in temperature-induced alterations of daytime sleep, potentially through signaling to DN1s, to which some anatomical connectivity has been demonstrated [53]. Elsewhere, CycA, Cdk1 and TARANIS interact to affect sleep in a wake-promoting pars lateralis (PL) population [54].
A final, often underappreciated ‘circuit’ member, are glial cells. Tools to selectively target glia adjoining specific neurons have largely been lacking. Nevertheless, astrocytes of the MB have been argued to affect homeostatic sleep, as Delta overexpression in MB neurons, or the intracellular domain of its receptor Notch in astroctyes can impair rebound sleep [55]. Additionally, the short-sleeping sleepless mutants are known to exhibit lower GABA levels, which is thought to result from upregulation of GABA transaminase in glia [56•].
Conclusion
In conclusion, multiple circuits of sleep–wake regulating neurons are found throughout the Drosophila brain. A clear, outstanding question is whether and how these disparate loci connect to control states, particularly given that any number of populations can mediate profound sleep–wake alteration. In fact, few studies [18] have attempted to link the currently known centers of sleep–wake circuitry.
Another major interest regards the interplay between baseline and homeostatic sleep regulation. A molecular or electrophysiological correlate of homeostatic sleep need has been described in at least three of the major circuits [20••,31,32••,35••], again prompting the question of connectivity. Further, several populations appear to function in daily as well as rebound sleep [20••,28,31], while others have a dedicated homeostatic role [35••,52•].
The Drosophila anatomy differs considerably from that of mammals [57], but the fly provides ultimate value in identifying signals integrated in sleep circuits and circuit principles that underlie regulation of the sleep state.
Acknowledgements
Research in the lab is supported by the Ellison Medical Foundation and the Glenn Foundation. AS is an investigator with the Howard Hughes Medical Institute. GA is also supported by an NIH T32 training grant (5T32HL007953–17).
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
References
- 1.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI: Rest in Drosophila is a sleep-like state. Neuron 2000, 25:129–138. [DOI] [PubMed] [Google Scholar]
- 2.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G: Correlates of sleep and waking in Drosophila melanogaster. Science 2000, 287:1834–1837. [DOI] [PubMed] [Google Scholar]
- 3.Crocker A, Sehgal A: Genetic analysis of sleep. Genes Dev 2010, 24:1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nall A, Sehgal A: Monoamines and sleep in Drosophila. Behav Neurosci 2014, 128:264. [DOI] [PubMed] [Google Scholar]
- 5.Griffith LC: Neuromodulatory control of sleep in Drosophila melanogaster: integration of competing and complementary behaviors. Curr Opin Neurobiol 2013, 23:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sehgal A, Mignot E: Genetics of sleep and sleep disorders. Cell 2011, 146:194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A: Identification of sleepless, a sleep-promoting factor. Science 2008, 321:372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi M, Yue Z, Kuryatov A, Lindstrom JM, Sehgal A: Identification of Redeye: a new sleep-regulating protein whose expression is modulated by sleep amount. Elife 2014, 3:e01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stavropoulos N, Young MW: Insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 2011, 72:964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo T-T, Dionne H, Abbott L, Axel R, Tanimoto H: The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 2014, 3:e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guérin G, Plaçais P-Y, Robie AA, Yamagata N, Schnaitmann C: Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. Elife 2014, 3:e04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joiner WJ, Crocker A, White BH, Sehgal A: Sleep in Drosophila is regulated by adult mushroom bodies. Nature 2006, 441:757–760. [DOI] [PubMed] [Google Scholar]
- 13.Pitman JL, McGill JJ, Keegan KP, Allada R: A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 2006, 441:753–756. [DOI] [PubMed] [Google Scholar]
- 14.Guo F, Yi W, Zhou M, Guo A: Go signaling in mushroom bodies regulates sleep in Drosophila. Sleep 2011, 34:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yi W, Zhang Y, Tian Y, Guo J, Li Y, Guo A: A subset of cholinergic mushroom body neurons requires go signaling to regulate sleep in Drosophila. Sleep 2013, 36:1809–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA: An internal thermal sensor controlling temperature preference in Drosophila. Nature 2008, 454:217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitamoto T: Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol 2001, 47:81–92. [DOI] [PubMed] [Google Scholar]
- 18.Cavanaugh DJ, Vigderman AS, Dean T, Garbe DS, Sehgal A: The Drosophila circadian clock gates sleep through time-of-day dependent modulation of sleep-promoting neurons. Sleep 2015, 39:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luan H, Peabody NC, Vinson CR, White BH: Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 2006, 52:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, Nitabach MN: Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr Biol 2015, 25:2915–2927. •• Along with the earlier Aso et al. publications, this study is one of the most detailed and comprehensive dissections of the mushroom body sleep–wake circuitry to date, examining KCs and MBONs, and showing molecular correlates of sleep need in the MBs.
- 21.Sitaraman D, Aso Y, Rubin GM, Nitabach MN: Control of sleep by dopaminergic inputs to the Drosophila mushroom body. Front Neural Circuits 2015:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nall AH, Shakhmantsir I, Cichewicz K, Birman S, Hirsh J, Sehgal A: Caffeine promotes wakefulness via dopamine signaling in Drosophila. Sci Rep 2016:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andretic R, Kim Y-C, Jones FS, Han K-A, Greenspan RJ: Drosophila D1 dopamine receptor mediates caffeine-induced arousal. Proc Natl Acad Sci 2008, 105:20392–20397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haynes PR, Christmann BL, Griffith LC: A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 2015, 4:e03868. • This study provides a thorough examination of DPM influence on the mushroom bodies and sleep, suggestive due to the otherwise prominent role of DPMs in memory.
- 25.Liu W, Guo F, Lu B, Guo A: Amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophys Res Commun 2008, 372:798–803. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Q, Joiner WJ, Sehgal A: A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol 2006, 16:1051–1062. [DOI] [PubMed] [Google Scholar]
- 27.Wolff T, Iyer NA, Rubin GM: Neuroarchitecture and neuroanatomy of the Drosophila central complex: a GAL4-based dissection of protocerebral bridge neurons and circuits. J Comp Neurol 2015, 523:997–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ: Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 2011, 332:1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K: Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 2012, 15:1516–1523. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN: Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol 2012, 22:2114–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donlea JM, Pimentel D, Miesenböck G: Neuronal machinery of sleep homeostasis in Drosophila. Neuron 2014, 81:860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pimentel D, Donlea JM, Talbot CB, Song SM, Thurston AJ, Miesenböck G: Operation of a homeostatic sleep switch. Nature 2016, 536:333–337. •• These authors describe an impressively detailed molecular mechanism by which the known sleep-promoting dFSB neurons switch between quiescent and active electrical states in relation to dopaminergic input.
- 33.Lee T, Luo L: Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 2001, 24:251–254. [DOI] [PubMed] [Google Scholar]
- 34. Kayser M, Yue Z, Sehgal A: A critical period of sleep for development of courtship circuitry and behavior in Drosophila. Science 2014, 344:269–274. • This study explores the phenomenon of enhanced sleep in young adult flies, establishing a mechanistic link to activity in the known wake-promoting DANs and the sleep-promoting dFSB.
- 35. Liu S, Liu Q, Tabuchi M, Wu MN: Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 2016, 165:1347–1360. •• This work uncovers a new population within the ellipsoid body which mediates homeostatic sleep and demonstrates both electrophysiological and plastic changes as correlating with sleep need.
- 36.Allada R, Chung BY: Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 2010, 72:605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar S, Sehgal A: Control of rest—activity behavior by the central clock in Drosophila. Mechanisms of Circadian Systems in Animals and Their Clinical Relevance. Springer; 2015:: 31–53. [Google Scholar]
- 38.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M: PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 2008, 60:672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R: The GABA A receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 2009, 19:386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shang Y, Griffith LC, Rosbash M: Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc Natl Acad Sci 2008, 105:19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou Y-T, Sharma VK, Holmes TC: Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 2008, 18:1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shang Y, Donelson NC, Vecsey CG, Guo F, Rosbash M, Griffith LC: Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 2013, 80:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S, Lamaze A, Liu Q, Tabuchi M, Yang Y, Fowler M, Bharadwaj R, Zhang J, Bedont J, Blackshaw S: WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 2014, 82:151–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo F, Yu J, Jung HJ, Abruzzi KC, Luo W, Griffith LC, Rosbash M: Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature 2016, 536:292–297. • These authors use optogenetics to show a sleep-promoting role of DN1 clock neurons, which inhibit the clock network through glutamate. Interesting connections between temperature, sex and clock cell activity are also reported.
- 45.Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, Nitabach MN: Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr Biol 2014, 24:2652–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Reiher W, Hermann-Luibl C, Sellami A, Cognigni P, Kondo S, Helfrich-Förster C, Veenstra JA, Wegener C: Allatostatin a signalling in Drosophila regulates feeding and sleep and is modulated by PDF. PLoS Genet 2016, 12:e1006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parisky KM, Rivera JLA, Donelson NC, Kotecha S, Griffith LC: Reorganization of sleep by temperature in Drosophila requires light, the homeostat, and the circadian clock. Curr Biol 2016, 26:882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foltenyi K, Greenspan RJ, Newport JW: Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 2007, 10:1160–1167. [DOI] [PubMed] [Google Scholar]
- 49.Crocker A, Shahidullah M, Levitan IB, Sehgal A: Identification of a neural circuit that underlies the effects of octopamine on sleep: wake behavior. Neuron 2010, 65:670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park S, Sonn JY, Oh Y, Lim C, Choe J: SIFamide and SIFamide receptor define a novel neuropeptide signaling to promote sleep in Drosophila. Mol Cells 2014, 37:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavanaugh DJ, Geratowski JD, Wooltorton JR, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A: Identification of a circadian output circuit for rest: activity rhythms in Drosophila. Cell 2014, 157:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Seidner G, Robinson JE, Wu M, Worden K, Masek P, Roberts SW, Keene AC, Joiner WJ: Identification of neurons with a privileged role in sleep homeostasis in Drosophila melanogaster. Curr Biol 2015, 25:2928–2938. • These authors conduct a screen for neurons involved in homeostatic sleep and aside from a novel population, find a surprising degree of disconnect between sleep loss and recovery.
- 53.Lamaze A, Öztürk-Çolak A, Fischer R, Peschel N, Koh K, Jepson JE: Regulation of sleep plasticity by a thermo-sensitive circuit in Drosophila. Sci Rep 2017:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afonso DJ, Liu D, Machado DR, Pan H, Jepson JE, Rogulja D, Koh K: TARANIS functions with Cyclin A and Cdk1 in a novel arousal center to control sleep in Drosophila. Curr Biol 2015, 25:1717–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ: Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Curr Biol 2011, 21:835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen W-F, Maguire S, Sowcik M, Luo W, Koh K, Sehgal A: A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol Psychiatry 2015, 20:240–251. • This study is one of the few early fly works to describe a glial contribution to sleep.sleepless flies are among the shortest sleeping mutants yet discovered, and have reduced GABA signaling due to GABA transaminase in the mitochondria of glia.
- 57.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE: Sleep state switching. Neuron 2010, 68:1023–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]


