Abstract
The concept of a digital twin came from the engineering, industrial, and manufacturing domains to create virtual objects or machines that could inform the design and development of real objects. This idea is appealing for precision medicine where digital twins of patients could help inform healthcare decisions. We have developed a methodology for generating and using digital twins for clinical outcome prediction. We introduce a new approach that combines synthetic data and network science to create digital twins (i.e. SynTwin) for precision medicine. First, our approach starts by estimating the distance between all subjects based on their available features. Second, the distances are used to construct a network with subjects as nodes and edges defining distance less than the percolation threshold. Third, communities or cliques of subjects are defined. Fourth, a large population of synthetic patients are generated using a synthetic data generation algorithm that models the correlation structure of the data to generate new patients. Fifth, digital twins are selected from the synthetic patient population that are within a given distance defining a subject community in the network. Finally, we compare and contrast community-based prediction of clinical endpoints using real subjects, digital twins, or both within and outside of the community. Key to this approach are the digital twins defined using patient similarity that represent hypothetical unobserved patients with patterns similar to nearby real patients as defined by network distance and community structure. We apply our SynTwin approach to predicting mortality in a population-based cancer registry (n=87,674) from the Surveillance, Epidemiology, and End Results (SEER) program from the National Cancer Institute (USA). Our results demonstrate that nearest network neighbor prediction of mortality in this study is significantly improved with digital twins (AUROC=0.864, 95% CI=0.857–0.872) over just using real data alone (AUROC=0.791, 95% CI=0.781–0.800). These results suggest a network-based digital twin strategy using synthetic patients may add value to precision medicine efforts.
Keywords: Digital twins, Precision medicine, Artificial intelligence, Synthetic data
1. Introduction to Digital Twins
The concept of a digital twin came from the engineering, industrial, and manufacturing domains and refers to the creation of virtual objects or machines that can inform the design and development of real objects (Grieves & Vickers 2017). The promise of this approach in manufacturing is to reduce costs, improve efficiency, reduce waste, and minimize variability among products (Attaran et al. 2023). This is accomplished by enumerating and evaluating design parameters of the digital twin of a physical product with some measurable outcome that can then be applied to manufacturing. Use cases in industry include product design, process design and optimization, supply chain management, preventive system maintenance, farm management, weather modeling, soil management, facility and operations design, construction, etc. (Attaran & Celik 2023). Consider the use case of monitoring weed pressure and crop growth (Verdouw et al. 2021). Data on crops, weeds, weather, and soil conditions are collected from crop sensors. These data are used to build a digital twin of the crops where parameters for a weeding machine can be enumerated and evaluated. Optimized parameters from the digital twin can then be put into practice for weed management with benefits including crop weight, size, and yield.
The successful use of digital twins in industry has opened the door for their use in medicine and healthcare where they represent virtual or simulated patients that could be used to inform health outcomes or treatment decision for real patients (Acosta et al. 2022). This idea of using digital twins in precision medicine has been explored for asthma management (Drummond et al. 2023), the treatment of immune-mediated diseases (Benson 2023), and dementia care (Wickramasinghe et al. 2022), for example. Despite the interest in this area, the development of computational methods and open-source software for creating and using digital twins has been slow to emerge. This is likely due to the industry focus on creating twins of mechanical objects using principles of physics and engineering that do not exist with enough detail to create simulated patients with molecular, cellular, physiological, and anatomical realness and appropriate environmental and societal context. Some of these challenges have been previously discussed (Benson 2023).
The goal of the present study was to create a computational methodology for generating digital twins based on synthetic patients rather than biophysics. The generation of synthetic data is becoming a mature field (Gonzales et al. 2023) and lends itself well to the digital twin strategy. The working hypothesis is that the correlation structure of clinical variables among patients can inform the creation of digital twins that represent unobserved individuals. In other words, patient relationships might be able to serve as a surrogate for biophysical realizations. The advantage of this surrogate approach is that it can be implemented and evaluated today while we wait for better and more complete biophysical models that could take decades to develop and validate.
We introduce here a new approach that combines synthetic data and network science to create digital twins (i.e. SynTwin) for precision medicine. Our approach starts by estimating the distance between all subjects based on their available features. We explore here several different distance metrics. Second, the distances are used to construct a network with subjects as nodes and edges defining distance less than the percolation threshold. Third, communities or cliques of subjects are identified using a Multilevel community detection algorithm. Fourth, a large population of synthetic patients or subjects are generated. Several synthetic data generators were evaluated. Fifth, digital twins are selected from the synthetic patient population that are within a given distance defining a subject community in the network. By design, the digital twins represent unobserved hypothetical patients with similar clinical profiles as their real patient counterparts. Finally, we compare and contrast community-based prediction of clinical endpoints using real subjects, digital twins, or both. This is compared to predictive performance using real patients outside the community as a baseline. We apply our synthetic digital twin (SynTwin) approach to predicting mortality in a population-based cancer registry (n=87,674) from the Surveillance, Epidemiology, and End Results (SEER) program from the National Cancer Institute (USA). Bootstrapping is used to assess the standard error of all performance metrics and to estimate 95% confidence intervals for hypothesis testing. Our results demonstrate that nearest network neighbor prediction of mortality in the SEER breast cancer data is significantly improved with digital twins. These results support a growing number of studies highlighting the benefit of synthetic data in other applications.
2. Methods
We describe here the data used and the detailed methods for the SynTwin approach.
2.1. Cancer Registry Data
We chose a population-based cancer registry from the Surveillance, Epidemiology, and End Results (SEER) program from the National Cancer Institute (USA) for this study due its large sample size and ease of access by simple registration with an email address to allow for reproducibility. We utilized SEER Stat Version 8.4.1 for data retrieval.
To extract patient data specifically for breast cancer, we applied the following filters:
Database name: Incidence - SEER Research Data, 17 Registries, Nov 2021 Sub (2000–2019) - Linked To County Attributes - Time Dependent (1990–2019) Income/Rurality, 1969–2020 Counties.
Additional filter criteria included:
Site recode ICD-O-3/WHO 2008 = ‘Breast’ AND Year of diagnosis = ‘2010’, ‘2011’, ‘2012’, ‘2013’, ‘2014’, ‘2015’ AND {Vital status recode (study cutoff used) = ‘Alive’ OR {Vital status recode (study cutoff used) = ‘Dead’ AND SEER cause-specific death classification = ‘Dead {attributable to this cancer dx}’}}
We chose to exclude data from the years 2015–2019 due to the significant imbalance observed within that period. Specifically, the data exhibited a notable disparity between the number of surviving patients and the number of deceased cases. More than 80% of the patients within that timeframe were still alive, rendering the dataset heavily skewed. Our criteria yielded 324,117 patient records. Removing redundant entries resulted in 231,930 records, consisting of 188,093 Alive cases and 43,837 Dead cases. Subsequently, we conducted a stratified sampling based on vital status to create a balanced dataset for prediction purposes. We retained all Dead cases (n=43,837) and randomly undersampled the same number of Alive cases (n=43,837). This process yielded a total of 87,674 records for our final dataset. We partitioned this sample into a training dataset (n=57,674) and a validation dataset of approximately 1/3 of the sample (n=30,000) to assess internal validity of the results. The training data was used to generate the digital twins while the validation dataset was held out for making predictions using the real patient data and their network and communities. The data processing steps are outlined by the flowchart in Figure 1.
Fig. 1.
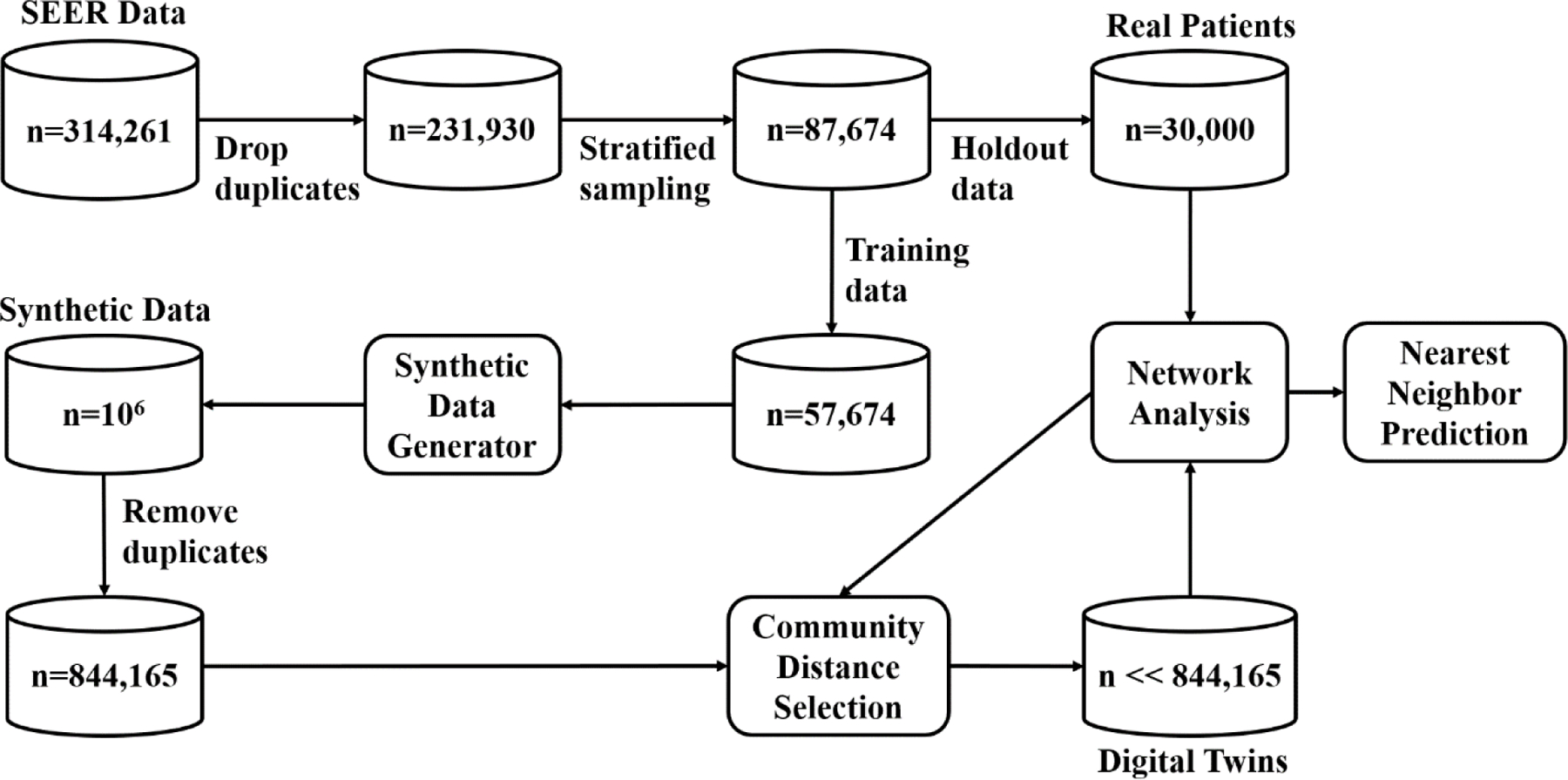
Flowchart for data processing and analysis.
Features included age, year of diagnosis, sex, race, ICDO3, tumor grade, laterality, primary site, survival in months, tumor sequence, diagnostic confirmation, ICCC site, combined summary stage, and vital status (Alive or Dead). The last feature was used as the clinical outcome of class variable for prediction.
2.2. Bootstrapping
A central goal of this study was to compare and contrast different methods for estimating patient distances, different methods for generating synthetic data, and different approaches to using digital twins to predict outcome. In order to generate a sampling distribution of all objective functions we carried out 1000-fold bootstrapping by sampling 90% of patients in the holdout or validation data with replacement in each community of size 10 or greater a total of 1000 times. Each performance measure was estimated using all 1000 replications to derive its empirical distribution. This allowed 95% confidence intervals to be estimated for all performance metrics. These were used for uncertainty quantification, statistical comparisons, and hypothesis testing.
2.3. The SynTwin Algorithm for Network-Based Generation of Digital Twins from Synthetic Data
We describe here our six-step algorithm for generating digital twins and using them for predicting mortality in the SEER data. This involves computing patient distances based on clinical features, constructing a network using distances based on the percolation threshold, identifying patient communities, generating synthetic patients, selection of digital twins, a nearest neighbor (i.e. within community) prediction of mortality.
2.3.1. Distance Measures
The first step is to estimate the distances between patients. We evaluated four different distance metrics. These include Euclidian, Manhattan, Cosine (Lee et al. 2015), and Gower (Gower 1971). Each has different strengths and weaknesses. For example, Gower is appealing because it is scale-invariant and works well with both discrete and continuous data. Further, as shown in the results, this distance measure yielded the best results.
2.3.2. Network Construction
The second step is to build a network with patients as nodes and edges with weights based on the estimated distances in the first step. To prevent an uninformative fully connected network, we used a percolation threshold equal to the first upward inflection point of the convex part of the sigmoid relationship between edge weight (X axis) and network size (Y axis) as an objective approach to filtering edges.
2.3.3. Community Detection
The third step is to detect communities of patients (i.e. cliques or modules) in the network. There are many different community detection algorithms for large networks. We selected the Multilevel algorithm (Blondel et al. 2008) for this study. This algorithm uses a heuristic for modularity optimization and is designed specifically for large networks. The Multilevel algorithm was shown to outperform other community detection algorithms available at the time and with better time complexity (Blondel et al. 2008). Further, a more recent study compared this algorithm with seven others on several graph benchmarks and showed that the Multilevel algorithm was best for both accuracy and time complexity (Yang et al. 2016). In our study, we varied the resolution parameter settings to maximize the number of communities with at least 10 subjects. This yielded between 11,000 and 19,000 communities across the four different distance metrics we investigated.
2.3.4. Synthetic Data Generation
The fourth step is to generate synthetic patients to be used as the population to select digital twins from. We evaluated three synthetic data generation algorithms. The first, categorical latent Gaussian process (CLGP), uses continuous latent variables to represent categorical variables that can then be modeled using a Gaussian process (Gal et al. 2015). Here, synthetic data can be generated by sampling from the posterior distribution of the latent variables. The second, mixture of product of multinomials (MPoM), uses a probabilistic model to generate synthetic data with similar statistical properties to the original data (Dunson & Xing 2009). The third, multi-categorical extension of a medical generative adversarial network (MC-MedGAN), uses two adversarial neural networks to generate synthetic data (Choi et al. 2017). Here, The first network learns to generate realistic synthetic data, and the second one attempts to distinguish between real and synthetic data generated by the first network. Autoencoders are used to transform the multivariate categorical data to continuous values, which are then used by the GAN to generate synthetic data.
All three of these methods were recently evaluated and compared (Goncalves et al. 2020). We used the following performance metrics highlighted in this study to evaluate each approach: pairwise correlation difference (PCD), log-cluster (LC), support coverage (SC), and cross-classification (CrCl). The PCD metric is computed as the Frobenius norm difference between Pearson correlation matrices of real and synthetic datasets. It measures how well a method captures the correlation between variables. The LC metric assesses the similarity in latent structure between real and synthetic datasets using k-means clustering. The SC metric quantifies the extent to which the variables support in real data is captured in synthetic data. It is calculated as the ratio of the cardinalities of number of levels (support) for each variable in real and synthetic data. CrCl assesses how accurately a synthetic dataset replicates the statistical dependence found in real data using a classifier.
We used the best hyperparameters reported for “small-set” in the study (Goncalves et al. 2020) to set up our synthetic data generation algorithms considering the smaller number of variables in our dataset. For CLGP we used 100 inducing points and 5-dimensional latent space. For MPoM we set the number of clusters (k) to 30, concentration parameter (α) to 10, Gibbs sampling steps to 10,000, and burn-in steps to 1,000. For MC-MedGAN we used a learning rate 1e-3 and batch size 100 samples. We applied L-2 regularization on the weights of the neural network with λ=1e-3 and set temperature parameter for Gumbel-Softmax trick to τ=0.666. The autoencoder part was built with a code size 64, two encoder layers (hidden size – 256 and 128), and two decoder layers (hidden size – 256 and 128). The GAN part consisted of one generator step with two generator layers (hidden size – 64 and 64) and two discriminator steps each with two discriminator layers (hidden size – 256 and 128). The autoencoder and the GAN were trained for 100 and 500 epochs, respectively.
2.3.5. Selection of Digital Twins
The fifth step is to select digital twins from a population of synthetic patients. For a synthetic twin to be a digital twin it must be within some distance of one or more real patients such that the clinical features can represent realistic unobserved measures and outcomes. For each community we selected those synthetic patients whose distances places them within that community. We refer to these virtual patients as digital twins of the real patients in the community. Only those digital twins in a community are used for prediction of mortality.
2.3.6. Prediction of Mortality
The final step is to use features from real patients and/or digital twins to predict mortality (Alive or Dead) using a majority vote using the study design described in the next section (2.4). This prediction strategy resembles k-nearest neighbor classification. We estimated six different classification performance measures for predicting mortality across 1000 bootstrapped samples of the holdout data sampled with replacement from each community with at least 10 patients. These included accuracy, balanced accuracy area under the receiver operating characteristic curve (AUROC), precision, recall, and F1. The mean of each performance metric across the 1000 bootstrapped datasets was reported along with the bootstrapped 95% confidence interval (CI).
2.4. Study Design and Analysis.
A central goal of this study is to evaluate whether digital twins add any value to predicting mortality beyond that provided by data from the real patients. To answer this question, we developed the following study design (Figure 2). Here, we evaluated prediction of mortality in target patients (black circle) using real patients (A), digital twins (B), real patients and digital twins (C), the closest digital twins equal to the number of real patients in the community (D), real patients and closest digital twins (E), and real patients outside the community (F) as a control for the value of considering communities. Here, each subject in a community alternate as the target patient in a leave-one-out style analysis.
Fig. 2.
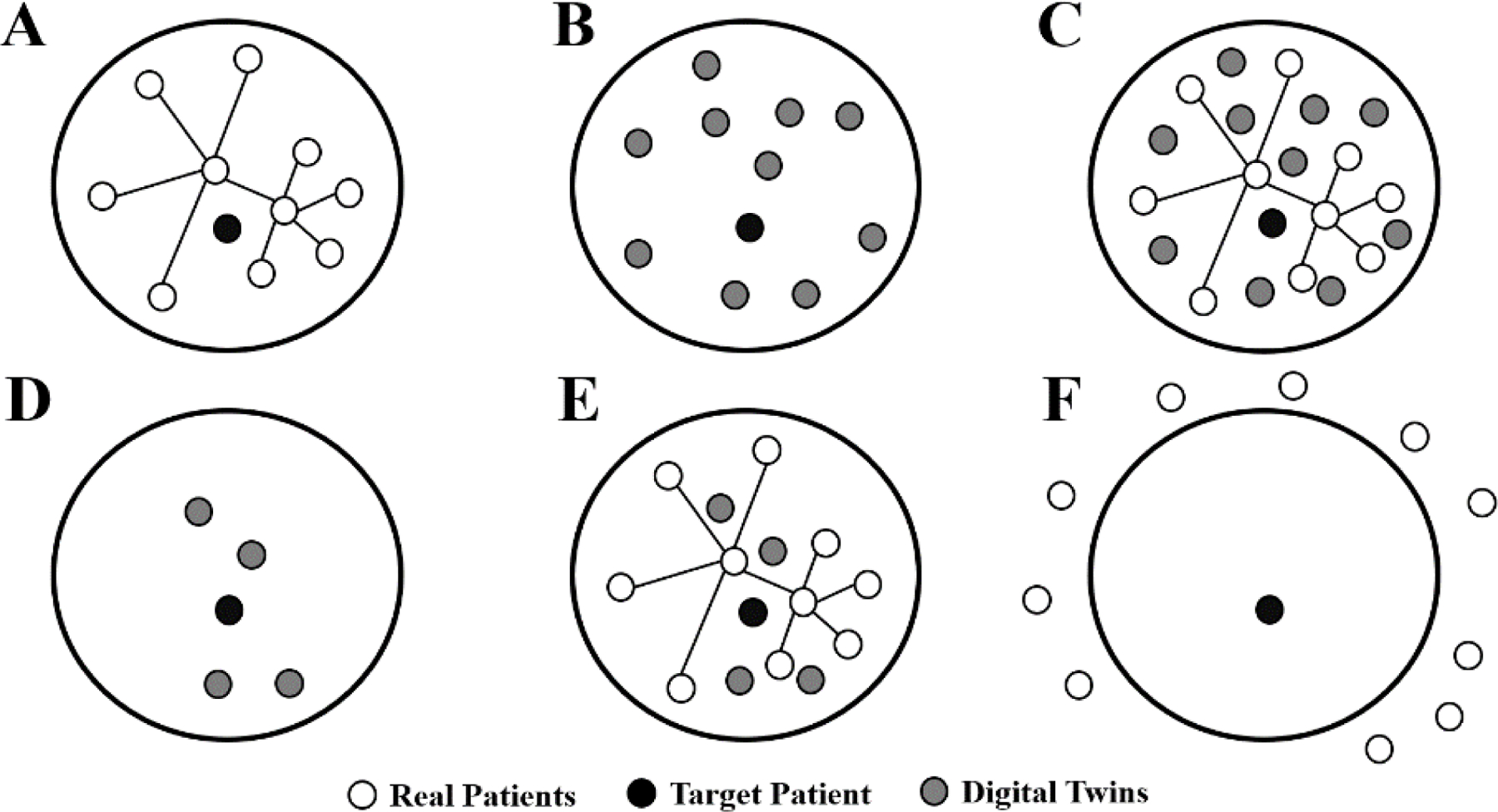
Study design for comparing outcome prediction using real patients and/or digital twins. The large circles represent a community within the patient network. Prediction of the target patient is carried out using real patients (A), digital twins (B), real patients and digital twins (C), the closest digital twins (D), real patients and closest digital twins (E), and real patients outside the community (F).
A total of one million synthetic patients were generated from the training data using the best synthetic data generation algorithm (MPoM). We predicted target patient mortality using the nearest neighbor majority vote classification method in the holdout or validation dataset. We estimated 95% confidence intervals for each of the classification performance metrics and statistically compared distance metrics, synthetic data generation algorithms, and study designs.
3. Results
Table 1 summarizes the performance metrics for the three synthetic data generators considered. Across all metrics, mixture of product of multinomials (MPoM) performed significantly better than the other two methods with nonoverlapping 95% confidence intervals. Consider for example that MPoM had a cross-classification (CrCl) of 0.982 indicating a very high degree of correlation between the same features in the real dataset and in the synthetic dataset. This was significantly higher than the CrCl for MC-MedGAN (0.759) and CLGP (0.645) with nonoverlapping confidence intervals when compared to MPoM. This was true for the other metrics. The only exception was the categorical latent Gaussian process (CLGP) for coverage which was comparable to MPoM. These results mirror a previous evaluation of these algorithms using the SEER data where MPoM outperformed the MC-MedGAN adversarial neural network approach (Goncalves et al. 2020). Therefore, we selected MPoM as our synthetic data generator and used it for the remainder of the study.
Table 1.
Comparison of synthetic data algorithms (columns) for four performance metrics (rows). Bolded metric values are significantly better than the others.
| CLGP |
MC-MedGAN |
MPoM |
||||
|---|---|---|---|---|---|---|
| Metric | Mean | 95% CI | Mean | 95% CI | Mean | 95% CI |
| CrCl | 0.645 | 0.533, 0.757 | 0.759 | 0.634, 0.884 | 0.982 | 0.941, 1.022 |
| LC | −1.545 | −1.594, −1.496 | −2.941 | −4.211, −1.672 | −5.191 | −6.146, −4.236 |
| SC | 1.000 | 0.999, 1.001 | 0.830 | 0.625, 1.036 | 0.989 | 0.978, 1.000 |
| PCD | 1.723 | 1.453, 1.992 | 2.772 | 1.007, 4.538 | 1.012 | 0.720, 1.305 |
Table 2 summarizes the AUROC for predicting mortality in the holdout or validation data for each of the four distance metrics and each of the six study designs (A-F, see Figure 1). Study designs D and E had significantly higher AUROCs than the others but were not significantly better than each other given overlapping confidence intervals. Unique to study designs D and E are the presence of digital twins selected to be close to the target patient being predicted. The performance of D and E was significantly higher for the Gower distance than Cosine, Euclidian, or Manhattan. Therefore, we are reporting the mean AUROCs for Gower distance. These patterns of significance were similar for accuracy, balanced accuracy, and the other performance metrics (tables not shown). For example, the Gower accuracies for D and E were 0.788 (95% CI=0.780–0.797) and 0.781 (95% CI=0.772–0.790), respectively. The Gower accuracy for just the real patients (A) in the community was 0.719 (95% CI=0.710–0728). The mean balanced accuracies were very similar for D (0.789), E (0.783), and A (0.721) suggesting that there were no biased accuracies due to imbalanced data. Thus, the accuracies associated with including close digital twins within communities was significantly higher than that for just real patients within communities.
Table 2.
Comparison of study design performance as measured by AUROC for each distance measure. Bolded metric values are significantly better than the others.
| Cosine |
Euclidean |
Gower |
Manhattan |
|||||
|---|---|---|---|---|---|---|---|---|
| Design* | mean | 95% CI | mean | 95% CI | mean | 95% CI | mean | 95% CI |
| A | 0.800 | 0.792, 0.808 | 0.807 | 0.800, 0.814 | 0.791 | 0.781, 0.800 | 0.800 | 0.792, 0.807 |
| B | 0.793 | 0.785, 0.801 | 0.799 | 0.792, 0.806 | 0.784 | 0.774, 0.794 | 0.792 | 0.784, 0.800 |
| C | 0.793 | 0.785, 0.801 | 0.798 | 0.791, 0.805 | 0.783 | 0.773, 0.793 | 0.791 | 0.783, 0.798 |
| D | 0.840 | 0.833, 0.847 | 0.848 | 0.842, 0.854 | 0.864 | 0.857, 0.872 | 0.852 | 0.845, 0.858 |
| E | 0.840 | 0.833, 0.847 | 0.845 | 0.839, 0.852 | 0.852 | 0.844, 0.860 | 0.846 | 0.839, 0.852 |
| F | 0.510 | 0.500, 0.521 | 0.512 | 0.503, 0.522 | 0.494 | 0.482, 0.507 | 0.485 | 0.475, 0.495 |
Real patients (A), digital twins (B), real patients and digital twins (C), closest digital twins (D), real patients and closest digital twins (E), and real patients outside the community (F).
Interestingly, the performance of A (real patients only), B (digital twins only), and C (real patients and digital twins) were not significantly different from one another across the different distance metrics including Gower. It is important to note that F (real patients outside the community) had an AUROC of approximately 0.50 as might be expected by chance given these patients have a distance that exceeds the percolation threshold and places them outside the community. Thus, the distance from the target patient being considered for prediction plays an important role in predictive accuracy and is highly relevant for precision medicine where context is a key consideration. An example network of real patients for three communities is shown in Figure 3 along with the corresponding digital patients.
Fig. 3.
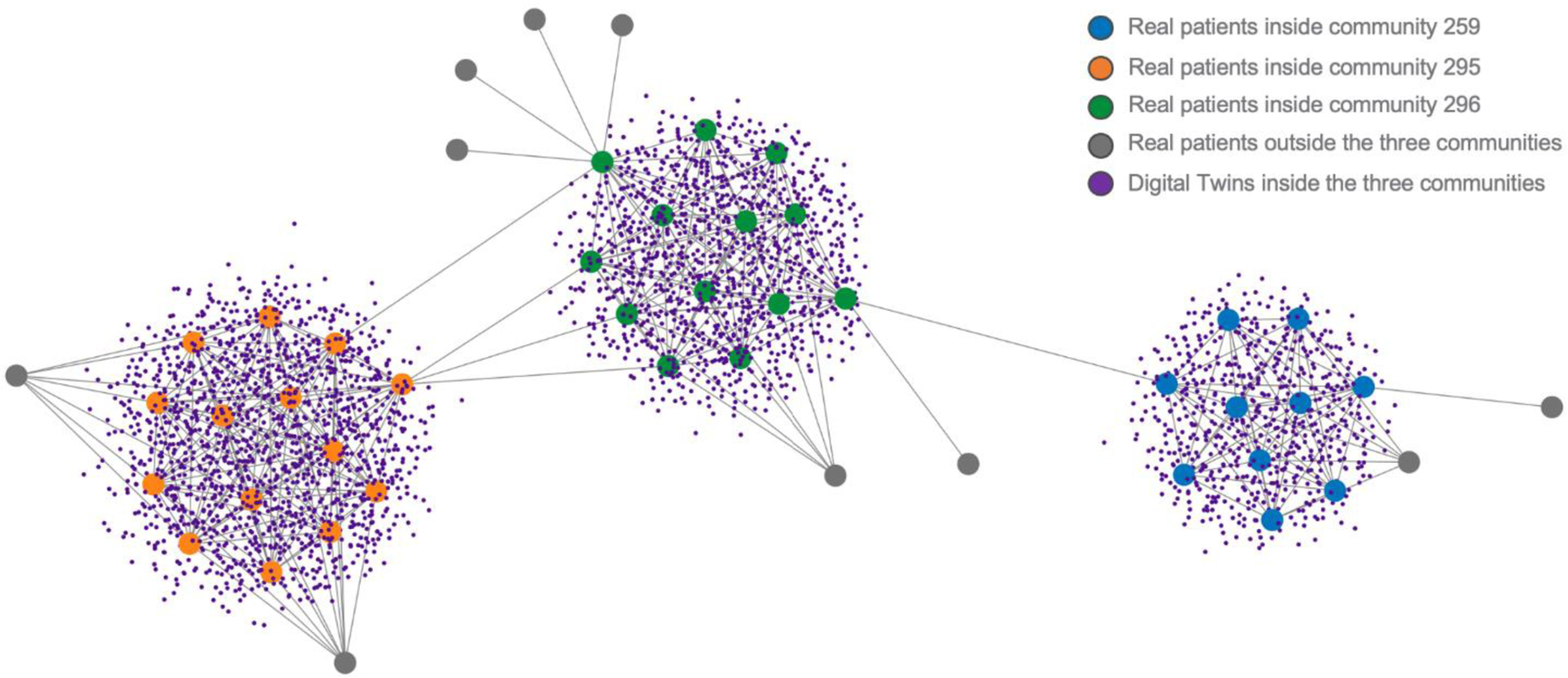
Section of the network showing three communities of real patients (orange, green, and blue circles). Also shown are the digital patients (small purple circles) and real patients outside the communities (grey circles).
4. Discussion
We have developed a new digital twin approach to improve the prediction of clinical endpoints. Our approach combines network science to model patient similarity and synthetic data generation to generate digital twins (SynTwin). Key to SynTwin is using patient similarity to synthesize nearby digital twins that represent hypothetical unobserved patients with clinical data correlations that are consistent with real patients. This distance-based approach is different than the digital twin approaches from industry that rely on well-known physical principles that govern a complex system (Attaran & Celik 2023; Attaran et al. 2023; Grieves & Vickers 2017). Biophysical properties governing health are not well known and are often only available for certain cellular or physiological processes. Indeed, simulating a single cell is quite challenging for a number of reasons including the lack of physics-based models (Thornburg et al. 2022). It is our working hypothesis that distance-based digital twins will be useful for informing patient outcomes above and beyond that provided by the observed clinical data. Indeed, our results suggest that generating and selecting digital twins close to the target patient whose outcome is being predicted significantly improves predictive performance above and beyond the real patients in the community. Choosing real patients outside the community for predicting target patients inside a community was not better than flipping a coin.
The generation and use of synthetic data for biomedical research is in and of itself not new. A recent review highlighted more than 70 published papers representing at least seven different use cases for synthetic data (Gonzales et al. 2023). Most of the use cases involve generating a synthetic dataset that can be used to avoid the privacy and security concerns of real data. For example, a synthetic dataset could be distributed to students to use for learning objectives without fear of identifying real patients. Other use cases involve using synthetic data to benchmark algorithms, evaluate information technology software, and public release of data. A very specific use case is to allow investigators to test a hypothesis without the need for Institutional Review Board (IRB) approval and the time it takes to retrieve data from an electronic health record which is a process that can take months depending on the complexity of the data and the wait time for available qualified personnel. Any interesting patterns found in the easily available synthetic data could then justify the time and expense of retrieving real data to confirm the finding before publication as has been suggested (Foraker et al. 2018). This approach was recently evaluated by comparing statistical and machine learning results obtained from real patient data and a synthetic derivative generated using a commercially available platform (Foraker et al. 2020). Similar results were seen when using a large integrated data resource (Foraker et al. 2021). In each case, the authors were able to draw the same conclusions from the analytical results using both real and synthetic datasets.
The use of synthetic data to generate digital twins was not mentioned in the review by Gonzales et al. (2023). However, using synthetic data to improve the sample size of a real dataset for improving predictive accuracy was specifically discussed. A study evaluating the addition of synthetic data to a real dataset showed that variance improved and five machine learning algorithms had improved prediction of heart disease (Aljaaf et al. 2016). The idea that synthetic data can improve machine learning performance has been observed in the image analysis domain. For example, a synthetic image generation approach using general adversarial networks (GANs) has been shown to improve image segmentation when the number of training examples is small (Thambawita et al. 2022). This approach may have clinical applications. For example, a recent study showed that synthetic colonoscopy images with polyps can improve the sensitivity of a deep learning neural network to detect polyps in real images (Adjei et al. 2022). This may be true in ophthalmology as well (You et al. 2022). Our observation that synthetic data may improve the performance of predictive accuracy is consistent with these studies. More studies are needed to validate this phenomenon.
Most synthetic data generation studies have focused on generating and using an entire synthetic dataset and checking to make sure the patterns detected by a machine learning algorithm are similar (Gonzales et al. 2023). Our SynTwin digital twin approach is different in the sense that we are using patient similarity and network community structure to select synthetic patients (i.e. twins) that can inform clinical outcome prediction. This is a more targeted approach that is much more consistent with the goals of precision medicine where treatment decisions and clinical outcomes are assessed in patient subgroups with similar characteristics. As such, this represents a fundamental shift in how synthetic data are used and may be more informative for clinical decision support.
Despite progress in this area, there are some possible limitations and challenges for moving forward. First, we applied our method to a dataset with a large sample size and a small number of features. On one hand, this was an ideal dataset to evaluate a new approach. Further, this dataset is publicly available, has been carefully curated, and has been well studied for understanding cancer risk and outcomes. However, the question remains of how the SynTwin approach will scale to hundreds or thousands of features or how it will behave when the synthetic data are generated from a dataset with small sample size. Further, the validation data was derived from the same cohort. Second, SynTwin is highly dependent on the community structure of the network. Not every patient is part of a community and prediction of outcomes in those patients may need to be performed using standard machine learning (ML) methods. Thus, a hybrid SynTwin-ML approach may need to be developed to make sure those patients are considered fully. Thirdly, it is of great interest to develop formal statistical inferential procedures to quantify the uncertainty of the subsequent analyses including estimation and prediction. Intuitively the generated digital twins should be weighted differently from the real patients in the precision of the downstream analyses. Finally, our implementation of SynTwin relied on bootstrapping to assign confidence intervals to performance metrics. This adds 1000-fold more computation time which might be prohibitive for larger datasets with more features. Future studies will need to balance the need for statistical inference with computing resources that are available. This study benefited from access to a 2000-core high-performance computing system to carry out all computations.
Precision medicine relies heavily on artificial intelligence and machine learning methods to develop models for predicting disease risk and patient outcomes in a manner that takes into account the uniqueness of the patient in question and other patients with similar profiles (Rajpurkar et al. 2022). The SynTwin digital twin strategy we presented here takes a step toward the use of synthetic data to augment the prediction of clinical outcomes by generating hypothetical unobserved patients to be used alongside real patients. The use of digital twins in medicine and biomedical research is in its infancy. We have a lot to learn from industrial uses of this approach and will need to develop new algorithms and software that consider the unique aspects of patients and their data. We agree with others who have speculated that digital twins will have a big impact on research and patient care but that new biophysical, computational, and statistical methods are needed (Acosta et al. 2022; Armeni et al. 2022; Attaran & Celik 2023; Attaran et al. 2023; Kamel Boulos & Zhang 2021).
5. Acknowledgments
This work was supported by NIH grants LM010098 and AG066833.
References
- Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. 2022. Multimodal biomedical AI. Nat Med 28(9):1773–84 [DOI] [PubMed] [Google Scholar]
- Adjei PE, Lonseko ZM, Du W, Zhang H, Rao N. 2022. Examining the effect of synthetic data augmentation in polyp detection and segmentation. Int J CARS 17(7):1289–1302 [DOI] [PubMed] [Google Scholar]
- Aljaaf AJ, Al-Jumeily D, Hussain AJ, Fergus P, Al-Jumaily M, Hamdan H. 2016. Partially Synthesised Dataset to Improve Prediction Accuracy. Intelligent Computing Theories and Application, pp. 855–66. Cham: Springer International Publishing [Google Scholar]
- Armeni P, Polat I, De Rossi LM, Diaferia L, Meregalli S, Gatti A. 2022. Digital Twins in Healthcare: Is It the Beginning of a New Era of Evidence-Based Medicine? A Critical Review. J Pers Med 12(8):1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran M, Attaran S, Celik BG. 2023. The impact of digital twins on the evolution of intelligent manufacturing and Industry 4.0. Adv Comput Intell 3(3):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran M, Celik BG. 2023. Digital Twin: Benefits, use cases, challenges, and opportunities. Decision Analytics Journal 6:100165 [Google Scholar]
- Benson M 2023. Digital Twins for Predictive, Preventive Personalized, and Participatory Treatment of Immune-Mediated Diseases. Arterioscler Thromb Vasc Biol 43(3):410–16 [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J. Stat. Mech 2008(10):P10008 [Google Scholar]
- Choi E, Biswal S, Malin B, Duke J, Stewart WF, Sun J. 2017. Generating Multi-label Discrete Patient Records using Generative Adversarial Networks. Proceedings of the 2nd Machine Learning for Healthcare Conference, pp. 286–305. PMLR [Google Scholar]
- Drummond D, Roukema J, Pijnenburg M. 2023. Home monitoring in asthma: towards digital twins. Curr Opin Pulm Med 29(4):270–76 [DOI] [PubMed] [Google Scholar]
- Dunson DB, Xing C. 2009. Nonparametric Bayes Modeling of Multivariate Categorical Data. Journal of the American Statistical Association 104(487):1042–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foraker R, Guo A, Thomas J, Zamstein N, Payne PR, et al. 2021. The National COVID Cohort Collaborative: Analyses of Original and Computationally Derived Electronic Health Record Data. J Med Internet Res 23(10):e30697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foraker R, Mann DL, Payne PRO. 2018. Are Synthetic Data Derivatives the Future of Translational Medicine? JACC: Basic to Translational Science 3(5):716–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foraker RE, Yu SC, Gupta A, Michelson AP, Pineda Soto JA, et al. 2020. Spot the difference: comparing results of analyses from real patient data and synthetic derivatives. JAMIA Open 3(4):557–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal Y, Chen Y, Ghahramani Z. 2015. Latent Gaussian Processes for Distribution Estimation of Multivariate Categorical Data. Proceedings of the 32nd International Conference on Machine Learning, pp. 645–54. PMLR [Google Scholar]
- Goncalves A, Ray P, Soper B, Stevens J, Coyle L, Sales AP. 2020. Generation and evaluation of synthetic patient data. BMC Medical Research Methodology 20(1):108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales A, Guruswamy G, Smith SR. 2023. Synthetic data in health care: A narrative review. PLOS Digital Health 2(1):e0000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower JC. 1971. A General Coefficient of Similarity and Some of Its Properties. Biometrics 27(4):857–71 [Google Scholar]
- Grieves M, Vickers J. 2017. Digital Twin: Mitigating Unpredictable, Undesirable Emergent Behavior in Complex Systems. In Transdisciplinary Perspectives on Complex Systems: New Findings and Approaches, eds. Kahlen F-J, Flumerfelt S, Alves A, pp. 85–113. Cham: Springer International Publishing [Google Scholar]
- Kamel Boulos MN, Zhang P. 2021. Digital Twins: From Personalised Medicine to Precision Public Health. Journal of Personalized Medicine 11(8):745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Maslove DM, Dubin JA. 2015. Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PLoS One 10(5):e0127428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurkar P, Chen E, Banerjee O, Topol EJ. 2022. AI in health and medicine. Nat Med 28(1):31–38 [DOI] [PubMed] [Google Scholar]
- Thambawita V, Salehi P, Sheshkal SA, Hicks SA, Hammer HL, et al. 2022. SinGAN-Seg: Synthetic training data generation for medical image segmentation. PLOS ONE 17(5):e0267976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg ZR, Bianchi DM, Brier TA, Gilbert BR, Earnest TM, et al. 2022. Fundamental behaviors emerge from simulations of a living minimal cell. Cell 185(2):345–360.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdouw C, Tekinerdogan B, Beulens A, Wolfert S. 2021. Digital twins in smart farming. Agricultural Systems 189:103046 [Google Scholar]
- Wickramasinghe N, Ulapane N, Andargoli A, Ossai C, Shuakat N, et al. 2022. Digital twins to enable better precision and personalized dementia care. JAMIA Open 5(3):ooac072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Algesheimer R, Tessone CJ. 2016. A Comparative Analysis of Community Detection Algorithms on Artificial Networks. Sci Rep 6(1):30750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You A, Kim JK, Ryu IH, Yoo TK. 2022. Application of generative adversarial networks (GAN) for ophthalmology image domains: a survey. Eye Vis (Lond) 9(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]


