Abstract
Streptococcus mutans JH1000 and its derivatives were previously shown (J. D. Hillman, K. P. Johnson, and B. I. Yaphe, Infect. Immun. 44:141–144, 1984) to produce a low-molecular-weight, broad-spectrum bacteriocin-like inhibitory substance (BLIS). The thermosensitive vector pTV1-OK harboring Tn917 was used to isolate a BLIS-deficient mutant, DM25, and the mutated gene was recovered by shotgun cloning in Escherichia coli. Sequence analysis of insert DNA adjacent to Tn917 led to the identification of four open reading frames including two (lanA and lanB) which have substantial homology to the Staphylococcus epidermidis structural gene (epiA) and a modifying enzyme gene (epiB) for biosynthesis of the lantibiotic epidermin, respectively. Although the BLIS activity could not be recovered from broth cultures, high yields were obtained from a solid medium consisting of Todd-Hewitt broth containing 0.5% agarose that was stab inoculated with JH1140 (a spontaneous mutant of JH1000 that produces threefold-elevated amounts of activity). Agar could not substitute for agarose. Chloroform extraction of the spent medium produced a fraction which yielded two major bands on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The faster-migrating band was absent in chloroform extracts of the mutant, DM25. The amino acid sequence of this band was determined by Edman sequencing and mass spectroscopy. The results showed that it is a lantibiotic, which we have named mutacin 1140, and that the sequence corresponded to that deduced from the lanA sequence. We observed a number of similarities of mutacin 1140 to epidermin and an S. mutans lantibiotic, B-Ny266, but it appears to have significant differences in the positions of its thioether bridges. It also has other unique features with regard to its leader sequence and posttranslational modification. A proposed structure for mutacin 1140 is presented.
Lactate dehydrogenase (LDH)-deficient mutants of mutans streptococci have been studied for their potential use in the replacement therapy of dental caries (10). Without LDH, fermentation of carbohydrates by this microorganism occurs via alternate pathways for pyruvate metabolism that yield significant amounts of neutral end products and smaller amounts of total acids (6). As a result, LDH-deficient mutants are less cariogenic. In addition to being less cariogenic, an effector strain for replacement therapy of dental caries must demonstrate superior colonization properties to displace indigenous mutans streptococcal strains which may be present in the host while preventing subsequent colonization by wild-type strains whenever the host is exposed to them.
Numerous studies (13, 14, 22, 25, 26) have documented the difficulty of introducing mutans streptococci into the mouths of humans, particularly if they already harbor an indigenous strain of this organism. We previously reported the isolation from a clinical sample of a strain of Streptococcus mutans called JH1000, which has unusually good colonization properties (9). This strain was found to produce a potent, broad-spectrum bacteriocin-like inhibitory substance (BLIS). In a deferred antagonism assay, the JH1000 BLIS was found to inhibit the growth of representative strains of S. salivarius, S. sanguis, S. oralis, S. mitis, S. pyogenes, Staphylococcus aureus, Lactobacillus salivarius, L. casei, Actinomyces israelii, A. naeslundii, and A. viscosus. In addition, virtually all 124 mutans streptococcal strains tested, including both fresh isolates and laboratory strains, were sensitive. Preliminary studies performed on crude BLIS preparations indicated that the active component was both small (molecular weight, <1,000) and proteinaceous.
Analysis of isogenic mutants demonstrated a good correlation between BLIS production and colonization potential in both a rodent model (9) and human subjects (8, 11). A mutant called JH1005, which produces ca. threefold-elevated levels of BLIS activity, could be recovered from saliva samples 3 years after brushing and flossing it onto the teeth of human subjects during a single, 3-min infection regimen (7, 8). During that period, the proportion of indigenous S. mutans cells recoverable from the same plaque samples decreased to undetectable levels. Based on these and other results, it was proposed that an LDH-deficient mutant of JH1005 would possess the necessary combination of properties, including low virulence and superior colonization potential, to serve as an effector strain in the replacement therapy of dental caries.
Previous, unreported attempts involving standard biochemical methods to isolate and characterize the JH1005 BLIS activity were unsuccessful, probably because its production is less reliable when cells are grown in liquid medium than when they are grown in solid medium. In this paper, we describe genetic methods which enabled us to identify the BLIS activity and to devise methods for its large-scale purification. It was found to be a lantibiotic with significant homology to epidermin produced by Staphylococcus epidermidis and to the recently reported lantibiotic from S. mutans Ny266 (18).
MATERIALS AND METHODS
Organisms and culture conditions.
S. mutans JH1005 and JH1140 are, respectively, ethyl methanesulfonate-induced and spontaneous mutants of JH1000 that show three- to fourfold-increased BLIS activity (9). Strain DM25 is a Tn917-induced BLIS-negative mutant of JH1005 (5). S. rattus BHT-2 (resistant to 1 mg of streptomycin sulfate ml−1 [9]) was used as a target strain in BLIS purification assays. Other bacterial strains are listed in Table 1. For routine cultivation, brain heart infusion (BHI) broth and agar (1.5%) (Difco Laboratories, Detroit, Mich.) were used. For determination of auxotrophic properties, the minimal medium of Carlsson (2) was used.
TABLE 1.
Spectrum of antibacterial activity of parent, mutant, and partially purified mutacin 1140 (fraction A)a
| Target strain | Zone of growth inhibition (mm)b for test strain or compound:
|
||
|---|---|---|---|
| JH1140 | DM25 | Fraction A | |
| Mutans streptococci | |||
| S. cricetus E49 | 11 | 0 | +c |
| S. rattus BHT-2 | 16 | 0 | + |
| S. mutans NG8 | 12 | 0 | + |
| S. mutans OMZ175 | 10 | 0 | + |
| S. mutans V100 | 11 | 0 | + |
| S. sobrinus SL1 | 10 | 0 | + |
| S. sobrinus ATCC 27352 | 8 | 0 | + |
| S. downeii ATCC 33748 | 17 | 0 | + |
| S. salivarius | |||
| ATCC 25975 | 17 | 0 | + |
| ATCC 27945 | 19 | 0 | + |
| S. mitis ATCC 19950 | 20 | 0 | + |
| S. sanguis | |||
| PC10 | 10 | 0 | + |
| PC13 | 10 | 0 | + |
| Challis | 8 | 0 | + |
| S. pyogenes | |||
| Manfredo | 18 | 0 | + |
| MC5 | 17 | 0 | + |
| Lactobacillus casei ATCC 7469 | 12 | 0 | + |
| Staphylococcus aureus Cowan | 16 | 0 | + |
Chloroform treatment of JH1140 culture liquor yielded a precipitate, which was dissolved in 50% ethanol to yield fraction A, containing partially pure mutacin 1140.
Diameter of zone of growth inhibition.
+, zone of growth inhibition was observed but not measured.
Tn917 mutagenesis and recovery of interrupted DNA.
Plasmid pTV1-OK is a repA(Ts) derivative of the Lactococcus lactis cryptic plasmid pWV01 for temperature-dependent replication in both S. mutans and Escherichia coli (5). It possesses a kanamycin resistance (Kanr) gene, which functions in both E. coli and S. mutans, and transposon Tn917, which confers erythromycin resistance (Emr) in streptococci and in E. coli MC1061. Transposon mutagenesis of strain JH1005 harboring pTV1-OK was performed by the method of Gutierrez et al. (5). After a temperature shift to eliminate the plasmid, Emr clones were selected on BHI agar containing 15 μg of the antibiotic ml−1. These were stab inoculated (25 stabs/plate) into the same medium without antibiotic. After incubation overnight in candle jars at 37°C, the plates were overlaid with 3 ml of top agar containing ca. 105 CFU of BHT-2 ml−1. Stabbed clones which failed to produce a zone of growth inhibition in the BHT-2 lawn in this deferred-antagonism assay were recovered and purified by streaking on medium containing erythromycin.
Chromosomal DNA from transposon mutant DM25 was digested with EcoRI, which does not cut in the transposon, and ligated to EcoRI-linearized and dephosphorylated pUC19 DNA as previously reported (5). Ligated DNA was used to transform E. coli MC1061, and recombinants were plated onto Luria-Bertani agar containing 100 μg of ampicillin ml−1. After 24 h at 37°C, the colonies were replica plated onto Luria-Bertani agar containing both ampicillin and 300 μg of erythromycin ml−1 and incubated for an additional 48 h. Colonies which arose were purified by streaking, and their plasmid DNA was isolated by a modified alkaline lysis-polyethylene glycol precipitation procedure (1). Insert DNA was sequenced with both Tn917- and pUC19-based primers (5) by the DNA Sequencing Core Facility of the Interdisciplinary Center for Biotechnology Research, University of Florida, using the Taq DyeDeoxy Terminator and DyePrimer cycle-sequencing protocols as described by Applied Biosystems, Inc. (Foster City, Calif.). The fluorescently labeled extension products were analyzed on an Applied Biosystems model 373A DNA sequencer. Homology searches of sequence databases were performed with the BLAST program from the National Centers for Biotechnology Information (Bethesda, Md.). Amino acid sequence alignments were generated with the CLUSTAL-W program and displayed with SeqVu v. 1.0.1 (Garvan Institute of Medical Research, Sydney, Australia).
Other genetic methods.
Southern analysis was performed with pTV1-OK as a probe and EcoRI-digested chromosomal DNA. The enhanced chemiluminescence gene detection system was used as specified by the manufacturer (Amersham International PLC, Little Chalfont, England). Transformation of S. mutans was carried out by the method of Perry and Kuramitsu (21). Other DNA manipulations were as described by Maniatis et al. (16).
Purification and characterization of the JH1140 BLIS.
Large-scale preparations of BLIS were performed with Todd-Hewitt broth (Difco) containing 0.5% SeaKem LE agarose (FMC BioProducts, Rockland, Maine). Batches of this medium (4 liters) in petri dishes were stab inoculated with a 10-prong replicator that was first stabbed into a BHI plate with a confluent lawn of JH1140. The plates were incubated in candle jars for 72 h at 37°C. The agar was scraped from the plates, aliquoted into centrifuge bottles, and frozen overnight at −20°C. The bottles were centrifuged at 4,000 × g for 60 min and then at 8,000 × g for 30 min at room temperature. The resulting supernatant was passed through Whatman no. 1 filter paper to remove agar fines. Chloroform (33.3 ml liter of culture supernatant−1) was added, and the mixture was vigorously agitated for 2 h with a magnetic stirrer. After overnight standing at room temperature, the aqueous phase was removed by aspiration. The remaining chloroform layer, containing a white precipitate, was centrifuged at room temperature for 8 min at 2,500 × g. The chloroform was decanted, and the precipitate was isolated, washed twice by centrifugation with 10 ml of chloroform, and dried under a stream of nitrogen in a 45°C water bath. A 1-ml volume of 50% ethanol was added to the resulting residue and vortexed vigorously for 15 s. Undissolved material was removed by centrifugation at 14,000 × g for 1 min at room temperature. The supernatant, which we called fraction A, was stored at 4°C.
A sample (300 μl) of fraction A was diluted 1:3 with distilled and deionized H2O and freeze-dried overnight in a Savant Speed Vac. The resulting residue was dissolved in 10 μl of Tricine-sodium dodecyl sulfate (SDS) sample buffer (Novex Experimental Technologies, San Diego, Calif.) and heated in a boiling-water bath for 5 min. After being cooled to room temperature, the sample was electrophoresed on an SDS-Tris-Tricine–10 to 20% polyacrylamide gradient Ready gel (Bio-Rad Laboratories, Hercules, Calif.) along with Mark 12 protein standards (Novex). Proteins were electroblotted at 90 V for 30 min onto an Immobilon P membrane (Millipore Corp., Bedford, Mass.) in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (pH 10) containing 20% methanol. The membranes were stained for 1 to 2 min with a solution containing 0.02% Coomassie brilliant blue R250, 40% methanol, and 5% acetic acid and destained for 4 to 5 min with 40% methanol–5% acetic acid. The appropriate band was then isolated for amino acid analysis and Edman degradation analysis, which were performed by the University of Florida Interdisciplinary Center for Biological Research Protein Chemistry Core. Sequencing also was performed by the alkaline ethanethiol derivatization method described by Meyer et al. (17) and modified for electroblotted samples on Immobilon P membranes. In this modification, ethanol was substituted for methanol in the reaction mixture. The membrane was carefully wetted with the reaction mixture to avoid dripping and was placed in a microcentrifuge tube under argon. The tube was incubated at 50°C for 1 h, and the band was applied to an ABI 494 protein sequencer operated under conditions of normal liquid-pulsed cycling. The lantibiotic nisin A (Sigma Chemical Co., St. Louis, Mo.) from Lactococcus lactis served as a control, and its analysis was carried out in parallel.
The purified polypeptide was analyzed by electrospray ionization mass spectroscopy (MS) on a PE Sciex API III Biomolecular mass analyzer. For detection of thioether and didehydro amino acids, the sample was vacuum dried and the residue was dissolved in 30 μl of a derivatization mixture composed of 1.4 M ethanethiol and 0.5 M NaOH in 46% aqueous ethanol as previously reported (20). The sample was then incubated for 60 min at 50°C under argon. The reaction was stopped by the addition of 2 μl of glacial acetic acid. Nisin A served as a control. Tandem mass spectrometry (MS/MS) was performed by selecting the parent ion having a particular m/z with the first quadrupole and allowing it to move into the second quadrupole, where it collided with argon atoms and fragments. These fragments were then analyzed with the third quadrupole.
Assay of mutacin 1140 activity.
Two methods were used to determine the presence of mutacin 1140 activity. In the quantitative method, samples (25 μl) of fractions to be tested for mutacin 1140 activity were serially twofold diluted in distilled and deionized H2O in 96-well microtiter plates. Top agar (BHI broth containing 0.75% agar) was melted and then cooled to 42°C in a water bath. An overnight culture of BHT-2 grown in BHI broth was added to the top agar to give a final concentration of ca. 105 CFU ml−1. A stock solution of streptomycin sulfate was also added to give a final concentration of 1 mg ml−1. Aliquots (200 μl) of the top agar were then added to the samples in the microtiter wells. After gelling, the plates were sealed with Mylar and incubated inverted at 37°C for 24 h. Growth of BHT-2 was determined for each well under ×10 magnification, and the activity titer for each sample tested was calculated as the reciprocal of the highest dilution which inhibited growth.
A deferred-antagonism assay was used to qualitatively test for the presence of mutacin 1140 activity. An overnight broth culture of the target strain was added to molten (42°C) top agar to a final concentration of ca. 105 CFU ml−1. A 3-ml sample was used to cover the surface of a THB plate. After gelling, 1 to 5 μl of the test sample was spotted on the surface and allowed to air dry. The plates were examined after overnight incubation at 37°C for zones of growth inhibition.
Nucleotide sequence accession number.
The GenBank/EMBL accession number for the nucleotide and deduced amino acid sequences of the genes described here is AF051560.
RESULTS
Genetic studies.
A culture of S. mutans JH1005, transformed with the Tn917-bearing, temperature-sensitive plasmid pTV1-OK, was shifted to 42°C to eliminate the plasmid. The frequency of transposition (Emr) was found to be ca. 10−4 cell−1, and the frequency of replicon fusion (integration of the entire plasmid into the chromosome; Emr Kanr) was ca. 10−6 cell−1. A total of 750 independent Tn917 transposon mutants of JH1005 were individually analyzed for BLIS production by determining their ability to inhibit the growth of the S. rattus target strain BHT-2 in the deferred-antagonism assay. One BLIS-negative mutant, DM25, was isolated in this fashion.
Besides the S. rattus strain used to identify DM25, the deferred-antagonism assay was used to show that this mutant could not inhibit the growth of other gram-positive bacterial species that are sensitive to the JH1005 BLIS (Table 1). No other phenotypic properties were associated with the genetic lesion in DM25. The mutant grew normally both aerobically and anaerobically in Carlsson’s minimal medium containing various carbon sources.
Southern analysis with pTV1-OK as a probe and EcoRI-digested chromosomal DNA indicated a single copy of Tn917 in the genome of DM25; no hybridization to chromosomal DNA of the parent strain, JH1005, was observed (data not shown). A backcross in which 10 μg of DM25 chromosomal DNA was used to transform JH1005 yielded two erythromycin-resistant clones. Both were found to be BLIS negative in the deferred-antagonism assay with BHT-2 as the target strain. These results suggested that a single, broad-spectrum BLIS is expressed by the parent and that its production was eliminated in the mutant by means of a single Tn917 insertion.
To identify the Tn917-inactivated gene associated with BLIS production in JH1005, marker rescue of the Tn917 erm gene and flanking chromosomal DNA 5′ and 3′ to the transposon insertion was performed as previously described (5). The pUC19-based plasmid that was obtained, pDM25 (ca. 10 kbp), had a total of 2,086 bp of flanking DNA. There are four significant open reading frames in this sequence, all of which are oriented in the same direction (Fig. 1). orfX is 162 bp and possesses no homology to any sequence in databases at the nucleotide or amino acid level. An intergenic region of 588 bp separates the end of orfX from the start of lanA. This region contains a large number of inverted repeats, some of which are shown. This region also contains streptococcal promoterlike −35 and −10 sequences and ribosome binding sites for the second open reading frame, lanA. BLAST analysis of lanA revealed significant homology to several lantibiotics, particularly epidermin, in databases. lanA encodes a polypeptide of 63 amino acid residues. At 27 bp after the end of lanA is the start of orfY, which also possesses no homology to database sequences. There are no apparent transcriptional or translational signal sequences upstream of orfY, but there is a strong inverted repeat that could serve as a rho-independent termination signal for lanA and that bridges the start of orfY. The site of Tn917 insertion is within orfY. There is also a stretch of 20 bases that exactly duplicates a portion of the lanA sequence, suggesting an evolutionary relationship. The start of the fourth open reading frame, lanB, is 133 bp downstream from the end of orfY. This intergenic region contains streptococcal promoterlike and ribosome binding sequences for transcription and translation initiation for lanB. This open reading frame is 555 bp and bears significant homology to epiB (24), a gene which encodes a modifying enzyme involved in the processing of the epidermin prepropeptide. lanB is predicted to encode a protein of 184 amino acids (21.6 kDa), which is considerably smaller than the predicted product of epiB.
FIG. 1.
lan gene cluster. (A) Diagram of the orientation and relationship of the four open reading frames and the site of Tn917 insertion (∇). (B) Nucleic acid sequence of the JH1005 DNA insert in pDM25, showing the deduced amino acid sequences encoded by the open reading frames, putative stem-loop structures, putative transcriptional and translational regulatory sequences, the site of cleavage of the prelantibiotic (↑), the partial gene duplication in lanA and orfY, and the site of Tn917 insertion (∇).
Purification and characterization of the JH1140 BLIS.
Although JH1005 and JH1140 make approximately threefold-elevated amounts of BLIS compared to their parent, JH1000, broth culture liquors of these strains did not contain reliably high levels of activity. Consequently, we attempted to purify the putative lantibiotic indicated by the previous studies by growing JH1140 on solid medium consisting of Todd-Hewitt broth containing 0.75% agar. Plates that were stab inoculated with JH1140 and incubated for 3 days showed large (>25-mm) zones of inhibition when overlaid with BHT-2. When the fluid phase of the agar was extracted by freeze-centrifugation and treated with chloroform, no activity was associated with the resulting precipitate, as would be expected for a lantibiotic (19). JH1140 produced comparable zones of growth inhibition on medium in which agarose (0.5%) was substituted for agar. Chloroform treatment of the fluid extracted from agarose-containing medium produced a precipitate which, when dissolved in 50% ethanol (fraction A), did contain large amounts of an activity that inhibited the growth of BHT-2 and other strains sensitive to the JH1140 BLIS (Table 1). This result suggested that agar contained a contaminant which bound the active BLIS component during the purification procedure and which was absent in agarose.
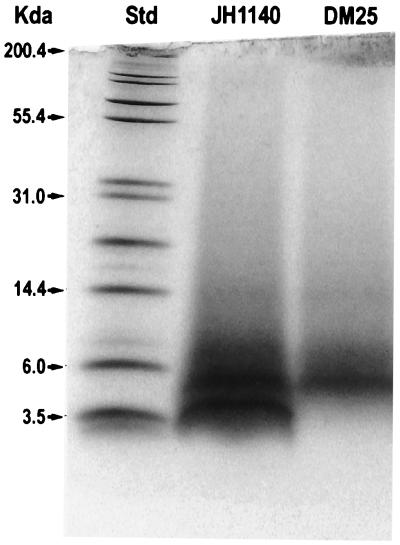
SDS-polyacrylamide gel electrophoresis (PAGE) analysis of fraction A using Tris-Tricine–10 to 20% polyacrylamide gels showed two major bands with apparent molecular weights of ca. 3,500 and 4,500 (Fig. 2). An identical preparation of the Tn917 mutant, DM25, failed to reveal the lower-molecular- weight band, indicating that this band was responsible for the BLIS activity. The bands were electrotransferred to an Immobilon P membrane, and the smaller band was shown by amino acid analysis to contain lanthionine and methyllanthionine residues with nisin as a control (data not shown). Edman degradation of this band stopped after two cycles, indicating that the subsequent residue was a didehydro residue typical of other lantibiotics (23). Treatment with alkaline ethanethiol allowed complete sequencing to be performed (Table 2). The results of this study were interpreted to confirm the primary amino acid sequence deduced from the nucleotide sequence of lanA. It also identified the site of cleavage of the leader sequence from the prepeptide (Fig. 1).
FIG. 2.
SDS-PAGE analysis of fraction A from the parent and mutant. Chloroform treatment of JH1140 and DM25 culture liquors yielded precipitates, which were dissolved in 50% ethanol to yield fraction A. Samples were electrophoresed on a Tris-Tricine-10 to 20% polyacrylamide gradient gel and stained with Coomassie brilliant blue. Std, protein molecular weight standard.
TABLE 2.
Edman sequencing of mutacin 1140 derivatized with ethanethiol
| Cycle | Residue predicted from DNA sequence | Identified residue |
|---|---|---|
| 1 | Phe | Phe |
| 2 | Lys | Lys |
| 3 | Ser | S-ECa |
| 4 | Trp | Trp |
| 5 | Ser | S-EC |
| 6 | Leu | Leu |
| 7 | Cys | S-EC |
| 8 | Thr | β-M-S-ECa |
| 9 | Pro | Pro |
| 10 | Gly | Gly |
| 11 | Cys | S-EC |
| 12 | Ala | Ala |
| 13 | Arg | Arg |
| 14 | Thr | β-M-S-EC |
| 15 | Gly | Gly |
| 16 | Ser | S-EC |
| 17 | Phe | Phe |
| 18 | Asn | Asn |
| 19 | Ser | S-EC |
| 20 | Tyr | Tyr |
| 21 | Cys | NDb |
| 22 | Cys | ND |
Thioethyl cysteine (S-EC) derived from ethanethiol treatment of lanthionine, 3-methyllanthionine, and 2,3-didehydroalanine; β-methylthioethyl cysteine (β-M-S-EC) derived from ethanethiol treatment of 3-methyllanthionine and 2,3-didehydrobutyrine according to the scheme of Meyer et al. (17).
ND, not detected.
Electrospray ionization MS indicated an Mr of 2,263.2 ± 0.01 by calculating the mass from doubly charged (m/z 1,132.63) and triply charged (m/z 755.42) molecular ions. This Mr was also confirmed by using a blotted peptide and a MALDI-TOF mass spectrometer. A combination of chemical modification and MS (20) confirmed the presence of modifications typical for lantibiotics as indicated by Edman sequencing. We detected six to eight modifications consisting of additions of ethanethiol molecules to the LanA peptide. The most prominent were the signals corresponding to six and seven modifications. These probably represent six dehydrations (four thioether bridges, two didehydro residues) and one oxidative decarboxylation at the C terminus, creating another double bond. The fully modified lanA product therefore has the predicted Mr for the unmodified propeptide minus 154 (6 × 18 [loss of water] and 46 [loss of HCOOH]), which is in perfect agreement with the detected Mr of 2,263.
The signal of the doubly charged molecular ion was so abundant that it permitted analyses by MS/MS and resulted in a series of daughter ion fragments of the molecule. The mass spectrum of daughter ions exhibited an unusual distribution, typical of lantibiotics studied by fast atom bombardment and thermospray MS (15). The spectrum below the m/z 1,133 of the parent ion had many signals indicative of overlapping peptide fragments. The region with mass above the m/z of the parent ion exhibited several signals separated by empty sections. These empty parts corresponded very well to the locations of thioether bridges in the molecule. The y series of signals (m/z 1,989, 1,920, 1,734, 1,450, and 1,367) corresponded to the molecular ion minus Phe and Lys residues, followed by deletions of didehydroalanine (dhA), Trp, Ser, Leu, Cys thioether ring, and didehydrobutyrine (dhB), respectively. The b series of ions (m/z 129, 276, and 345) confirmed the N-terminal sequence as Phe-Lys-dhA. m/z 1,467 and 1,524 are likely to represent the N-terminal fragment cleaved before and after Gly at position 15. The m/z 1,192 would thus be derived from m/z 1,467 by removal of the N-terminal Phe and Lys. These data confirmed the results of Edman sequencing and allowed us to assign some of the posttranslational modifications. Ser in position 3 and Thr in position 8 appear to be dehydrated. Ser in position 5 is the partner for Cys in position 7. Also, Cys in position 11 is likely to bridge with Thr in position 14, but this finding needs confirmation. The C-terminal part contains two thioether bridges, and their assignment will have to be resolved, most probably by nuclear magnetic resonance spectroscopy.
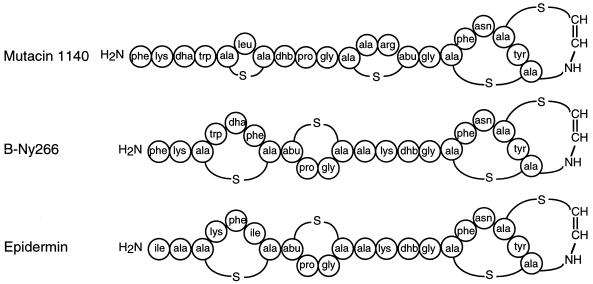
Based on these results, we have proposed a tentative structure for mutacin 1140 (Fig. 3). This molecular structure has a predicted molecular weight of 2,263, which agrees precisely with results obtained by MS analysis.
FIG. 3.
Proposed structure of mutacin 1140 and comparison to B-Ny266 and epidermin. Abbreviations: dha, 2,3-didehydroalanine; dhb, 2,3-didehydrobutyrine; abu, 2-aminobutyric acid.
DISCUSSION
Because strain JH1005 is poorly transformable, it was not possible to create a library by conventional insertional inactivation methods to screen for BLIS mutants. We have previously described the construction and successful testing of the thermosensitive vector, pTV1-OK harboring Tn917, as a tool for random mutagenesis in poorly transformable strains of S. mutans (5) and other streptococci. Previously, we reported the isolation of a Tn917-induced BLIS-negative mutant which proved to be defective in formyl-tetrahydrofolate synthetase (3). In the present study, we isolated another BLIS-negative mutant, DM25, that allowed us to identify the BLIS activity as a lantibiotic called mutacin 1140.
Marker rescue of the DM25 Tn917 insertion yielded a pUC19-based plasmid containing a total of 2,086 bp of DNA 5′ and 3′ to the transposon. Four potential open reading frames were identified, two of which (orfX and orfY) had no homology to database sequences. The remaining two open reading frames had extensive homology to the structural gene and a gene encoding a modifying enzyme for the S. epidermidis lantibiotic epidermin.
The BLIS-negative phenotype of DM25 cannot be clearly explained by the location of Tn917 in orfY, which has no apparent ribosome binding site. If orfY is expressed, it may be from a nonstandard start site upstream or downstream of the indicated ATG, or there may be a nonstandard ribosome binding site. Expression of orfY may occur in the absence of a ribosome binding site by ribosomal hopping, as described by Farabaugh (4), from lanA, the end of which is only 27 bp removed from the start of orfY. Polar effects of Tn917 on lanB could be invoked, even though this gene appears to have its own promoter based on sequence analysis. It remains to be determined whether lanB is part of the lanA transcript or is transcribed separately.
A large number of BLIS activities from a variety of species have been difficult to purify and characterize owing largely to their poor production in liquid medium. We have preliminary data (27) which indicate that, in the case of mutacin 1140, this is due in part to inhibition of transcription of the structural gene by the mature lantibiotic. We hypothesize that diffusion of the lantibiotic away from stab-inoculated cells growing on solid medium permits a greatly extended period of synthesis before a threshold inhibitory concentration is reached.
However, recovery of mutacin 1140 from solid medium in practical amounts was not possible until we replaced standard cultivation agar with agarose. Comparable inhibition zone sizes were observed when an S. rattus target strain was overlaid on either agar or agarose plates stabbed with JH1140. Hence, agar did not affect the synthesis or stability of mutacin 1140 but must have bound it in a fashion that prevented its extraction into chloroform without altering its biological activity. We did not examine the possibility of eluting mutacin 1140 from agar, which might have served as a practical purification step. Rather, we found that substitution of agarose for agar permitted the recovery of mutacin 1140 from freeze-centrifugation extracts where it constituted one of only two major protein bands as seen on polyacrylamide gels. Perhaps as many as 20 additional proteins were also present in small or trace amounts.
The amino acid sequence of purified mutacin 1140 as determined by Edman analysis and MS corresponded to that deduced from the lanA sequence. The observed molecular weight of 2,263 is considerably larger than that predicted in previous studies (9), in which mutacin 1140 was found to pass through dialysis membranes with a molecular weight cutoff of 1,000. Also, the migration of mutacin 1140 in SDS-PAGE analysis was slower than expected based on its known molecular weight (Fig. 2). We believe that both of these apparent discrepancies are because mutacin 1140, like other lantibiotics (23), possesses a somewhat rigid, rod-like structure that would enhance its ability to move through dialysis membrane pores and retard its electrophoretic mobility.
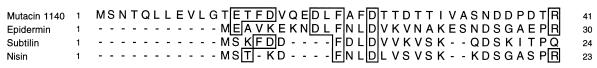
The deduced primary amino acid sequence of the lanA leader sequence showed limited homology to the leader sequences of other reported lantibiotics (Fig. 4). However, 16 of the 22 amino acids that form the mature mutacin 1140 are identical to the epidermin sequence and 2 others have conserved homology (Fig. 3). Based on the proposed structure of the S. mutans lantibiotic B-Ny266 (18), mutacin 1140 is probably more closely related to this molecule. Since Edman sequencing of ethanethiol-treated lantibiotics does not permit assignment of a position for Cys and Ser residues involved in the formation of lanthionine, and in the absence of genetic data, the precise primary amino acid sequence and secondary structure of B-Ny266 are unknown. However, it is possible that B-Ny266 differs from mutacin 1140 in as few as two positions, i.e., Phe versus Leu at residue 6 and Lys versus Arg at position 13. Both of these differences can occur as a result of single-base changes in the lanA nucleotide sequence.
FIG. 4.
Multiple amino acid sequence alignment of the leader sequences of mutacin 1140 and other lantibiotics. Sequence homology alignment was performed with the CLUSTAL-W program. Mutacin 1140 residues that are identical to other aligned lantibiotic residues are indicated by boxes.
The proposed secondary structure of mutacin 1140 differs significantly in the N-terminal half of the molecule from that proposed for B-Ny266 and from the established structure for epidermin (Fig. 3). Our MS data strongly support the smaller and more widely separated ring structures as depicted. Differences in the ring structures of the C-terminal half of the molecules may also exist, but our MS data were not able to provide a definitive structure for this portion of mutacin 1140. Additional data from nuclear magnetic resonance spectroscopy analysis are being gathered to answer this question. The variation between mutacin 1140 and B-Ny266 or epidermin in the positions of their thioether bridges and didehydro amino acids probably reflects differences in the specificities of the modifying enzymes, although it is also possible that the several differences in their amino acid sequences are responsible. It would be of interest to put the lanA structural gene into an appropriate Ny266 or S. epidermidis background to clarify this point.
A novel feature of the proposed mutacin 1140 structure is that the Cys-derived “half” of the methyllanthionine (residues 11 to 14) is located on the N-terminal side of the diamino acid while the Thr-derived half is located toward the C terminus. This has not been observed previously among the type A lantibiotics (23). Also, the deduced lanA polypeptide does not show a consensus sequence surrounding the point of cleavage between the leader peptide and the propeptide (Fig. 3 and 4). Unlike other reported lantibiotics, which contain hydrophobic amino acids in positions −4 and +2 and Pro in position −2, mutacin 1140 contains Pro, Lys, and Thr, respectively. This finding differs from results for previously reported lantibiotics and suggests the possibility that processing of mutacin 1140 differs from that of other known lantibiotics.
We have discovered by Edman degradation following ethanethiol derivatization that the other major protein present in chloroform precipitates of JH1140 culture liquors has a sequence that is identical to the faster-migrating band, which we know from the above studies to be the mature lantibiotic mutacin 1140. No additional residues were found at either the amino- or the carboxy-terminal end of the molecule. This result has been confirmed by MALDI-TOF MS of the blotted band, which showed a peak with a molecular weight of 2,263, along with several other minor contaminant peaks. This observation is difficult to explain: the slower band may be a partially or wholly unmodified or differently modified molecule, reflecting the Tn917 insertion which knocks out expression of one or more enzymes involved in processing the prepropeptide. While this would be expected to result in differences in apparent molecular weight by SDS-PAGE analysis, we would also expect differences in the Edman sequencing, which were not observed. We anticipate that further MS analyses will help to resolve this question.
Mutant analysis of JH1000 provided the first strong evidence for the role of a BLIS activity in promoting the colonization of a susceptible host by a producer strain (9). The identification here that the JH1000 BLIS is a lantibiotic accords well with our proposed scheme to use a JH1000 derivative as an effector strain for replacement therapy of dental caries. Nisin, the prototype lantibiotic, has low toxicity (12) and has been used widely for food preservation. Other lantibiotics are also being developed for use in clinical treatment of infections and other applications.
ACKNOWLEDGMENTS
We thank Marion Kirk, Lori Coward, and Stephen Barnes, University of Alabama, for mass spectrometric measurements and the Interdisciplinary Center for Biological Research, University of Florida, for expert technical assistance.
This work was supported in part by NIH grant DEO4529 and by a University of Florida Research and Technology grant.
REFERENCES
- 1.Applied Biosystems, Inc. High-quality template DNA for Taq cycle sequencing using Dye-Deoxy terminators: an improved preparation procedure. User bulletin 18. October 1991. Foster City, Calif: Applied Biosystems, Inc.; 1991. [Google Scholar]
- 2.Carlsson J. Nutritional requirements of Streptococcus mutans. Caries Res. 1970;4:305–320. doi: 10.1159/000259653. [DOI] [PubMed] [Google Scholar]
- 3.Crowley P J, Gutierrez J A, Hillman J D, Bleiweis A S. Genetic and physiologic analysis of a formyl-tetrahydrofolate synthetase mutant of Streptococcus mutans. J Bacteriol. 1997;179:1563–1572. doi: 10.1128/jb.179.5.1563-1572.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farabaugh P J. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez J A, Crowley P J, Brown D P, Hillman J D, Youngman P, Bleiweis A S. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillman J D, Andrews S W, Dzuback A L. Acetoin production by wild-type strains and a lactate dehydrogenase-deficient mutant of Streptococcus mutans. Infect Immun. 1987;55:1399–1402. doi: 10.1128/iai.55.6.1399-1402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillman J D, Andrews S W, Painter S, Stashenko P. Adaptive changes in a strain of Streptococcus mutans during colonisation of the human oral cavity. Microb Ecol Health Dis. 1989;2:231–239. [Google Scholar]
- 8.Hillman J D, Dzuback A L, Andrews S W. Colonization of the human oral cavity by a Streptococcus mutans mutant producing increased bacteriocin. J Dent Res. 1987;66:1092–1094. doi: 10.1177/00220345870660060101. [DOI] [PubMed] [Google Scholar]
- 9.Hillman J D, Johnson K P, Yaphe B I. Isolation of a Streptococcus mutans strain producing a novel bacteriocin. Infect Immun. 1984;44:141–144. doi: 10.1128/iai.44.1.141-144.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hillman J D, Socransky S S. Replacement therapy for the prevention of dental disease. Adv Dent Res. 1987;1:119–125. doi: 10.1177/08959374870010010301. [DOI] [PubMed] [Google Scholar]
- 11.Hillman J D, Yaphe B I, Johnson K P. Colonization of the human oral cavity by a strain of Streptococcus mutans. J Dent Res. 1985;64:1272–1274. doi: 10.1177/00220345850640110301. [DOI] [PubMed] [Google Scholar]
- 12.Hurst A. Nisin. Adv Appl Microbiol. 1981;27:85–123. [Google Scholar]
- 13.Jordan H V, Englander H R, Engler W O, Kulczyk S. Observations on the implantation and transmission of Streptococcus mutans in humans. J Dent Res. 1972;51:515–518. doi: 10.1177/00220345720510024501. [DOI] [PubMed] [Google Scholar]
- 14.Krasse B, Edwardsson S, Svensson I, Trell L. Implantation of caries-inducing streptococci in the human oral cavity. Arch Oral Biol. 1967;12:231–236. doi: 10.1016/0003-9969(67)90042-8. [DOI] [PubMed] [Google Scholar]
- 15.Liptak M, Vekey K, van Dongen W D, Heerma W. Fast atom bombardment mass spectrometry of some lantibiotics. Biol Mass Spectrom. 1994;23:701–706. doi: 10.1002/bms.1200231109. [DOI] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Meyer H E, Heber M, Eisermann B, Korte H, Metzger W, Jung G. Sequence analysis of lantibiotics: chemical derivatization procedures allow a fast access to complete Edman degradation. Anal Biochem. 1994;223:185–190. doi: 10.1006/abio.1994.1571. [DOI] [PubMed] [Google Scholar]
- 18.Mota-Meira M, Lacroix C, LaPointe G, Lavoie M C. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett. 1997;410:275–279. doi: 10.1016/s0014-5793(97)00425-0. [DOI] [PubMed] [Google Scholar]
- 19.Novak J, Caufield P W, Miller E J. Isolation and biochemical characterization of a novel lantibiotic mutacin from Streptococcus mutans. J Bacteriol. 1994;176:4316–4320. doi: 10.1128/jb.176.14.4316-4320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Novak J, Kirk M, Caufield P W, Barnes S, Morrison K, Baker J. Detection of modified amino acids in lantibiotic peptide mutacin II by chemical derivatization followed by electrospray ionization mass spectroscopic analysis. Anal Biochem. 1996;236:358–360. doi: 10.1006/abio.1996.0181. [DOI] [PubMed] [Google Scholar]
- 21.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruangsri P, Orstavik D. Effect of the acquired pellicle and of dental plaque on the implantation of Streptococcus mutans on tooth surfaces in man. Caries Res. 1977;11:204–210. doi: 10.1159/000260269. [DOI] [PubMed] [Google Scholar]
- 23.Sahl H-G, Jack R W, Bierbaum G. Lantibiotics: biosynthesis and biological activities of peptides with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 24.Schnell N, Engelke G, Augustin J, Rosenstein R, Ungermann V, Gotz F, Entian K D. Analysis of genes involved in the biosynthesis of lantibiotic epidermin. Eur J Biochem. 1992;204:57–68. doi: 10.1111/j.1432-1033.1992.tb16605.x. [DOI] [PubMed] [Google Scholar]
- 25.Svanberg M, Krasse B. Oral implantation of saliva-treated Streptococcus mutans in man. Arch Oral Biol. 1981;26:197–201. doi: 10.1016/0003-9969(81)90130-8. [DOI] [PubMed] [Google Scholar]
- 26.Tanzer J M, Krasse B, Svanberg M. Conditions for implantation of Streptococcus mutans mutant 805 in adult human mouths. J Dent Res. 1982;61:334. [Google Scholar]
- 27.Wojciechowski, L., and J. D. Hillman. Unpublished data.