Abstract
Here, we describe the “Obelisks,” a previously unrecognised class of viroid-like elements that we first identified in human gut metatranscriptomic data. “Obelisks” share several properties: (i) apparently circular RNA ~1kb genome assemblies, (ii) predicted rod-like secondary structures encompassing the entire genome, and (iii) open reading frames coding for a novel protein superfamily, which we call the “Oblins”. We find that Obelisks form their own distinct phylogenetic group with no detectable sequence or structural similarity to known biological agents. Further, Obelisks are prevalent in tested human microbiome metatranscriptomes with representatives detected in ~7% of analysed stool metatranscriptomes (29/440) and in ~50% of analysed oral metatranscriptomes (17/32). Obelisk compositions appear to differ between the anatomic sites and are capable of persisting in individuals, with continued presence over >300 days observed in one case. Large scale searches identified 29,959 Obelisks (clustered at 90% nucleotide identity), with examples from all seven continents and in diverse ecological niches. From this search, a subset of Obelisks are identified to code for Obelisk-specific variants of the hammerhead type-III self-cleaving ribozyme. Lastly, we identified one case of a bacterial species (Streptococcus sanguinis) in which a subset of defined laboratory strains harboured a specific Obelisk RNA population. As such, Obelisks comprise a class of diverse RNAs that have colonised, and gone unnoticed in, human, and global microbiomes.
Keywords: RNA metaviromics, Viroid, Hepatitis Delta Virus, Human Microbiome
Introduction
RNA viruses (Riboviria) are in part defined by their encoding of their own replicative polymerases, a feature that can be leveraged for homology-based viral discovery 1-5. By contrast, viroids 6,7 and Hepatitis Delta-like viral (HDV) ‘satellites’ 8 (Supplementary Figure 1) co-opt eukaryotic host RNA polymerases for their replication, resulting in some of biology's smallest known genomes (viroids: ~350 nt; Delta: ~1.7 kb). These streamlined genomes define the working limits of biological information transfer 9,10, and their simplicity raises the question of why, compared to Riboviria, there are so few known examples of viroids and similar agents. Recently, enquiries based on protein similarity have uncovered new Delta-like agents 2,11. Likewise, viroids, which lack any protein-coding capacity, are beginning to be surveyed at a larger scale based in part on circular genome maps and the presence of ribozyme-like features. These searches have led to an expanded family of known viroid-like RNAs and a revision of earlier models that their distribution is limited to plants 12-14. As such, these studies have already shifted virological paradigms, leaving open the possibility that an even broader category of viroid-like elements are present in living systems that might have been overlooked due to a lack of detectable similarity to known viroids and HDV family members.
The human gut microbiome (hGMB) is experimentally attractive for discovery of novel genetic agents. Indeed, metagenomic and metatranscriptomic 15 profiling of the hGMB has yielded new insights into prokaryotic, viral 16-18, and plasmid 19 ecology To this end, we developed a reference-free bioinformatic approach (VNom) to identify novel viroid-like elements. We initially applied VNom to published Integrative Human Microbiome Project (iHMP) data 20 resulting in the identification of a new class of hGMB-colonising RNA agents, which we term ‘Obelisks’. Obelisks form a distinct phylogenetic group restricted to RNA datasets and lack any evident homology to characterised genomes or viromes. Obelisk RNA reads assemble into ~1000 nt circles, which are predicted to fold into rod-like RNA secondary structures and code for at least one member of a novel “Oblin” protein superfamily. We further found that a subset of Obelisks harbour Obelisk-specific hammerhead ribozyme motifs. Querying 5.4 million public sequencing datasets, we identified 29,959 distinct Obelisks (90 % ID threshold) present across ~220,000 datasets representing diverse ecosystems beyond the hGMB. Amongst the datasets with clear Obelisk representatives, we identified a definitive Obelisk-Host pair, with Streptococcus sanguinis acting as a replicative host. Lastly, we surveyed Obelisks in five published human oral and gut microbiome studies from 472 donors, finding an estimated ~9.7 % donor prevalence within these datasets, with an apparent anatomy-specific Obelisk distribution.
Results
A novel, human microbiome-associated viroid-like RNA
Viroids and Delta viruses are in part typified by their single stranded, circular genomes, both of which are molecular features that can be detected in strand specific RNA-seq. To search for such features in microbiome RNA sequencing (RNA-seq) datasets, we created a bioinformatic tool, VNom (see VNom, and Supplementary Figure 2), and applied it to microbiome RNA-seq datasets (see Initial Obelisk identification). In particular, we chose an iHMP human stool dataset 20 for its strand-specific RNA-seq, its longitudinal nature (regular sampling over ~1 year), and its cohort size (104 donors), qualities well suited for identifying persistent hGMB colonists.
We next filtered VNom-nominated RNAs to retain contigs with no evident homology to the NCBI BLAST (nt or nr) databases 21 (see Initial Obelisk identification). One class of 15 related (<2 % sequence variation, Supplementary Table 1), 1164 nt RNAs stood out with their extended predicted secondary structure reminiscent of HDV and Pospiviroidae (Figure 1b, Supplementary Figures 1a-b and 2b). Owing to a strong predicted rod-like secondary structure, we term this group of RNAs Obelisk-alpha (Obelisk-α, “Obelisk_000001” in Supplementary Table 1). At 1164 nt in length, the rod-like secondary structure was striking because typical mRNA sequences are not predicted to readily fold in this manner (as evidenced by the efforts required to maximise the degree of “rod-ness” in mRNA vaccines 22). Based on open reading frame (ORF) predictions, Obelisk-α has the capacity to code for two proteins (202 and 53 amino acids [aa]). Both open reading frames (ORFs) lack evident nucleotide or protein sequence homology when querying a number of reference databases (NCBI nt, nr, or CDD 23, Pfam 24). Tertiary structure protein alignment yielded similar negative results (see Protein tertiary structure prediction). As such, we chose new names, terming these two proteins Oblin-1 and Oblin-2, respectively. We specifically note that despite some similar characteristics between Obelisk-α and HDV (apparently circular, predicted highly structured RNA genome, and ability to code for at least one ~200 aa ORF, Supplementary Figure 1a), there is no evident sequence homology at the RNA or protein level or structural homology at the protein level between Obelisks and HDV. In further contrast to HDV, whose large hepatitis Delta antigen (L-HDAg) occurs on one strand of the extended HDV predicted secondary structure (Supplementary Figure 1a), the Obelisk-α Oblin-1 encoding region is largely self-complementary within the open reading frame, forming a ~300 base pair hairpin making up half of the predicted Obelisk-α RNA secondary structure. Obelisk-α sequences were found to occur in 7 of the 104 iHMP donors (Table 1) with donors A-C exhibiting consistent prevalence for over 200 days (Figure 1d, note: positive donors are renamed for brevity, with original donor alias equivalences in Table 1). Further, Obelisk-α sequences were found to largely cluster together based on donor identity, when grouped by sequence variation (Figure 1e). We noted some co-clustering of sequences between donors (A and E in cluster 3, and D and G in cluster 5); this co-clustering could be explained by either transient prevalence or by library cross contamination, as each minor member of such clusters was both low prevalence (few positive timepoints) and low abundance (low counts in positive timepoints) (see Table 1). Regardless of the source of the relatively rare cross-sample reads, Obelisk-α appears to persist within human donors, with each donor appearing to harbour their own distinct ‘strain.’ Lastly, in companion DNA-seq data from this project, no detectable Obelisk reads are found (Table 2). Taken together, these findings are consistent with Obelisk-α representing an as of yet uncharacterised RNA element with viroid-like features that occurs in human stool, further comprised of subspecies that persist in individual donors over time.
Figure 1. Obelisk alpha has a predicted extensive secondary structure and appears to colonise and speciate within the human gut.
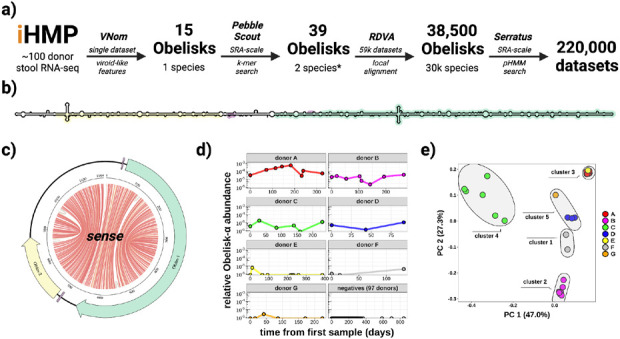
a) overview of the iterative approach taken in Obelisk discovery, (see methods) b) schematic of the predicted sense consensus secondary structure derived from all non-redundant, 1164 nt Obelisk-αs found using SRA-scale k-mer matching (PebbleScout). Predicted open reading frames (ORFs) 1 and 2 (green/yellow), and Shine-Delgarno sequences (purple) shown, c) “jupiter” plot of Obelisk-α coloured as in b), chords illustrate predicted basepairs (basepair probabilities grey, 0.1, to red, 1.0) d) Obelisk-α relative read abundance for six donors (A-G); sequence data from in Lloyd-Price et al., 2019 and time in days from first sample. e) Principal component analysis of sequence variation seen in Obelisk-α reads in Lloyd-Price et al., 2019 (the initial iHMP dataset), grouped by k-means clustering with 5 centres, coloured as in d).
Table 1. Figure 1d metadata.
Donors with at least one Obelisk-alpha read from Lloyd-Price et al. 2019, with their SRA accession code, alias as used in Figure 1d, disease state, relative (to total reads per sample) Obelisk-α read abundance (see Methods), date of sampling, and original donor numeric ID.
| SRA (Lloyd-Price et al. 2019) |
alias | clinical classification |
Obelisk-alpha relative cont |
sample date | donor |
|---|---|---|---|---|---|
| SRR5949245 | donor A | Healthy Donor | 3.22E-05 | 2015-08-31 | d_2077 |
| SRR5950275 | donor A | Healthy Donor | 1.53E-04 | 2015-11-12 | d_2077 |
| SRR5950410 | donor A | Healthy Donor | 2.55E-04 | 2015-12-23 | d_2077 |
| SRR5950468 | donor A | Healthy Donor | 4.06E-04 | 2016-01-21 | d_2077 |
| SRR5950280 | donor A | Healthy Donor | 5.71E-04 | 2016-03-01 | d_2077 |
| SRR5950352 | donor A | Healthy Donor | 2.75E-05 | 2016-04-19 | d_2077 |
| SRR5950308 | donor A | Healthy Donor | 1.19E-04 | 2016-04-26 | d_2077 |
| SRR5950313 | donor A | Healthy Donor | 5.50E-05 | 2016-07-21 | d_2077 |
| SRR5950297 | donor B | Ulcerative Colitis | 1.97E-05 | 2015-11-12 | d_6038 |
| SRR5950395 | donor B | Ulcerative Colitis | 2.62E-05 | 2015-12-01 | d_6038 |
| SRR5950415 | donor B | Ulcerative Colitis | 1.21E-05 | 2016-01-05 | d_6038 |
| SRR5950446 | donor B | Ulcerative Colitis | 2.45E-05 | 2016-02-23 | d_6038 |
| SRR5950283 | donor B | Ulcerative Colitis | 6.46E-06 | 2016-03-01 | d_6038 |
| SRR5950351 | donor B | Ulcerative Colitis | 2.31E-06 | 2016-04-02 | d_6038 |
| SRR5950317 | donor B | Ulcerative Colitis | 2.18E-05 | 2016-05-23 | d_6038 |
| SRR5950257 | donor B | Ulcerative Colitis | 3.78E-05 | 2016-08-04 | d_6038 |
| SRR5949129 | donor C | Crohn's Disease | 3.61E-06 | 2015-04-28 | d_2068 |
| SRR5949216 | donor C | Crohn's Disease | 1.88E-05 | 2015-05-27 | d_2068 |
| SRR5949201 | donor C | Crohn's Disease | 2.34E-06 | 2015-07-21 | d_2068 |
| SRR5950374 | donor C | Crohn's Disease | 4.46E-06 | 2015-09-15 | d_2068 |
| SRR5950426 | donor C | Crohn's Disease | 0 | 2015-10-05 | d_2068 |
| SRR5950333 | donor C | Crohn's Disease | 1.28E-05 | 2015-12-15 | d_2068 |
| SRR5949407 | donor D | Crohn's Disease | 5.09E-06 | 2014-09-16 | d_5001 |
| SRR5949179 | donor D | Crohn's Disease | 1.37E-06 | 2014-10-29 | d_5001 |
| SRR5949326 | donor D | Crohn's Disease | 1.12E-05 | 2014-12-16 | d_5001 |
| SRR5949222 | donor E | Ulcerative Colitis | 0 | 2015-06-15 | d_2071 |
| SRR5949120 | donor E | Ulcerative Colitis | 6.33E-06 | 2015-06-26 | d_2071 |
| SRR5949586 | donor E | Ulcerative Colitis | 0 | 2015-08-19 | d_2071 |
| SRR5950375 | donor E | Ulcerative Colitis | 0 | 2015-09-16 | d_2071 |
| SRR5950383 | donor E | Ulcerative Colitis | 0 | 2016-01-19 | d_2071 |
| SRR5963925 | donor E | Ulcerative Colitis | 0 | 2016-01-27 | d_2071 |
| SRR5950464 | donor E | Ulcerative Colitis | 0 | 2016-02-02 | d_2071 |
| SRR5950281 | donor E | Ulcerative Colitis | 0 | 2016-02-29 | d_2071 |
| SRR5950429 | donor E | Ulcerative Colitis | 0 | 2016-07-06 | d_2071 |
| SRR5949469 | donor F | Crohn's Disease | 1.09E-06 | 2014-08-13 | d_2025 |
| SRR5949284 | donor F | Crohn's Disease | 0 | 2014-08-28 | d_2025 |
| SRR5949607 | donor F | Crohn's Disease | 4.24E-06 | 2014-12-23 | d_2025 |
| SRR5949267 | donor G | Ulcerative Colitis | 0 | 2015-06-23 | d_3034 |
| SRR5949148 | donor G | Ulcerative Colitis | 0 | 2015-07-08 | d_3034 |
| SRR5949435 | donor G | Ulcerative Colitis | 2.58E-06 | 2015-08-04 | d_3034 |
| SRR5950377 | donor G | Ulcerative Colitis | 0 | 2015-09-17 | d_3034 |
| SRR5950332 | donor G | Ulcerative Colitis | 0 | 2015-12-11 | d_3034 |
| SRR5950460 | donor G | Ulcerative Colitis | 0 | 2016-02-02 | d_3034 |
Table 2. Obelisk read counts from paired metagenomic (DNA) and metatranscriptomic (RNA) samples.
Paired metagenomic DNA and metatranscriptomic RNA Kraken2 read counts (Bracken-corrected, see methods) for Lloyd-Price et al. 2019 20 and Maghini and Dvorak et al. 2023 79 for Obelisks -α and -β. Means taken over donors (aliased as in Table 6) and total read counts also reported. Note that no reads mapping to either Obelisk were found in the DNA datasets, suggesting an RNA-only Obelisk lifestyle. For full Kraken2 and Bracken outputs, see Data Availability.
| study | donor | mean α RNA |
sdv α RNA |
mean β RNA |
sdv β RNA |
mean RNA reads |
sdv RNA reads |
mean α DNA |
sdv α DNA |
mean β DNA |
sdv β DNA |
mean DNA reads |
sdv DNA reads |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lloyd-Price et al. 2019 | A | 2310 | 3180 | 1 | 1 | 13372847 | 8928775 | 0 | 0 | 0 | 0 | 12434953 | 7135963 |
| Lloyd-Price et al. 2019 | B | 120 | 75 | 0 | 0 | 6471339 | 1941900 | 0 | 0 | 0 | 0 | 8624205 | 2706287 |
| Lloyd-Price et al. 2019 | C | 59 | 85 | 0 | 0 | 7465073 | 3571490 | 0 | 0 | 0 | 0 | 7861229 | 2987857 |
| Lloyd-Price et al. 2019 | D | 35 | 36 | 0 | 0 | 5991299 | 1931426 | 0 | 0 | 0 | 0 | 11930216 | 3223559 |
| Lloyd-Price et al. 2019 | E | 5 | 13 | 0 | 0 | 10205726 | 4505426 | 0 | 0 | 0 | 0 | 10182743 | 4150798 |
| Lloyd-Price et al. 2019 | F | 14 | 6 | 0 | 0 | 7261820 | 2583126 | 0 | 0 | 0 | 0 | 9293180 | 3041056 |
| Lloyd-Price et al. 2019 | G | 3 | 4 | 0 | 0 | 7544276 | 3692007 | 0 | 0 | 0 | 0 | 8342623 | 2564388 |
| Maghini and Dvorak et al. 2023 | 1 | 8080 | 5500 | 319 | 246 | 24534988 | 11197080 | 0 | 0 | 0 | 0 | 59395503 | 52132729 |
| Maghini and Dvorak et al. 2023 | 2 | 0 | 0 | 0 | 0 | 17277835 | 6182452 | 0 | 0 | 0 | 0 | 63203793 | 31663431 |
| Maghini and Dvorak et al. 2023 | 3 | 0 | 0 | 0 | 0 | 14125535 | 9993287 | 0 | 0 | 0 | 0 | 35879627 | 10380606 |
| Maghini and Dvorak et al. 2023 | 4 | 0 | 0 | 0 | 0 | 15983644 | 5232547 | 0 | 0 | 0 | 0 | 19114616 | 10718757 |
| Maghini and Dvorak et al. 2023 | 5 | 24 | 42 | 0 | 0 | 18237767 | 5281905 | 0 | 0 | 0 | 0 | 38104238 | 18496583 |
| Maghini and Dvorak et al. 2023 | 6 | 0 | 0 | 0 | 0 | 19476656 | 3667861 | 0 | 0 | 0 | 0 | 44924227 | 19547694 |
| Maghini and Dvorak et al. 2023 | 7 | 0 | 0 | 0 | 0 | 18639827 | 6428812 | 0 | 0 | 0 | 0 | 75407696 | 72530680 |
| Maghini and Dvorak et al. 2023 | 8 | 4 | 6 | 0 | 0 | 18449872 | 6842782 | 0 | 0 | 0 | 0 | 27950881 | 10870547 |
| Maghini and Dvorak et al. 2023 | 9 | 1 | 1 | 0 | 0 | 15639556 | 9328742 | 0 | 0 | 0 | 0 | 42868581 | 32749586 |
| Maghini and Dvorak et al. 2023 | 10 | 12417 | 1740 | 1 | 2 | 13021384 | 4204129 | 0 | 0 | 0 | 0 | 20243492 | 8732203 |
Public data are replete with Obelisk-like elements
Using Obelisk-α as a starting point, 21 additional full-length examples of Obelisk-α (<4 % sequence variation, Supplementary Table 1) were found in 7 datasets using a k-mer search (PebbleScout 25) of ~3.2 million “metagenomic” annotated sequence read archive (SRA) datasets. All 7 datasets were human-derived metatranscriptome (metagenomic RNA) BioProjects (Table 3, see Obelisk homologue detection in additional public data); 0 sequences were found in metagenomic DNA samples. The repeated finding of Obelisk-α in disparate BioProjects supported the notion that Obelisk-α is a bona fide biological entity. Based on the prevalence of Obelisk-α in these human microbiome transcriptome datasets (Table 3), we investigated the possibility that additional Obelisks might be present in such data (as identified by both VNom and Oblin-1 protein similarity). This search ultimately lead to the discovery of Obelisk-β (“Obelisk_000002” in Supplementary Table 1), a 1182 nt, likely hGMB-resident, Obelisk-like RNA with similar characteristics to Obelisk-α (circular assembly map, rod-like predicted secondary structure, absence in paired DNA sequence) and low-but-evident protein sequence similarity to Oblin-1 (~38 % protein similarity and pairwise mean BLASTp E-value: 5.2x10−14). Thus, both Obelisks appear to be Oblin-1-encoding elements. Analysis of the Oblin-2 homology at this stage was limited by the short size of the proteins – nonetheless both Obelisk-α and Obelisk-β encode second proteins of ~50 amino acids rich in helix-forming residues (Supplementary Figure S3a/c). Next, utilising the uniqueness of the Obelisk-α/β Oblins-1 and -2 as Obelisk-specific hallmark sequences, we searched over 12 trillion contigs in the RNA deep virome assemblage (RDVA), a database of assembled metatranscriptomes 13,26 (see Obelisk homologue detection in additional public data), yielding over 38,500 Oblin-encoding RNA assemblies. Following this search, the smaller Obelisk-α/β proteins were determined to likely be Oblin-2 homologues (~31 % protein similarity and mean BLASTp E-value against the Oblin-2 consensus sequence: 2.5x10−6). Ultimately, by insisting on evidence of apparent circularity, we created a stringent subset of 7,202 clustered Obelisks (1,744 clusters at 80 % nucleotide identity) as a conservative database for future studies (Supplementary Table 1). Building from these RDVA hits, we then queried ~5.4 million SRA datasets for distant Oblin-1 and -2 homology using Serratus 2 (applying an inclusion threshold from earlier Serratus projects, see Serratus), yielding over 220,000 putatively Obelisk-positive datasets. From these datasets, we followed up on the 4,505 datasets with confident Oblin-1 hits (see Serratus). These searches suggest that Obelisk-like elements are found globally (Figure 3c), with a possible bias in reported datasets towards mammalian microbiome-related origins (Figure 3b), and represent a distinct, diverse group of phylogenetically related RNA-based elements (Figure 3a).
Table 3. varied human stool metatranscriptomes are positive for Obelisk-alpha.
Obelisk-alpha positive bioProjects identified by PebbleScout (see methods)
| bioProject | human stool study details |
|---|---|
| PRJNA398089 | longitudinal - part of iHMP: where Obelisk-alpha was first identified |
| PRJNA940499 | testing stool storage conditions: where Obelisk-beta was first identified |
| PRJNA407499 | longitudinal - typhoid vaccine challenge trial |
| PRJNA354235 | longitudinal - part of the Health Professionals Follow-up Study |
| PRJNA600008 | secondary bile acid intestinal inflammation |
| PRJNA541981 | pre immunotherapy treatment in melanoma patients |
| PRJNA338184 | longitudinal - enterotoxigenic Escherichia coli challenge |
| PRJNA306874 | longitudinal - part of Inflammatory Bowel Disease Multi-omics Data (IBDMDB) |
| PRJNA389280 | longitudinal - part of iHMP |
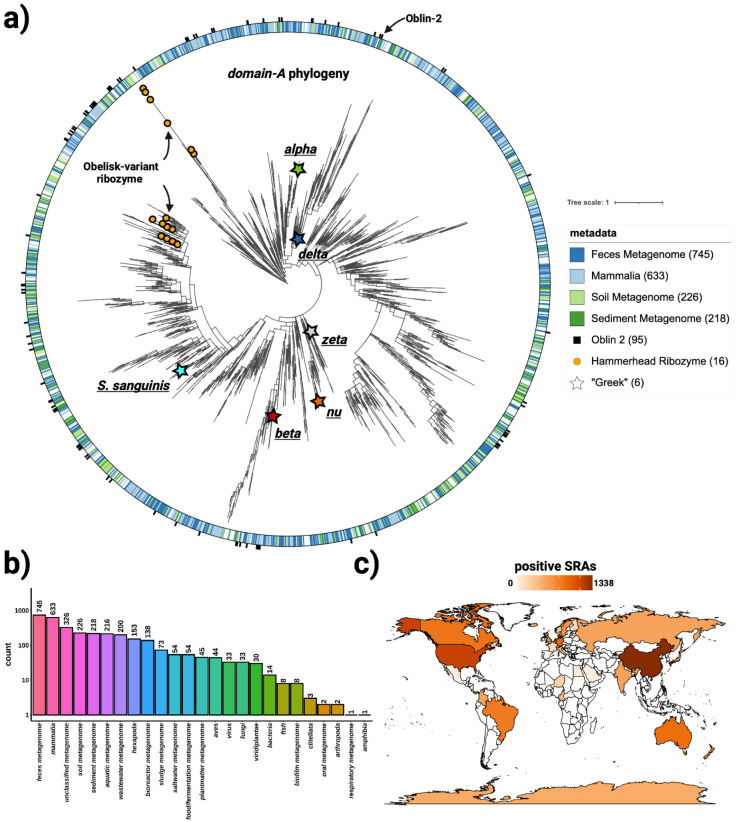
Figure 3. Obelisks form their own globally distributed phylogenetic group.
a) a maximum likelihood, midpoint-rooted, phylogenetic tree (see methods) constructed from a non-redundant set of 3265 Serratus and RDVA domain-A sequences, with RDVA genomes positive for Obelisk-variant self cleaving Hammerhead Type III ribozymes illustrated as orange circles on leaves, and the top four known classes of SRA “host” metadata depicted as the colour band (see legend), and with per-RDVA-genome co-occurrence of Oblin-2 (based on blastp hits against the Oblin-2 consensus) illustrated as the outer ring (black studs). Leaves that correspond to domain-A sequences from Figure 4 are illustrated with stars. b) Counts of non de-replicated SRA datasets used to construct a) sorted by their “host” metadata; we note that “host” metadata likely fails to account other organisms’ genetic material that was sequences alongside the “host” (e.g. signals from these hosts’ microbiomes maybe be detected in tandem). c) Counts of non de-replicated SRA datasets used to construct a) arranged by sample geolocation (where known) illustrated on a world map (darker orange = more SRA datasets contributed to a)). We note that SRA counts are not expected to correlate with true geo-/ecological prevalence, but are still indicative of global presence.
An oral commensal bacterium, Streptococcus sanguinis, serves as one Obelisk host
The task of identifying specific host-agent pairings from metagenomic data presented a number of challenges. Most samples with Obelisk homologues that were retrieved from the various searches were from metatranscriptomic samples derived from complex mixtures such as highly biodiverse microbiome and waste water samples (Figure 3b). As such, the potential host(s) of Obelisk elements were not immediately clear. While correlation and co-occurrence based methods for inferring potential hosts are possible 3,4,16, concerns about their statistical validity and interpretability 27-29 motivated a more direct strategy for Obelisk host identification. Consequently, we combed the Serratus results for Obelisk-like elements found in limited-complexity samples, such as defined monoculture and/or co-cultures. This search yielded a set of independent sequencing datasets from Streptococcus sanguinis (strain SK36), a commensal bacterium of the healthy human oral microbiome 30. Several RNA-seq datasets (Table 4) from S. sanguinis strain SK36 contained an Oblin-1 coding Obelisk-like sequence (see Streptococcus sanguinis bioinformatics). These datasets evidenced a well-defined RNA element which we refer to as “Obelisk-S.s” (“Obelisk_000003” in Supplementary Table 1). This RNA has the hallmark features of an Obelisk: a characteristic length (1137 nt) circular assembly with an obelisk-shaped predicted RNA secondary structure; genome similarity to Obelisks-α and -β (41 and 35 % nucleotide identity, respectively) and an Oblin-1 homologue (α and β: 33 % protein similarity, and mean pairwise E-values of 5.2x10−5 and 4.5x10−7, respectively). Unlike the other two Obelisks, however, it lacks a predicted Oblin-2 homologue (Supplementary Figure S2a/d). Overall, based on sequence homology, the predicted genomic secondary structure, the Oblin-1 tertiary structure, and the Obelisk-characteristic Oblin-1 self-complementarity (Supplementary Figure S2d), this RNA element is a bona fide Obelisk. Further, the robust co-occurrence of S. sanguinis SK36 with Obelisk RNA-seq reads (Table 4), positions S. sanguinis SK36 as a model system for future Obelisk characterisation.
Table 4. multiple Streptococcus sanguinis SK36 datasets contain Obelisk reads.
A curated set of Streptococcus sanguinis SK36 monoculture or low complexity bioProjects identified by scanRabbit (see methods) that contain Obelisk reads produced from differing institutions over differing years and thus suggestive of an S. sanguinis SK36 - Obelisk relationship.
| bioProject | composition | year | submitting institution |
|---|---|---|---|
| PRJNA270301 | SK36 monoculture | 2014 | Kyungpook National University |
| PRJNA632881 | SK36 / deletion mutants monoculture | 2020 | Virginia Commonwealth University |
| PRJNA731039 | SK36 / deletion mutants monoculture | 2021 | Virginia Commonwealth University |
| PRJNA862079 | SK36 / deletion mutants monoculture | 2022 | University of Florida |
| PRJNA862955 | SK36 and other Streptococci co-culture | 2022 | Ohio State University |
Structural prediction indicates a novel globular domain characteristic of Oblin-1 proteins
Due to the lack of obvious protein sequence homology in existing, non-Obelisk databases, we performed protein tertiary structure predictions in an attempt to identify both shared predicted structural elements, and homology through tertiary structure similarity searches. Owing to Oblin-1 and -2's previously unrecorded nature and apparent monophyly, we avoided automated multiple sequence alignment construction during conventional tertiary structure prediction using ColabFold (an implementation of AlphaFold2) 31,32 and instead opted for custom RDVA alignments (see Protein tertiary structure prediction). This yielded a folding prediction of Oblin-1 (mean per-residue confidence estimate, μ-pLDDT ± standard deviation, of 83.8 ± 13.4, where 70-90 pLDDT values are “a generally good backbone prediction” 33 and higher is better) with a more confidently predicted N-terminal “globule” (μ-pLDDT of 90.1 ± 8.7, Figure 2a). ColabFold was not able to confidently place the two flanking backbones between the first and last predicted alpha helices and the rest of the Oblin-1 sequence (Supplementary Figure 4a), and owing to heterogeneity in the last alpha helix's placement across predictions (see Data Availability), the “globule” was further focused on. The “globule” was predicted to form a consistent fold (Figure 4): a three alpha helix bundle (two smaller alpha helices co-axially aligned along the larger alpha helix) partially wrapping over a semi-orthogonal four alpha helix bundle - all bookended with a two strand beta sheet “clasp” (Figure 2b). Interestingly, no confident fold was predicted for the largest conserved region in Oblin-1 (Supplementary Figure 4), termed domain-A, (Figure 2a-b - magenta). Suggestive of an anion binding function, this 18 amino acid stretch is enriched for positively charged residues (arginine, histidine, and lysine) with the Obelisk-α domain-A containing five arginines, three histidines, and a lysine residue (50 % of domain-A, Figure 2b and Protein homology bioinformatics). Additionally, a “GYxDxG” motif appears prominently in domain-A. If Oblin-1, represents a new class of RNA binding protein, ColabFold may miss the fold of domain-A due to the absence of its client RNA ligand.
Figure 2. Obelisks encode putatively well-folded proteins.
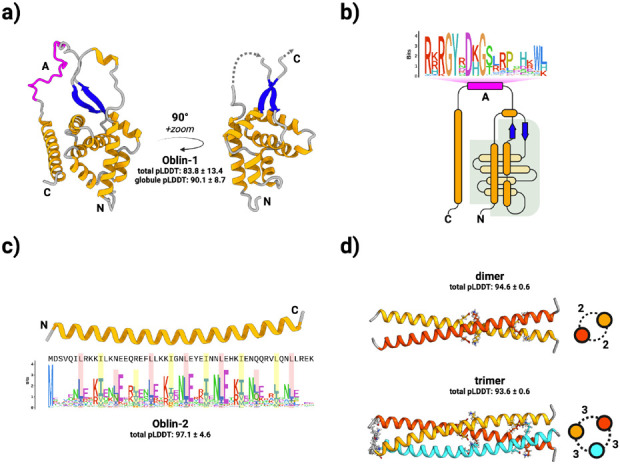
a) Obelisk open reading frame 1 (Oblin-1) is predicted (total mean-pLDDT ± SD = 83.8 ± 13.4, see methods) to fold into a stereotyped N-terminal “globule” formed of a three alpha helix (orange) bundle partially wrapping around an orthogonal four helix bundle, capped with a beta sheet “clasp” (blue, globule mean-pLDDT = 90.1 ± 8.7), joined by an intervening region harbouring the conserved domain-A (magenta) with no predicted tertiary structure, to an arbitrarily placed C-terminal alpha helix. “Globule” emphasised on the right. b) a to-scale (secondary structure) topological representation of Oblin-1 with the “globule” shaded in grey, and the domain-A emphasised with this bit-score sequence logo (see methods). c) Obelisk Oblin-2 is confidently predicted (mean-pLDDT = 97.1 ± 4.6 ) to fold into an alpha helix which appears to be a leucine zipper. Sequence logo of an “i+7” leucine spacing emphasised in red, with hydrophobic “d” position residues emphasised in yellow (expanded in Supplementary Figure 4b). d) homo-multimer predictions of Obelisk-alpha Oblin-2. top: dimer (mean-pLDDT = 94.6 ± 0.6), bottom: trimer (mean-pLDDT = 93.6 ± 0.6). Side-on representations of homomultimers shown with numbers of inter-helix salt-bridges (see Supplementary Figure 5).
Figure 4. Obelisks form a self-consistent set.
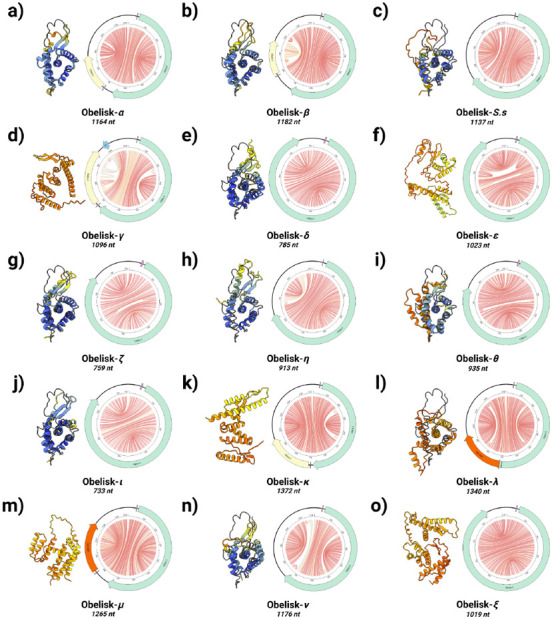
Predicted Obelisk secondary structures depicted as “jupiter” plots where chords represent predicted basepairs (coloured by basepair probability from 0, grey, to 1, red, see methods) with predicted open reading frames (ORFs, preceded by predicted Shine-Delgarno sequences, purple) depicted: Oblin-1 (green), Oblin-2 (yellow, based on blastp hits against the Oblin-2 consensus), and “2ndORF” (orange). Obelisk-ɣ’s suggested CRISPR spacer match illustrated in light blue. ColabFold predictions of Oblin-1 tertiary “globule” structures built with ad hoc multiple sequence alignment (MSA) construction (coloured cartoons) superimposed over the RDVA-derived MSA prediction for Obelisk-α where possible (black line, Figure 2a, see methods). Prediction confidence (pLDDT) shown as cartoon colouring as in Supplementary Figure 3. Greek letter key: α : alpha, β : beta, ɣ : gamma, δ : delta, ε : epsilon, ζ : zeta, η : eta, θ : theta, ι: iota, κ : kappa, λ : lambda, μ : mu, ν : nu, and ξ : xi.
Oblin-2 modelling suggests a leucine zipper alpha helix
Oblin-2 modelling with ColabFold resulted in a high-confidence prediction (μ-pLDDT of 97.1 ± 4.6) that this protein forms a solitary alpha helix (Figure 2c). In the RDVA consensus, the Oblin-2 alpha helix consists of a leucine zipper motif (see Protein conservation and phylogenetics), with the characteristic “i+7” spacing of leucines at the “a” position; another hydrophobic residue (leucine or isoleucine) with “i+7” spacing at the “d” position; and complementary charged residues (glutamic acid and lysine or arginine) at the “e” and “g” positions, respectively 34 (Supplementary Figure 4b). Based on μ-pLDDT, ColabFold predicts that Oblin-2 might be able to homo-multimerize as a dimer (μ-pLDDT of 94.6 ± 0.6), or a trimer (μ-pLDDT of 93.6 ± 0.6) with a coiled-coil forming with 2 or 3 inter-helix salt bridges per helix pair, respectively (Figure 2d, and Supplementary Figure 5a-b). Although conceivable, a higher order Oblin-2 homo-tetramer is less well supported by ColabFold (μ-pLDDT of 65.3 ± 7.9, Supplementary Figure 5c). Leucine zippers typically act as multimerization motifs that bring together other protein domains such as the DNA-binding basic leucine zipper domain (bZIP) 35. Oblin-2 does not appear to include any other sequence motifs (e.g. a non-zipper poly-basic patch similar to bZIP proteins), suggesting potential function as a homo-multimer, or as a binding partner to other host leucine zippers.
A subtype of Obelisks bear ribozyme signatures of a viroid-like replication mechanism
Viroids of the family Avsunviroidae and HDV code for self-cleaving ribozymes used in their respective replicative cycles 6,8 (Supplementary Figure 1a,c), and previous bioinformatic studies have found self-cleaving ribozymes in candidate viroid-like genomes 12-14. Upon querying for Hammerhead type-III ribozyme-coding Obelisks, we identified 23 initial hits and noticed that these ribozymes slightly differed from the reference covariance model (Rfam: RF00008). Therefore, we constructed an “Obelisk-variant Hammerhead type-III” ribozyme (ObV-HHR3) covariance model (Supplementary Figure 6b, see RNA homology bioinformatics), yielding 339 total Obelisks containing HHRs in the RDVA set with stringent similarity (35 clustered at 80 % identity in Supplementary Table 1 - “ObV-HHR3” column). These “HHR-Obelisks” are similarly rod-shaped, ~1 kb in length, and code for diverged Oblin-1 proteins (20.6 % identity and 31.7 % similarity to the Obelisk-α Oblin-1) that are similarly largely self-complementary (Supplementary Figure 6a), do not code for Oblin-2, but do include an unrelated “smaller ORF.” Additionally, some Obelisks appear to include a bidirectional pair of ObV-HHR3 ribozymes (Supplementary Figure 6a), a feature used by Avsunviroidae, HDV, and ambiviruses for their rolling-circle replicative cycles. For the subset of ObV-HHR3 ribozyme-containing Obelisks, ColabFold predicts a “globule” fold (total μ-pLDDT of 76.8 ± 20.1, and “globule” μ-pLDDT of 88.3 ± 8.6, Supplementary Figure 6c, Supplementary Figure 7), that is similar to the non-HHR Oblin-1 model but with additional specific tertiary structure features. Namely, the beta-sheet “clasp” region is expanded by an extra sheet as well as some small alpha helices, and the C-terminal alpha helix is predicted to be shorter (Supplementary Figure 6d). Additionally, the domain-A region appears to be diverged in the ObV-HHR3 class, yet still exhibits the positive residue skew as well as the “GYxDxG” protein motif also found in non-HHR-Obelisks (Supplementary Figure 6d). These subset-specific features, and the correlation with HHR co-occurrence, suggest that at least HRR-Obelisks may replicate via a viroid-like mechanism, with Oblin-1 and/or Oblin-2 as potential cofactors.
A phylogeny of Oblin-1’s domain-A provides evidence for in-family evolution and places ribozyme-baring Obelisks in distinct clades
Following the RDVA and Serratus searches, an initial Obelisk phylogeny spanning diverse sampling sites (Figure 3b) from around the globe (Figure 3c) was constructed using domain-A as a marker sequence (see Protein homology bioinformatics and Protein conservation and phylogenetics, Figure 3a). This domain-A phylogeny was sufficient to partially explain the distribution of ObV-HHR3-bearing Obelisks, which segregate tightly into two clades (Figure 3a - orange circles), implying both an evolutionary relationship between Obelisk genome processing and domain-A, as well as two different evolutionary paths for domain-A ‘speciation’ within the presence of ObV-HHR3. Additionally, this phylogeny indicates that the human microbiome-associated Obelisks (Figure 3a - stars) are widely distributed, implying a complex intersection between human and Obelisk biology. However, the co-occurrence of Oblin-2 (Figure 3a - black studs), and the sampling site of origin (Figure 3a - coloured band), are not adequately explained by this domain-A phylogeny, suggesting either multiple gains or losses of such features over the course of Obelisk evolution or recombination events that would confound the construction of a simple tree.
Absence of captured Obelisk matches among available CRISPR spacer datasets
Searches through CRISPR spacer databases offer an opportunity to deduce past associations between specific mobile genetic elements and potential cellular prokaryotic hosts 4,14. We applied a conservative k-mer matching approach (see Obelisk spacer analysis) to gauge the extent to which Obelisks appear to be sampled by the CRISPR spacer arrays, using a dataset of 29,857,318 spacers predicted by the Joint Genome Institute's (JGI's) IMG/M database 36. Ultimately, only one spacer locus out of ~140,000 initially mapping spacers confidently mapped to an >1000 nt Obelisk-like contig which we term Obelisk-“gamma” (Obelisk-ɣ, Supplementary Figure 8, “Obelisk_000004” in Supplementary Table 1). This mapping could suggest that Obelisk-ɣ has previously infected the Alphaproteobacterium Bombella mellum, however, the Obelisk that this spacer maps to deviates from the “rod-like” nature seen in other Obelisks (Figure 4 - “jupiter” plots), suggestive of a chimeric misassembly. While Obelisk-ɣ does resemble other Obelisks (1096 nt, mostly rod-shaped, and contains both Oblin-1 and Oblin-2 homologues - Supplementary Figure 8c), the assembly appears to contain an unpaired, “frayed” end. The coincidence of the spacer mapping to the “frayed” end, and the fact that only one mapping was found (out of ~39,000 RDVA Obelisks) casts some doubt on the validity of this mapping. As such, using this spacer mapping approach, we have no evidence to date that CRISPR systems interact with Obelisks, but that if they do, these interactions appear to be rare events or make use of a CRISPR system that has not been appended to the IMG/M database. Alternatively, the surveyed Obelisks, and the methods used to identify them, might have serendipitously been biassed against identifying CRISPR-interacting Obelisks.
Obelisks are prevalent in tested human microbiomes
Next, we sought to roughly estimate the prevalence of Obelisks in human gut and oral microbiomes by searching five datasets (three gut, two oral, Table 5) spanning 472 human donors primarily from North America (due to representational bias on the SRA). 25 donors (5.3 %) were identified as positive for Obelisks -α, -β, or -S.s, and a further 21 donors (4.4 %) appeared to be positive for novel Obelisks (Supplementary Figure 9), for a total of 9.7 % Obelisk-positivity (see Surveying for Obelisks in human data). Upon separating by microbiome source, 6.6 % (29 donors) of gut microbiome, and 53 % (17 donors) of oral microbiome samples contained Obelisks. These data therefore implicate the oral microbiome as a reservoir of Obelisks with more than half of the donors positive for such elements, though this could also be explained by an idiosyncrasy of the major oral dataset (Belstrøm and Constancias et al. 2021 112) that contributes to this count. Ultimately, 11 new, distinct, full-length Obelisks were identified upon examining the Obelisk-positive donors without Obelisk -α, -β, or -S.s homology - which we name “delta” through “xi” (see Surveying for Obelisks in human data, Figure 4, “Obelisk_000005” through “Obelisk_000015” in Supplementary Table 1). Obelisks “alpha,” “beta,” “epsilon,” “zeta,” and “eta” were restricted to gut microbiome samples (Obelisk-ε was found in one oral sample), whereas Obelisks “S. sanguinis,” and “theta” through “xi” were primarily orally restricted (Obelisk-S.s was found in one stool sample) - indicating an anatomical specificity of Obelisks despite the oral-gastric connection. These studies used different library preparation strategies (Table 5) and show varying Obelisk sensitivity as a function of read depth (Supplementary Figure 9 - scale bars), consistent with the technical expectation that not all metatranscriptomic sequencing workflows would be equally good at detecting Obelisks. This raises the question of a potential technological blind-spot to these (and similar) elements with some protocols. In any case, the observed values certainly represent a lower bound, and these data point to Obelisks being a non-negligible member of the tested adult oral and gut microbiomes. By their public nature, these datasets lack complete donor medical metadata; this lack and the relatively small sample size leave the investigation of correlations between Obelisk prevalence (and abundance) and the health of human hosts for future studies.
Table 5. human oral and stool metatranscriptome datasets queried for Obelisks.
Human metatranscriptomic datasets used for Supplementary Figure 9 with sampled niche and library preparation method indicated.
| bioProject | study | niche | RNA-seq library method |
|---|---|---|---|
| PRJNA398089 | Lloyd-Price et al. 2019 20 | stool | Modified RNAtag-seq |
| PRJNA940499 | Maghini and Dvorak et al. 2023 79 | stool | Illumina stranded total rna prep |
| PRJNA812699 | Jacobs et al. 2023 112 | stool | Viomega |
| PRJNA678453 | Belstrøm and Constancias et al. 2021 37 | oral | Illumina TruSeq Stranded mRNA |
| PRJNA917314 | Tong et al. 2023 113 | oral | NEBNext Small RNA-seq |
Discussion
The RNA viroid/sub-viral component of the biosphere is beginning to be estimated 12-14, but sequence-matching-based strategies, though potent for RNA viral discovery 1-5, are blind to previously unnoticed classes of agents. Here, we applied a generic molecular-feature-focused search strategy (VNom) to identify viroid-like RNAs in public RNA-seq datasets. We ultimately focused on a large monophyletic group of viroid-like elements that we term Obelisks. A single clear Obelisk-host pairing (S. Sanguinis SK36 - Obelisk-S.s) indicates that Obelisks can be a component of bacterial cells; while we don't know the “hosts” of other Obelisks, it is reasonable to assume that at least a fraction may be present in bacteria.
Obelisk genomes are predicted to fold into conspicuous “rod-like” secondary structures, with a largely self-complementary and conserved Oblin-1 ORF that accounts for at least half of the circular sequence assembly. Oblin-1 itself is predicted to fold into a stereotyped “globule” tertiary structure (Figure 4) with its most conserved motif, domain-A, lacking a confident tertiary structure prediction (Figure 2a). Furthermore, the presence of a subset of hammerhead ribozyme-bearing Obelisks with distinct Oblin-1 features (Supplementary Figure 6), and an interplay with domain-A evolution (Figure 3a) suggests an Oblin-ribozyme functional relationship, perhaps in viroid-like rolling-circle or rolling hairpin 37 replication. We note that conservative ribozyme detection thresholds were used in this work, leaving open the possibility that a larger diversity of ribozymes could be present in the Obelisks, including potentially novel self-cleaving ribozymes. As such, the exact interplay between Obelisk genome processing (via ribozymes) and Oblins-1, -2, and others is currently unknown.
Obelisks appear to be globally distributed (Figure 3c) and are a constituent member of the human oral and gut microbiomes, occurring in ~10 % of human donors in five assayed human metatranscriptomic studies (Table 5, Supplementary Figure 9). Of particular interest, we note one oral microbiome study showing a ~50 % Obelisk prevalence (Supplementary Figure 9d). We also note that observed Obelisk prevalence is likely to be quite dependent on the population in question, sampling scheme, type and depth of sequencing, and other features. Lastly, a specific Obelisk strain, Obelisk-α, appears to persist and speciate within microbiomes of human donors (Figure 1d-e). The prevalence and apparent novelty of these elements implies more is yet to be learned about their interplay with microbial and human life.
Constructing a full Obelisk phylogenetic tree with explanatory power proved difficult (Figure 3a). This is likely due to several factors including the fact that Obelisks appear to be under selection for a highly basepaired genomic coding region that must also code for stereotyped protein fold (Figure 4). Classical phylogenetic tools cannot account for evolutionary signals from non-position-independent RNA secondary structure constraints 38, consistent with the complexities in estimating trees from such families 39. Further, recent advances in protein tertiary structure prediction may now allow for protein structure based phylogenetic reconstruction that may be tolerant of greater sequence divergence 40,41. As such, definitive phylogenetic work on Obelisks might benefit from future tools that incorporate both evolutionary signals from RNA secondary structure conservation, and from structural alignment of predicted Oblin-1 “globule” tertiary structures. Lastly, the Serratus approach taken for large-scale Obelisk discovery was run using homology models built from sequences initially homologous to Obelisk-α (see Protein homology bioinformatics), and thresholds derived from RNA viral discovery campaigns 2, so while a mammalian sample-origin bias is seen (Figure 3b), this could be explained by an auto-correlation based on the mammalian origin of Obelisk-α, potentially confounded by the choice of RNA viral discovery threshold. Due to this aforementioned bias, as well as a lack of a systematised method for discovery, it should be noted that the breadth of Obelisk diversity reported in this study could be an underestimate. Further, while we focused on Obelisks, their prevalence and diversity suggest that similar, unrelated viroid-like RNAs are likely widespread and waiting to be discovered in public sequencing data.
The observation that distinct subsets of Obelisks appear to occur in human oral versus gastric sites, an anatomic specificity that mirrors the site-specificity of human microbiomes 42 (Supplementary Figure 9), supports the notion that Obelisks might include colonists of said human microbiomes. Building on this, donor-specific factors such as diet or lifestyles therefore likely play a role in Obelisk (re-)colonisation and retention. Further, given that Streptococcus sanguinis is a commensal of the healthy human oral microbiome 43, but also a causative agent of bacterial endocarditis 44, study of the implied S. sanguinis-Obelisk-S.s relationship might begin to reveal the relevance of Obelisks to the natural oral niche and potentially to human health, as well as offer a tractable model system to study Obelisk molecular biology. With 15 exemplar Obelisk sequences (Figure 4), an “Obelisk blueprint” arises: a ~730-1340 nt apparently circular RNA; with an extended “rod-like”, largely symmetrical predicted RNA secondary structure (Figure 4 - “jupiter” plots); an Oblin-1 homologue whose RNA sequence is largely self-complementary (which ColabFold predicts occupies a “globule-like” tertiary structure in 9 of 15 examples, Figure 4 - tertiary structures); and an occasionally present second, smaller protein (e.g. Oblin-2).
Many questions arise about the Obelisks. Does their transmission involve a separate, more complex, infectious agent (like HDV)? Do they primarily spread via virus-like particles, or cytoplasmically like viroids? Are Obelisks plasmid-like in that they can co-exist, and in some cases, contribute to host adaptability and fitness? Like viroids and HDV, do Obelisks replicate via rolling circle replication using a co-opted host RNA polymerase? What roles do the apparently circular Obelisk genome topology and the evidently conserved Obelisk genomic secondary structure play in the Obelisk lifecycle? Is Oblin-1 an RNA binding protein, and how does domain-A factor into its function? Does Oblin-2 act as a competitive inhibitor of host leucine zippers, as a multimerizing element, and/or can it interact with Oblin-1? How do Obelisks that lack Oblin-2 complement its function(s)? What role do the Obelisk-specific self-cleaving ribozymes play, and how do they interact with the Oblin proteins? How do Obelisks affect their host, and are they largely a deleterious or beneficial element to harbour? And what impact, if any, does harbouring an Obelisk have on ‘meta’-host physiology, is Obelisk positivity predictive of human health states?
Lastly, Obelisks do not closely resemble any existing mobile genetic elements, raising the question of their appropriate designation. Throughout this work, Obelisks have been referred to as ‘viroid-like’, drawing comparisons to viroids and HDV. However, viroids are in-part defined by their non-coding nature 6,7, and HDV-like elements are defined by homology to the large hepatitis Delta antigen (L-HDAg, and in the case of HDV, human tropism and a satellite relationship to Hepatitis B virus) 8. By virtue of their predicted coding capacity, which does not resemble L-HDAg, Obelisks are then neither strictly viroids, nor Delta-like elements. The predicted self-complementarity of the Oblin-1 further deviates from L-HDAg, likely imposing a set of unique evolutionary constraints (protein tertiary structure in addition to RNA secondary structure), that are not experienced by viroids and HDV-like elements. We therefore propose these proteins be referred to as “Oblins”. Viruses are already ill-defined, with sub-viral agents (such as viroids and HDV) being defined within the then more nebulous ‘perivirosphere’ 45, but part of ‘sub-virality’ is the implication of virus-like behaviour, either in transmission (e.g. via virions), in host impact (e.g. a pathology), or in replication (e.g. a co-opting viral replication machinery). Currently, it is not possible to assign transmission mode, host impact, or replication mode of Obelisks, suggesting that these elements might not even be ‘viral’ in nature and might more closely resemble “RNA plasmids”. As such, we propose that the term “Obelisk” be used to refer to these agents as they are distinct from other sub-viral satellites 46, viroids, and HDV.
Methods
VNom
VNom (pronounced venom, short for “Viroid Nominator”) was written to sequentially filter, in a homology-independent manner, for contigs with molecular features consistent with viroid-like biology from de novo assembled stranded RNA-seq data, namely: apparent circularity, and the co-occurrence of both positive- and negative-sense strands within a given sample (Supplementary Figure 2). As an input, VNom can take in any De Bruijn graph assembled contigs from stranded RNA-seq data; however, VNom is optimised to work on the output from rnaSPAdes 47. Initially, apparent circularity is inferred by identifying perfect k-mer repeats between the start and end of a contig: a previously exploited 2,12,48 sequence feature produced from circular De Bruijn graphs which are in turn produced from repetitive or circular transcripts during assembly. These apparently circular contigs are further de-concatenated into apparent unit-length, monomeric sub-sequences if a regular repetition of the identified k-mer is found, as is analogously done in 14. The resulting apparently circular contigs are then clustered with circUCLUST 49 and clusters containing at least one apparent sense and one antisense contig are kept (as inferred by k-mer counting). Any previously filtered out contigs that produce strong global alignments (usearch -usearch_global) 50 to these resulting sense-antisense clusters are then re-introduced where any clusters with now mutual contigs are merged. Local alignment (usearch -usearch_local) is then used to resolve and annotate any new multi-unit-length contigs into monomeric sub-sequences, and any sub-unit-length sequences into fragments. Finally, the resulting clusters are all “phased” to the same circular permutation using the multiple sequence aligner MARS 51. VNom is freely available at github.com/Zheludev/VNom.
Initial Obelisk identification
Stranded RNA-seq data were fetched from the SRA 52 using fasterq-dump 53, adapter and quality filtered using fastp (--average_qual=30 --n_base_limit=0 --cut_front --cut_tail) 54, and de novo assembled with rnaSPAdes (default settings). Viroid-like sequences were identified using VNom (-max 2000 -CF_k 10 -CF_simple 0 -CF_tandem 1 -USG_vs_all 1).
Obelisk RNA was initially identified in a longitudinal dataset of human stool stranded metatranscriptomics from the Integrative Human Microbiome Project (iHMP) 20. All paired-end RNA-seq datasets were downloaded (104 donors), trimmed, and assembled as described. Contigs were then grouped by donor ID and passed through VNom. The 2306 resulting VNom-nominated sense contigs were then queried manually for apparent lack of nucleotide, or protein-coding homology to the NCBI nt/nr (see later in this paragraph). Amongst these, we chose a sequence with striking predicted RNA secondary structure (high degree of basepairing, by eye, RNAfold -p -d2 --noLP --circ) 55. Obelisk RNAs were also manifest when VNom nominated contigs were passed through the following pipeline: the sense contigs were queried against a custom database (see Data Availability) of self-cleaving ribozymes (CMscan, default settings, keeping any, including likely spurious, hits) 56, these resulting 196 contigs were then assayed against the NCBI nt database (11 Oct 2021, blastn, default settings) 21,57, and contigs that yielded no hits, or whose best (by E-value) hits aligned to less than 40 % of contig's length were kept. These resulting 20 contigs were then queried against the NCBI nr database (8 Nov 2021, blastx, default settings), similarly keeping sub-40 % alignment length best hits, yielding 11 contigs, of which 5 had a unit length of 1164 nt (one contig was 1166nt) - suggesting a common class of RNA. These were later defined as the Obelisk RNAs. Similarly, blastn/p filtering the 2306 sense contigs but without the CMscan step yielded 107 contigs, 8 of which were over 1000 nt in length, comprising the 6 Obelisk RNAs. Lastly, running blastn on all the iHMP contigs against the 6 Obelisk RNAs resulted in a final total of 15 unique Obelisk RNA sequences.
Taxonomic classification
Taxa from length-filtered reads (fastp, as above with --length_required 75) were classified using Kraken2 (default settings) 58 against the Phanta 59 database, modified with non-redundant Obelisk-α/β sequences using KrakenGrafter 60, followed by Bayesian re-estimation using Bracken (-r 75) 61, lastly taxon counts were combined using Bracken2OTU 60, summing any samples that came from the same donor on the same day (indicative of split sequencing lanes).
Obelisk-α positive length-filtered read datasets, were assessed for sequence diversity relative to a fixed, arbitrarily chosen Obelisk-α reference 62. Namely, single nucleotide polymorphisms (SNPs) and small structural variants were measured by aligning reads (bwa-mem2, default settings) 63 to the reference, followed by deduplication (picard, MarkDuplicates) 64, and detection freebayes (--ploidy 1 --pooled-discrete --pooled-continuous) 65. SAMtools 66 and bamaddrg 67 were used throughout. Principal component analysis (PCA) on the resulting vcf file was computed using SNPRelate (snpgdsPCA) 68, as described in 69, clusters were identified by kmeans (centers = 5) 70.
Obelisk homologue detection in additional public data
Close Obelisk-α homologues were identified in the Short Read Archive (SRA) 52 using PebbleScout (“Metagenomic” database, default settings) 25, a recently released tool that efficiently queries ~3.2 million (mid 2022) raw sequencing data for exact 42 k-mer matches. 9 metatranscriptome BioProjects (comprising 34 short read datasets) were identified (PBSscore > 65) with close (~1 % nucleotide divergence) matches to Obelisk-α, of which 3 were part of iHMP or its predecessor 71, 5 were from other human stool studies 72-76, and 1 was from a fox gut autopsy 77. Using the VNom pipeline (see above), 21 datasets (from 7 BioProjects) yielded full length Obelisk-α sequences, all from human hGMB studies (Table 3).
Finding Obelisk-α homologues in studies separate from the iHMP lent support to these RNA elements being legitimate biological entities. Further, one Obelisk-α homologue was found in a study from our own institution 76, suggesting that Obelisk-like RNAs could be locally present. Emboldened by this, we solicited hGMB stranded RNA-seq data from the local academic community and identified closely related Obelisk-α homologues in a dataset that at the time had not been uploaded onto the SRA (now available at PRJNA940499: donors D01 - both Obelisks -α and -β; and D10 - just Obelisk-α) 78. Further, within this dataset we identified a diverged Obelisk-like sequence with similar: length (1182 nt), lack of apparent homology to reference databases, predicted obelisk-like secondary structure, and two ORFs but with low homology to Obelisk-α. In comparison to Obelisk-α, this new “Obelisk-β” had a 41.30 % nucleotide sequence identity, and 23.42/38.29 % and 18.75/31.25 % on the amino acid level identities/similarities for ORFs 1 and 2, respectively (see below, Supplementary Figure 3a/c).
Owing to their apparent sequence novelty, the Obelisk-α/β Oblin-1 and -2 protein sequences were next used as hallmark sequences specific to Obelisk-like RNAs - analogous to the use of RNA-dependant RNA polymerase (RdRP) hallmark sequences in RNA viral discovery 1-5. To identify divergent Obelisk-like elements, we searched the RNA Deep Virome Assemblage (RDVA, v0.2) 13,26, a collection of 58,557 assemblies of ~12.5 trillion contigs, with diamond (--very-sensitive) 79 using Obelisk-α/β Oblin -1 and -2 protein sequences deduplicated at 90 % sequence identity (UCLUST, default settings) as queries 50. This resulted in 38,545 sub-5000 nt hits which when de-replicated, circularly clustered (circUCLUST) into 29,859 and 19,808 clusters at 90 % and 75 % nucleotide sequence identity, respectively (see Data Availability). A conservative database of 7,202 Obelisks was built by keeping assemblies with a CircleFinder (VNom defaults) implied circularity, with each genome “phased” to 50 nt from the start codon of its largest predicted ORF (prodigal, -p meta). This database was clustered (circUCLUST) into 1,744 80 % identity clusters which were then sub-clustered at 95 % identity (Supplementary Table 1). The assemblies were then named based on these nested clusterings. A naming convention is proposed with the following pattern “Obelisk_X_Y_Z” where “X” refers to the 80 % cluster ordinate, “Y” to the 95 % cluster ordinate, and “Z” as a unique identifier within the 95 % cluster. The first 15 80 % ordinates are defined as the Obelisks depicted in Figure 4, the next 10 80 % ordinates are defined as the remaining letters in the Greek alphabet (omicron through omega). As such, the centroid Obelisk-α sequence that is also the centroid of the first 95 % sub-type is defined as “Obelisk_000001_000001_000001”.
Serratus
Extending from the RDVA search, a larger breadth of public datasets (5,470,176 runs) was next assessed for diverged Obelisk-like sequence presence. Profile hidden Markov models (pHMMs) of ORFs 1 and 2 were derived from the RDVA hits (see below) and used as queries in the Serratus architecture 2, an optimised, cloud-based pipeline for efficiently identifying sequencing reads that align to pHMMs. By looking for pHMM matches, Serratus is able to find more distantly related Obelisk-like sequences where k-mer match searches (e.g. PebbleScout) would fail, but at a considerable computational expense. Datasets were defined as a Serratus hit if at least one read aligned (E-value <1x10−4) to either Oblin-1 or Oblin-2. Of the resulting 949,810 non-redundant SRA hits, 215,398 datasets were selected by filtering with a virus-presence score (≥25, explained in github.com/ababaian/serratus/wiki/.summary-Reports) which attempts to predict ORF de novo assembly success, ultimately yielding 1,499 datasets containing both Oblin-1 and Oblin-2, 3,006 containing only Oblin-1, and 213,891 containing only Oblin-2. Per hit SRA, high confidence ORF mapping reads were then de novo assembled using rnaSPAdes (default settings) yielding Obelisk “micro-assemblies”. This Serratus run was conducted along with other pHMM queries, meaning that de novo assembly happened in aggregate with all other hits, as such, diamond (--very-sensitive) was used to extract Oblin-1/-2 micro-assembly protein sequences.
Protein homology bioinformatics
To probe the deep sequence diversity of Oblins 1 and 2, corresponding single domain profile hidden Markov models (pHMMs) were individually constructed from the RDVA hits using an iterative approach: A multiple sequence alignment (MSA) from the initial PebbleScout set was computed using Muscle5 (default settings) 80, from which an initial pHMM was computed using HMMbuild (default settings) 81. Each genome in the RDVA non-redundant 90 % sequence identity cluster centroid set was doubled in length using SeqDoubler 60 and ORFs were predicted using Prodigal (-p meta) 82. ORFs with predicted N- or C- terminal truncation were omitted and a non-redundant set was kept (usearch -fastx_uniques) 50. This ORF database was queried against (HMMsearch, default settings) the initial pHMM and hits with global E-values lower than 1x10−15 for Oblin-1 or 1x10−8 for Oblin-2 were kept. HMMalign (--trim) and MSACleaner (-ref from the PebbleScout set and -fxn 0.01) 60 were used recursively (until no new sequences were omitted) to filter the constituent MSA sequences to omit sequences that contributed large indels relative to the initial pHMM. A new pHMM was computed and the HMMsearch (on the remaining ORFs), HMMalign (without --trim), and MSACleaner steps were repeated once. This resulting MSA was filtered by sequence length FASTACleanUp (-lower 150 for Oblin-1, -lower 40 for Oblin-2) 60 and a final pHMM was computed. msaconverter 83 was used throughout. There were no overlapping sequences between the resulting Oblin-1 and -2 pHMMs.
A contiguous alignment block of 18 amino acids was noticed in the resulting Oblin-1 pHMM (Obelisk-α: 152-RRRGYKDHGSRRFPHEVH-169) and was selected as a marker sequence, terming it domain-A. Because the Serratus Oblin-1 micro-assemblies may include some that are not full-length (wrt Oblin-1), further aggregation from the Serratus data utilised a search for similarity to domain-A. To incorporate the Serratus results, an initial 503 sequence domain-A alignment was extracted from the RDVA pHMM (and later used with K-mer Rabbit, below) and a new pHMM was constructed (HMMbuild, default settings). A length sorted (seqkit sort -l -r), non-redundant (usearch -fastx_uniques) set of Serratus Oblin-1 micro-assemblies was then iteratively queried with an ever-rebuilt domain-A pHMM: keeping HMMsearch (default settings) hits with E-values lower than 1x10−4, intermediate MSAs were re-built (HMMalign --trim) relative to the previous iteration and sequences with at least 8 amino acids (seqkit seq -g -m 8) were kept, next, the resulting sequences were re-aligned to the current pHMM and a new pHMM was built, lastly, all <1x10−4 E-value hits were omitted and a new iteration was started. A finalised Serratus-inclusive domain-A pHMM was constructed with 30,686 sequences after 12 cycles. This process was repeated for two other less well-conserved domains, domain-B (Obelisk-α: 96-CLTSKSGMLNFLEDTTLY-113), and domain-C (Obelisk-α: 53-RSKKDLLALAIISWWLEE-70), with 5076 and 5103 resulting sequences, respectively. Domains -B/-C were not studied further in this work.
Protein tertiary structure prediction
For initial, monomeric tertiary structure prediction, RDVA pHMM MSAs were re-aligned (Muscle5, default settings) relative to ORFs-1/2 from Obelisk-α and used with ColabFold (v1 .5.2-patch) 32 implementation of AlphaFold2 (default settings, no amber, no dropout) 31. The HHblits suite was used to convert between fasta and a3m MSA formats 84. Tertiary structure homology was assessed using the Phyre2 (default settings) 85, Dali (PDB Search) 86, FoldSeek (all databases, 3Di/AA and TM-align scoring) 87, and the Clustered AlphaFold Database 88 webservers (see Data Availability). For all other tertiary structure predictions, ColabFold was used with mmseqs2 uniref env for MSA generation. For 9 in 15 predictions, including Obelisks -α, -β, and -S.s, this yielded qualitatively similar “globule” predictions (Figure 4 - tertiary fold predictions). An equivalent 73 sequence MSA was constructed for Oblin-1 homologues from ribozyme-baring Obelisks (see RNA homology bioinformatics) by first filtering any Prodigal-predicted proteins for length (seqkit seq -m 200 -M 250), aligning the resulting sequences (Muscle5), and manually removing any sequences that appeared to disrupt the MSA. ColabFold v1.5.3 was used for ribozyme-baring Oblin-1 protein tertiary fold predictions and Obelisk-nu.
Protein conservation and phylogenetics
Oblin-1/-2 conservation analysis was conducted on Obelisk-α-relative a3m alignments against the BLOSUM62 substitution matrix 89 using msaConservationScore (gapVsGap = 0) 90 and the Biostrings package 91. The Oblin-2 sequence logo was constructed using ggseqlogo 92, and a consensus sequence was generated with msaConsensusSequence (upperlower, thresh = 20,0).
Owing to the micro-assembly used in the Serratus search, phylogenetic analysis was limited to the highly conserved domain-A. To ensure a domain-A phylogenetic tree encompassed the observed sequence diversity from ribozyme-baring Obelisks, the underlying multiple sequence alignment (MSA) construction started with an iterative pHMM construction approach similar to method used to build the initial Oblin-1 pHMM. First Oblin-1 homologues from ribozyme-baring Obelisks (see RNA homology bioinformatics) were queried (HMMsearch --max, E-value ≤ 1x10−8) against the initial Oblin-1 pHMM, yielding only sequences homologous to domain-A. These sequences were re-aligned (Muscle5) and an initial ribozyme-associated domain-A pHMM was built. This ribozyme-associated pHMM was then iteratively built upon with successive rounds of similarity searches (HMMsearch --max, E-value ≤ 1x10−8) against the RDVA’s ribozyme-baring Obelisk’s predicted proteins followed by re-alignment with Muscle5. Once no new sequences were found, the cycle was continued at an E-value threshold of 1x10−5. This resulting ribozyme-associated MSA was then re-aligned to the initial Oblin-1 MSA (HMMalign, default settings) and the alignment column corresponding to domain-A was manually excised, and re-aligned (Muscle5). The entirety of the full-length predicted proteins from the RDVA were then similarly iteratively queried but at a E-value threshold of 1x10−4, and without an intermediate Muscle5 step. The converged alignment was then re-aligned with Muscle5 (Super5) and similarly iteratively queried against the Serratus micro-assemblies, keeping the best hit per micro-assembly until convergence. The resulting 46,884 total domain-A sequences were finally re-aligned with Muscle5 (Super5). This MSA was then deduplicated, and optimised using CIAlign 93 to remove insertions (minimum size 1, minimum 0.05 %), to crop divergent sequences (minimum identity proportion 0.01, minimum non-gap proportion 0.5, buffer size 4), and to remove any resulting sequences shorter than or equal to 16 aa. A final round of deduplication yielded a 3265 non-redundant sequence domain-A no-gap alignment of 17 aa (Supplementary Table 2).
A maximum likelihood phylogenetic tree was then constructed from this 17 aa alignment using iqtree 94 The LG+G4 substitution model (testnewonly) was selected (ModelFinder 95) based on a consensus between the Akaike and Bayesian Information Criteria. Tree construction was run with 33,000 UFBoot bootstraps 96, Nearest Neighbour Interchange optimization, and 33,000 SH-like approximate likelihood ratio tests (-B 33000 -bnni -alrt 33000). The resulting tree was plotted using iTOL 97.
ScanRabbit
For rapidly searching smaller, locally-held datasets for novel Obelisk homologues, we developed a second tool, ScanRabbit, which focuses on a short segment of any multiple sequence alignment. ScanRabbit was run using the position-specific-scoring matrix (PSSM) based on the multiple sequence alignment used to build the Oblin-1 profile hidden Markov model (see above) from the RDVA hits corresponding to Domain-A. ScanRabbit accelerates searches on local hardware through direct bitwise conversion of the PSSM to a local bitwise scoring that can be applied to the raw binary representation of RNA-seq reads, and a just-in-time compiler PyPy 98. ScanRabbit is available on GitHub at github.com/FireLabSoftware/ScanRabbit.
Obelisk spacer analysis
The presence of Obelisks in known prokaryotic CRISPR spacer arrays was assessed using a conservative k-mer matching approach. Namely, the RDVA Obelisk dataset was queried against predicted CRISPR spacers in the Joint Genome Institute (JGI) IMG/M spacer database (May 2023) 36. To estimate a lower length bound on matching noise, a parallel analysis was conducted on “reversed” (not reverse complemented) Obelisk sequences. Initially, RDVA Obelisk sequences were searched against the IMG/M spacer database using blastn (default settings), only keeping perfect matches with no gaps or mismatches (k-mers) - the longest k-mer match between a given spacer/Obelisk pairing was kept. Next, all kept spacers containing any 12-mer match to common Illumina sequencing adaptors were omitted using KmerCatcher (default settings) 60. For each remaining spacer, the information content was estimated 99 by comparing how efficiently the compression algorithm zip (-9) 100 could “deflate” a given spacer - a larger length normalised deflation indicates a less complex spacer sequence that is less likely to be unambiguously mapped to a specific (Obelisk) sequence. The repetitive content of each spacer was also assessed using etandem (-minrepeat 4, -maxrepeat 15, -threshold 2) 101. Spacers with a length normalised deflation less than 1.0 percent per nucleotide were kept (137,667 forward, 118,411 reverse), these spacers also qualitatively had a low etandem score though this metric was not used for filtration (Supplementary Figure 8a). Next, only the 23 forward spacers longer than the maximum length of the reverse spacers (25 nt) were kept as any mappings below this threshold would be indistinguishable from noise (reverse-mapping, Supplementary Figure 8b). Lastly, the corresponding Obelisks mapping to these spacers were minimum length filtered to 1000 nt (seqkit seq -m 1000), resulting in two contigs. Only one of these contigs gave blastn (default settings, NCBI webserver, August 2023) a largely (~95 %) unknown sequence with a singular ~45 nt sequence mostly showing up in high G+C Gram-positive bacteria and cyanobacteria (consistent with a CRISPR spacer array, see Data Availability). This largely unknown 1096 nt contig was found to encode (prodigal -p meta) homologues of both Oblin-1 and Oblin-2 (HMMsearch, default settings, against the RDVA pHMMs), and is predicted to fold (see below) into an obelisk-like RNA secondary structure (Supplementary Figure 8c) - features consistent with being an Obelisk which we term Obelisk-“gamma” (Obelisk-ɣ). Two spacers were found to map to Obelisk-ɣ, both from the same Bombella mellum genome (RefSeq GCF_014048465.1) 102 - these spacers (which differ by one extra nucleotide) were found at the same putative CRISPR locus but predicted in the IMG/M database with two different tools (PILER-CR and CRT) 103,104, as such, this is likely one spacer. Obelisk-ɣ's predicted secondary structure is not as “rod-like” as other Obelisks (Figure 4 - “jupiter” plots), with the spacer mapping to the “frayed” end; additionally, the spacer mapping position coincides with the locus identified by blastn; and lastly CircleFinder (VNom default settings) did not identify a start-end k-mer repeat indicative of a circular genome. The Obelisk-ɣ Oblin-1 was also not predicted (see above) to fold into the characteristic “globule” fold (Figure 4 - tertiary fold predictions), though the discriminatory power of this is unclear and so ignored. These features suggest that the Obelisk-ɣ genome might be mis-assembled, with the putative spacer mapping sequence arising from a chimeric assembly. As such, this conservative approach to CRISPR spacer mapping was not able to unambiguously identify any Obelisk relationships to CRISPR spacer arrays as we currently recognise them.
Identity and similarity measurements
Unless otherwise stated, all nucleotide identity, and protein identity and similarity measurements were computed by first building a pairwise alignments Muscle5 (default settings) of “phased” genomes (as below) followed by calculation with Ident and Sim (default settings) 105.
RNA homology bioinformatics
Figure 1b, Figure 4, Supplementary Figure 3b, and Supplementary Figure 6a RNA secondary structures were predicted using RNAalifold (-p, -r, -d2, --noLP, --circ) 107 on the non-redundant (usearch -fastx_uniques), 1164 nt long, PebbleScout set of the above “phased” Obelisk-α sequences, split by genome polarity, using a Muscle5 (default settings) derived MSA. Supplementary Figures 1a and 2c-d secondary structures were predicted on singular genomes using RNAfold (-p -r -d2 --noLP --circ). RNA secondary structures were illustrated using circlize 108 for “jupiter” plots, and R2R 109 for “skeleton” diagrams. Conserved RNA element (e.g. ribozymes) coordinates in Supplementary Figure 1 were identified using CMscan (--rfam --cut_ga) against the Rfam 14.6 database 110.
23 Obelisk-encoded hammerhead type-III ribozyme homologous sequences were initially identified (CMsearch) using the RF00008 reference covariance model against the 90 % identity-clustered (circUCLUST), sequence-doubled (SeqDoubler) RDVA dataset, using stringent cutoffs for confident (E-value ≤ 1x10−5), full-length (--notrunc) hits, keeping only the best hit per Obelisk genome. An Obelisk-specific, “Obelisk-variant hammerhead type-III” (ObV-HHR3) covariance model (CM) was constructed using an iterative approach: an initial CM was constructed using the 23 hit sequences by aligning them against RF00008 (CMalign, default settings), optimising the alignment using CaCoFold 111 (R-scape: -s, --cacofold, --rna), and finally building (CMbuild, default settings), and calibrating (CMcalibrate, default settings) the CM. Using this initial CM as a starting point, the sequence-doubled RDVA dataset was iteratively passed through the CMsearch, CMalign, CaCoFold, CMbuild, and CMcalibrate pipeline, each time only keeping the best, non-truncated, E-value ≤ 1x10−5 hits (one hit per Obelisk genome) and additively appending them to the CM, subtracting the hits from the RDVA set as they were found, until no new hits were found. Ultimately, a 178 sequence ObV-HHR3 was constructed with 15 significantly covarying positions identified (Supplementary Figure 6b). When re-querying (CMsearch, --no-trunc) the full RDVA dataset with this finalised CM at an E-value ≤ 1x10−4, 339 Obelisk genomes were identified. The ObV-HHR3 column in Supplementary Table 1 was annotated with CMsearch, --no-trunc, ≤ 1x10−5 on sequence-doubled genomes.
Streptococcus sanguinis bioinformatics
In an attempt to identify Obelisk-like elements that had been serendipitously sequenced in isolation with their putative cellular host(s), Oblin-1 positive filtered Serratus hits were screened for potentially low biodiversity experimental designs such as defined co-culture, single-cell RNA-seq, and isolate culture. As such, isolate RNA-seq experiments of Streptococcus sanguinis (strain SK36, a commensal of the human oral microbiome) stood out (Table 4). Upon further investigation (using CircleFinder from VNom), a 1137 nt, obelisk-shaped RNA coding only for Oblin-1 was identified. This so-called “Obelisk-S.s” exhibited 40.65 % and 35.47 % nucleotide sequence identity with Obelisk-α and Obelisk-β, respectively, and 19.92/33.47 % and 21.05/32.71 % Oblin-1 amino acid identity and similarity to Obelisk-α and Obelisk-β, respectively (see above, Supplementary Figure 3a/d). Additionally, Obelisk-S.s was further found in human oral microbiome samples (Table 5, Supplementary Figure 9), and by comparing isolate cultures from different growth media, S. sanguinis was determined to be the likely cellular host as opposed to Obelisk-S.s being a contamination from complex media.
Surveying for Obelisks in human data
The prevalence of Obelisks in five human microbiome datasets (three gastric, hGMB, and two oral, hOMB, Table 5 and Table 6) was (re-)evaluated after both Obelisks -α, -β, and -S.s were identified, and the RDVA pHMMs were constructed. For human gut metatranscriptome data, the 104 iHMP donors 20, and the 10 “ZF” donors from the dataset where Obelisk-β was found 79 were reanalysed; additionally, 326 new donor samples from an irritable bowel syndrome study 112 were queried, for a total of 440 hGMB donors analysed. For human oral metatranscriptome data, 22 (50/50 healthy/case) donors from a Dutch cohort studying periodontitis 37, and 10 healthy donors from a oral extracellular vesicles study 113 were queried for a total of 32 hOMB donors analysed. To identify more diverged Obelisk elements, a pHMM mapping approach was taken - similarly to Serratus. Namely, each dataset's trimmed reads (as before) were translated in all six frames (seqkit -f 6 -F) and assessed for Oblin-1 homology using HMMsearch (default settings) against the RDVA Oblin-1 pHMM. Donors with greater than or equal to 10 translated reads (averaging over per-donor replicates, time points, or sampling locations if present) mapping with an E-value less than or equal to 1x10−5 were counted as true Oblin-1 hits. Additionally, these trimmed reads were assessed for Obelisk -α, -β, and -S.s presence using a modified Phanta Kraken2 and Bracken database constructed as before incorporating all non-redundant Obelisk -α, -β, and -S.s sequences (only the previous Obelisk-α positive iHMP datasets were re-assessed in this way). Across these five datasets, 21 donors were identified as positive for Obelisk homologues (>10 HMMsearch hits) but negative for Obelisks -α, -β, or -S.s (<10 Kraken2 hits), additionally, 25 donors were identified as positive for Obelisks -α, -β, or -S.s (>10 Kraken2 hits, Supplementary Figure 9). The presence of pHMM-mapping reads in the absence of k-mer reads suggested the existence of new Obelisks, as such, these 21 donors' datasets were assessed for new Obelisks. Briefly, these donor's trimmed reads were assembled as before, keeping any contigs with Oblin-1 homology (HMMsearch, --max, E-value ≤1x10−5), and then selecting for apparently circular contigs with CircleFinder (default VNom settings). These selected contigs were next assessed for Oblin-2 coding capacity (prodigal -p meta, followed by HMMsearch and blastn versus the Oblin-2 RDVA pHMM, and Obelisk-α Oblin-2 sequence and consensus, respectively E-value ≤1x10−4), and obelisk-like secondary structure as before. Clustering all resulting and previously identified contigs (circUCLUST -id 0.8), 11 new full-length Obelisks were identified, which we named “delta” through “xi” (“Obelisk_000005” to “Obelisk_000015” in Supplementary Table 1, Figure 4). “Delta,” “epsilon,” “zeta,” and “eta” were found in the hGMB datasets and all remaining Obelisks were found in the Dutch hOMB dataset - indicating a human sampling site specificity to Obelisk species. Of these 11 Obelisks, eight apparently only code for an Oblin-1 homologue, Obelisk-“kappa” codes for an Oblin-2 homologue, and Obelisks -“lambda” and -“mu” code for a second ORF similar in size to Oblin-2 but with no obvious homology (which we term the “2ndORF” as more study is needed to determine if this is actually a bona fide new ORF). Four of these new Obelisks’ (“epsilon,” “kappa,” “mu,” and “xi”) Oblin-1 sequences were not predicted to fold (as before) into the otherwise characteristic “globule” tertiary structure (Figure 4 - tertiary fold predictions). These new Obelisks span between 733 nt (Obelisk- “iota”) to 1372 nt (Obelisk- “kappa”). Considering these new Obelisk sequences, as well as donors which did not yield full-length Obelisk candidates, Obelisks appear to occur in 9.5 % of the human donors assayed (6.6 % of hGMB samples, and 53 % of hOMB samples) and describe a wider breadth of characteristics that Obelisks seem to be able to possess (length and coding capacity).
Table 6. Supplementary Figure 9 donor aliases.
Human donor aliases used for brevity in this study (Supplementary Figure 9) and their corresponding nomenclature from their original studies. Note, datasets from Maghini and Dvorak et al. 2023 79 and Tong et al. 2023 113 were used without re-assigning new aliases.
| study | alias to study nomenclature key |
|---|---|
| Lloyd-Price et al. 2019 20 | A:2077, B:6038, C:2068, D:5001, E:2071, F:2025, G:3034, H:4016, I:4022 |
| Jacobs et al. 2023 112 | A:A6405, B:A6673, C:1634K, D:A6089, E:1596C, F:A7010, G:A6942, H:A6889, I:A6419, J:A6797, K:A6083, L:A6974, M:A6131, N:A6739, O:A6810, P:A6745, Q:1558C |
| Belstrøm and Constancias et al. 2021 37 | A:K2, B:K3, C:MP8, D:MP10, E:K11, F:K10, G:MP11, H:K9, I:MP1, J:K6, K:MP4, L:MP2, M:MP9, N:K7, O:K5 |
Supplementary Material
Acknowledgements
This work is dedicated to the memory of mentor and friend, Paul Berg.
This work was funded by: Stanford Graduate Fellowship (INZ); University of Valencia Margarita Salas Fellowship MS21-067 (MJLG); Generalitat Valenciana Grant PROMETEO CIPROM/2022/21 (MDLP); Ministry of Economics and Competitivity of Spain-FEDER grant PID2020-116008GB-I00 (MDLP); Canadian Institutes of Health Project Grant PTJ-496709 (AB); Computing resources were provided by the University of British Columbia Community Health and Wellbeing Cloud Innovation Centre, powered by AWS (AB); NIH Grant R01AI148623 (National Institute of Allergy and Infectious Diseases, NIAID) (ASB); NIH Grant R01AI143757 (NIAID) (ASB); Convergence Grant 3.1416 (Stand up 2 Cancer) (ASB); Paul Allen Distinguished Investigator Award (ASB); and NIH Grant R35GM130366 (National Institute of General Medical Sciences) (AZF).
We are grateful to the greater biology community for open data sharing. Some figures were arranged in BioRender.
Data availability
Code and tabular summaries are available at the Stanford Digital Repository (purl.stanford.edu/wb363nt3637).
Bibliography
- 1.Shi M., Lin X.-D., Tian J.-H., Chen L.-J., Chen X., Li C.-X., Qin X.-C., Li J., Cao J.-P., Eden J.-S., et al. (2016). Redefining the invertebrate RNA virosphere. Nature 540, 539–543. 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 2.Edgar R.C., Taylor J., Lin V., Altman T., Barbera P., Meleshko D., Lohr D., Novakovsky G., Buchfink B., Al-Shayeb B., et al. (2022). Petabase-scale sequence alignment catalyses viral discovery. Nature 602, 142–147. 10.1038/s41586-021-04332-2. [DOI] [PubMed] [Google Scholar]
- 3.Zayed A.A., Wainaina J.M., Dominguez-Huerta G., Pelletier E., Guo J., Mohssen M., Tian F., Pratama A.A., Bolduc B., Zablocki O., et al. (2022). Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science 376, 156–162. 10.1126/science.abm5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neri U., Wolf Y.I., Roux S., Camargo A.P., Lee B., Kazlauskas D., Chen I.M., Ivanova N., Zeigler Allen L., Paez-Espino D., et al. (2022). Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell 185, 4023–4037.e18. 10.1016/j.cell.2022.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Olendraite I., Brown K., and Firth A.E. (2023). Identification of RNA Virus–Derived RdRp Sequences in Publicly Available Transcriptomic Data Sets. Molecular Biology and Evolution 40, msad060. 10.1093/molbev/msad060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Serio F., Li S.-F., Matoušek J., Owens R.A., Pallás V., Randles J.W., Sano T., Verhoeven J.Th.J., Vidalakis G., Flores R., et al. (2018). ICTV Virus Taxonomy Profile: Avsunviroidae. Journal of General Virology 99, 611–612. 10.1099/jgv.0.001045. [DOI] [PubMed] [Google Scholar]
- 7.Di Serio F., Owens R.A., Li S.-F., Matoušek J., Pallás V., Randles J.W., Sano T., Verhoeven J.Th.J., Vidalakis G., Flores R., et al. (2020). ICTV Virus Taxonomy Profile: Pospiviroidae. Journal of General Virology 102, 001543. 10.1099/jgv.0.001543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Magnius L., Taylor J., Mason W.S., Sureau C., Dény P., Norder H., and ICTV Report ConsortiumYR 2018 (2018). ICTV Virus Taxonomy Profile: Deltavirus. Journal of General Virology 99, 1565–1566. 10.1099/jgv.0.001150. [DOI] [PubMed] [Google Scholar]
- 9.Eigen M. (1971). Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 58, 465–523. 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 10.Gago S., Elena S.F., Flores R., and Sanjuán R. (2009). Extremely High Mutation Rate of a Hammerhead Viroid. Science 323, 1308–1308. 10.1126/science.1169202. [DOI] [PubMed] [Google Scholar]
- 11.Bergner L.M., Orton R.J., Broos A., Tello C., Becker D.J., Carrera J.E., Patel A.H., Biek R., and Streicker D.G. (2021). Diversification of mammalian deltaviruses by host shifting. Proceedings of the National Academy of Sciences 118, e2019907118. 10.1073/pnas.2019907118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg C.E., Olzog V.J., Eckert I., and Weinberg Z. (2021). Identification of over 200-fold more hairpin ribozymes than previously known in diverse circular RNAs. Nucleic Acids Research 49, 6375–6388. 10.1093/nar/gkab454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forgia M., Navarro B., Daghino S., Cervera A., Gisel A., Perotto S., Aghayeva D.N., Akinyuwa M.F., Gobbi E., Zheludev I.N., et al. (2023). Hybrids of RNA viruses and viroid-like elements replicate in fungi. Nat Commun 14, 2591. 10.1038/s41467-023-38301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee B.D., Neri U., Roux S., Wolf Y.I., Camargo A.P., Krupovic M., Simmonds P., Kyrpides N., Gophna U., Dolja V.V., et al. (2023). Mining metatranscriptomes reveals a vast world of viroid-like circular RNAs. Cell. 10.1016/j.cell.2022.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tisza M.J., and Buck C.B. (2021). A catalog of tens of thousands of viruses from human metagenomes reveals hidden associations with chronic diseases. Proceedings of the National Academy of Sciences 118, e2023202118. 10.1073/pnas.2023202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutilh B.E., Cassman N., McNair K., Sanchez S.E., Silva G.G.Z., Boling L., Barr J.J., Speth D.R., Seguritan V., Aziz R.K., et al. (2014). A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5, 4498. 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camarillo-Guerrero L.F., Almeida A., Rangel-Pineros G., Finn R.D., and Lawley T.D. (2021). Massive expansion of human gut bacteriophage diversity. Cell 184, 1098–1109.e9. 10.1016/j.cell.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahlman S., Avellaneda-Franco L., Kett C., Subedi D., Young R.B., Gould J.A., Rutten E.L., Gulliver E.L., Turkington C.J.R., Nezam-Abadi N., et al. (2023). Temperate gut phages are prevalent, diverse, and predominantly inactive. Preprint at bioRxiv, 10.1101/2023.08.17.553642 10.1101/2023.08.17.553642. [DOI] [Google Scholar]
- 19.Fogarty E.C., Schechter M.S., Lolans K., Sheahan M.L., Veseli I., Moore R., Kiefl E., Moody T., Rice P.A., Yu M.K., et al. (2023). A highly conserved and globally prevalent cryptic plasmid is among the most numerous mobile genetic elements in the human gut. Preprint at bioRxiv, 10.1101/2023.03.25.534219 10.1101/2023.03.25.534219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Kelly C., Kim S., et al. (2022). Database resources of the national center for biotechnology information. Nucleic Acids Research 50, D20–D26. 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H., Zhang L., Lin A., Xu C., Li Z., Liu K., Liu B., Ma X., Zhao F., Jiang H., et al. (2023). Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 621, 396–403. 10.1038/s41586-023-06127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu S., Wang J., Chitsaz F., Derbyshire M.K., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Marchler G.H., Song J.S., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48, D265–D268. 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Research 49, D412–D419. 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiryev S.A., and Agarwala R. (2023). Indexing and searching petabyte-scale nucleotide resources. Preprint at bioRxiv, 10.1101/2023.07.09.547343 10.1101/2023.07.09.547343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin V., Ravichandran G., Ha K., Kinoshita B.P., and Babaian A. (2022). RNA Deep Virome Assemblage. GitHub. https://github.com/ababaian/serratus/wiki/RNA-Deep-Virome-Assemblage. [Google Scholar]
- 27.Gloor G.B., Macklaim J.M., Pawlowsky-Glahn V., and Egozcue J.J. (2017). Microbiome Datasets Are Compositional: And This Is Not Optional. Frontiers in Microbiology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coenen A.R., and Weitz J.S. (2018). Limitations of Correlation-Based Inference in Complex Virus-Microbe Communities. mSystems 3, e00084–18. 10.1128/mSystems.00084-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirano H., and Takemoto K. (2019). Difficulty in inferring microbial community structure based on co-occurrence network approaches. BMC Bioinformatics 20, 329. 10.1186/s12859-019-2915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caufield P.W., Dasanayake A.P., Li Y., Pan Y., Hsu J., and Hardin J.M. (2000). Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infect Immun 68, 4018–4023. 10.1128/IAI.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirdita M., Schütze K., Moriwaki Y., Heo L., Ovchinnikov S., and Steinegger M. (2022). ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeepMind, and EMBL-EBI (2022). AlphaFold Protein Structure Database: Frequently Asked Questions. https://alphafold.ebi.ac.uk/faq. [Google Scholar]
- 34.O’Shea E.K., Lumb K.J., and Kim P.S. (1993). Peptide “Velcro”: design of a heterodimeric coiled coil. Curr Biol 3, 658–667. 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 35.Sinden R.R. (1994). CHAPTER 8 - DNA–Protein Interactions. In DNA Structure and Function, Sinden R. R., ed. (Academic Press; ), pp. 287–325. 10.1016/B978-0-08-057173-7.50013-4. [DOI] [Google Scholar]
- 36.Chen I.-M.A., Chu K., Palaniappan K., Ratner A., Huang J., Huntemann M., Hajek P., Ritter S., Varghese N., Seshadri R., et al. (2021). The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Res 49, D751–D763. 10.1093/nar/gkaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belstrøm D., Constancias F., Drautz-Moses D.I., Schuster S.C., Veleba M., Mahé F., and Givskov M. (2021). Periodontitis associates with species-specific gene expression of the oral microbiota. NPJ Biofilms Microbiomes 7, 76. 10.1038/s41522-021-00247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tattersall P., and Ward D.C. (1976). Rolling hairpin model for replication of parvovirus and linear chromosomal DNA. Nature 263, 106–109. 10.1038/263106a0. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen J.S., Forsberg R., Meyer I.M., and Hein J. (2004). An evolutionary model for protein-coding regions with conserved RNA structure. Mol Biol Evol 21, 1913–1922. 10.1093/molbev/msh199. [DOI] [PubMed] [Google Scholar]
- 40.Patiño-Galindo J.Á., González-Candelas F., and Pybus O.G. (2018). The Effect of RNA Substitution Models on Viroid and RNA Virus Phylogenies. Genome Biol Evol 10, 657–666. 10.1093/gbe/evx273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moi D., Bernard C., Steinegger M., Nevers Y., Langleib M., and Dessimoz C. (2023). Structural phylogenetics unravels the evolutionary diversification of communication systems in gram-positive bacteria and their viruses. Preprint at bioRxiv, 10.1101/2023.09.19.558401 10.1101/2023.09.19.558401. [DOI] [Google Scholar]
- 42.Caroline Puente-Lelievre Ashar J. Malik, Douglas Jordan, Ascher David, Baker Matthew, Allison Jane, Anthony M Poole Daniel Lundin, Fullmer Matthew, Bouckaert Remco, et al. (2023). Tertiary-interaction characters enable fast, model-based structural phylogenetics beyond the twilight zone. bioRxiv, 2023.12.12.571181. 10.1101/2023.12.12.571181. [DOI] [Google Scholar]
- 43.Kennedy M.S., and Chang E.B. (2020). The microbiome: composition and locations. Prog Mol Biol Transl Sci 176, 1–42. 10.1016/bs.pmbts.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu P., Alves J.M., Kitten T., Brown A., Chen Z., Ozaki L.S., Manque P., Ge X., Serrano M.G., Puiu D., et al. (2007). Genome of the Opportunistic Pathogen Streptococcus sanguinis. J Bacteriol 189, 3166–3175. 10.1128/JB.01808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mylonakis E., and Calderwood S.B. (2001). Infective Endocarditis in Adults. New England Journal of Medicine 345, 1318–1330. 10.1056/NEJMra010082. [DOI] [PubMed] [Google Scholar]
- 46.Koonin E.V., Dolja V.V., Krupovic M., and Kuhn J.H. (2021). Viruses Defined by the Position of the Virosphere within the Replicator Space. Microbiology and Molecular Biology Reviews 85, e00193–20. 10.1128/MMBR.00193-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Symons R.H. (1991). The intriguing viroids and virusoids: what is their information content and how did they evolve? Mol Plant Microbe Interact 4, 111–121. 10.1094/mpmi-4-111. [DOI] [PubMed] [Google Scholar]
- 48.Bushmanova E., Antipov D., Lapidus A., and Prjibelski A.D. (2019). rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 8, giz100. 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin Y., Xu T., Lin W., Jia Q., He Q., Liu K., Du J., Chen L., Yang X., Du F., et al. (2020). Reference-free and de novo Identification of Circular RNAs. Preprint at bioRxiv, 10.1101/2020.04.21.050617 10.1101/2020.04.21.050617. [DOI] [Google Scholar]
- 50.Edgar R.C. (2022). circuclust. github.com/rcedgar/circuclust [Google Scholar]
- 51.Edgar R.C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.Ayad L.A.K., and Pissis S.P. (2017). MARS: improving multiple circular sequence alignment using refined sequences. BMC Genomics 18, 86. 10.1186/s12864-016-3477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leinonen R., Sugawara H., and Shumway M. (2011). The Sequence Read Archive. Nucleic Acids Res 39, D19–D21. 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.NCBI, S.T.D.T. (2022). SRA Toolkit. Version 3.0.0. [Google Scholar]
- 55.Chen S., Zhou Y., Chen Y., and Gu J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., and Hofacker I.L. (2011). ViennaRNA Package 2.0. Algorithms for Molecular Biology 6, 26. 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nawrocki E.P., and Eddy S.R. (2013). Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935. 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., and Madden T.L. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421. 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood D.E., Lu J., and Langmead B. (2019). Improved metagenomic analysis with Kraken 2. Genome Biology 20, 257. 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinto Y., Chakraborty M., Jain N., and Bhatt A.S. (2023). Phage-inclusive profiling of human gut microbiomes with Phanta. Nat Biotechnol, 1–12. 10.1038/s41587-023-01799-4. [DOI] [PubMed] [Google Scholar]
- 61.Zheludev I.N. (2022). FireTools. github.com/Zheludev/FireTools. [Google Scholar]
- 62.Lu J., Breitwieser F.P., Thielen P., and Salzberg S.L. (2017). Bracken: estimating species abundance in metagenomics data. PeerJ Comput. Sci. 3, e104. 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 63.Jeffares D. (2019). Workshop 4: Calling and filtering SNPs and indels. https://www-users.york.ac.uk/~dj757/popgenomics/workshop4.html. [Google Scholar]
- 64.Vasimuddin Md., Misra S., Li H., and Aluru S. (2019). Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. In 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS), pp. 314–324. 10.1109/IPDPS.2019.00041. [DOI] [Google Scholar]
- 65.Broad I. (2022). Picard Tools - By Broad Institute. http://broadinstitute.github.io/picard/. [Google Scholar]
- 66.Garrison E., and Marth G. (2012). Haplotype-based variant detection from short-read sequencing. Preprint at arXiv, 10.48550/arXiv.1207.3907 10.48550/arXiv.1207.3907. [DOI] [Google Scholar]
- 67.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garrison E. (2022). ekg/bamaddrg. [Google Scholar]
- 69.Zheng X., Levine D., Shen J., Gogarten S.M., Laurie C., and Weir B.S. (2012). A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328. 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.adel922 (2019). Working with VCF files and Trees. https://rpubs.com/adel922/560260. [Google Scholar]
- 71.R, C.T. (2022). R: A language and environment for statistical computing. (R Foundation for Statistical Computing; ). [Google Scholar]
- 72.Abu-Ali G.S., Mehta R.S., Lloyd-Price J., Mallick H., Branck T., Ivey K.L., Drew D.A., DuLong C., Rimm E., Izard J., et al. (2018). Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol 3, 356–366. 10.1038/s41564-017-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petersen L.M., Bautista E.J., Nguyen H., Hanson B.M., Chen L., Lek S.H., Sodergren E., and Weinstock G.M. (2017). Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 5, 98. 10.1186/s40168-017-0320-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Brady A., Jones C., Song Y., Darton T.C., Jones C., Blohmke C.J., Pollard A.J., Magder L.S., Fasano A., et al. (2018). Compositional and Functional Differences in the Human Gut Microbiome Correlate with Clinical Outcome following Infection with Wild-Type Salmonella enterica Serovar Typhi. mBio 9, e00686–18. 10.1128/mBio.00686-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richter T.K.S., Michalski J.M., Zanetti L., Tennant S.M., Chen W.H., and Rasko D.A. (2018). Responses of the Human Gut Escherichia coli Population to Pathogen and Antibiotic Disturbances. mSystems 3, e00047–18. 10.1128/mSystems.00047-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peters B.A., Wilson M., Moran U., Pavlick A., Izsak A., Wechter T., Weber J.S., Osman I., and Ahn J. (2019). Relating the gut metagenome and metatranscriptome to immunotherapy responses in melanoma patients. Genome Medicine 11, 61. 10.1186/s13073-019-0672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sinha S.R., Haileselassie Y., Nguyen L.P., Tropini C., Wang M., Becker L.S., Sim D., Jarr K., Spear E.T., Singh G., et al. (2020). Dysbiosis-Induced Secondary Bile Acid Deficiency Promotes Intestinal Inflammation. Cell Host & Microbe 27, 659–670.e5. 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campbell S.J., Ashley W., Gil-Fernandez M., Newsome T.M., Giallonardo F.D., Ortiz-Baez A.S., Mahar J.E., Towerton A.L., Gillings M., Holmes E.C., et al. (2020). Red fox viromes across an urban-rural gradient. Preprint at bioRxiv, 10.1101/2020.06.15.153858 10.1101/2020.06.15.153858. [DOI] [Google Scholar]
- 79.Maghini D.G., Dvorak M., Dahlen A., Roos M., Kuersten S., and Bhatt A.S. (2023). Quantifying bias introduced by sample collection in relative and absolute microbiome measurements. Nat Biotechnol, 1–11. 10.1038/s41587-023-01754-3. [DOI] [PubMed] [Google Scholar]
- 80.Buchfink B., Xie C., and Huson D.H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 81.Edgar R.C. (2022). Muscle5: High-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat Commun 13, 6968. 10.1038/s41467-022-34630-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eddy S.R. (2011). Accelerated Profile HMM Searches. PLOS Computational Biology 7, e1002195. 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hyatt D., Chen G.-L., LoCascio P.F., Land M.L., Larimer F.W., and Hauser L.J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meng G. (2022). msaconverter. github.com/linzhi2013/msaconverter [Google Scholar]
- 85.Remmert M., Biegert A., Hauser A., and Söding J. (2012). HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9, 173–175. 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 86.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., and Sternberg M.J.E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10, 845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holm L. (2022). Dali server: structural unification of protein families. Nucleic Acids Research 50, W210–W215. 10.1093/nar/gkac387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.van Kempen M., Kim S.S., Tumescheit C., Mirdita M., Gilchrist C.L.M., Söding J., and Steinegger M. (2022). Foldseek: fast and accurate protein structure search. Preprint at bioRxiv, 10.1101/2022.02.07.479398 10.1101/2022.02.07.479398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hernandez I.B., Yeo J., Jänes J., Wein T., Varadi M., Velankar S., Beltrao P., and Steinegger M. (2013). Clustering predicted structures at the scale of the known protein universe. Preprint at bioRxiv, 10.1101/2023.03.09.531927 10.1101/2023.03.09.531927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Henikoff S., and Henikoff J.G. (1992). Amino acid substitution matrices from protein blocks. Proceedings of the National Academy of Sciences 89, 10915–10919. 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bodenhofer U., Bonatesta E., Horejš-Kainrath C., and Hochreiter S. (2015). msa: an R package for multiple sequence alignment. Bioinformatics 31, 3997–3999. 10.1093/bioinformatics/btv494. [DOI] [PubMed] [Google Scholar]
- 92.Pagès H., Aboyoun P., Gentleman R., DebRoy S., Carey V., Delhomme N., Ernst F., Lakshman A., O’Neill K., Obenchain V., et al. (2023). Biostrings: Efficient manipulation of biological strings. Version 2.68.1 (Bioconductor version: Release (3.17)). 10.18129/B9.bioc.Biostrings 10.18129/B9.bioc.Biostrings. [DOI] [Google Scholar]
- 93.Wagih O. (2017). ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics 33, 3645–3647. 10.1093/bioinformatics/btx469. [DOI] [PubMed] [Google Scholar]
- 94.Tumescheit C., Firth A.E., and Brown K. (2022). CIAlign: A highly customisable command line tool to clean, interpret and visualise multiple sequence alignments. PeerJ 10, e12983. 10.7717/peerj.12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., and Lanfear R. (2020). IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution 37, 1530–1534. 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., and Jermiin L.S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14, 587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., and Vinh L.S. (2018). UFBoot2: Improving the Ultrafast Bootstrap Approximation. Molecular Biology and Evolution 35, 518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Letunic I., and Bork P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49, W293–W296. 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.PyPy, T. (2019). PyPy. https://www.pypy.org/. [Google Scholar]
- 100.Avinery R., Kornreich M., and Beck R. (2019). Universal and Accessible Entropy Estimation Using a Compression Algorithm. Phys. Rev. Lett. 123, 178102. 10.1103/PhysRevLett.123.178102. [DOI] [PubMed] [Google Scholar]
- 101.Katz P. (1989). ZIP. (PKWare). [Google Scholar]
- 102.Rice P., Longden I., and Bleasby A. (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16, 276–277. 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 103.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44, D733–745. 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Edgar R.C. (2007). PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformatics 8, 18. 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bland C., Ramsey T.L., Sabree F., Lowe M., Brown K., Kyrpides N.C., and Hugenholtz P. (2007). CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 8, 209. 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stothard P. (2000). The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28, 1102, 1104. 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 107.Bernhart S.H., Hofacker I.L., Will S., Gruber A.R., and Stadler P.F. (2008). RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics 9, 474. 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gu Z., Gu L., Eils R., Schlesner M., and Brors B. (2014). circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 109.Weinberg Z., and Breaker R.R. (2011). R2R - software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12, 3. 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., Griffiths-Jones S., Toffano-Nioche C., Gautheret D., Weinberg Z., et al. (2021). Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Research 49, D192–D200. 10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rivas E. (2020). RNA structure prediction using positive and negative evolutionary information. PLOS Computational Biology 16, e1008387. 10.1371/journal.pcbi.1008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jacobs J.P., Lagishetty V., Hauer M.C., Labus J.S., Dong T.S., Toma R., Vuyisich M., Naliboff B.D., Lackner J.M., Gupta A., et al. (2023). Multi-omics profiles of the intestinal microbiome in irritable bowel syndrome and its bowel habit subtypes. Microbiome 11, 5. 10.1186/s40168-022-01450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tong F., Tang G., and Wang X. (2023). Characteristics of Human and Microbiome RNA Profiles in Saliva. RNA Biol 20, 398–408. 10.1080/15476286.2023.2229596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saldanha J.A., Thomas H.C., and Monjardino J.P. (1990). Cloning and sequencing of RNA of hepatitis delta virus isolated from human serum. Journal of General Virology 71, 1603–1606. 10.1099/0022-1317-71-7-1603. [DOI] [PubMed] [Google Scholar]
- 115.Gross H.J., Domdey H., Lossow C., Jank P., Raba M., Alberty H., and Sänger H.L. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273, 203–208. 10.1038/273203a0. [DOI] [PubMed] [Google Scholar]
- 116.Bussière F., Ouellet J., Côté F., Lévesque D., and Perreault J.P. (2000). Mapping in Solution Shows the Peach Latent Mosaic Viroid To Possess a New Pseudoknot in a Complex, Branched Secondary Structure. Journal of Virology 74, 2647–2654. 10.1128/JVI.74.6.2647-2654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson A.D. (2010). An extended IUPAC nomenclature code for polymorphic nucleic acids. Bioinformatics 26, 1386–1389. 10.1093/bioinformatics/btq098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code and tabular summaries are available at the Stanford Digital Repository (purl.stanford.edu/wb363nt3637).