Abstract
Culture filtrate proteins from Mycobacterium tuberculosis induce protective immunity in various animal models of tuberculosis. Two molecular mass regions (6 to 10 kDa and 24 to 36 kDa) of short-term culture filtrate are preferentially recognized by Th1 cells in animal models as well as by patients with minimal disease. In the present study, the 24- to 36-kDa region has been studied, and the T-cell reactivity has been mapped in detail. Monoclonal antibodies were generated, and one monoclonal antibody, HYB 71-2, with reactivity against a 29-kDa antigen located in the highly reactive region below the antigen 85 complex was selected. The 29-kDa antigen (CFP29) was purified from M. tuberculosis short-term culture filtrate by thiophilic adsorption chromatography, anion-exchange chromatography, and gel filtration. In its native form, CFP29 forms a polymer with a high molecular mass. CFP29 was mapped in two-dimensional electrophoresis gels as three distinct spots just below the antigen 85 complex component MPT59. CFP29 is present in both culture filtrate and the membrane fraction from M. tuberculosis, suggesting that this antigen is released from the envelope to culture filtrate during growth. Determination of the N-terminal amino acid sequence allowed cloning and sequencing of the cfp29 gene. The nucleotide sequence showed 62% identity to the bacteriocin Linocin from Brevibacterium linens. Purified recombinant histidine-tagged CFP29 and native CFP29 had similar T-cell stimulatory properties, and they both elicited the release of high levels of gamma interferon from mouse memory effector cells isolated during the recall of protective immunity to tuberculosis. Interspecies analysis by immunoblotting and PCR demonstrated that CFP29 is widely distributed in mycobacterial species.
Tuberculosis (TB) remains a major global health problem and is the most frequent cause of death from a single infectious agent (40). The seriousness of the problem is reflected by the fact that the World Health Organization declared the TB pandemic a global emergency situation in 1993. In the light of the inconsistent efficacy imparted by Mycobacterium bovis bacillus Calmette-Guérin (BCG) (14), the development of an improved TB vaccine is a very high international research priority.
Immunity to TB is mediated by the cellular branch of the immune system, and it has been demonstrated that Mycobacterium tuberculosis culture filtrate proteins are strongly recognized by T cells involved in protection against TB (3, 26). Given as experimental subunit vaccines, culture filtrate proteins promote efficient acquired cellular resistance against the disease (3, 27, 31). Single protective antigens could be included in a future vaccine, as either a subunit or a DNA vaccine or in the form of recombinant BCG expressing these proteins. However, until recently, very limited information on single antigens recognized by T cells was available. By fractionation of extracellular M. tuberculosis proteins into narrow molecular mass fractions we have previously identified two regions that are strongly recognized and stimulate proliferation and gamma interferon (IFN-γ) production in T cells isolated in the first phase of infection from various species (4, 6, 12, 29). In the low-molecular-mass fraction (5 to 12 kDa), ESAT-6 (for 6-kDa early secretory antigenic target) has been identified as the key target (4). The other mass region (24 to 36 kDa) comprises several well-characterized culture filtrate proteins, including T-cell antigens in the antigen 85 complex (4), which consists of the three proteins, MPT44, MPT45, and MPT59. The present study was undertaken to identify novel T-cell antigens in the 24- to 36-kDa region. This investigation led to the identification and purification of a 29-kDa M. tuberculosis protein, CFP29, which is strongly recognized by mouse memory effector cells. This antigen was characterized, and the corresponding gene was cloned and sequenced. The immunological activity of native as well as recombinant, histidine-tagged CFP29 was evaluated.
MATERIALS AND METHODS
Bacterial strains and media.
Short-term culture filtrate (ST-CF), which is highly enriched in extracellular M. tuberculosis proteins, was produced in modified Sauton medium (5). Other culture filtrates from mycobacterial species were prepared as described previously (2). Chromosomal DNAs from mycobacterial species were obtained as described in detail elsewhere (39).
For PCR procedures, cloning, and recombinant expression, the Escherichia coli K-12 DH5α strain, the cloning vector pMCT6, and standard protocols were used (32). pMCT6 is an expression plasmid containing unique restriction sites allowing the construction of in-frame fusions with a leader peptide containing a stretch of eight histidine residues (15). The mRNA for the peptide is transcribed from the tac promoter and translated from a plasmid-encoded translational start site. The plasmid also encodes the Lac repressor to ensure tight control of gene expression.
The bacteriocin assay was carried out with bacterial strains from the Weihenstephan Strain Collection. The Listeria monocytogenes strains were grown on tryptose agar slants, stored at 4°C, and subcultured for 24 h at 30°C in 10 ml of tryptose broth (Merck, Darmstadt, Germany). The Mycobacterium diernhoferi strain was cultured in brain heart infusion broth (Merck) at 30°C for 72 h.
Animals and experimental infections.
Female C57BL/6J and BALB/c mice were purchased from Bomholtegaard (Ry, Denmark). Memory immune C57BL/6J mice were generated as previously described (7). Briefly, mice received a primary infection with 5 × 104 CFU of M. tuberculosis via the lateral tail vein, after which they were treated with isoniazid (Merck, Rahway, N.J.) and rifabutin (Farmatalia Carlo Erba, Milan, Italy) in their drinking water for 2 months to clear the infection. The mice were rested for a period of 4 to 6 months before challenge with 106 CFU of bacteria intravenously, and the animals were sacrificed on day 4 postinfection.
Protein purification.
Narrow molecular mass fractions of ST-CF were obtained by the multielution method (6). Briefly, 6.5 mg of ST-CF was separated by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the gel was preequilibrated in the elution buffer (25 mM 3-[cyclohexylamino]-1-propanesulfonic acid [CAPS], 37 mM ammonia, pH 10.2). The gel was electroeluted in a Whole Gel Elutor (Bio-Rad, Richmond, Calif.) for 20 min at 40 V, followed by reversal of the current for 10 s. Thirty fractions of approximately 3 ml were harvested from the unit.
For purification of CFP29, ammonium sulfate was added to concentrated ST-CF to obtain a final concentration of 1.5 M ammonium sulfate. The soluble proteins were subsequently subjected to thiophilic adsorption chromatography (30) on an Affi-T gel column (Kem-En-Tec, Copenhagen, Denmark). Proteins were eluted by a linear 1.5 to 0 M gradient of ammonium sulfate, fractions containing CFP29 were concentrated, and the buffer was changed to 10 mM Tris-HCl (pH 8.5) by ultrafiltration. Further purification was performed on a Mono Q HR 5/5 column (Pharmacia, Uppsala, Sweden) equilibrated with 10 mM Tris-HCl (pH 8.5) and eluted with a linear gradient of sodium chloride (0 to 0.75 M). As the final purification step, gel filtration was performed on a Superose 6 HR 10/30 column (Pharmacia) equilibrated with phosphate-buffered saline (PBS), pH 7.4. Standard proteins were aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa) (Pharmacia). SDS-PAGE followed by silver staining and immunoblot analysis with monoclonal antibody (MAb) HYB 71-2 was used to detect CFP29 during the purification procedures.
For generation of subcellular fractions of M. tuberculosis, bacteria were lysed in a French pressure cell and fractionated into a cell wall fraction, a membrane fraction, and a cytosol fraction by differential centrifugation as previously described (34). Further fractionation of the membrane proteins was performed by extraction and phase separation with 4% (wt/vol) precondensed Triton X-114 (22).
Purified MPT51, MPT59, and MPT64 proteins were kind gifts from S. Nagai (Osaka City University Medical School, Osaka, Japan).
The Brevibacterium linens Linocin preparation was obtained from sterile filtered culture supernatant by ultrafiltration and ultracentrifugation (37).
Protein concentrations were determined by the micro-bicinchoninic acid method (Pierce Europe, Oud-Beijerland, The Netherlands).
MAbs.
BALB/c mice were immunized four times at 2-week intervals. MAbs were generated by immunization with SDS-PAGE-purified 26- to 29-kDa proteins from ST-CF in adjuvant R-730 (RIBI ImmunoChem Research Inc., Hamilton, Mont.) (first and second immunizations) or in aluminum hydroxide gel adjuvant (2.0% Alhydrogel; Superfos Biosector, Kvistgård, Denmark) (third immunization and boosting). Spleens were isolated, and spleen cells were fused with the P3-X63 myeloma cell line. Supernatants from wells with visible growth were tested for antibodies by immunoblotting of SDS-PAGE-separated ST-CF. The subclass of the MAb HYB 71-2 was determined with an RPN29 mouse monoclonal antibody isotyping kit (Amersham, Buckinghamshire, United Kingdom) according to the manufacturer’s instructions. MAb HBT12, reacting with PstS, has been described previously (24).
SDS-PAGE and 2-DE.
Electrophoresis experiments were performed in the Protean IIxi system (Bio-Rad). Standard SDS-PAGE was done with 10 to 20% gradient gels (16 by 16 by 0.075 cm) as described by Laemmli (21). The gels were either silver stained (10) or transferred to nitrocellulose (Schleicher and Schuell, Dassel, Germany) as previously described (34). For immunoblot analysis, the nitrocellulose membranes were incubated with mouse MAbs and then with alkaline phosphatase-labeled rabbit anti-mouse antibodies (D314; DAKO, Glostrup, Denmark). Nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate was used as a substrate.
For the separation of proteins for sequencing, Tricine SDS-PAGE was carried out in 12% gels as described elsewhere (33). After electrophoresis, the gels were blotted to Problott polyvinylidene difluoride (PVDF) membranes (Applied Biosystems, Foster City, Calif.) by semidry electroblotting in 10 mM CAPS–10% methanol, pH 11. PVDF membranes were stained with 0.1% Coomassie blue R-250 in 40% methanol–1% acetic acid and destained in 50% methanol.
Two-dimensional electrophoresis (2-DE) in polyacrylamide gels was carried out as described previously (18), except that in the first dimension, Nonidet P-40 was replaced by Tween 80. The first-dimension isoelectric focusing tube gels (14 by 0.15 cm) contained Biolyt 4/6 and Biolyt 5/7 (2:3) (Bio-Rad). After the first dimension, samples were separated by SDS-PAGE in 10 to 20% gradient gels or by Tricine SDS-PAGE in 12% gels (16 by 16 by 0.10 cm). The pI scale was calibrated by measuring the pHs of pieces of isoelectric focusing gels soaked in Milli Q water.
For glycan detection, protein samples were loaded on SDS-polyacrylamide gels. After blotting to nitrocellulose membranes, the Immun-Blot kit for glycoprotein detection was applied according to the instructions of the manufacturer (Bio-Rad). Creatinase (Boehringer Mannheim, Mannheim, Germany) and ovalbumin (Bio-Rad) (0.2 and 1 μg of each protein) were included as negative and positive glycan controls, respectively.
N-terminal sequencing.
Separated proteins were blotted to PVDF membranes after Tricine SDS-PAGE. The proteins of interest were excised and subjected to N-terminal sequence analysis by automated Edman degradation with a Procise 494 sequencer (Applied Biosystems) as described by the manufacturer. The EMBL and SWISSPROT databases were searched with the TFASTA and FASTA algorithms (28), respectively.
Cloning and DNA sequencing.
Based on the homology to the DNA sequence of the B. linens lin gene (38), the following degenerate primers for PCR cloning of the cfp29 gene were constructed: 5′-CCCGGCTCGAGAACCTYTACCGCGACCTYGCYCC and 5′-GGGCCGGATCCGAYGCYGCGTCCTTYACYGGXTGCCA (where Y is G or C and X is T or C). These primers are derived from nucleotides 6 to 30 and 301 to 327, respectively, of the lin gene.
A DNA fragment of 322 nucleotides was obtained by PCR amplification of M. tuberculosis H37Rv chromosomal DNA with these primers, purified by using Spinn-X columns (Costar, Cambridge, Mass.), and sequenced with a cycle sequencing kit (Pharmacia). The obtained sequence was used for a homology search of the Sanger Centre M. tuberculosis database (http://www.sanger.ac.uk/Projects/M_tuberculosis/blast_server.shtml) by using the Blast program. On cosmid CY444 in the database, a sequence that was identical to the sequence of the amplified DNA fragment was identified. This sequence on cosmid CY444 was contained within a 795-bp open reading frame, which was PCR amplified, cloned into a BglII- and NcoI-digested pMCT6 expression vector, and sequenced.
All primers used for cloning and sequencing were synthesized at the Statens Serum Institut with an ABI-391 DNA synthesizer (Applied Biosystems).
Histidine-tagged recombinant CFP29 was expressed and purified by metal ion affinity chromatography on an Ni2+-Iminodiacetic acid-epoxy-activated Sepharose 6B fast-flow column (Sigma Chemical Co., St. Louis, Mo.) in the presence of 8 M urea (36). Fractions were analyzed by SDS-PAGE, and those fractions containing recombinant CFP29 were pooled and applied to a 1-ml Resource Q column (Pharmacia) equilibrated with 10 mM Tris-HCl–3 M urea (pH 8.5) and eluted with a linear gradient of sodium chloride (0 to 1 M). Fractions containing recombinant CFP29 were pooled and dialyzed in 25 mM HEPES, pH 8.0. The lipopolysaccharide content in the recombinant CFP29 preparation was determined by the Limulus amoebocyte lysate clot test (8) to be <0.2 ng/μg of protein, and this concentration had no influence on cellular activity.
Interspecies analysis.
One microliter of chromosomal DNA (∼20 ng/μl) was used as template for PCR with the cfp29 gene-specific primers 5′-GGACGTTCAAGCGACACATCGCCG and 5′-CAGCACGAACGCGCCGTCGATGGC, giving rise to 572-bp DNA fragments. The annealing temperature was 55°C, and the number of cycles was 30. PCR products were analyzed on 1% agarose gels. As positive controls for the PCRs, identical PCRs were set up with 16S rRNA gene primers that give rise to a 1,018-bp DNA fragment with chromosomal DNAs from all the mycobacterial strains tested (11). All chromosomal DNA preparations were positive, as determined by the amplification of the 1,018-bp PCR fragment. Chromosomal DNA preparations that were negative with the cfp29-specific primers were tested again under less stringent conditions by lowering the annealing temperature to 45°C and increasing the number of PCR cycles to 35.
Bacteriocin assay.
The spot-on-the-lawn method (9) was used to study the inhibitory effects of the sterile protein samples. To prepare the plates, 0.1 ml of a liquid culture of the indicator organism was inoculated into 6.5 ml of tryptose-soft agar (tryptose broth with 8 g of agar/liter), mixed, and poured into petri dishes (diameter, 10 cm). After solidification of the agar, 10 μl each of sterile filtered ST-CF (4.5 mg/ml), native CFP29 (40 μg/ml), and B. linens Linocin (100 μg/ml) was applied on the agar. The agar plates were incubated at 30°C, and possible inhibition zones were detected by using Henry’s lumination (17). The plates were checked for inhibition zones after 24, 48, and 72 h of incubation.
Cellular assays.
Spleen lymphocytes were isolated from memory immune mice during the recall of protective immunity as previously described (13). Briefly, cells pooled from three mice were cultured in microtiter wells in a volume of 200 μl of RPMI 1640 medium supplemented with β-mercaptoethanol, penicillin-streptomycin, glutamine, and fetal calf serum. Recombinant mouse interleukin-2 (2.5 U/ml; GenZyme, Cambridge, Mass.) was added to all cultures. ST-CF and purified proteins to be tested were all buffer exchanged into compatible buffers before analysis and used at various concentrations (0.5 to 8 μg/ml) in cultures. Supernatants were harvested after 72 h of incubation for detection of IFN-γ by enzyme-linked immunosorbent assay.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. Y12820.
RESULTS
Investigation of the highly stimulatory 24- to 36-kDa region of ST-CF.
The recall of long-lived memory immunity in the mouse model is characterized by rapid recruitment and activation of memory effector cells directed predominantly to molecules in the 5- to 12- and 24- to 36-kDa regions of ST-CF (4). To identify novel antigens in the 24- to 36-kDa region, ST-CF proteins were separated by SDS-PAGE and divided into narrow fractions by the multielution method, giving rise to six fractions from the selected region (Fig. 1). These fractions were used to stimulate T cells isolated from the spleens of mice during the recall response. Fractions 3 and 4 contained members of the antigen 85 complex as major components, and as expected, stimulation with these fractions elicited a prominent release of IFN-γ (4). However, large amounts of IFN-γ were also released in response to fraction 2, indicating that T-cell antigens in the 26- to 29-kDa region were also present (Fig. 1).
FIG. 1.
IFN-γ release from mouse memory effector cells in response to narrow molecular mass fractions of ST-CF. (A) SDS-PAGE, followed by silver staining, of ST-CF and the panel of fractions (lanes 1 to 6). Numbers at the left indicate molecular masses in kilodaltons, and the bands representing the HYB 71-2 antigen (HYB 71-2Ag), MPT51, MPT59, and MPT64 in ST-CF are indicated. (B) The fractions generated were used to stimulate mouse memory effector cells (obtained at day 4 of M. tuberculosis challenge) in culture, and the release of IFN-γ was measured. All values represent means from triplicate analyses. Standard errors of the mean were not above 10% of the mean.
To investigate this molecular mass region, MAbs were generated by immunization of BALB/c mice with the fraction in RIBI adjuvant followed by boosting in aluminum hydroxide. The HYB 71-2 MAb (immunoglobulin M class) showed strong reactivity to a ST-CF band of 29 kDa located in the highly active fraction 2 and was selected for thorough analysis.
Purification of a 29-kDa antigen.
ST-CF proteins were loaded on an Affi-T gel column for thiophilic adsorption chromatography. Immunoblot analysis with HYB 71-2 demonstrated that the reactive molecule was eluted in the ammonium sulfate concentration range of 0.44 to 0.31 M. The relevant fractions were pooled and applied to a Mono Q column, and this step was very efficient for separation of the 29-kDa protein from other ST-CF proteins; fractions of almost pure 29-kDa protein eluted at between 400 and 450 mM sodium chloride. Final purification was performed by gel filtration on a Superose 6 column equilibrated with PBS, pH 7.4. The reactive molecule eluted near the void volume before any of the marker proteins. In the gel filtration step, minor contaminating bands were removed and the buffer was changed to PBS, which was compatible with the cellular assays. As judged by SDS-PAGE, an at least 90% pure preparation was obtained by this protocol, which provided 40 μg of the purified protein from 400 mg of ST-CF (Fig. 2). The 29-kDa protein was designated CFP29.
FIG. 2.
Fractions from the different steps used in the purification of CFP29 analyzed by SDS-PAGE followed by silver staining (A) and by immunoblot analysis with HYB 71-2 (B). Lanes: 1, ST-CF; 2, fraction obtained from the Affi-T gel column; 3, fraction obtained after anion-exchange chromatography and gel filtration. Numbers at the left are molecular masses in kilodaltons.
The CFP29 band was excised from a PVDF membrane after Tricine SDS-PAGE, and microsequencing revealed two N-terminal sequences: MNNLYRDLAPVTEAAWAEI and NNLYRDLAPVTEAAWAEI. The finding of these two sequences indicated that the N-terminal methionine in the mature protein is partially cleaved off.
2-DE analysis of CFP29.
2-DE analysis of purified CFP29 revealed a cluster of three distinct spots of the same molecular mass, and all three spots were recognized by HYB 71-2 in immunoblot analysis (Fig. 3A). Tricine SDS-–2-DE analysis was performed, and the spots representing CFP29 were excised from the PVDF membrane for sequence analysis. For all three spots, the same two sequences were detected after six cycles: MNNLYR and NNLYRD. This finding confirms the partial cleavage of the N-terminal methionine and suggests that all three spots represent the same gene product.
FIG. 3.
2-DE analysis. (A) Immunoblot analysis of the purified CFP29 with HYB 71-2. The three individual spots reacting with HYB 71-2 are marked by arrowheads. (B) Silver-stained gel of ST-CF. The positions of individual proteins are indicated. Numbers at the left of each panel are molecular masses in kilodaltons.
Figure 3B shows the pattern obtained in 2-DE of ST-CF proteins. The labeled spots were identified by 2-DE immunoblot analysis or by addition of individual purified proteins to samples of ST-CF followed by 2-DE analysis. The three spots representing CFP29 were mapped just below MPT59 in the pH range of 5.0 to 5.2.
To investigate whether glycosylation could be the explanation of the heterogeneity observed in 2-DE gels, a glycan detection kit was used to examine CFP29 for glycosylation. After SDS-PAGE, electroblotting to nitrocellulose, and labeling for glycan, 0.2 μg of the positive control (ovalbumin) from this kit was clearly identified as a glycoprotein; however, no staining of the CFP29 band (1 to 5 μg was applied) was observed (data not shown), indicating that CFP29 is not glycosylated.
Distribution of CFP29 in subcellular fractions.
Many M. tuberculosis culture filtrate antigens are derived from the envelope, from which they are gradually released (5). ST-CF and subcellular fractions of M. tuberculosis were analyzed for the presence of CFP29 and PstS (Table 1). CFP29 was found in ST-CF only in small quantities but was highly enriched in the membrane fraction. In agreement with earlier studies, the PstS homolog, the lipidated 38-kDa protein recognized by HBT12, was located predominantly in the membrane and cell wall fractions. The association between CFP29 and the membrane was evaluated by Triton X-114 phase separation of the cell membrane fraction. CFP29 preferably partitioned in the aqueous phase, whereas the majority of PstS was detected in the detergent phase, in agreement with previously published data (41).
TABLE 1.
Subcellular localization of CFP29 and PstS
| Subcellular fractiona | Reactivityb with protein (MAb):
|
|
|---|---|---|
| CFP29 (HYB 71-2) | PstS (HBT12) | |
| ST-CF | + | + |
| Cell wall | − | ++ |
| Cytosol | − | − |
| Membrane | +++ | +++ |
| Membrane, aq. | +++ | + |
| Membrane, det. | − | +++ |
aq. and det., aqueous and detergent phases, respectively, obtained after Triton X-114 phase separation. The same quantity of total protein was applied for all samples, except for the aqueous and detergent phases, which in total represented the same protein content as the other fractions.
Values from immunoblot analysis: −, no reactivity, +, weak reactivity, ++, intermediate reactivity, +++, strong reactivity.
The presence of CFP29 in the membrane fraction and ST-CF suggests that it may be gradually released from the envelope to the extracellular milieu during growth.
Characterization of the cfp29 gene.
Database searches for homology to the N-terminal sequence of CFP29 in EMBL and SWISSPROT by the TFASTA and FASTA algorithms, respectively, identified a B. linens protein, M18 Linocin (accession no. X93588) with 74% identity to the 19 N-terminal amino acids of CFP29. Linocin, encoded by the lin gene, is a potent bacteriocin with activity against Listeria species and other gram-positive bacteria (37).
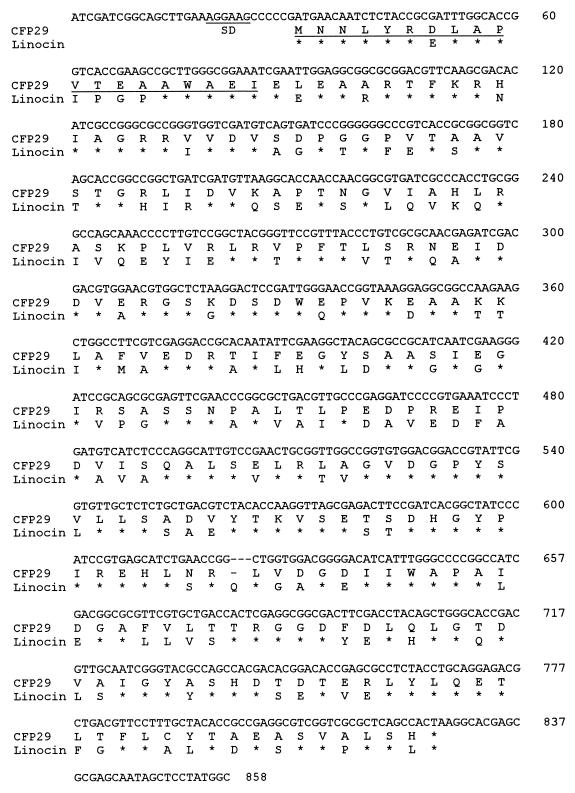
Because of this high degree of sequence identity, the lin gene sequence was used to design two degenerate PCR primers. By PCR, a DNA fragment was obtained with these primers and M. tuberculosis chromosomal DNA as the template. The sequence of the PCR fragment was identified in a 795-bp open reading frame on cosmid CY444 in the M. tuberculosis genome sequencing database at the Sanger Centre, Cambridge, United Kingdom. This 795-bp open reading frame has 62% identity to the lin gene sequence of B. linens, and the 5′ end translates into a sequence that shows 100% identity to the 19 N-terminal amino acids of CFP29. We therefore conclude that the 795-bp open reading frame corresponds to the gene encoding CFP29. This open reading frame was PCR amplified, cloned into the pMCT6 expression vector, and sequenced. The determined nucleotide sequence of the open reading frame, named cfp29, and the deduced sequence of 265 amino acids are shown in Fig. 4. The open reading frame is preceded by a putative Shine-Dalgarno-like sequence, 5′-GGAAGC-3′, centered 8 bp upstream of the ATG start codon. The nucleotide sequence of the cfp29 gene was completely identical to that of the 795-bp open reading frame identified on cosmid CY444 from the M. tuberculosis genome sequencing database. The start of the deduced amino acid sequence is identical to the N-terminal sequence of the mature CFP29, indicating that CFP29 has no signal sequence, and no consensus signal sequence was identified for the cfp29 gene by computer-assisted analysis (25).
FIG. 4.
Nucleotide sequence of the amplified PCR fragment and deduced amino acid sequence of CFP29. The putative Shine-Dalgarno (SD) site and the amino acid sequence confirmed by N-terminal sequencing are underlined. Alignment of the deduced amino acid sequence of CFP29 and that of B. linens Linocin is shown. A single gap was inserted to improve alignment. Identical amino acid residues are indicated by asterisks.
Comparison of Linocin and CFP29.
The deduced amino acid sequence of CFP29 shows 58% identity to that of Linocin (Fig. 4). The deduced molecular mass of CFP29 is 28,860 Da, and the calculated pI is 4.70. In comparison, the calculated mass of Linocin is 28,596 Da, and the pI is 4.41.
The observed similarities point to the existence of a Linocin homolog in M. tuberculosis. Linocin exists in its native form as a polymer with a high molecular mass (>2,000 kDa) (37). Like Linocin, CFP29 elutes close to void volume before any of the marker proteins on a Superose 6 gel filtration column, indicating that the molecular mass of CFP29 in its native form is at least 700 kDa.
Bacteriocin activity was therefore examined by spotting samples of native proteins on agar containing the indicator strains. Twenty-four Listeria monocytogenes strains with different sensitivities to inhibitory effects from Linocin were investigated. A single mycobacterial species, M. diernhoferi, was also included as indicator strain, since this species has been reported to be inhibited by a not-yet-defined molecule from M. tuberculosis (35). Linocin inhibited the growth of all L. monocytogenes indicator strains, as expected, whereas no inhibition of M. diernhoferi by Linocin was observed (data not shown). None of the indicator strains were inhibited by ST-CF or CFP29.
Interspecies analyses.
The presence of CFP29 in different mycobacterial species was examined at the protein level with HYB 71-2 by immunoblot analysis of mycobacterial culture filtrates. A 29-kDa band reacting with HYB 71-2 was observed in all culture filtrates tested (Table 2).
TABLE 2.
Presence of the CFP29 protein and gene in different species and strains of mycobacteria
| Species and strain | Sourcea | Presence of:
|
|
|---|---|---|---|
| Proteinb | Genec | ||
| M. tuberculosis | |||
| H37Rv (ATCC 27294) | ATCC | + | + |
| H37Ra (ATCC 25177) | ATCC | + | + |
| M. africanum patient isolate | SSI | + | + |
| M. bovis | |||
| AN5 (MNC 433) | SSI | + | + |
| BCG Danish 1331 | SSI | + | + |
| M. avium ATCC 15769 | ATCC | + | − |
| M. terrae ATCC 15755 | ATCC | + | + |
| M. marinum ATCC 927 | ATCC | + | + |
| M. szulgai patient isolate | SSI | + | + |
| M. scrofulaceum ATCC 19275 | ATCC | + | − |
| M. diernhoferi ATCC 19340 | ATCC | + | + |
ATCC, American Type Culture Collection; SSI, Statens Serum Institut mycobacterium collection.
The protein was identified in culture filtrates by immunoblot analysis with HYB 71-2.
The presence of the gene was investigated by PCR.
At the genetic level, PCR analysis with cfp29-specific primers demonstrated that the cfp29 gene is present in all species with the exception of Mycobacterium avium and Mycobacterium scrofulaceum (Table 2). DNAs from these two species were tested under less stringent conditions, but no PCR product could be detected. Since a HYB 71-2-reactive 29-kDa band is present in both species, the lack of PCR product may be explained by polymorphism in the region corresponding to the primers or by the presence of a shared epitope in a different protein with the same mobility in SDS-PAGE as CFP29.
Immunological activity of CFP29.
CFP29 is present in only small amounts in ST-CF, so to obtain CFP29 for immunological studies, recombinant histidine-tagged CFP29 was expressed and purified to homogeneity as described in Materials and Methods.
The T-cell recognition of purified native and recombinant CFP29 was evaluated and compared to those of other culture filtrate antigens from the 24- to 36-kDa region. Memory effector cells isolated from mice during the recall of protective immunity were stimulated in vitro with different concentrations of CFP29, MPT59, MPT51, and MPT64, and the cellular responses were compared with the response to ST-CF (Table 3). All antigens except MPT64 were recognized and gave rise to a dose-dependent release of IFN-γ in the cell cultures. As previously reported, MPT59 is a very prominent target molecule in this region (4), and MPT59 gave rise to almost the same cellular reactivity as ST-CF (Table 3). The CFP29 preparations also gave rise to high levels of IFN-γ, whereas MPT51 elicited only a limited release of IFN-γ. The native and recombinant CFP29 samples gave rise to similar dose-response curves, with increasing release of IFN-γ in the dose range of 0.5 to 8 μg/ml and the same level of IFN-γ at the highest concentration (Table 3).
TABLE 3.
Mouse IFN-γ response to ST-CF and antigens from the 24- to 36-kDa regiona
| Antigen | Concn (μg/ml) | IFN-γ release (ng/ml)b |
|---|---|---|
| ST-CF | 8.0 | 24.36 (0.22) |
| 2.0 | 24.09 (0.33) | |
| 0.5 | 11.12 (0.24) | |
| nCFP29c | 8.0 | 12.22 (0.13) |
| 2.0 | 3.74 (0.12) | |
| 0.5 | 1.20 (0.01) | |
| rCFP29d | 8.0 | 11.43 (0.18) |
| 2.0 | 9.37 (0.18) | |
| 0.5 | 1.61 (0.17) | |
| MPT51 | 8.0 | 2.33 (0.12) |
| 2.0 | 0.73 (0.03) | |
| 0.5 | 0.78 (0.03) | |
| MPT59 | 8.0 | 19.30 (0.55) |
| 2.0 | 19.37 (0.15) | |
| 0.5 | 10.67 (0.37) | |
| MPT64 | 8.0 | <0.05 |
| 2.0 | <0.05 | |
| 0.05 | <0.05 |
The responses were measured with memory effector T cells isolated from the spleen 4 days after challenge with M. tuberculosis.
Expressed as mean values from triplicate analyses performed with cells pooled from three mice. Numbers in parentheses are standard errors of the means.
nCFP29, native CFP29.
rCFP29, recombinant CFP29.
DISCUSSION
It is now considered a fact that M. tuberculosis culture filtrate contains targets for protective T cells, and in animal models, experimental vaccines based on culture filtrate proteins induce protective immunity (3, 27).
In the present study, we describe a novel T-cell antigen, CFP29, from the highly stimulatory 24- to 36-kDa region of M. tuberculosis ST-CF. This region contains antigens recognized strongly by memory effector cells in mouse and guinea pig models of TB (4, 16). Furthermore, this region is strongly recognized by Th1 cells in human TB patients with minimal disease (12). Although CFP29 is present in ST-CF in very limited amounts, we were able to isolate enough native CFP29 for a partial biochemical characterization and demonstration of its potent T-cell-stimulatory properties. In cellular assays, native as well as recombinant CFP29 was clearly identified as a T-cell antigen triggering the release of large quantities of IFN-γ from memory effector cells isolated during the recall of protective immunity in the mouse model of TB infection. However, given the complexity of the reactive 26- to 29-kDa fraction in SDS-PAGE, our identification of CFP29 does not exclude that other, as-yet-unidentified T-cell antigens may also contribute to the prominent release of IFN-γ in response to this fraction.
It has been suggested that the extracellular proteins most relevant for novel vaccines are the ones appearing in abundant quantities in culture filtrates (19). Since intracellular growth of M. tuberculosis in macrophages induces a significant change in protein expression compared to extracellular growth in culture medium (1, 23), the search for new M. tuberculosis antigens should not be biased by their relative quantities in vitro. The identification of the highly reactive T-cell antigens CFP29 and ESAT-6 (34), both of which are present in small amounts in culture filtrates, demonstrates that important antigens may be overlooked if only the most abundant molecules are characterized. For both antigens, immunoblot analysis with specific MAbs has proven to be a sensitive technique for identification and isolation of proteins which constitute less than 1% of the total protein quantity.
Investigation of subcellular fractions of M. tuberculosis showed that CFP29 is present in the membrane and culture filtrates but not in the cytosol fraction. This leads to the conclusion that CFP29 is exported and probably gradually released from the envelope, making it available for immune recognition in the early stage of infection. The localization of CFP29 emphasizes the relevance of defining antigens from various subcellular fractions of M. tuberculosis, and this matter is currently being pursued as part of our attempts to identify new M. tuberculosis T-cell antigens.
A consensus signal sequence is absent from the cfp29 gene, a phenomenon also observed for other extracellular mycobacterial proteins, like ESAT-6 and superoxide dismutase (34, 42), suggesting the existence of signal sequence-independent mechanisms for translocation of proteins across the bacterial membrane. The partitioning of CFP29 in the aqueous phase indicates a peripheral association of the protein with the membrane, and in agreement with this finding, no transmembrane segments in the amino acid sequence of CFP29 were identified by the algorithm of Klein et al. (20).
2-DE analysis of CFP29 showed three different spots in the pI range of 5.0 to 5.2 with the same N-terminal sequence. The cause of the heterogenity of CFP29 observed in 2-DE is not known, but it could be explained by deamidation, oxidation of amino acids, or posttranslational modifications such as phosphorylation. Our data suggested that glycosylation was not a possible explanation.
CFP29 is widely distributed in mycobacterial species, and its expression was demonstrated in culture filtrates from pathogenic species as well as nonpathogenic species of environmental origin, suggesting that CFP29 could play an important physiological role in mycobacteria. Comparison of the amino acid sequences, calculated pIs, and molecular masses of CFP29 and B. linens Linocin indicated that CFP29 is a mycobacterial homolog of Linocin, and it was therefore relevant to investigate whether CFP29 possesses bacteriocin activity. Twenty-four L. monocytogenes strains and M. diernhoferi were tested for growth inhibition in the presence of purified native CFP29 or ST-CF. However, no inhibition was observed with any of the strains tested. CFP29 may not share this property with Linocin, due to either significant differences in the amino acid sequences or lack of posttranslational modifications which may be important for the bacteriocin activity. Alternatively, CFP29 may have lost its activity during the purification process.
In this regard, M. tuberculosis is known to possess bacteriocin activity against several rapidly growing mycobacterial strains, including M. diernhoferi (35). The inhibitory activity was sensitive to heat treatment and proteinase digestion, implying that the responsible molecule is of proteinaceous origin. To our knowledge, no detailed characterization of the molecule responsible for this mycobacteriocin activity has been published. However, the expression of a 29-kDa protein which reacts with HYB 71-2 in the rapidly growing, nonpathogenic species M. diernhoferi further supports that CFP29 is not the molecule responsible for this inhibition observed by Takeya and Tokiwa (35). It therefore still remains to be established which physiological role CFP29 may play in mycobacteria.
Work is in progress to test the recognition of recombinant CFP29 by human TB patients in different stages of disease. The results obtained in the mouse model of TB infection are encouraging and suggest that CFP29 should be added to the list of important T-cell antigens derived from culture filtrates.
ACKNOWLEDGMENTS
This investigation received financial support from the WHO Global Programme for Vaccines and Immunization, The Danish Research Center for Biotechnology. The European Community (project no. TS*/CF94/0113), and the Hofmansgave Foundation.
We are grateful to Marie Olesen, Jette Pedersen, Vita Skov, Tove Slotved Simonsen, Birgitte Smedegaard, Mette Paulli Andersen, Bente Isbye, and Anne Blicher for excellent technical assistance. We thank Martin Elhay for critical reading of the manuscript.
REFERENCES
- 1.Alavi M R, Affronti L F. Induction of mycobacterial proteins during phagocytosis and heat shock: a time interval analysis. J Leukocyte Biol. 1994;55:633–641. doi: 10.1002/jlb.55.5.633. [DOI] [PubMed] [Google Scholar]
- 2.Andersen Å B, Ljungqvist L, Hasløv K, Bentzon M W. MPB 64 possesses ′tuberculosis-complex′-specific B- and T-cell epitopes. Scand J Immunol. 1991;34:365–372. doi: 10.1111/j.1365-3083.1991.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 3.Andersen P. Effective vaccination of mice against Mycobacterium tuberculosis infection with a soluble mixture of secreted mycobacterial proteins. Infect Immun. 1994;62:2536–2544. doi: 10.1128/iai.62.6.2536-2544.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen P, Andersen Å B, Sørensen A L, Nagai S. Recall of long-lived immunity to Mycobacterium tuberculosis infection in mice. J Immunol. 1995;154:3359–3372. [PubMed] [Google Scholar]
- 5.Andersen P, Askgaard D, Ljungqvist L, Bennedsen J, Heron I. Proteins released from Mycobacterium tuberculosis during growth. Infect Immun. 1991;59:1905–1910. doi: 10.1128/iai.59.6.1905-1910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersen P, Heron I. Simultaneous electroelution of whole SDS-polyacrylamide gels for the direct cellular analysis of complex protein mixtures. J Immunol Methods. 1993;161:29–39. doi: 10.1016/0022-1759(93)90195-d. [DOI] [PubMed] [Google Scholar]
- 7.Andersen P, Heron I. Specificity of a protective memory immune response against Mycobacterium tuberculosis. Infect Immun. 1993;61:844–851. doi: 10.1128/iai.61.3.844-851.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baek L. New, sensitive rocket immunoelectrophoretic assay for measurement of the reaction between endotoxin and Limulus amoebocyte lysate. J Clin Microbiol. 1983;17:1013–1020. doi: 10.1128/jcm.17.6.1013-1020.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry A L. Procedure for testing antibiotics in agar media: theoretical considerations. In: Lorian V, editor. Antibiotics in laboratory medicine. Baltimore, Md: Williams & Wilkins; 1980. pp. 1–23. [Google Scholar]
- 10.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 11.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boesen H, Jensen B N, Wilcke T, Andersen P. Human T-cell responses to secreted antigen fractions of Mycobacterium tuberculosis. Infect Immun. 1995;63:1491–1497. doi: 10.1128/iai.63.4.1491-1497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandt L, Oettinger T, Holm A, Andersen Å B, Andersen P. Key epitopes on the ESAT-6 antigen recognized in mice during the recall of protective immunity to Mycobacterium tuberculosis. J Immunol. 1996;157:3527–3533. [PubMed] [Google Scholar]
- 14.Fine, P. E. 1989. The BCG story: lessons from the past and implications for the future. Rev. Infect. Dis. 11(Suppl. 2):S353–S359. [DOI] [PubMed]
- 15.Harboe M, Malin A S, Dockrell H S, Wiker H G, Ulvund G, Holm A, Jørgensen M C, Andersen P. B-cell epitopes and quantification of the ESAT-6 protein of Mycobacterium tuberculosis. Infect Immun. 1998;66:717–723. doi: 10.1128/iai.66.2.717-723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasløv K, Andersen A, Nagai S, Gottschau A, Sørensen T, Andersen P. Guinea pig cellular immune responses to proteins secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:804–810. doi: 10.1128/iai.63.3.804-810.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry B S. Dissociation in the genus Brucella. J Infect Dis. 1933;52:374–402. [Google Scholar]
- 18.Hochstrasser D F, Harrington M G, Hochstrasser A-C, Miller M J, Merril C R. Methods for increasing the resolution of two-dimensional protein electrophoresis. Anal Biochem. 1988;173:424–435. doi: 10.1016/0003-2697(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz M A. A new TB vaccine. Immunologist. 1997;5:15–20. [Google Scholar]
- 20.Klein P, Kanehisa M, Delisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1987;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 22.Lee B-Y, Hefta S A, Brennan P J. Characterization of the major membrane protein of virulent Mycobacterium tuberculosis. Infect Immun. 1992;60:2066–2074. doi: 10.1128/iai.60.5.2066-2074.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee B-Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Invest. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljungqvist L, Worsaae A, Heron I. Antibody responses against Mycobacterium tuberculosis in 11 strains of inbred mice: novel monoclonal antibody specificities generated by fusions, using spleens from BALB.B10 and CBA/J mice. Infect Immun. 1988;56:1994–1998. doi: 10.1128/iai.56.8.1994-1998.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Orme I M, Miller E S, Roberts A D, Furney S K, Griffin J P, Dobos K M, Chi D, Rivoire B, Brennan P J. T lymphocytes mediating protection and cellular cytolysis during the course of Mycobacterium tuberculosis infection. Evidence for different kinetics and recognition of a wide spectrum of protein antigens. J Immunol. 1992;148:189–196. [PubMed] [Google Scholar]
- 27.Pal P G, Horwitz M A. Immunization with extracellular proteins of Mycobacterium tuberculosis induces cell-mediated immune responses and substantial protective immunity in a guinea pig model of pulmonary tuberculosis. Infect Immun. 1992;60:4781–4792. doi: 10.1128/iai.60.11.4781-4792.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollock J M, Andersen P. Predominant recognition of the ESAT-6 protein in the first phase of infection with Mycobacterium bovis in cattle. Infect Immun. 1997;65:2587–2592. doi: 10.1128/iai.65.7.2587-2592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porath J, Maisano F, Belew M. Thiophilic adsorption—a new method for fractionation. FEBS Lett. 1985;185:306–310. doi: 10.1016/0014-5793(85)80928-5. [DOI] [PubMed] [Google Scholar]
- 31.Roberts A D, Sonnenberg M G, Ordway D J, Furney S K, Brennan P J, Belisler J T, Orme I M. Characteristics of protective immunity engendered by vaccination of mice with purified culture filtrate protein antigens of Mycobacterium tuberculosis. Immunology. 1995;85:502–508. [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 34.Sørensen A L, Nagai S, Houen G, Andersen P, Andersen Å B. Purification and characterization of a low-molecular-mass T-cell antigen secreted by Mycobacterium tuberculosis. Infect Immun. 1995;63:1710–1717. doi: 10.1128/iai.63.5.1710-1717.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeya K, Tokiwa H. Bacteriocin-typing of Mycobacterium tuberculosis. Am Rev Respir Dis. 1974;109:304–305. doi: 10.1164/arrd.1974.109.2.304. [DOI] [PubMed] [Google Scholar]
- 36.Theisen M, Vuust J, Gottschau A, Jepsen S, Høgh B. Antigenicity and immunogenicity of recombinant glutamate-rich protein of Plasmodium falciparum expressed in Eschericia coli. Clin Diagn Lab Immunol. 1995;2:30–34. doi: 10.1128/cdli.2.1.30-34.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdés-Stauber N, Scherer S. Isolation and characterization of Linocin M18, a bacteriocin produced by Brevibacterium linens. Appl Environ Microbiol. 1994;60:3809–3814. doi: 10.1128/aem.60.10.3809-3814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valdés-Stauber N, Scherer S. Nucleotide sequence and taxonomical distribution of the bacteriocin gene lin cloned from Brevibacterium linens M18. Appl Environ Microbiol. 1996;62:1283–1286. doi: 10.1128/aem.62.4.1283-1286.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. The occurence and stability of insertion sequences in Mycobacterium tuberculosis complex strains; evaluation of LS-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. Tuberculosis control and research strategies for the 1990s: memorandum from a WHO meeting. Bull W H O. 1992;70:17–21. [PMC free article] [PubMed] [Google Scholar]
- 41.Young D B, Garbe T R. Lipoprotein antigens of Mycobacterium tuberculosis. Res Microbiol. 1991;142:55–65. doi: 10.1016/0923-2508(91)90097-t. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Lathigra R, Garbe T, Catty D, Young D. Genetic analysis of superoxide dismutase, the 23 kilodalton antigen of Mycobacterium tuberculosis. Mol Microbiol. 1991;5:381–391. doi: 10.1111/j.1365-2958.1991.tb02120.x. [DOI] [PubMed] [Google Scholar]