Summary
Plant defence against pathogens generally occurs at the expense of growth and yield. Uncoupling the inverse relationship between growth and defence is of great importance for crop breeding, while the underlying genes and regulatory mechanisms remain largely elusive. The exocytosis complex was shown to play an important role in the trafficking of receptor kinases (RKs) to the plasma membrane (PM). Here, we found a Cucumis sativus exocytosis subunit Exo70B (CsExo70B) regulates the abundance of both development and defence RKs at the PM to promote fruit elongation and disease resistance in cucumber. Knockout of CsExo70B resulted in shorter fruit and susceptibility to pathogens. Mechanistically, CsExo70B associates with the developmental RK CsERECTA, which promotes fruit longitudinal growth in cucumber, and contributes to its accumulation at the PM. On the other side, CsExo70B confers to the spectrum resistance to pathogens in cucumber via a similar regulatory module of defence RKs. Moreover, CsExo70B overexpression lines showed an increased fruit yield as well as disease resistance. Collectively, our work reveals a regulatory mechanism that CsExo70B promotes both fruit elongation and disease resistance by maintaining appropriate RK levels at the PM and thus provides a possible strategy for superior cucumber breeding with high yield and robust pathogen resistance.
Keywords: cucumber, Exo70B, receptor kinases, fruit length, disease resistance
Introduction
In nature, plants constantly encounter a variety of pathogens, and they have evolved sophisticated defence mechanisms to against pathogens. However, prolonged defence often leads to growth inhibition and yield penalty (Huot et al., 2014). On the other hand, the rapid growth of plants is usually accompanied by increased susceptibility to pathogens (He et al., 2022). One prevailing explanation for this phenomenon is energy competition, in which the limited resource drives the trade‐off between growth and defence (Coley et al., 1985; Herms and Mattson, 1992). Despite recent studies showed that genetic manipulation of single proteins, such as transcription factors (TFs) and aquaporins (AQPs), or of different genes simultaneously can make plants grow well and defend better (He et al., 2022; Lu et al., 2022; Wang et al., 2018; Xu et al., 2022), the underlying genes and regulatory mechanisms of growth‐defence balance remain largely elusive.
Receptor‐like kinases (RLKs) are encoded by expanded gene families and can act as receptors, co‐receptors, and regulatory partners. They play vital roles in plant growth, development, and adaptation to diverse environmental conditions (Liang and Zhou, 2018; Morris and Walker, 2003; Shiu and Bleecker, 2001a,b). Many RLKs function as immune receptor kinases (RKs) to sense conserved molecular patterns derived from microbes or plants during potential pathogen infection to activate plant defence (Couto and Zipfel, 2016; Gust et al., 2017). For example, the FLAGELLIN SENSING 2 (FLS2) and ELONGATION FACTOR‐TU (EF‐Tu) RECEPTOR (EFR) are the receptors of a conserved 22 amino acid epitope (flg22) and elf18 derived from bacterial flagellin and elongation factor, respectively (Gómez and Boller, 2000; Zipfel et al., 2004, 2006). Similarly, LysM‐RKs LYSINE MOTIF RECEPTOR KINASE5 (LYK5) senses fungal chitin and activates downstream immune responses (Cao et al., 2014). Other RKs have been shown to recognize endogenous peptides to regulate plant development. For example, two different RKs including CLAVATA1 (CLV1) and RECEPTOR‐LIKE PROTEIN KINASE 2 (RPK2) perceive the secreted peptide CLAVATA3 (CLV3) to limit shoot apical meristem size (Clark et al., 1997; Kinoshita et al., 2010). A disulphated pentapeptide phytosulfokine (PSK) plays a ubiquitous role in plant growth and development (Matsubayashi, 2014), which perceived by the receptor PSKR in carrot and Arabidopsis (Amano et al., 2007; Matsubayashi et al., 2002). Moreover, ERECTA (ER) senses the secreted cysteine‐rich peptides of EPF/EPFL family to regulate many biological processes including longitudinal growth of aboveground organs and reproductive development (Shpak, 2013; Shpak et al., 2003). The appropriate abundance of RKs at plasma membrane (PM) is critical for signal recognition and transduction. Nonetheless, only a few of regulators that function in the trafficking of RKs were identified in plants. For instance, the ER‐resident proteins RETICULAN LIKE PROTEIN B1/2 (RTNLB1/2) and RABA1b regulate the trafficking of de novo‐synthesized FLS2 out of the endoplasmic reticulum and trans‐Golgi network (TGN) to the PM, respectively (Choi et al., 2013; Lee et al., 2011).
Recently, exocyst complex has been shown to play an important role in maintenance of the abundance of cargos at PM (Saeed et al., 2019). The exocyst was initially identified in yeast and contains eight protein subunits: Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84. It mediates the tethering process of secretory vesicles to the PM (TerBush et al., 1996). In particular, Sec3 and Exo70 work as vital tethers that guarantees the first contact between vesicles and PM, followed by the soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor (SNARE) mediates membrane‐fusion (Saeed et al., 2019). In plants, Exo70 has the most copies and is broadly involved in multiple biological processes (Chong et al., 2010; Zárský et al., 2009). In Arabidopsis, Exo70A1 has been reported to be involved in the secretion of PIN‐FORMED (PIN) transporters to the PM and polar auxin transport and then regulates the apical dominance and organ morphogenesis (Drdová et al., 2013; Synek et al., 2006). Exo70H4 mediates the secretion of a stress‐induced callose synthase powdery mildew resistant 4 (PMR4) to PM and functions in the development of inner cell wall layer (Kulich et al., 2015, 2018). Beyond that, Exo70B1 and Exo70B2 are the vital regulators of pattern‐triggered immunity (PTI) response, exo70B1–3 and exo70B2 mutants both displayed more susceptibility to Pseudomonas syringae pv. tomato (Pto) strain DC3000 (Stegmann et al., 2012, 2013). A recent study showed that Exo70B1 and Exo70B2 function in trafficking FLS2 to the PM, possibly by tethering the FLS2‐containing vesicles to the targeted PM, thereby regulating plant defence response in Arabidopsis (Wang et al., 2020a). Similarly, OsExo70B1 interacts with the RK‐chitin elicitor receptor kinase 1 (CERK1), the co‐receptor of chitin, to resist the Magnaporthe oryzae (M. oryzae) in rice (Hou et al., 2020). Above these suggested that Exo70B mainly regulates plant resistance via trafficking the RKs to the PM. However, whether Exo70B regulates developmental RKs in a similar mode remains largely unknown.
Cucumber (Cucumis sativus L.) is an economically important vegetable crop, which bears pepo fruits harvested immaturely at 8–18 days after anthesis (DAA) (Pan et al., 2017; Che et al., 2023), but its production is substantially affected by various diseases (Weng and Wehner, 2017). The most important pathogens that affect cucumber include the Fusarium oxysporum f. sp. Cucumerinum Owen (Foc) and Pseudomonas syringae pv. lachrymans (Psl), which cause fusarium wilt (also known as cancer) and angular leaf spot in cucumber, respectively (Chand and Walker, 1964; Dong et al., 2019). In this study, we showed a CsExo70B positively regulates both fruit elongation and disease resistance. Further analyses revealed that CsExo70B associates with multiple RKs, including CsERECTA, CsFLS2 and CsLYK5 and promotes their accumulation at the PM. More importantly, overexpression of CsExo70B resulted in an increase of cucumber fruit yield and disease resistance. Together, our findings provide a strategy to enhance cucumber yield and resistance.
Results
CsExo70B positively regulates fruit elongation in cucumber
Previous studies showed that Exo70B members are involved in immunity by regulating the accumulation of RK‐FLS2 at PM in Arabidopsis (Wang et al., 2020a). To explore whether Exo70B regulates plant development by associating with RKs, we identified the Exo70B in cucumber (CsaV3_1G036490) and named as CsExo70B hereinafter (Liu et al., 2023). CRISPR/Cas9 method was used to generate Csexo70b knockout mutants. Two homozygous mutants were obtained. The Csexo70b‐5 # allele has 29 bp deletion and 3 bp replacement, and Csexo70b‐4 # allele has 131 bp deletion. Both mutations resulted in premature stop codon at 136Arg and 156Tyr, respectively (Figure 1a, Figure S1a). There were no off‐target loci detected in both mutants (Table S1). Interestingly, we found that the fruit length was significantly reduced in Csexo70b lines at all observed stages including anthesis, 10 DAA, and 30 DAA (Figure 1b–d). The fruit length of Csexo70b‐5 # and Csexo70b‐4 # mutants have a 21.9% and 29.45% reduction at anthesis, 36.37% and 35.3% reduction at 10 DAA, and 25.6% and 26.8% reduction at 30 DAA compared to that of wild type (WT) plants in the corresponding periods (Figure 1e–g). Moreover, the microscopic structures of longitudinal sections of the fruit pericarp at 10 DAA were examined for WT and Csexo70b mutants. We found that the cell size of Csexo70b mutants was 15.2% smaller, and the cell number was also reduced 19.4% compared to that in WT plants. These results suggested that CsExo70B promotes fruit elongation by stimulating both cell expansion and cell division (Figure 1h–k). Furthermore, the Csexo70b mutants also displayed slightly dwarfed plant height, smaller male flowers and seeds, as well as increased carpels in cucumber (Figure S1b–h), indicating that CsExo70B plays an important role in cucumber development.
Figure 1.
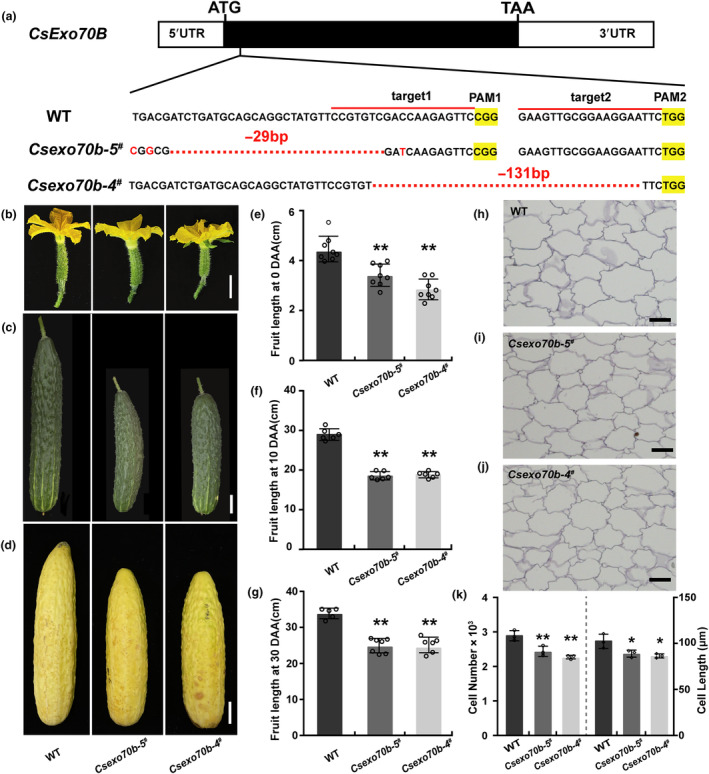
Construction and characterization of the Csexo70b mutants in cucumber. (a) CRISPR/Cas9‐mediated mutations of CsExo70B gene. The single‐guide RNAs (sgRNAs) are marked in red line, and protospacer‐adjacent motif (PAM) sites are highlighted in yellow. ‘−29’ and ‘−131’‐bp indicate frame shifts in Csexo70b mutant lines. (b–d) Morphology of ovaries at anthesis (b), fruits at 10 days after anthesis (DAA) (c), and 30 DAA (d) in wild type (WT) and Csexo70b mutants, scale bar = 1 cm (b) or 3.5 cm (c, d). (e–g) Statistical analyses of ovary length shown in (b) and fruit length observed in (c) and (d). Values are means ± SD, n ≥ 5. (h–j) Longitudinal pericarp sections of fruits at 10 DAA in WT and Csexo70b mutants, scale bar = 100 μm. (k) The cell number and cell length quantification of fruit pericarp in WT and Csexo70b mutants along a longitudinal axis. Values are means ± SD, n = three individual fruits. Significant differences relative to WT are indicated by asterisks (*P < 0.05; and **P < 0.01, one‐way ANOVA).
CsExo70B directly interacts with CsERECTA and affects its accumulation at PM
Considering the roles of CsExo70B in multiple aspects of development, we explored whether CsExo70B interacts with some development‐related RKs using firefly split‐luciferase complementation (luc) assay (Figure S2). Very interestingly, we found CsExo70B strongly associated with CsERECTA (Figure S2), a founding RK regulating silique length in Arabidopsis (Pillitteri et al., 2007; Shpak et al., 2003). To investigate the function of CsERECTA in cucumber, we also used CRISPR/Cas9 system to knock out CsERECTA. Two mutant alleles Cserecta‐2 # (with 1 bp insertion and 4 bp deletion) and Cserecta‐12 # (with 1 bp and 3 bp deletions) were obtained. Both mutations resulted in premature stop, generating truncated proteins of 31 amino acids and 39 amino acids, respectively (Figure 2a, Figure S3a), and no off‐target loci were detected (Table S1). As expected, Cserecta mutants exhibited significantly shorter fruits from anthesis to 30 DAA (Figure 2b–d). The average fruit length of Cserecta‐2 # and Cserecta‐12 # mutants had a 40.8% and 36.15% reduction at anthesis, 51.28% and 51.94% reduction at 10 DAA, and both 40% reduction at 30 DAA compared to that in WT (Figure 2e–g). Further study showed that cell size and cell number was significantly decreased in Cserecta mutants (Figure 2h–k), suggesting that CsERECTA also positively regulates fruit length by promoting cell expansion and cell division. In addition, Cserecta mutants also resulted in similar phenotypes to Csexo70b mutants, including dwarfed plants, and reduced size of male flowers and seeds (Figure S3b–i).
Figure 2.
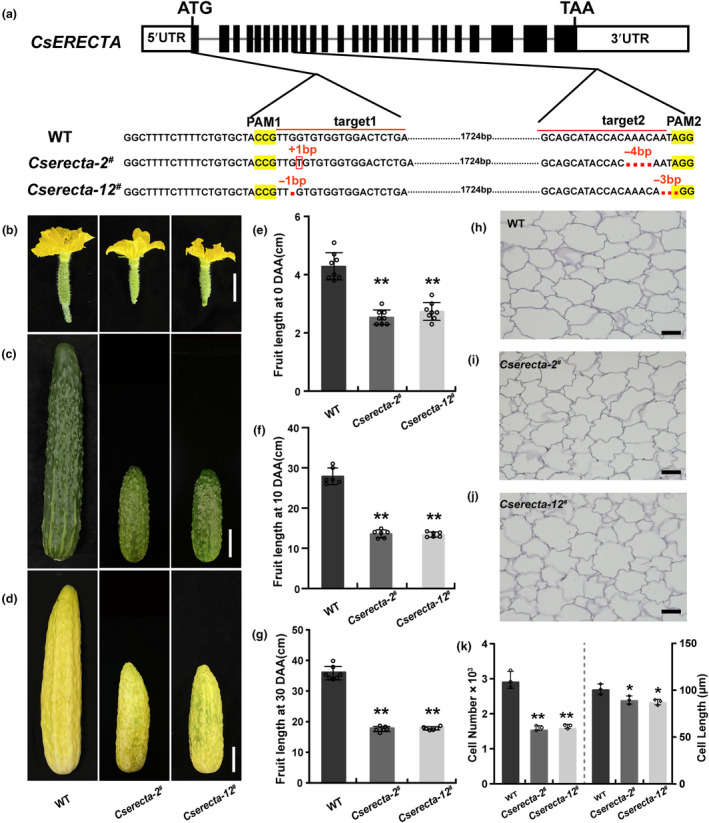
Phenotypic characterization of CsERECTA knockout lines in cucumber. (a) Mutant alleles of Cserecta. The red‐lined sequences in the WT denote the regions targeted by sgRNA. The Cserecta‐2 # allele has 1‐bp insertion and 4‐bp deletion, and the Cserecta‐12 # allele has 1‐bp and 3‐bp deletions. (b–d) The fruit morphology of WT and Cserecta mutants. Pictures were collected at anthesis (b), 10 DAA (c) and 30 DAA (d). Scale bar = 2 cm in (b), 3.5 cm in (c), and 5 cm in (d). (e–g) The length quantification of fruits in (b–d). Data are shown as means ± SD; n ≥ 6. Significant differences relative to WT are indicated by asterisks (**P < 0.01, one‐way ANOVA). (h–j) Longitudinal sections of fruit pericarp at 10 DAA of WT and Cserecta mutants. Scale bar = 100 μm. (k) Statistical analyses of cell number and cell length phenotypes along a longitudinal axis. Data are shown as means ± SD; n = three individual fruits. Statistical significances were determined by one‐way ANOVA (*P < 0.05; **P < 0.01).
The same phenotypes of Csexo70b and Cserecta promoted us to test if CsExo70B regulates CsERECTA accumulation at PM. First, we sought to verify that CsExo70B interacts with CsERECTA using multiple assays. As shown in Figure 3a and Figure S4, the split‐luc and the bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana (N. benthamiana) indicated that CsExo70B interacted with CsERECTA, but not CsBAK1. The CsExo70B‐CsERECTA interaction was also detected using co‐immunoprecipitation (co‐IP) assays when a FLAG‐tagged CsERECTA was co‐expressed with CsExo70B‐HA in N. benthaminana (Figure 3b). Co‐IP experiments using stable CsExo70B‐HA transgenic plant roots which expressing with CsERECTA‐FLAG further confirmed the CsExo70B‐CsERECTA interaction in cucumber plants (Figure 3c).
Figure 3.
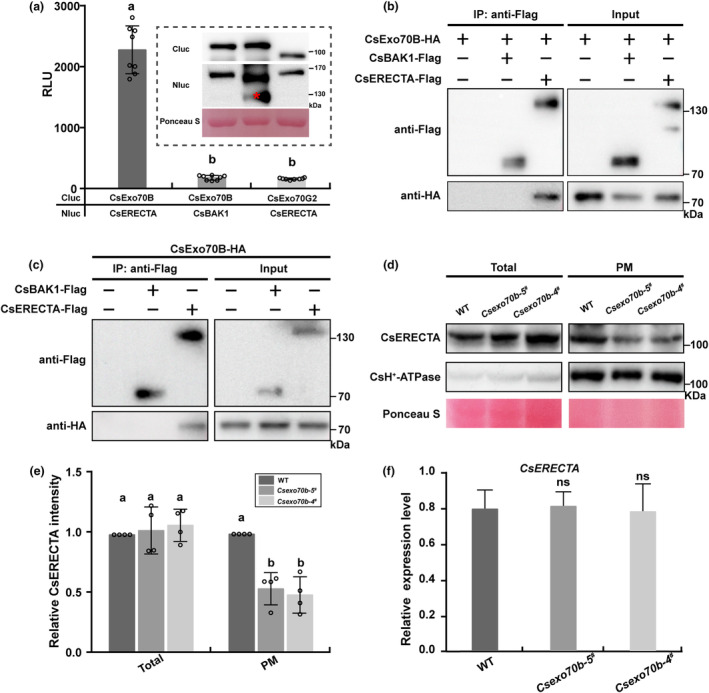
CsExo70B interacts with CsERECTA and contributes to its accumulation at plasma membrane (PM). (a) Firefly split‐luciferase complementation (LUC) assay showed CsExo70B interacts with CsERECTA. The indicated constructs were expressed in Nicotiana benthamiana (N. benthamiana) leaves, and the relative LUC activity was recorded by a luminometer. Immunoblot showing the Cluc and Nluc fusion proteins, as determined by anti‐Cluc and anti‐HA antibodies, respectively. RLU, relative luminescence unit. Data are represented as means ± SD, n = 8. Different lowercase letters indicated statistically significant differences (P < 0.01, one‐way ANOVA analysis with Tukey's test). (b) Co‐immunoprecipitation (IP) of CsExo70B and CsERECTA in N. benthamiana leaves. These indicated constructs were expressed in N. benthamiana leaves, and co‐IP assay was performed by anti‐FLAG antibody. CsBAK1‐FLAG was used as a negative control. (c) Co‐IP assay between CsExo70B and CsERECTA in cucumber roots. The CsERECTA‐FLAG was expressed in hairy roots of stable CsExo70B‐HA overexpression lines using Agrobacterium rhizogenes mediated transformation, and co‐IP assay was performed by anti‐FLAG antibody. (d) Immunoblotting analysis of endogenous CsERECTA levels in different fractions. CsH+‐ATPase was used as PM protein marker. Total and PM proteins were extracted from ovaries and detected by anti‐CsERECTA antibody. Equal loading was demonstrated by Ponceau Staining (Ponceau S) of Rubisco. (e) Statistical analysis of the relative CsERECTA levels from four independent replicates. Numbers indicate relative protein band density of CsERECTA normalized to CsH+‐ATPase, and the data are means ± SD, n = 4. Different lowercase letters indicate statistically significant differences (P < 0.05, one‐way ANOVA analysis with Tukey's test). (f) Transcript level of CsERECTA in WT and Csexo70b mutants. Data are means ± SD, n = 3 (ns, no significant difference, one‐way ANOVA).
To further investigate if CsExo70B regulates protein accumulation of CsERECTA at the PM, we developed peptide‐specific antibodies recognizing CsERECTA and CsH+‐ATPase. The specificity of CsERECTA antibody was confirmed by Western blot using the lysates from WT and Cserecta mutants and N. benthamiana leaves (Figure S5a,b), CsH+‐ATPase antibody specificity was also confirmed in cucumber (Figure S5c). Total proteins were extracted from ovaries and PM fractions were separated, followed by CsERECTA detection with specific antibody. In the Csexo70b mutants, less CsERECTA was detected in the PM fraction compared to WT, but there were no significant differences of total CsERECTA protein between Csexo70b mutants and WT (Figure 3d,e). Additionally, the transcripts of CsERECTA were not significantly different between WT and Csexo70b plants (Figure 3f), indicating that the reduced CsERECTA accumulation at PM in Csexo70b mutants is not a consequence of lower total levels. These results support the notion that CsExo70B specifically regulates the protein accumulation of CsERECTA at PM, thereby promoting fruit elongation in cucumber.
In addition, our results showed that CsExo70B also interacted with CsCLV1 in N. benthamiana (Figure S2). Consistent with the phenotype of Csexo70b mutants (Figure S1d,g), which has been reported that CsCLV1 mutation leads to increased carpels in cucumber (Cheng et al., 2022), we speculated that CsExo70B may also regulate CsCLV1‐mediated carpel development in cucumber. Together, these results demonstrate that CsExo70B associates with multiple RKs to regulate development in cucumber.
CsExo70B regulates fruit elongation through CsERECTA‐mediated signal pathway in cucumber
To further confirm that CsExo70B and CsERECTA act in the same pathway to regulate fruit elongation in cucumber, we generated the Csexo70b‐5 # Cserecta‐2 # double mutant by crossing the Csexo70b‐5 # and Cserecta‐2 # . During the anthesis, the fruit length of Csexo70b‐5 # Cserecta‐2 # double mutants was indistinguishable from those of Cserecta‐2 # mutants, but significantly shorter than Csexo70b‐5 # mutants and WT lines (Figure 4a,c). The length of fruit at 10 DAA in Csexo70b‐5 # Cserecta‐2 # was slightly reduced, but still similar to that in Cserecta‐2 # (Figure 4b,d), indicating that genetically CsERECTA acts downstream of CsExo70B.
Figure 4.
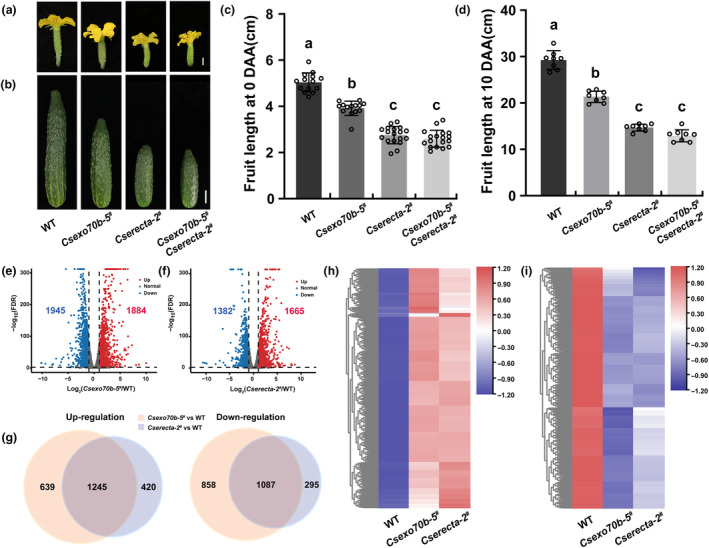
CsExo70B and CsERECTA co‐regulate the fruit elongation in cucumber. (a, b) The morphology of fruit at anthesis (a) and 10 DAA (b) of WT, Csexo70b‐5 #, Cserecta‐2 # and Csexo70b‐5 # Cserecta‐2 #. Scale bar = 1 cm in (a) and 3 cm in (b). (c, d) Quantification of fruit length in the indicated lines shown in (a) and (b). Values are means ± SD (n ≥ 7). The different lowercase letters indicate significant differences (P < 0.05) by one‐way ANOVA analysis with Turkey's test. (e, f) Transcriptome analysis for fruits at 3 DAA of WT, Csexo70b‐5 # and Cserecta‐2 # mutants. Volcano diagrams showing the differentially expressed genes (DEGs) in Csexo70b‐5 # (e) and Cserecta‐2 # (f) relative to WT (Fold change ≥ 2.0, FDR < 0.01). Down‐regulated and up‐regulated genes were appeared in bule and red, and grey means normal genes. (g) Veen diagrams showing the overlapped DEGs of Csexo70b‐5 # and Cserecta‐2 # mutants compared with WT, yellow and blue represent the DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants, respectively. (h, i) Heat map showing the different expression patterns of shared DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants. The data for each sample are averaged from three biological repeats.
In addition, we performed RNA‐seq analyses for fruits of WT, Csexo70b‐5 # , and Cserecta‐2 # plants. A total of 3829 differentially expressed genes (DEGs) were detected in Csexo70b‐5 # mutants and 3047 DEGs in Cserecta‐2 # mutants relative to WT (Fold change ≥ 2.0, FDR < 0.01) (Figure 4e,f). Among the 1665 up‐regulated genes in Cserecta‐2 # mutants, 1245 genes were overlapped with DEGs in Csexo70b‐5 # mutants. Similarly, 1087 down‐regulated genes in Cserecta‐2 # mutants were overlapped with DEGs in Csexo70b‐5 # mutants (Figure 4g), suggesting that the majority (76.5%) of transcripts were co‐regulated by CsExo70B and CsERECTA. A heat map was generated to further show the expression patterns of these common DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants (Figure 4h,i), which participated in multiple signalling pathways such as plant‐pathogen interaction and plant hormone signal transduction (Figures S6 and S7, Tables S2 and S3). These data further support the notion that CsExo70B regulates fruit elongation through CsERECTA‐mediated signal pathway in cucumber.
CsExo70B is required for resistance to fungal and bacterial pathogens
Previous studies showed that Exo70B members are required for plant immunity in Arabidopsis and rice (Hou et al., 2020; Wang et al., 2020a). Thus, we sought to dissect the disease resistance role of CsExo70B in cucumber. Firstly, we examined chitin‐ and flg22‐induced ROS production in Csexo70b mutants. When treated with chitin, Csexo70b mutants accumulated 60%–70% less ROS compared with WT (Figure 5a). Similarly, flg22‐induced ROS production was also reduced to 50%–60% in Csexo70b mutants compared with WT (Figure 5b).
Figure 5.
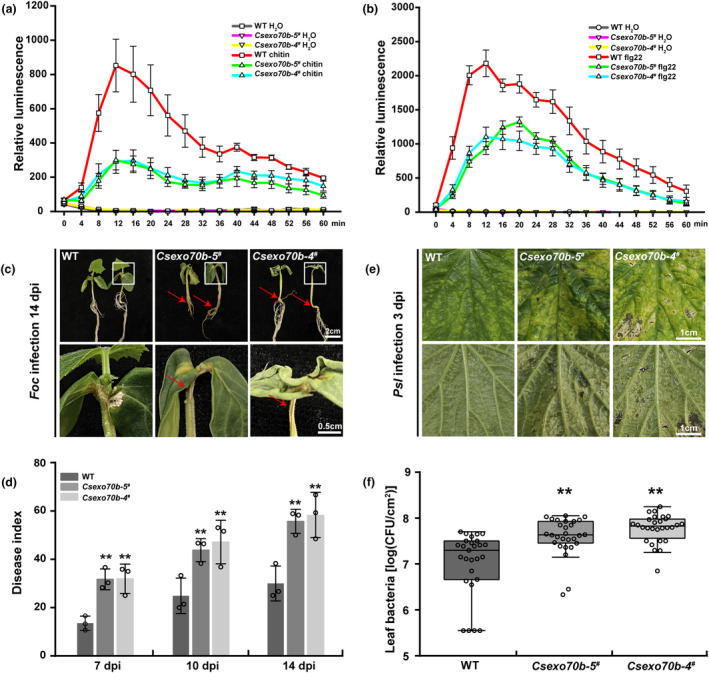
Knockout of CsExo70B compromises pathogen resistance in cucumber. (a, b) CsExo70B knockout lines exhibited reduced reactive oxygen species (ROS) production triggered by flg22 (a) or chitin (b). Data represent the means ± SD (n = 6). (c) CsExo70B knockout lines are more susceptible to Fusarium oxysporum f. sp. Cucumerinum Owen (Foc). Images were taken at 14 days post‐inoculation (dpi), and the red arrows indicated wilted hypocotyls and stems. (d) Disease index (DI) in WT and CsExo70B knockout lines shown in (c). Data are means ± SD (n = 3). (e) The disease resistance phenotypes of CsExo70B knockout lines after inoculation with Pseudomonas syringae pv. lachrymans (Psl). The cucumber plants with two true leaves were spray‐inoculated with Psl, and images were taken at 3 dpi. Scale bar = 1 cm. (f) CsExo70B knockout lines displayed increased susceptibility to Psl. Bacterial population in the leaf was measured 3 days after spray inoculation. Values are means of log (colony‐forming units [CFU] cm−2 leaf tissue) ± SD (n ≥ 28). Statistical significances relative to WT are indicated by asterisks (**P < 0.01, one‐way ANOVA).
To further confirm the function of CsExo70B in cucumber disease resistance, Csexo70b mutants and WT were firstly challenged with fungal vascular pathogen Foc using radicle dipping method (Figure S8a). We observed the severity of disease symptoms in both WT and Csexo70b mutants increased steadily with increasing days of infection. The disease symptom progression in Csexo70b mutants was earlier than that in WT plants. In Csexo70b mutants, minor disease symptoms firstly appeared at 7 days post‐inoculation (dpi), severe wilting symptoms subsequently appeared in stem vascular and cotyledon at 10 dpi and 14 dpi, while in WT plants, only minor disease symptoms were observed until 10 dpi to 14 dpi (Figure S8b,c, Figure 5c). Consistently, the quantification of disease index (DI) and the percentage of disease grade in Csexo70b lines further confirmed this phenotype (Figure 5d, Figure S8d), indicating that the resistance to Foc in Csexo70b mutants is greatly reduced. Next, the Csexo70b and WT seedlings were spray‐inoculated with bacterial leaf pathogen Psl. Three days after inoculation, more disease spots were observed in the Csexo70b‐5 # and Csexo70b‐4 # than WT, and the bacterial populations in the Csexo70b‐5 # and Csexo70b‐4 # mutants were approximately 2.6‐ to 3.6‐fold greater than that in WT (Figure 5e,f). These results imply that CsExo70B positively regulates resistance to fungal and bacterial pathogens in cucumber.
CsExo70B associates with immune RKs and affects their abundance at the PM
The requirement of CsExo70B in RK signalling and resistance to bacterial and fungal pathogens promotes us to test whether CsExo70B similarly regulates immune RK abundance at the PM in cucumber. The well‐known immune RKs FLS2 and LYK5, which perceive flg22 of bacterial flagellin and fungal chitin, respectively (Cao et al., 2014; Zipfel et al., 2004), were selected for further study. Multiple assays, including firefly Split‐luc and BiFC assays in N. benthamiana leaves (Figure 6a,b, Figure S4), and Co‐IP assays in N. benthamiana leaves (Figure 6c,d) and cucumber roots (Figure 6e,f), all showed that CsExo70B associated with CsLYK5 and CsFLS2 in vivo. To study if CsExo70B affects immune RK accumulation at PM in cucumber, we also developed peptide‐specific antibodies recognizing CsLYK5 and CsFLS2. Unfortunately, only antibody of CsLYK5 was available (Figure S5d,e). Our results showed that CsLYK5 levels at PM in Csexo70b‐5 # and Csexo70b‐4 # were obviously reduced than that in WT, but the total protein and transcription levels of CsLYK5 and CsFLS2 were not changed in Csexo70b mutants (Figure 6g–i, Figure S9a), indicating that CsExo70B similarly affects immune RK accumulation at the PM, thus regulating disease resistance in cucumber.
Figure 6.
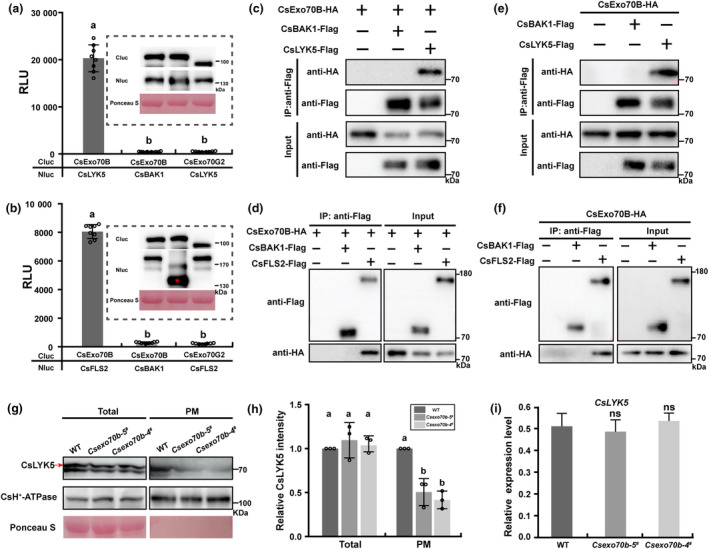
CsExo70B physically interacts with immune RKs and contributes to their accumulation at PM. (a, b) LUC assays indicated CsExo70B interacts with CsLYK5 (a) and CsFLS2 (b). The LUC assays were performed in N. benthamiana leaves by Agrobacterium‐mediated transient expression of the indicated constructs. The relative luciferase activities were measured using a luminometer. Data are shown as means ± SD; n = 8. Different lower‐case letters indicate statistically significant differences (P < 0.001, one‐way ANOVA analysis with Tukey's test). (c, d) CsExo70B associates with CsLYK5 (c) and CsFLS2 (d) in N. benthamiana. These indicated constructs were transiently expressed in N. benthamiana leaves, and co‐IP was performed by anti‐FLAG antibody. CsBAK1‐FALG was used as a negative control. (e, f) CsExo70B interacts with CsLYK5 (e) and CsFLS2 (f) in cucumber roots. The indicated constructs were expressed in hairy roots of stable CsExo70B‐HA overexpression lines, and co‐IP assays were performed with anti‐FLAG antibody. (g) Immunoblotting analysis of CsLYK5 in WT and CsExo70B knockout lines. Total proteins were extracted from true leaves and subjected to protein fractionation. CsH+‐ATPase was used as PM protein control. (h) Quantified analysis of the relative CsLYK5 levels shown in (g). The CsH+‐ATPase protein was used as control. Data represent means ± SD of three independent experiments. Different lowercase letters indicate statistically significant differences (P < 0.05, one‐way ANOVA analysis with Tukey's test). (i) The levels of CsLYK5 transcript were normal in WT and Csexo70b mutants. Data are means ± SD, n = 3 (ns, no significant difference, one‐way ANOVA).
Overexpression of CsExo70B enhances fruit yield and disease resistance in cucumber
The aforementioned results supported the notion that CsExo70B simultaneously maintains the abundance of developmental and immune RKs at the PM, regulating development and disease resistance in cucumber. To further test whether CsExo70B overexpression leads to higher yield and stronger defence, three stable overexpression transgenic lines were obtained. qRT‐PCR results showed that the transcript levels of CsExo70B in CsExo70B‐OE‐3#, CsExo70B‐OE‐4# and CsExo70B‐OE‐6# were 5.3‐, 7.1‐ and 8.0‐fold greater than WT. Immunoblot assay confirmed that CsExo70B‐HA protein was accumulated in all CsExo70B‐OE transgenic lines (Figure 7a). Compared to WT, CsExo70B‐OE lines exhibited significantly longer fruits of both commercial and physiological maturity periods (Figure 7b,c, Figure S10a). We further explored fruit yield per plant in WT and CsExo70B‐OE plants and observed 11.6%–22.9% higher yield in CsExo70B‐OE lines than that in WT (Figure 7d). In addition, we detected significantly larger male flower and higher plant height in CsExo70B‐OE lines (Figure S10b,c). These results suggest that CsExo70B‐overexpressing increases fruit yield in cucumber.
Figure 7.
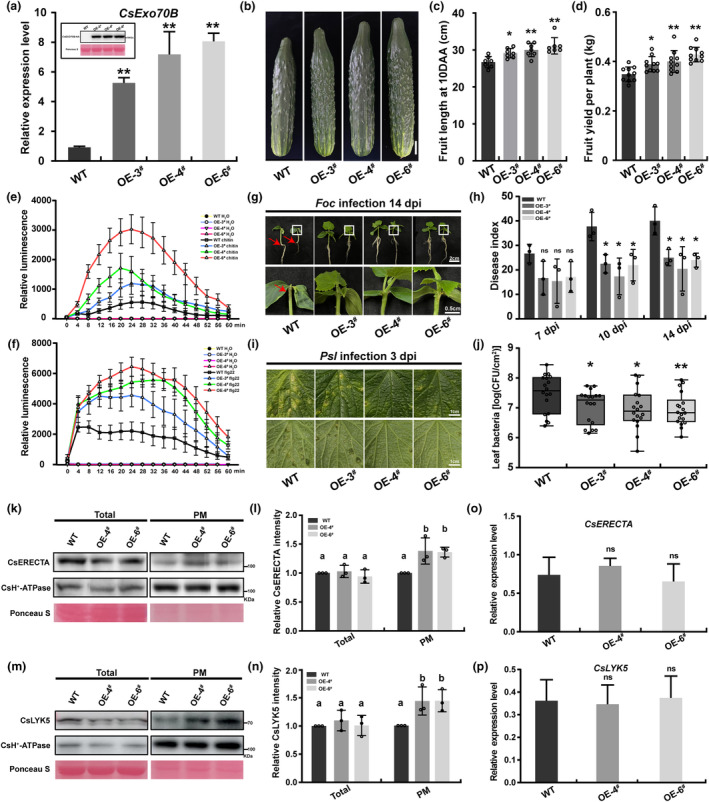
Overexpression of CsExo70B enhances fruit yield and resistance to pathogens in cucumber. (a) The identification of CsExo70B‐OE lines. Transcripts of CsExo70B were detected by qRT‐PCR. Data are shown as means ± SD (n = 3). Statistical significances were determined by one‐way ANOVA (**P < 0.01). The gels shown the protein levels of CsExo70B detected by anti‐HA immunoblot. Equal loading was demonstrated by Ponceau S of Rubisco. (b) Morphology of cucumber fruit at 10 DAA of WT and CsExo70B‐OE lines. Scale bar = 3 cm. (c) The length of fruits at 10 DAA shown in (b) was quantified. Data are shown as means ± SD, n = 11 (*P < 0.05; **P < 0.01, one‐way ANOVA). (d) Fruit yield per plant in WT and CsExo70B‐OE lines. Data represent as mean ± SD, n = 10 individual plants (*P < 0.05, **P < 0.01, one‐way ANOVA). (e, f) chitin (e) and flg22 (f) triggered ROS production in CsExo70B‐OE lines. Data represent the means ± SD (n = 8). (g) The seedling morphology of WT and CsExo70B‐OE lines at 14 dpi with Foc. White boxes indicate ‘zoomed‐in’ regions. (h) The DI of WT and CsExo70B‐OE lines shown in (g). Data represent the means ± SD, n = 3. (*P < 0.05, one‐way ANOVA). (i, j) CsExo70B‐OE lines confer resistance to Psl. The cucumber plants with two true leaves were spray‐inoculated with Psl, and the disease resistance phenotypes were collected at 3 dpi. Values are means of log ([CFU] cm−2 leaf tissue) ± SD; n = 18. Significant differences relative to WT are indicated by asterisks (*P < 0.05; **P < 0.01 one‐way ANOVA). (k) Immunoblotting analysis of CsERECTA levels in WT and CsExo70B‐OE lines. Ponceau S was used as protein loading control. (l) Quantified analysis of relative CsERECTA levels in WT and CsExo70B‐OE lines. The relative protein band density of CsERECTA was normalized to that of CsH+‐ATPase. Data represent means ± SD of three independent experiments. Different lower‐case letters indicate statistically significant differences (P < 0.05, one‐way ANOVA analysis with Tukey's test). (m) Immunoblotting analysis of CsLYK5 levels in CsExo70B‐OE lines. Total proteins and PM proteins were extracted from leaves and detected by anti‐CsLYK5 antibody. (n) Quantified analysis of the relative CsLYK5 levels shown in (m). The CsH+‐ATPase levels were used as normalization standard. Data are presented as mean ± SD of three independent experiments. Different lowercase letters indicate statistically significant differences (P < 0.05, one‐way ANOVA analysis with Tukey's test). (o, p) Transcript analyses of CsERECTA (o) and CsLYK5 (p) in CsExo70B‐OE lines. Data are means ± SD, n = 3 (ns, no significant difference, one‐way ANOVA).
On the other side, we found chitin‐ and flg22‐induced ROS production was greatly increased in CsExo70B transgenic plants, especially in CsEXO70B‐OE‐4# and CsEXO70B‐OE‐6# lines (Figure 7e,f). When CsExo70B‐OE lines and WT were simultaneously infected by a higher concentration of Foc spore suspension, WT plants displayed obvious stem disease symptoms, and a few of them gradually showed cotyledon wilting at 14 dpi, while CsExo70B‐OE lines exhibited mild disease symptoms with no wilted cotyledons until 14 dpi (Figure 7g). Consistently, the DI of all the CsExo70B‐OE lines was lower that of WT at 10 dpi and 14 dpi (Figure 7h). Similarly, CsExo70B‐OE plants supported reduced Psl bacterial growth and disease spot area compared to WT (Figure 7i,j), indicating that CsExo70B overexpression improves pathogen resistance in cucumber.
Immunoblot analyses were further performed to detect the PM levels of RKs in CsExo70B‐OE‐4# and CsExo70B‐OE‐6# lines. Consistently, increased CsERECTA and CsLYK5 accumulation were observed at the PM in CsExo70B‐OE lines (Figure 7k–n), and the expression of CsERECTA, CsLYK5, and CsFLS2 was unchanged in CsExo70B‐OE lines (Figure 7o,p, Figure S9b).
Overall, these findings further support that CsExo70B promotes cucumber fruit elongation and disease resistances by RK levels at PM, providing a possible strategy for breeding cucumber with high yield and enhanced defence.
Discussion
When infected by pathogens, plants activate defence responses quickly and properly, but frequently penalize growth and development. Recent studies showed that the growth and defence ‘trade‐off’ can be uncoupled via genetic manipulations (Campos et al., 2016; Lu et al., 2022; Wang et al., 2014, 2018). In this study, we found that CsExo70B promotes both yield and disease resistance by regulating RKs homeostasis at PM.
Firstly, we revealed a novel function of CsExo70B in cucumber development. Previous evidence showed that the exo70B1 mutant is smaller compared to WT in Arabidopsis when grown under short days for 5 weeks and exhibited hypersensitive response‐like cell death and enhanced disease resistance to various pathogens (Zhao et al., 2015), subsequent study found that these phenotypes of exo70B1 was caused by constitutive activation of an atypical intracellular immune receptor TIR‐NBS2 (TN2) (Zhao et al., 2015). Thus, the development defect of exo70B1 mutant in Arabidopsis is due to constitutive immune activation, and Exo70B was not considered as a real developmental regulator. Interestingly, the cucumber exo70b mutants displayed shorter fruits and slightly reduced plant height (Figure 1, Figure S1). However, it seems that the developmental defects were not caused by constitutive immune activation, because no cell death was observed in Csexo70b mutants (Figure S11a), and the expression of pathogenesis‐related genes CsPR1 and CsPR2 also exhibited normal in Csexo70b mutants (Figure S11b,c). Besides, Csexo70b mutants showed more susceptible rather than resistance to pathogens (Figure 5). In addition, previous study reported that there are only two genes encoding TIR‐NBSs in cucumber, and both of them have low similarity to TN2 in sequences (Wan et al., 2013). Therefore, our evidences demonstrated that CsExo70B directly regulates cucumber development.
Due to Exo70B has been linked to the trafficking of immune RKs to the PM in previous studies (Hou et al., 2020; Wang et al., 2020a). The short fruit phenotype of Csexo70b mutants reminds us of ERECTA, which mutation led to short siliques in Arabidopsis (Pillitteri et al., 2007; Shpak et al., 2003). As expected, the fruit length and plant height were greatly reduced in Cserecta mutants generated by CRISPR/Cas9 system (Figure 2, Figure S3). Our extensive analyses found that CsExo70B interacts with CsERECTA and regulates its accumulation at the PM in fruit tissue (Figure 3). Consistently, the fruit length of the Csexo70b Cserecta double mutant is similar to that of Cserecta mutant rather than Csexo70b mutant (Figure 4a–d), indicating that CsERECTA acted genetically downstream of CsExo70B. Notably, the short‐fruit phenotype of Cserecta was more severe than that of Csexo70b (Figures 1, 2 and 4), which is plausible considering that CsExo70B regulating CsERECTA's accumulation at the PM, thus knockout of CsExo70B exhibited only partial phenotype of Cserecta. Moreover, the RNA‐seq analysis demonstrated that 76.5% transcripts were co‐regulated by CsExo70B and CsERECTA (Figure 4e–i), indicating that CsExo70B and CsERECTA regulate fruit elongation in the same pathway in cucumber. Among the down‐regulated DEGs, YUCC4 and other auxin‐ and cell division‐responsive factors were significantly decreased (Figure S12), implying that the shorter fruits in Csexo70b and Cserecta mutants may be mediated by plant hormone pathways. Furthermore, CsExo70B interacts with CsCLV1 (Figure S2), which mutation leads to an increased carpel number in cucumber (Cheng et al., 2022). The increased carpel number was also observed in Csexo70b mutants (Figure 1d,g). It is speculated that CsExo70B may regulate carpel number via modulating CLV1 accumulation at the PM in cucumber.
Secondly, previous studies have shown that Exo70B1 and Exo70B2 are involved in resistances to fungal and bacterial pathogens, which cause leaf disease in rice and Arabidopsis (Hou et al., 2020; Pecenková et al., 2011). Our analyses found that cucumber Csexo70b mutant exhibited more susceptible to Psl infection in leaves (Figure 5e,f). Not only that, Csexo70b mutant also displayed reduced resistance to vascular pathogen Foc (Figure 5c,d), which causes wilt in cucumber, indicating cucumber resistance to leaf and vascular pathogens both depends on CsExo70B. Similarly, CsExo70B also interacts with immune RKs CsFLS2 and CsLYK5 and promotes their accumulation at the PM in cucumber (Figure 6). Consistent with the reduced immune RK levels at the PM, the RK‐mediated ROS production was compromised in Csexo70b mutants (Figure 5a,b). Together, these results suggest CsExo70B provides a broad‐spectrum resistance to different pathogens through regulating the accumulation of immune RKs at the PM.
Moreover, we found that the cucumber plants with higher yield and stronger defence can be achieved simultaneously by overexpression of CsExo70B (Figure 7), indicating that CsExo70B acts as an integral component to promote both fruit elongation and defence response in cucumber. However, the molecular mechanism underlying CsExo70B regulates growth‐defence tradeoffs is unknown. Several plausible mechanisms may explain the switch of growth to defence. One possibility is that CsExo70B may function in an organ‐ or tissue‐specific manner. Our results showed that CsERECTA was specifically highly expressed in fruit, while both CsFLS2 and CsLYK5 were significantly lower expressed in fruit than other tissues (Figure S13). When infected by pathogens, the expression levels of CsExo70B (Liu et al., 2023) and immune RKs (CsFLS2 and CsLYK5) were significantly up‐regulated, but the expression of CsERECTA was markedly down‐regulated in roots and leaves rather than fruits (Figures S14 and S15). So, one possibility is that CsExo70B may preferentially promote CsERECTA accumulation in fruit to regulate fruit elongation; while CsExo70B may have a prior choice to traffic more immune RKs to PM to activate stronger defence in roots and leaves upon pathogen attack. In addition, previous study showed IPA1 promotes disease resistance and yield by activating different genes through reversible phosphorylation (Wang et al., 2018). So, another possibility that cannot be ruled out is CsExo70B may also have different post‐translational modification states before and after pathogen infection, and select to traffic different RKs to enhance fruit length and disease resistance.
Collectively, the data presented here reveal a regulatory mechanism that CsExo70B positively regulates fruit elongation and disease resistances via fine‐tuning the PM accumulation of CsERECTA and CsLYK5 in cucumber (Figure 8). In field, cucumber is often challenged by various pathogens, resulting in reduced production. In the future, fine tuning the expression level of Exo70B will provide a promising breeding strategy with high yield and robust defence in cucumber and other species.
Figure 8.
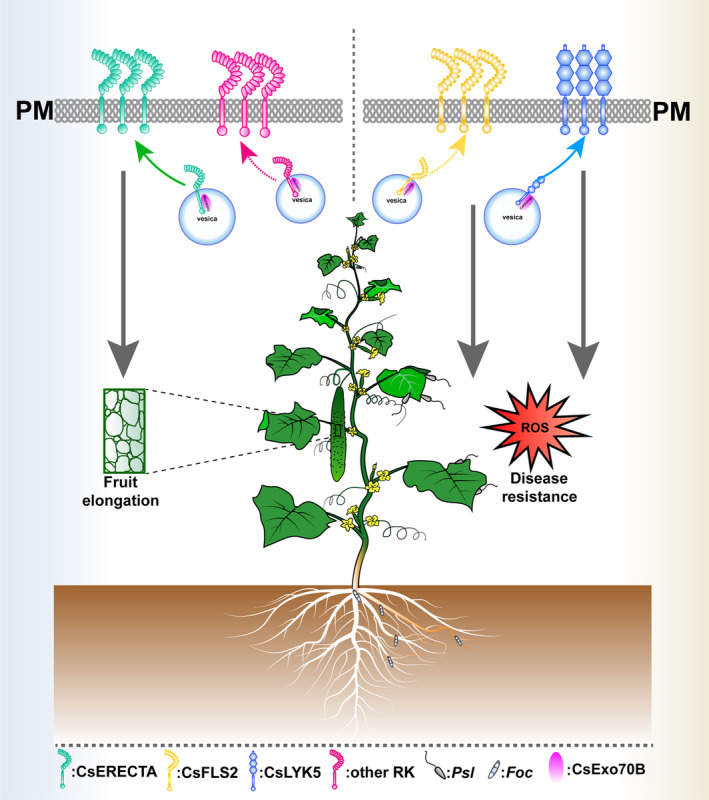
The working model for CsExo70B promotes fruit elongation and pathogen resistance in cucumber. In cucumber, the exocyst subunit CsExo70B interacts with the developmental RK CsERECTA and maintains the level of CsERECTA at PM, promoting fruit elongation. On the other hand, CsExo70B also associates with immune RKs CsFLS2 and CsLYK5 and contributes their accumulation at the PM for resistance to pathogens.
Experimental procedures
Plant materials and growth conditions
Cucumber (Cucumis sativus L.) inbred line XTMC was used in this study for genetic transformation, pathogen infection assays, and expression analysis. The cucumber seedlings at three true‐leaf stage were transplanted to the greenhouse with standard water and fertilizer management and pest control in China Agricultural University, Beijing. The N. bethamiana was grown in a growth chamber set at 24 °C with 16‐h‐light/8‐h‐dark, and the 5‐week‐old plants were used for protein–protein interaction analysis.
Cucumber transformation
To generate Csexo70b and Cserecta mutants using the CRISPR/Cas9 system, the single‐guide RNA (sgRNA) target sites of CsExo70B and CsERECTA were selected by the sgRNA design web (http://crispr.hzau.edu.cn/CRISPR2/). The corresponding guide RNAs were cloned into pKSE402G, which contained a green fluorescent protein (GFP) reporter (Hu et al., 2017; Xing et al., 2014). To generate CsExo70B overexpression transgenic plants, the full‐length coding sequence of CsExo70B without termination codon was first cloned into PUC19‐35S‐HA vector, and then, the full length of CsExo70B‐HA was cloned into pCAMBIA1305.4 vector, which contained a 35S‐GFP expression cassette and a 35S‐GUS expression cassette, SmaI and BstEII sites were selected for replacing GUS to CsExo70B‐HA. All the resulting constructs were transferred into the Agrobacterium tumefaciens strain EHA105, and the cucumber transformation was performed as described previously (Hu et al., 2017).
All positive T0 transgenic plants were selected by GFP reporter. The CsExo70B and CsERECTA mutated lines were genotyped by PCR using gene‐specific primers, and the CsExo70B overexpression transgenic lines were detected by immunoblot using anti‐HA antibodies. All primers used in cucumber transformation are shown in Table S4.
Measurements of the cell length and cell number
The cucumber fruits at 10 DAA were fixed in 3.7% FAA (50 mL Ethanol, 5 mL Acetic acid, 10 mL 37% Formaldehyde, 35 mL Ultrapure H2O) solution. 1‐cm‐thick slices from cucumber fruit pericarp were embedded, sectioned, dewaxed, and stained with toluidine blue. Finally, the sections were observed under a microscope (Olympus DP72, Japan), and the cell length and cell number along a longitudinal axis were quantified using the ImageJ software.
ROS production assays
ROS assays were performed in cucumber plants as described previously in Arabidopsis (Zhang et al., 2007) with some modification. Leaf discs were collected from the second true leaves of cucumber seedlings (two discs per leaf), placed into the white 96‐well plate with 200 μL sterile water overnight and then removed the water and treated with 100 nM flg22 or 0.2 mg mL−1 chitin in 200 μL of buffer containing 100 μM luminol and 10 μg mL−1 horseradish peroxidase. Luminescence was immediately detected with a luminometer (Tecan Infinite F200).
Pathogen infection assays
Pseudomonas syringae pv. lachrymans (Psl) were cultured in King's medium B (King et al., 1954) at 28 °C. Leaves of the cucumber seedlings at two‐three true leaf stage were sprayed with Psl at 1 × 108 CFU mL−1, containing 0.017% Silwet L‐77, and bacterial number in leaves was determined 3 days post‐inoculation (dpi). One leaf discs served as one replicate, and each data point represented at least 18 replicates from at least 6 plants for each genotype (Zhang et al., 2010).
Fusarium oxysporum f. sp. Cucumerinum Owen (Foc) stain was grown on potato dextrose agar (PDA) (Difco™) for 7–10 days at 27 °C, then the hyphae were transferred into Armstrong Fusarium liquid Medium on a shaker (25 °C; 150 rpm; 5 days) for sporulation. The spores were harvested and then adjusted to an inoculated concentration of 1 × 106 spores/mL for Csexo70b mutants and 5 × 106 spores/mL for CsExo70B‐OE lines. The cucumber seedlings with 1 cm radicles were inoculated with the spore suspension for 1 h. Finally, the infected seedlings were planted in sterilized soil at 25 °C and 80% relative humidity with a 16‐h‐light/8‐h‐dark photoperiod. The severity of disease symptoms of each cucumber seedling was graded from 0 to 4, where 0 = no symptoms, 1 = cotyledons or stems lose lustre with mild symptoms, 2 = one cotyledon wilted, 3 = two cotyledons and stem severely wilted, 4 = the whole plant wilted and died. The disease index (DI) was recorded and calculated using a previously described method with minor modifications (Wang et al., 2020b), and all infection experiments were repeated three to four times.
RNA extraction and qRT‐PCR
Total RNA was isolated using the Eastep® Super Total RNA Extraction Kit (Promega) according to the manufacturer's instructions, and 1.2 μg total RNA was reverse‐transcripted using a FastKing gDNA Dispelling RT SuperMix Kit (Tiangen). All qRT‐PCR assays were performed using TB Green® Premix Ex Taq™ II (Takara) in a CFX384 Real‐Time PCR System (BIO‐RAD). The cucumber Ubiquitin gene (CsaV3_5G031430) was used as a reference, and the relative expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Three biological replicates and three technical replicates were performed (Wan et al., 2010).
For determination of the expression levels of CsERECTA, CsFLS2 and CsLYK5 in indicated genotypes, RNA was extracted from 15‐day‐old cucumber seedlings. For analyses of Psl‐inducible gene expression, cucumber seedlings at two‐three true leaf stage were spray‐inoculated with Psl, and leaves were collected at 0, 1, 2, 3 dpi for RNA extraction. For analyses of Foc‐inducible gene expression, roots were collected at 0, 2, 4, and 6 dpi by Foc for RNA extraction. All primer sequences for qRT‐PCR are listed in Table S4.
Split‐luciferase complementation assays
The CDSs of CsExo70B and CsExo70G2 were cloned into pCAMBIA1300‐CLuc vector, and the CDSs of CsERECTA, CsFLS2, CsLYK5, CsBAK1, CsFERONIA and CsCLV1 without stop codons were amplified and inserted into pCAMBIA1300‐HA‐NLuc vector, respectively. These above constructs were then transferred into the A. tumefaciens strain GV3101 and co‐infiltrated into N. benthamiana leaves as described previously (Zhou et al., 2018). After 40–48 h infiltration, the relative luciferase activity was recorded in a 96‐well plate using a luminometer (Tecan Infinite F200) as described previously (Zhou et al., 2018). These Cluc‐ and Nluc‐fusion proteins were detected by anti‐Cluc (Sigma‐Aldrich) anti‐HA (CWBIO) antibodies, respectively.
Co‐immunoprecipitation (IP) assays
Full‐length coding sequence without stop codon of CsExo70B was cloned into pCAMBIA1300‐35S‐HA, and CsERECTA, CsFLS2, CsLYK5 and CsBAK1 were cloned into pCAMBIA1300‐35S‐FLAG vector. For Co‐IP assays in N. benthamiana leaves, these resulting plasmids were transferred into the A. tumefaciens strain GV3101 and transient expressed as previously described (Shi et al., 2013; Zhou et al., 2018). For co‐IP assays in cucumber, these constructs including CsERECTA‐FLAG, CsFLS2‐FLAG, CsLYK5‐FLAG and CsBAK1‐FLAG were transferred into the A. rhizogenes strain K599 and infiltrated the hypocotyl of transgenic 35S:CsExo70B‐HA plants to induce hairy roots for protein extraction as previously described (Yao et al., 2023). Total protein was extracted with the protein extraction buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 1 mM EDTA [pH 8.0], 1 mM DTT, 0.05% (v/v)Triton X‐100, 1× protease inhibitor cocktail [Roche]) and incubated with agarose conjugated anti‐FLAG antibody (Sigma‐Aldrich) for 1–2 h at 4 °C. The beads were washed 5–6 times with the protein extraction buffer. The bound protein was eluted with 0.5 mg/mL 3× FLAG (Sigma‐Aldrich) for 0.5 h at 4°C and separated by SDS‐PAGE and detected using anti‐HA (CWBIO) and anti‐FLAG (Sigma‐Aldrich) antibodies.
Bimolecular fluorescence complementation (BiFC) assays
Full‐length coding sequences of CsExo70B, CsERECTA, CsFLS2 and CsLYK5 without stop codons were cloned and inserted into pSPYCE‐35S and pSPYNE‐35S vectors carrying the N‐terminal/C‐terminal half of Yellow Fluorescent Protein (YFP) expression fragment (Walter et al., 2004). These constructs were transiently expressed in N. benthamiana leaves and were performed as previously described (Zhou et al., 2018), YFP signals were imaged by confocal imaging (Olympus BX51, Japan) under 488 nm excitation wavelength at 72 h post‐infiltration. The primers used are listed in Table S4.
Antibody production
CsERECTA, CsLYK5 and CsH+‐ATPase specific antibodies were produced against the peptides CTKRQPSDRPTMHE, TRAKGMDSKIDKNM and IKNEAVDLEHIPIE by GenScript (www.genscript.com), respectively. Specificity of CsERECTA antibody was tested using Cserecta knockout mutants and N. benthamiana expressing CsERECTA‐FLAG, and CsLYK5 antibody was detected by cucumber total protein or CsLYK5‐FLAG protein expressed in N. benthamiana, and CsH+‐ATPase antibody was verified by cucumber total protein and plasma membrane protein.
Plasma membrane (PM) protein fractionation and immunoblotting assays
The PM protein was extracted from cucumber fresh true leaves or ovaries using a Minute™ Plant Plasma Membrane Protein Isolation Kit (Invent Biotechnologies) according to the instruction and operation manual. Immunoblotting analysis of endogenous levels of CsERECTA, CsLYK5 and CsH+‐ATPase was detected with synthetic endogenous antibodies.
RNA sequencing and data analysis
For RNA‐seq analysis, ovaries at 3 DAA of WT, Csexo70b‐5 # and Cserecta‐2 # mutants were collected for RNA extraction and subjected to RNA‐Seq analysis. Three ovaries were collected from different plants as one biological repeat, and three biological repeats were performed. RNA library construction and sequencing were performed on the Illumina HiSeq X‐ten platform by Biomarker Technologies Corporation (www.biomarker.com.cn).
Clean read pairs were mapped to reference cucumber genome (Chinese long v.3.0). DEGs were identified via fragments per kilobase of transcript per million mapped fragments (FPKM) on the BMKCloud online platform (Fold change ≥ 2.0, FDR < 0.01), the heat maps were generated via TBtools Software (Chen et al., 2020).
Statistical analysis
All experiments were repeated at least three times. Data were analysed on GraphPad Prism9 with two‐tailed Student's t test for comparison between two samples, and one‐way ANOVA Tukey's test for pairwise comparisons among multiple samples.
Conflict of interest
The authors declare no competing interests.
Accession numbers
Sequence data in this article can be found in Cucurbit Genomics Database (http://www.cucurbitgenomics.org/); all the accession numbers of genes are listed in Table S5.
Author contributions
X.Z., Z.Z. and L.L. designed experiments. L.L. performed the majority of the experiments and analysed the data. L.L., Z.Z. and X.Z. wrote this manuscript. J.C. C.G., S.W., Y.X., L.H., W.S. and J.Z. provided experimental assistances. Z.Z., Z.W. and X.L. provided technical guidance. M.L., C.L. and L.W. checked this manuscript. All authors discussed the results and contributed to this manuscript.
Supporting information
Figure S1 Genotyping and phenotypic analysis of Csexo70b mutants.
Figure S2 CsExo70B interacts with CsERECTA and CsCLV1 in N. benthamiana plants.
Figure S3 Knockout of CsERECTA resulted in dwarfed plants, smaller male flowers and seeds in cucumber.
Figure S4 The bimolecular fluorescence complementation (BiFC) assays showing CsExo70B interacts with CsERECTA, CsFLS2 and CsLYK5.
Figure S5 Detection of the specificity of CsERECTA, CsLYK5 and CsH+‐ATPase antibodies.
Figure S6 KEGG enrichment analyses of common up‐regulated DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants.
Figure S7 KEGG enrichment analyses of common down‐regulated DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants.
Figure S8 Knockout of CsExo70B promotes Fusarium oxysporum f. sp. Cucumerinum Owen (Foc) infection in cucumber.
Figure S9 The expression level of CsFLS2 was normal in Csexo70b mutants and CsExo70B‐OE lines.
Figure S10 Phenotypic analysis of CsExo70B overexpression lines.
Figure S11 The phenotypes of Csexo70b mutants.
Figure S12 Heat map showing the expression pattern of genes involved in auxin‐related and cell division‐related pathway.
Figure S13 Expression patterns of CsERECTA, CsFLS2 and CsLYK5 in cucumber different tissues.
Figure S14 Pathogen‐induced RK expression in cucumber.
Figure S15 The expression of CsExo70B and RKs was normal in fruits infected by pathogen.
Table S1 List of off‐target analyses of CsExo70B and CsERECTA.
Table S4 List of primers used in this study.
Table S5 List of gene accession numbers used in this study.
Table S2 List of common up‐regulated DEGs in Csexo70b and Cserecta mutants.
Table S3 List of common down‐regulated DEGs in Csexo70b and Cserecta mutants.
Acknowledgements
We would like to thank Prof. Sanwen Huang for providing CRISPR/Cas9 vector and Prof. Xueyong Yang for giving pCAMBIA1305.4 vector, Prof. Xuezheng Wang for providing the greenhouse, and Prof. Zhiming Wu for Pseudomonas syringae pv. lachrymans (Psl) strain. The wok was supported by grants from the Natural Science Foundation of China (32025033, 31930097 and 32070286), the 2115 Talent Development Program of China Agricultural University, Expert Workstation in Yunnan Province (202205AF150021), and Pinduoduo‐China Agricultural University Research Fund (PC2023B01002).
Contributor Information
Xiaolan Zhang, Email: zhxiaolan@cau.edu.cn.
Zhaoyang Zhou, Email: zyzhou@cau.edu.cn.
References
- Amano, Y. , Tsubouchi, H. , Shinohara, H. , Ogawa, M. and Matsubayashi, Y. (2007) Tyrosine‐sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis . Proc. Natl. Acad. Sci. USA, 104, 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos, M.L. , Yoshida, Y. , Major, I.T. , de Oliveira Ferreira, D. , Weraduwage, S.M. , Froehlich, J.E. , Johnson, B.F. et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth–defense tradeoffs. Nat. Commun. 7, 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, Y. , Liang, Y. , Tanaka, K. , Nguyen, C.T. , Jedrzejczak, R.P. , Joachimiak, A. and Stacey, G. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin‐induced complex with related kinase CERK1. Elife, 3, e03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand, J.N. and Walker, J.C. (1964) Inheritance of resistance to angular leaf spot of cucumber. Phytopathology, 54, 51–53. [Google Scholar]
- Che, G. , Song, W.Y. and Zhang, X.L. (2023) Gene network associates with CsCRC regulating fruit elongation in cucumber. Vegetable Research, 3, 7. [Google Scholar]
- Chen, C. , Chen, H. , Zhang, Y. , Thomas, H.R. , Frank, M.H. , He, Y. and Xia, R. (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant, 13, 1194–1202. [DOI] [PubMed] [Google Scholar]
- Cheng, F. , Song, M. , Zhang, M. , Cheng, C. , Chen, J. and Lou, Q. (2022) A SNP mutation in the CsCLAVATA1 leads to pleiotropic variation in plant architecture and fruit morphogenesis in cucumber (Cucumis sativus L.). Plant Sci. 323, 111397. [DOI] [PubMed] [Google Scholar]
- Choi, S.W. , Tamaki, T. , Ebine, K. , Uemura, T. , Ueda, T. and Nakano, A. (2013) RABA members act in distinct steps of subcellular trafficking ofthe FLAGELLIN SENSING2 receptor. Plant Cell, 25, 1174–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, Y.T. , Gidda, S.K. , Sanford, C. , Parkinson, J. , Mullen, R.T. and Goring, D.R. (2010) Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 185, 401–419. [DOI] [PubMed] [Google Scholar]
- Clark, S.E. , Williams, R.W. and Meyerowitz, E.M. (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis . Cell, 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Coley, P.D. , Bryant, J.P. and Chapin, F.S. (1985) Resource availability and plant antiherbivore defense. Science, 230, 895–899. [DOI] [PubMed] [Google Scholar]
- Couto, D. and Zipfel, C. (2016) Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Dong, J. , Xu, J. , Xu, X. , Xu, Q. and Chen, X. (2019) Inheritance and quantitative trait locus mapping of Fusarium wilt resistance in cucumber. Front. Plant Sci. 10, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drdová, E.J. , Synek, L. , Pečenková, T. , Hála, M. , Kulich, I. , Fowler, J.E. , Murphy, A.S. et al. (2013) The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis . Plant J. 73, 709–719. [DOI] [PubMed] [Google Scholar]
- Gust, A.A. , Pruitt, R. and Nurnberger, T. (2017) Sensing danger: key to activating plant immunity. Trends Plant Sci. 22, 779–791. [DOI] [PubMed] [Google Scholar]
- Gómez, G.L. and Boller, T. (2000) FLS2: an LRR receptor‐like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis . Mol. Cell, 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- He, Z. , Webster, S. and He, S.Y. (2022) Growth‐defense trade‐offs in plants. Curr. Biol. 32, R634–R639. [DOI] [PubMed] [Google Scholar]
- Herms, D.A. and Mattson, W.J. (1992) The dilemma of plants: to grow or defend. Q. Rev. Biol. 67, 283–335. [Google Scholar]
- Hou, H. , Fang, J. , Liang, J. , Diao, Z. , Wang, W. , Yang, D. , Li, S. et al. (2020) OsExo70B1 positively regulates disease resistance to Magnaporthe oryzae in rice. Int. J. Mol. Sci. 21, 7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, B. , Li, D. , Liu, X. , Qi, J. , Gao, D. , Zhao, S. , Huang, S. et al. (2017) Engineering non‐transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 system. Mol. Plant, 10, 1575–1578. [DOI] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomerya, B.L. and He, S.Y. (2014) Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant, 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kinoshita, A. , Betsuyaku, S. , Osakabe, Y. , Mizuno, S. , Nagawa, S. , Stahl, Y. , Simon, R. et al. (2010) RPK2 is an essential receptor‐like kinase that transmits the CLV3 signal in Arabidopsis . Development, 137, 3911–3920. [DOI] [PubMed] [Google Scholar]
- Kulich, I. , Vojtíková, Z. , Glanc, M. , Ortmannová, J. , Rasmann, S. and Žárský, V. (2015) Cell wall maturation of Arabidopsis trichomes is dependent on exocyst subunit EXO70H4 and involves callose deposition. Plant Physiol. 168, 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich, I. , Vojtíková, Z. , Sabol, P. , Ortmannová, J. , Neděla, V. , Tihlaříková, E. and Žárský, V. (2018) Exocyst subunit EXO70H4 has a specific role in callose synthase secretion and silica accumulation. Plant Physiol. 176, 2040–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H.Y. , Bowen, C.H. , Popescu, G.V. , Kang, H.G. , Kato, N. , Ma, S. , Dinesh‐Kumar, S. et al. (2011) Arabidopsis RTNLB1 and RTNLB2 reticulon‐like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell, 23, 3374–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. and Zhou, J.M. (2018) Receptor‐like cytoplasmic kinases: central players in plant receptor kinase–mediated signaling. Annu. Rev. Plant Biol. 69, 267–299. [DOI] [PubMed] [Google Scholar]
- Liu, L. , Gu, C.H. , Zhang, J.H. , Guo, J.Y. , Zhang, X.L. and Zhou, Z.Y. (2023) Genome‐wide analysis of exocyst complex subunit Exo70 gene family in cucumber. Int. J. Mol. Sci. 24, 10929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu, K. , Chen, X. , Yao, X. , An, Y. , Wang, X. , Qin, L. , Li, X. et al. (2022) Phosphorylation of a wheat aquaporin at two sites enhances both plant growth and defense. Mol.Plant. 15, 1772–1789. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. (2014) Posttranslationally modified small‐peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413. [DOI] [PubMed] [Google Scholar]
- Matsubayashi, Y. , Ogawa, M. , Morita, A. and Sakagami, Y. (2002) An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science, 296, 1470–1472. [DOI] [PubMed] [Google Scholar]
- Morris, E.R. and Walker, J.C. (2003) Receptor‐like protein kinases: the keys to response. Curr. Opin. Plant Biol. 6, 339–342. [DOI] [PubMed] [Google Scholar]
- Pan, Y. , Liang, X. , Gao, M. , Liu, H. , Meng, H. , Weng, Y. and Cheng, Z. (2017) Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 130, 573–586. [DOI] [PubMed] [Google Scholar]
- Pecenková, T. , Hála, M. , Kulich, I. , Kocourková, D. , Drdová, E. , Fendrych, M. , Toupalová, H. et al. (2011) The role for the exocyst complex subunits Exo70B2 and Exo70H1 in the plant‐pathogen interaction. J. Exp. Bot. 62, 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri, L. , Bemis, S. , Shpak, E. and Torii, K. (2007) Haploinsufficiency after successive loss of signaling reveals a role for ERECTA‐family genes in Arabidopsis ovule development. Development, 134, 3099–3109. [DOI] [PubMed] [Google Scholar]
- Saeed, B. , Brillada, C. and Trujillo, M. (2019) Dissecting the plant exocyst. Curr. Opin. Plant Biol. 52, 69–76. [DOI] [PubMed] [Google Scholar]
- Shi, H. , Shen, Q. , Qi, Y. , Yan, H. , Nie, H. , Chen, Y. , Zhao, T. et al. (2013) BR‐SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis . Plant Cell, 25, 1143–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, S.H. and Bleecker, A.B. (2001a) Plant receptor‐like kinase gene family: diversity, function, and signaling. Sci. STKE, 2001, re22. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H. and Bleecker, A.B. (2001b) Receptor‐like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA, 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak, E.D. (2013) Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 55, 1238–1250. [DOI] [PubMed] [Google Scholar]
- Shpak, E.D. , Lakeman, M.B. and Torii, K.U. (2003) Dominant‐negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine‐rich repeat receptor–like kinase signaling pathway that regulates organ shape. Plant Cell, 15, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann, M. , Anderson, R.G. , Ichimura, K. , Pecenkova, T. , Reuter, P. , Žársky, V. , McDowell, J.M. et al. (2012) The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP‐triggered responses in Arabidopsis . Plant Cell, 24, 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmann, M. , Anderson, R.G. , Westphal, L. , Rosahl, S. , McDowell, J.M. and Trujillo, M. (2013) The exocyst subunit Exo70B1 is involved in the immune response of Arabidopsis thaliana to different pathogens and cell death. Plant Signal. Behav. 8, e27421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synek, L. , Schlager, N. , Eliás, M. , Quentin, M. , Hauser, M.T. and Zárský, V. (2006) AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development. Plant J. 48, 54–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush, D.R. , Maurice, T. , Roth, D. and Novick, P. (1996) The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae . EMBO J. 15, 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Walter, M. , Walter, M. , Chaban, C. , Schütze, K. , Batistic, O. , Weckermann, K. , Näke, C. et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wan, H. , Yuan, W. , Bo, K. , Shen, J. , Pang, X. and Chen, J. (2013) Genome‐wide analysis of NBS‐encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS‐encoding genes in Cucurbitaceae crops. BMC Genomics, 14, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, H. , Zhao, Z. , Qian, C. , Sui, Y. , Malik, A.A. and Chen, J. (2010) Selection of appropriate reference genes for gene expression studies by quantitative real‐time polymerase chain reaction in cucumber. Anal. Biochem. 399, 257–261. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. et al. (2018) A single transcription factor promotes both yield and immunity in rice. Science, 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Liu, N. , Gao, C. , Cai, H. , Romeis, T. and Tang, D. (2020a) The Arabidopsis exocyst subunits EXO70B1 and EXO70B2 regulate FLS2 homeostasis at the plasma membrane. New Phytol. 227, 529–544. [DOI] [PubMed] [Google Scholar]
- Wang, P. , Zhou, L. , Jamieson, P. , Zhang, L. , Zhao, Z. , Babilonia, K. , Shao, W. et al. (2020b) The cotton wall‐associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell, 32, 3978–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cheng, X. , Shan, Q. , Zhang, Y. , Liu, J. , Gao, C. and Qiu, J.L. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. [DOI] [PubMed] [Google Scholar]
- Weng, Y. and Wehner, T.C. (2017) Cucumber gene catalogue 2017. In: Cucurbit Genetic Cooperative ( Grumet, R. , Crosby, K. , Wehner, T. , Myers, J. , Behera, T. , eds), pp 17–29. Charleston: Vegetable Laboratory. [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Wang, X. , Liu, J. , Jia, A. , Xu, C. , Deng, X.W. and He, G. (2022) Natural variation in the transcription factor REPLUMLESS contributes to both disease resistance and plant growth in Arabidopsis . Plant Commun. 3, 100351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, X. , Li, H. , Nie, J. , Liu, H. , Guo, Y. , Lv, L. , Yang, Z. et al. (2023) Disruption of the amino acid transporter CsAAP2 inhibits auxin‐mediated root development in cucumber. New Phytol. 239, 639–659. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Li, W. , Xiang, T. , Liu, Z. , Laluk, K. , Ding, X. , Zou, Y. et al. (2010) Receptor‐like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe, 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zhang, J. , Shao, F. , Li, Y. , Cui, H. , Chen, L. , Li, H. , Zou, Y. et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP‐induced immunity in plants. Cell Host Microbe, 1, 175–185. [DOI] [PubMed] [Google Scholar]
- Zhao, T. , Rui, L. , Li, J. , Nishimura, M.T. , Vogel, J.P. , Liu, N. , Liu, S. et al. (2015) A truncated NLR protein, TIR‐NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 11, e1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z. , Bi, G. and Zhou, J.M. (2018) Luciferase complementation assay for protein‐protein interactions in plants. Curr. Protoc. Plant Biol. 3, 42–50. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. and Boller, T. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
- Zárský, V. , Cvrcková, F. , Potocký, M. and Hála, M. (2009) Exocytosis and cell polarity in plants – exocyst and recycling domains. New Phytol. 183, 255–272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Genotyping and phenotypic analysis of Csexo70b mutants.
Figure S2 CsExo70B interacts with CsERECTA and CsCLV1 in N. benthamiana plants.
Figure S3 Knockout of CsERECTA resulted in dwarfed plants, smaller male flowers and seeds in cucumber.
Figure S4 The bimolecular fluorescence complementation (BiFC) assays showing CsExo70B interacts with CsERECTA, CsFLS2 and CsLYK5.
Figure S5 Detection of the specificity of CsERECTA, CsLYK5 and CsH+‐ATPase antibodies.
Figure S6 KEGG enrichment analyses of common up‐regulated DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants.
Figure S7 KEGG enrichment analyses of common down‐regulated DEGs in Csexo70b‐5 # and Cserecta‐2 # mutants.
Figure S8 Knockout of CsExo70B promotes Fusarium oxysporum f. sp. Cucumerinum Owen (Foc) infection in cucumber.
Figure S9 The expression level of CsFLS2 was normal in Csexo70b mutants and CsExo70B‐OE lines.
Figure S10 Phenotypic analysis of CsExo70B overexpression lines.
Figure S11 The phenotypes of Csexo70b mutants.
Figure S12 Heat map showing the expression pattern of genes involved in auxin‐related and cell division‐related pathway.
Figure S13 Expression patterns of CsERECTA, CsFLS2 and CsLYK5 in cucumber different tissues.
Figure S14 Pathogen‐induced RK expression in cucumber.
Figure S15 The expression of CsExo70B and RKs was normal in fruits infected by pathogen.
Table S1 List of off‐target analyses of CsExo70B and CsERECTA.
Table S4 List of primers used in this study.
Table S5 List of gene accession numbers used in this study.
Table S2 List of common up‐regulated DEGs in Csexo70b and Cserecta mutants.
Table S3 List of common down‐regulated DEGs in Csexo70b and Cserecta mutants.


