Abstract
Equine infectious anemia virus (EIAV) is a lentivirus with in vivo cell tropism primarily for tissue macrophages; however, in vitro the virus can be adapted to fibroblasts and other cell types. Tropism adaptation is associated with both envelope and long terminal repeat (LTR) changes, and findings strongly suggest that these regions of the genome influence cell tropism and virulence. Furthermore, high levels of genetic variation have been well documented in both of these genomic regions. However, specific EIAV nucleotide or amino acid changes that are responsible for cell tropism changes have not been identified. A study was undertaken with the highly virulent, macrophage-tropic strain of virus EIAVwyo to identify LTR changes associated with alterations in cell tropism. We found the stepwise generation of a new transcription factor binding motif within the enhancer that was associated with adaptation of EIAV to endothelial cells and fibroblasts. An LTR that contained the new motif had enhanced transcriptional activity in fibroblasts, whereas the new site did not alter LTR activity in a macrophage cell line. This finding supports a previous prediction that selection for new LTR genetic variants may be a consequence of cell-specific selective pressures. Additional investigations of the EIAVwyo LTR were performed in vivo to determine if LTR evolution could be detected over the course of a 3-year infection. Consistent with previous in vivo findings, we observed no changes in the enhancer region of the LTR over that time period, indicating that the EIAVwyo LTR was evolutionarily stable in vivo.
Genetic variation is a hallmark of retroviruses that is believed to be a consequence of the absence of proofreading activity of the virally encoded enzyme reverse transcriptase. Selective forces that act to alter, limit, or diversify the viral population are well documented (42). For instance, human immunodeficiency virus (HIV) variants in response to antiviral drugs or neutralizing antibodies rapidly arise (5, 11, 30). Sequence variation throughout the HIV long terminal repeat (LTR) between and within different clades of virus is documented; however, in general, nucleotide variation is limited within the HIV enhancer/promoter proximal region that usually consists of two NF-κB and three Sp1 sites (52). In contrast to HIV, the LTR enhancer region of equine infectious anemia virus (EIAV) is one of the most variable regions of the EIAV genome in tissue culture isolates and different field isolates (6, 25, 36, 43). A study that examined seven in vivo isolates demonstrated that 45% of the nucleotide positions within the LTR enhancer varied between the isolates whereas little to no variation was found in the rest of the LTR (36). Similar levels of enhancer variation can be found in different tissue culture isolates. These enhancer region alterations included point mutations, insertions, and deletions in the roughly 90-bp region (33).
EIAV LTR sequences influence the tropism of the virus (43). While EIAV expression in macrophages utilizes a repertoire of transcription factors to initiate transcription that includes the macrophage-specific factor PU.1, EIAV expression in fibroblasts is driven by a series of other, more generic transcription factors such as AML-1, Oct-1, and ATF-1 (32, 34). In some cases, the enhancer motifs that are occupied in either macrophages or fibroblasts physically overlap and transcription factor motifs that optimize LTR expression in a macrophage-specific manner may not be present in a fibroblast-specific strain of virus and vice versa (36). These observations have led us to propose that in tissue culture, alterations in the EIAV enhancer region may result from cell-specific utilization of different transcription factors (33). Consistent with this, long-term adaptation in tissue culture has been frequently associated with changes in the LTR enhancer (6, 43). While adaptation of EIAV to new cell types in tissue culture, such as fibroblasts, has been a useful approach to facilitate the maintenance of virus stocks and study the molecular biology of the virus (20, 21), fibroblast tropism of the virus is associated with loss of both in vivo virulence (41) and the ability of the adapted strain to replicate in macrophages (7). Serial passages of fibroblast-tropic strains in horses can reconstitute mild virulence (40, 41). The LTR enhancer regions of these mildly virulent readapted strains differ from that found in both highly virulent strains of virus, as well as fibroblast-tropic strains (36).
To understand the evolutionary selection of LTR changes associated with changes in cell tropism, the LTR enhancer region within a stock of EIAV was monitored during passage through primary macrophages, endothelial cells, and fibroblasts. Serial passage through these three types of cells was performed since this approach has been used in the past to generate tissue culture-adapted EIAV (7, 28) and resulting tissue culture-adapted viral populations are found to have highly variable sequences present in the LTR enhancer region (6). Sequence evolution was observed in the LTR, but not the adjacent TM region, over the course of our passage study. The LTR sequence that was fixed within our virus population contained a new transcription factor binding motif. The altered LTR increased the transcription capacity in fibroblasts, providing direct evidence that the LTR enhancer evolves in parallel with cell tropism changes. Additionally, we investigated whether LTR enhancer evolution was detected in vivo in an experimental EIAV infection that was monitored over 3 years as the viral infection changed from acutely viremic to inapparent. In this in vivo study, the consensus sequence of the EIAV LTR U3 did not change over this time course, suggesting that rapid evolution of the LTR of virulent strains of EIAV does not occur in vivo.
MATERIALS AND METHODS
Primary cells and cell lines. Monocyte-derived macrophages (MDMs) were purified from peripheral equine blood from EIAV-seronegative horses as previously described (32). MDMs were maintained in high-glucose DMEM with 15% horse serum and 15% fetal calf serum (FCS) and penicillin/streptomycin (pen/strep). Approximately 106 MDMs were plated in each well of a 48-well cluster and infected with EIAV within 24 h of cell isolation. Umbilical cord endothelial cells (UVECs) were isolated from equine umbilical cords by trypsinization as previously described (35). UVECs were expanded, and aliquots of cells were frozen. Thawed UVECs were maintained in high-glucose DMEM with 40% FCS and pen/strep and passaged twice each week.
Three cell lines that support EIAV replication were used in these studies. Canine fibroblastic line Cf2Th (ATCC CRL1430) was used for reporter gene assays. DH82 cells (ATCC CRL10389), a canine histiocytic sarcoma cell line, were also used in reporter gene assays. Equine dermal fibroblasts (ED) (ATCC CCL57) were used in the cell tropism studies. All cell lines were maintained in high-glucose DMEM with 10% FCS and pen/strep.
EIAV inoculum.
The inoculum used for the in vitro and in vivo studies was plasma obtained from an initial fever episode from an EIAV (Wyoming strain)-infected horse (horse 2078) as previously described (3, 39). Stocks of EIAVwyo2078 plasma contained approximately 108 horse infectious doses of virus/ml as previously described (3, 39).
Reverse transcriptase (RT) PCR amplification of EIAV sequences.
Virus particles were pelleted from equine plasma, serum, or tissue culture medium at an average relative centrifugal force of 76,000 × g for 1 h at 4°C. mRNA was extracted from particles by particle lysis, followed by mRNA binding to an oligo(dT) column using the Fast Track kit (Invitrogen). The column was washed extensively, and mRNA was eluted from the column. Negative-strand cDNA was synthesized from viral mRNA using either EIAV U3-specific antisense primer MunIC′ (AGTGCCCAATTGTCAG) or a random primer supplied by Invitrogen. From the cDNA, a 450-bp product consisting of the 3′ terminus of TM and LTR U3 was amplified with Taq polymerase using 35 cycles of 92°C for 30s, 50°C for 30s, and 72°C for 1.5 min, followed by a 4-min extension at 72°C on the last cycle using primers 7333 (CCCAAGAAGGAACTCTCGCT) and MunIC′. All amplifications were performed in duplicate or triplicate to obtain independent amplification replicates.
Evolutionary analysis of EIAV clones.
Amplified sequences were ligated into pGEM-T (Promega), and competent Escherichia coli JM109 cells were transformed with the ligation mixture. Clones that produced appropriately sized restriction fragments were sequenced in both the sense and antisense directions using T7 and the Sp6 primers present within the pGEM-T vector. Six to 22 individual clones were analyzed from each in vitro passage or in vivo time point. From each time point, clones obtained from at least two separate amplifications were included in the analysis.
EIAV sequences were trimmed to remove the primer sequences and any plasmid DNA sequences. The clones amplified from each passage were aligned using the algorithm ClustalW multiple alignment program in the software package BioEdit (16, 53). Estimation of effective population size was done using the program FLUCTUATE (22). Nucleotide diversity (51) (using the Kimura 2 parameter model) and divergence (1) (using the Kimura 2 parameter model) were done using the program MEGA2 (23). The software package DNASP 3.51 was also used to perform additional evolutionary tests (49). Substitution rate was calculated by dividing the total number of substitutions in the population at a certain time point compared to the inoculum consensus sequence by the total number of nucleotide positions at the time point. The resultant number was divided by the number of generations the virus had undertaken from the beginning of the study. The number of viral generations was estimated by dividing the total number of days in passage by the viral generation time as previously described (50). Trees were created using MEGA version 2.1 (23) using the neighbor-joining method with the Kimura 2 parameter model for nucleotide substitution (1).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as previously described (32). Nuclear extract (NE) from MDM, endothelial, and ED cells were generated by the procedure of Dignam et al. and stored in modified buffer D (50 mM HEPES [pH 8.0], 80 mM KCl, 1 mM MgCl2, 0.5 mM dithiothreitol, 15% glycerol, 0.5 mM phenylmethylsulfonyl fluoride) at −80°C until use (14). Sense and antisense synthetic oligonucleotides containing LTR nucleotide sequences −91 to −75 relative to the start of transcription were designed with 5′ overhanging ends. 32P-labeled and unlabeled blunt-ended oligonucleotides were generated by fill-in reactions using the Klenow fragment of E. coli DNA polymerase and the addition of appropriate deoxynucleotides. Twenty thousand counts per minute of 32P-labeled probe was used in each EMSA reaction, as well as 5 to 10 μg of NE. Blunt-ended competitor oligonucleotides were used as noted in Results. Binding reactions in 80 mM KCl and 1 mM MgCl were performed at room temperature for 15 to 30 min, and bands were separated on a 1× Tris/borate/EDTA-6% acrylamide gel at 200 V. Gels were fixed, dried, and visualized by autoradiography.
Generation of LTR/reporter gene constructs and chloramphenicol acetyltransferase (CAT) assays.
The LTR was amplified from proviral sequences present in tissue from an experimentally infected horse. Primers 7606 Xho (GGTTTCTCGAGGGGTTTTATAAATG) and Xba 323C′ (TCTAGAGTAGGATCTCGAACA) were used for amplification, which allowed us to clone the full-length LTR into a modified pCATBasic (Promega) vector containing XhoI and XbaI sites. Mutagenesis of the EIAVwyo LTR/CAT plasmid was performed to generate EIAVfibrowyo LTR/CAT that contained two nucleotide changes at positions −82 and −84 relative to the start of transcription. Using the QuikChange strategy for mutagenesis (Stratagene), sense (GCTAGGCAACTAACCTGCAATAACCGGAAGTTCCTCAATATAGTTCCGCATTTGTG) and antisense (CACAAATGCGGAACTATATTGAGGAACTTCCGGTTATTGCAGGTTAGTTGCCTAGC) primers were synthesized that contained the two nucleotide changes. Mutagenesis was performed by 12 rounds of amplification, followed by DpnI digestion to cleave parental sequences. Colonies were isolated and DNA sequencing was performed to verify the incorporation of the mutations into the LTR.
The LTR/CAT constructs were transfected into either DH82 or Cf2Th cells using Gene Porter (Gene Therapy Systems) as previously described (37). One microgram of LTR/CAT plasmid, 0.5 μg of eTat plasmid (13), and 0.5 μg of cytomegalovirus-driven β-galactosidase (β-gal) plasmid were transfected in triplicate. Cells were lysed 40 to 48 h posttransfection, and cell lysates were normalized for β-gal activity. Cell lysates were analyzed for CAT activity using [14C]chloramphenicol as previously described (38). Transfections were performed at least three times in each cell line, and CAT values represent the mean and standard error of the percent acetylation/0.1 mU of β-gal activity/h.
Analysis of TM/LTR U3 sequences in experimentally infected horses.
One thousand horse infectious units (HIU) of EIAVwyo2078 was inoculated into pony 524. Details of the clinical course of EIAV infection of pony 524 have been previously described (3, 39). Following experimental inoculation of EIAVwyo, serum and plasma samples from pony 524 were collected and frozen at −80°C at each febrile spike, as well as periodically over the course of the 3-year infection. Plasma samples from days 12, 39, 67, 201, 216, 289, 385, 437, 567, 832, and 878 postinfection (p.i.) were thawed and ultracentrifuged at an average relative centrifugal force of 76,000 × g for 1 h at 4°C to pellet virus. RNA was extracted from virions as described above, and RT-PCR was performed using the oligonucleotides and conditions described above for the in vitro study. Viral sequences were analyzed as described for the in vitro passage study.
LTR U3 sequences from three additional horses that were infected with EIAVwyo2078 were analyzed. Horse 2084 was inoculated with 105 HIU of EIAVwyo2078 and was necropsied during the first acute viremic episode at day 15 p.i. Horse 2085 was also inoculated with 105 HIU of EIAVwyo2078 and progressed through a typical acute and chronic EIA profile with severe acute fever and thrombocytopenia from day 7 to day 30, followed by persistent thrombocytopenia and viremia in the absence of a recurrent fever. Horse 2085 was necropsied on day 124 p.i. Horse 586 was inoculated with 101 HIU of EIAVwyo2078 and had both an acute and three chronic fever episodes accompanied by thrombocytopenia. At necropsy on day 386 p.i., horse 586 was afebrile and had normal platelet counts. Total RNA was extracted from samples of spleen with a commercial RNA extraction kit (Trizol Reagent, Life Technologies) in accordance with the manufacturers directions, and cDNA was synthesized with random hexamer primers. The LTR U3 sequences were then PCR amplified with the 7333 and MunIC′ primers as described above. The amplicons were cloned into the pCR2.1 sequencing vector (TOPO TA cloning kit; Invitrogen), and strands from a single clone each were sequenced in both directions by automated dideoxy DNA methods.
RESULTS
Generation of EIAV with altered cell tropism.
To determine if LTR U3 region changes correlated with the expansion of EIAV cell tropism, a virulent, macrophage-tropic strain of EIAV (EIAVwyo2078) that was obtained during an initial viremic episode of an experimentally infected horse was serially passaged onto MDMs, endothelial cells, and fibroblasts (Fig. 1a). A 450-bp fragment of the EIAV genome was amplified. A total of 437 bp were analyzed, encompassing 217 bp of gp45 or TM, 17 untranslated nucleotides following the stop codon of env, and the LTR U3. This region was monitored for changes over the course of passage (Fig. 1b).
FIG. 1.
(A) Protocol used to alter the cell tropism of EIAV. A highly virulent, macrophage-tropic strain of EIAV (EIAVwyo) was used to infect primary MDMs. Supernatants were collected and passaged into fresh MDMs nine additional times. Concurrently, unsuccessful attempts were made to passage EIAVwyo stocks into primary equine endothelial cells (UVECs) and equine fibroblast cells (ED cells). Upon 10 MDM passages, viral stocks replicated in UVECs but the stocks still did not replicate in ED cells. Following 10 passages in UVECs, stocks were able to replicate in ED cells. 3′ TM and the LTR U3 sequences were RT-PCR amplified at passages noted and were analyzed as described below. (B) The 450-bp region of the EIAV genome amplified. This region encompassed the last 217 bp of TM, a 17-nucleotide spacer, and the 202 bp of the LTR U3.
A well-characterized stock of EIAV (Wyoming strain) (EIAVwyo2078) was used as the starting point in this in vitro study and is referred to as the inoculum. Cloning and sequencing of RT-PCR-amplified fragments of the TM/LTR U3 sequences present in the EIAVwyo2078 stock demonstrated that the inoculum contained little sequence variation in both the 3′ terminus of the TM and the LTR U3. Of the 17 clones of the inoculum analyzed, 9 out of 7,174 positions were variable, for an overall variance of 0.125% (supplemental Fig. S1A).
To determine if the level of nucleotide variation we observed within the inoculum differed from the error rate generated by Taq polymerase during the amplification, we determined the specific error rate for our primers under our amplification condition using a molecular clone of EIAV (p29A). p29A was amplified using the same primers and conditions used in the evolution study, and the amplified DNA was cloned into pGEM-T. Fourteen clones were sequenced. Seven errors in a total of 5,966 nucleotides were observed, indicating a rate of 0.117% for our amplification conditions. Since similar levels of variation were observed in our amplified clones that were generated from either the inoculum stock or from a molecular clone, we concluded that this region of the genome present in the EIAVwyo2078 stock contained similar levels of variation to that of a virus stock generated from a molecular clone.
To initiate the study, freshly isolated MDM cultures were infected in duplicate with approximately 108 HIU of EIAVwyo2078. Supernatants were collected daily, and supernatant RT assays were performed to monitor the infection. MDM supernatants from the first in vitro passage were negative for RT activity, so the collected supernatants were pooled and ultracentrifuged and the pellet was resuspended and blind passaged onto fresh MDMs. Supernatant RT positivity was detectable in the second and all subsequent MDM passages. Supernatants containing the greatest quantity of RT activity (usually day 5, 6, or 7 p.i.) were used as viral stocks for each subsequent round of MDM infection. A total of 10 MDM passages were performed.
UVEC and ED cells were infected with equivalent quantities of the EIAVwyo2078 inoculum in parallel with the first round of MDM infection. As with the initial infection of MDMs, passage of these stocks onto UVEC or ED cells did not produce RT-positive supernatants. However, unlike MDM cultures, two subsequent rounds of ultracentrifuge-concentrated supernatant passage onto either UVECs or fibroblasts did not result in detectable virus replication (data not shown), indicating that the EIAVwyo2078 stock did not replicate in the UVEC or ED populations. The ability of a stock of EIAVwyo to replicate in MDMs, but not endothelial cells or fibroblasts, is consistent with previously reported findings (46).
From the 10th passage of virus in MDMs (macP10 virus), RT-positive supernatants were collected and ultracentrifuged to concentrate the virus and added to either UVEC or ED cells. MacP10 virus replicated on UVECs as detected by both supernatant RT positivity and viral antigen immunostaining of infected UVEC cultures (data not shown). In contrast, macP10 virus replication was not detected on ED cells in parallel repeated attempts. Virus was passaged 10 times on UVECs. Peak RT activity within the supernatants arose more slowly in UVEC cultures than in MDMs with peak activity between days 10 and 14 p.i. Supernatants from the 10th UVEC passage (endoP10 virus) were passaged onto ED cell cultures, and supernatant RT positivity was detected in the first and all subsequent ED cell passages. Five additional rounds of passage in ED were performed.
Evolution of TM and LTR U3 sequences during virus passage.
As with the inoculum, the 3′ TM/LTR U3 region of the viral genome was analyzed for variation over the course of passage. This region was RT-PCR amplified from RT-positive supernatants from MDM passages 3, 6, and 10, UVEC passages 1, 3, 6, and 10, and ED passages 1, 3, and 6 (Fig. 1b). Two or three independent amplifications of each time point were performed. Amplified DNA was cloned and sequenced. One hundred thirty-three clones were analyzed over the course of the cell passages (supplemental Fig. S1A shows all of the sequences utilized in this study), and 101 unique clones were identified. Thirty-five clones varied in both TM and the LTR U3 region, 36 clones varied only in TM, and 30 clones varied only in the U3. Single nucleotide changes were primarily responsible for the genetic variation, although nucleotide insertions and deletions were also observed. Variation present in the population during the passages was analyzed by investigating both the quasispecies diversity within each passage and the divergence of the passage populations from the EIAVwyo2078 inoculum (Fig. 2a and b). Similar trends in variation were observed within the LTR U3 and TM. The relative homogeneity of the inoculum sequences diversified and diverged during macrophage passage 3 to passage 6. This was followed by restriction of genetic diversity and divergence occurring at macP10. Thereafter, diversity remained relatively constant during the passages in endothelial cells and fibroblasts. Divergence also occurred at a relatively constant rate within the population of TM sequences over the remaining passages, with somewhat greater divergence observed in the LTR sequences.
FIG. 2.
Analysis of the quasispecies populations over the course of virus passage. (a) Diversity plot demonstrating the genetic variation present within a population. (b) Divergence plot showing percent change in the quasispecies population from that of the inoculum population. (c) Substitutions/site/generation analysis in each passage. (d) Ratio of nonsynonymous to synonymous changes found in the carboxy-terminal 72 amino acids of TM.
Estimates of the substitution rate/site/generation showed a high rate of mutation during early macrophage passages (Fig. 2c). The substitution rate spiked during the sixth passage in MDM but leveled off subsequently, remaining relatively constant for the remainder of the study. Consistent with the trends found in substitution rates, analysis of the ratio of nonsynonymous to synonymous substitutions of amino acids in the TM demonstrates that in early macrophage passages more nonsynonymous changes are evident in the population than synonymous changes, suggesting that selection of genetic diversification of the population is occurring (Fig. 2d) (15). This trend was followed by an absence of amino acid change at macP10. Subsequent passages fluctuated between diversification of the protein sequence (NS/S ∼2) and purification or restriction of the sequence. In terms of sequence evolution, these findings suggest that an initial diversifying selection occurred that was acting upon the small initial population, followed by stochastic mechanisms directing evolution as the virus was passaged into endothelial cells and fibroblasts. Similar patterns of early, rapid genetic diversification have been previously observed in HIV-infected individuals (26).
Fixation of the sequence changes within the population.
While nucleotide diversity and divergence were observed in the carboxy TM terminus, this variation was not fixed in the population. As a result, the consensus sequence of this region of TM remained unchanged over the course of the experiment (Fig. 3a). The absence of change as the cell tropism of the virus occurred indicated that changes in the carboxy TM terminus are not required for alterations in EIAV cell tropism. Consistent with the TM sequence variation not being fixed within the population, rooted trees of the TM sequence were highly branched and short (trees are available in supplemental Fig. S2).
FIG. 3.
Consensus sequences of the LTR U3 change as the cell tropism of the virus changes, but the carboxy terminal TM sequences do not. (A) Consensus deduced amino acid sequences of the TM. (B) Consensus nucleotide sequences for the LTR U3 region. Inoc, inoculum.
In contrast to the TM sequences, four nucleotide changes within the LTR U3 were fixed in the population (Fig. 3b). An initial change of C to T was observed at position −123 (C-123T) relative to the start of transcription in the macP3 quasispecies population. Four out of 12 clones contained the change. The C-123T change was found in 100% of the clones in the macP6 population and remained present and predominant in the population throughout the remainder of the study. A second nucleotide change from T to G at position −85 (T-85G) was first observed in the endoP1 population, where 3 of 15 clones contained the change. In endoP3 and all subsequently isolated sequences, the T-85G change was present in all of the clones. By endoP10, a T-to-A transition at position −83 (T-83A) was fixed in the population. In the final ED cell passage (fibroP6), half of the clones also contained a G-to-C change at position −45. The G-to-C change at position −45 has been previously identified in some fibroblast-tropic EIAV isolates (43).
Nucleotide changes that were fixed within the population resulted in introduction of a novel transcription factor binding site.
LTR changes that we observed with alteration of cell tropism of the virus were assessed for their impact on LTR function. We chose to look at the first three changes that took place in the LTR (C-123T, T-85G, and T-83A) since the functional implication of the fourth change (G-45C) has been explored previously (43). To determine if the upstream C-to-T change at nucleotide −123 altered levels of LTR activity, we introduced a C-123T mutation into the EIAVwyo LTR and into an EIAVwyo LTR that contained T-85G and T-83A. The C-123T change in DH82 (macrophages) or Cf2Th (fibroblasts) did not alter the transcriptional activity of either LTR in either cell type (data not shown). Thus, it was concluded that the −123 substitution did not impact the activity of the LTR.
Next, we explored the binding and transcriptional consequences of the nucleotide changes at positions −85 and −83. The two changes produced a DNA sequence (−89AACCGGA−83) that resembled an AML (PEA-2 or CBP) binding motif (AACCGCA) in six out of seven positions (Fig. 4a). AML motifs have previously been identified in numerous tissue culture isolates of EIAV (8, 9, 34) but not in virulent strains of EIAV such as EIAVwyo (36). In most EIAV isolates that contain an AML site, the site is located at positions −98 to −92, slightly upstream to the location of the mutations noted here. However, a more promoter-proximal AML site resides at −89 to −83 in several Wyoming-derived tissue culture isolates that contain two AML motifs (55). To determine if the mutations at positions −85 and −83 resulted in the gain or loss of a transcription factor binding motif during viral passage, EMSAs were performed using 32P-labeled oligonucleotides containing the EIAVwyo sequence AACCTGT (Wyo), the intermediate AACCGGT motif (Wyoendo) that contains the T-85G change, or the AACCGGA (Wyofibro) motif found in endoP10 and the fibroblast-passaged virus (Fig. 4b). In all three sets of oligonucleotides used in these EMSA studies, the oligonucleotides contained changes (from TTCCTCAA to TTCCGGGA) that eliminated the downstream Ets (PU.1) binding site. We have previously found that when PU.1 protein is present in NEs, binding to the Ets site masks binding to adjacent sites (data not shown). Thus, mutations that have been shown to eliminate PU.1 binding (32) were introduced into the Ets site. Using NEs from equine endothelial cells, equine fibroblasts (ED cells) or a macrophage cell line, DH82, a band was retarded within the gel with both Wyo and Wyoendo probes; however, both specific and nonspecific cold competitor oligonucleotides competed for the binding to equivalent levels, indicating that the binding was nonspecific (data not shown). Specific binding was observed to a 32P-labeled Wyofibro oligonucleotide using all three NEs, as demonstrated by the ability of specific competitor to eliminate binding but the inability of equivalent concentrations of nonspecific competitor to compete (Fig. 4c and d). Unlabeled competitor Wyo oligonucleotide did not compete for Wyofibro binding, and competitor Wyoendo oligonucleotide was able to compete slightly at the highest concentration of competitor. The binding pattern observed with macrophage NE differed from that observed with endothelial or fibroblast NE with a series of additional more rapidly migrating bands (Fig. 4d). Interestingly, competitor Wyo and Wyoendo oligonucleotides did compete to some degree for these faster-migrating bands, suggesting that macrophage NEs contain proteins that bind to a site present in all three oligonucleotides (Wyo, Wyoendo, and Wyofibro).
FIG. 4.
Alternations in the EIAV enhancer generate a new transcription factor binding motif. (A) Alignment of several in vivo and in vitro EIAV enhancer regions identifying the motifs frequently found within the EIAV enhancer. (B) Sequences of oligonucleotides used as probes and competitors in EMSAs. (C) Binding of endothelial and fibroblast (ED cell) NEs to the WyoFibro probe. Increasing concentrations of unlabeled Wyofibro competitor (Comp.) oligonucleotide eliminated binding of endothelial NE, whereas similar molar concentrations of the Wyo oligonucleotide were unable to compete. The highest concentrations of the Wyoendo oligonucleotide competed slightly for binding. The three right-hand lanes show a darker exposure of the ED NE binding. (D) Binding of fibroblast and macrophage NEs to the WyoFibro probe. Approximately 200-fold excesses of competitor oligonucleotides (27.5 to 30 pmol) were used to compete for binding. The arrow in panel C indicates specific band retardation.
To define the new motif, a series of competing mutant oligonucleotides were tested against the Wyofibro probe (Fig. 5a). The inability of mutant oligonucleotides 2 to 5 to compete for the retarded bands indicated that the nucleotides CCGGAAGT were important for the DNA/protein interactions (Fig. 5b). This sequence had similarities to both an AML site and an Ets core motif. However, neither oligonucleotides (105 and 111) that contained a consensus AML motif nor oligonucleotides (124 and 163) that contained a core Ets motif were able to compete for binding (Fig. 5c). Our studies suggest that the evolution of the enhancer has generated a binding site not previously recognized within the EIAV LTR.
FIG. 5.
The binding motif of the site generated during tissue culture passage is CCGGAAGT. (A) Sequences of unlabeled oligonucleotides that were mutated at different locations across the Wyofibro oligonucleotide were used as competitors (Comp.) in EMSAs. (B) EMSA competition studies using the mutant Wyofibro oligonucleotides as competitors and UVEC NEs. (C) Competition EMSAs using oligonucleotides containing a consensus AML site (oligonucleotides 105 and 111) (34) or an Ets site that had been shown previously to bind to Ets transcription factor PU.1 (oligonucleotide 124 or 163) (32). ED NEs were used in this EMSA. All competitor oligonucleotides (27.5 −30 pmol) were in approximately 200-fold excess over the probe.
New U3 motif enhanced LTR activity in fibroblasts.
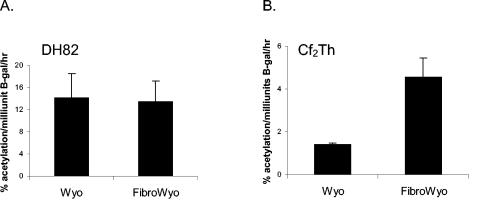
To determine if the new binding site resulted in altered transcription activity of the LTR in macrophages or fibroblasts, the site was introduced into an EIAVwyo LTR/CAT construct, generating EIAVwyofibro. In the macrophage cell line, DH82, the EIAVwyo LTR and the EIAVwyofibro LTRs had indistinguishable levels of activity in the presence of EIAV Tat (Fig. 6). In contrast, in fibroblasts the EIAVwyofibro LTR had higher levels of LTR activity than the EIAVwyo LTR, with an average of 3.25-fold higher activity than the EIAVwyo LTR in the presence of Tat. In total, these findings indicated that changes at LTR positions −85 and −83 generated a new binding motif that enhanced the transcriptional activity of the LTR in fibroblasts but not in macrophages.
FIG. 6.
The new enhancer motif results in enhanced LTR activity in fibroblasts but not in macrophages. LTR/CAT constructs were transfected into either DH82 or Cf2Th cells, and cell lysates were analyzed for CAT activity at 40 h posttransfection.
The LTR enhancer alterations were not observed in an experimentally infected pony.
Our in vitro passage results indicated that LTR U3 sequences evolve as the cell tropism of the virus changes. A previous study from our lab had demonstrated that LTR U3 sequences isolated from healthy, EIAV-seropositive horses differed from LTRs found in EIAV-infected horses that were acutely viremic at necropsy (36). Furthermore, LTRs from some healthy, seropositive horses did not contain transcription factor binding motifs that are important for transcription in macrophages. Our in vitro passage results in combination with the results from the earlier in vivo study suggested the possibility that LTR evolution might be observed over the course of in vivo EIAV infection as the infection evolved from frankly viremic to subclinical. To investigate this, pony 524 was experimentally inoculated with 103 HIU of EIAVwyo2078 and the infection was monitored for approximately 3 years for fever and thrombocytopenia (Fig. 7a) (3, 39). Pony 524 was frankly viremic with accompanying fever and thrombocytopenia during an initial episode of EIAV and chronically cycled with viremia over subsequent months. Fever episodes abated during year 2, but the animal became chronically thrombocytopenic with ongoing detectable viremia. Several fever spikes were detected as late as day 799 p.i., and the pony was necropsied on day 878, when the pony was weakly viremic and afebrile and had normal platelet counts. Virion RNA was amplified by RT-PCR from plasma samples taken 12 times over the course of the infection using primers 7333 and MunIC′ that were used in our in vitro study that amplified the 3′ terminus of TM and the LTR U3 (arrows in Fig. 7a denote LTR sampling times). As was observed in the in vitro passage study, the relatively homogeneous inoculum population of LTR U3 sequences expanded rapidly during the initial fever episode (day 12), with high rates of nucleotide substitutions (Fig. 7b) (supplemental Fig. S1B contains all of the sequences isolated at each passage). While diversity remained high over the course of the infection, by day 67, despite ongoing viremic episodes, substitution rates dropped to levels reported for HIV and divergence from the inoculum sequence remained lower than the diversity throughout the infection. Interestingly, changes in the analysis parameters did not appear to correlate with the disease state (thrombocytopenia or fever) of the infected pony.
FIG. 7.
The consensus sequence of the EIAVwyo LTR U3 does not change over long term experimental infections of horses. (A) Clinical course of experimentally infected Pony 524. The bars indicate time points when elevated body temperatures were observed. Platelet counts/μl of blood over the course of infection are also shown (♦). Arrows at the top indicate time points where TM/LTR sequences were sampled in this study. (B) Evolutionary analysis of the pony 524 TM/LTR U3 clones demonstrated high rates of substitution (▴) during early infection but not at later time points. Nucleotide diversity and divergence fluctuate over the course of the infection. (C) Consensus sequence of the LTR U3 from pony 524 at 12 time points p.i. Bolded nucleotides denote sequence changes from the consensus sequence. (D) Consensus sequence of the LTR U3 in four EIAVwyo-infected horses at time of necropsy. All horses were infected with a Wyo2078 stock. Horse 2084 was necropsied at 15 days p.i., when acutely febrile. Horse 2085 was necropsied at 124 days p.i., following acute viremia and chronic thrombocytopenia. Horse 586 was necropsied at 586 days p.i., following acute viremia and three subsequent fever episodes. Horse 586 and pony 524 were afebrile and had normal platelet counts at necropsy. Bolded bases denote nucleotides that differ from that of the consensus (Con) sequence. Inoc, inoculum.
Despite the nucleotide diversity that was observed over the course of the infection, the consensus sequence of the TM (data not shown) and the LTR U3 did not evolve (Fig. 7c). LTR U3 consensus sequences for days 67 and 567 differed slightly from the consensus sequences present at other time points, but these differences did not predominate long term in the quasispecies population. Rooted tree analysis also demonstrated an absence of evolution of the sequences (supplemental Fig. S3). Consistent with the stability of the EIAVwyo LTR U3 in pony 524 over a 3-year period, the LTR U3 consensus sequences RT-PCR amplified from plasma from three other experimentally EIAVwyo2078-inoculated horses were found to be identical to that of pony 524 and the inoculum consensus sequence (Fig. 7d). These findings indicate that while LTR U3 variation can be observed in field isolates and in vitro isolates (6, 36), detectable in vivo evolution of LTR U3 sequences from a virulent Wyoming strain of virus was not evident. This observation is consistent with previous findings (47). Furthermore, the absence of change within the consensus sequence suggests that while the LTR U3 population may diversify at points during the infection, the consensus sequence seems to be highly suited for the environment and cell type infected during this longitudinal study.
DISCUSSION
Previous studies have retrospectively identified alterations in EIAV LTR enhancer sequences that are associated with long-term passage in macrophages or leukocyte cultures (56) or fibroblasts (6, 25, 27, 43). These changes include loss and gain of transcription factor binding motifs (36). Some studies have identified extensive EIAV LTR enhancer changes including multiple nucleotide insertions or deletions (27, 57), whereas other studies have found more modest changes (6, 43). LTR alterations that are associated with fibroblast tropism have been correlated with the alteration of LTR activity in a cell-specific manner (43, 57); however, other reports suggest that the LTR changes found in tissue culture-derived isolates are not associated with cell-specific activity in vitro (25).
This study was undertaken to prospectively investigate the changes in the LTR U3 that occur as cell tropism of the virus is altered. We investigated intra- and interpassage variation, as well as changes in consensus sequences over time. High levels of nucleotide variation were observed as measured by nucleotide diversity and divergence in early macrophage passages in both the 3′-terminal TM sequences and the LTR U3. These levels were about five times higher than that reported for HIV in a single-cycle analysis (29, 31). However, it should be noted that genetic variation studies on the equivalent region of the HIV genome have not been extensively explored and levels of variation can be influenced by the region of the genome under study (24). Concomitant with a burst of genetic diversification in macrophages, a C-to-T change upstream from the enhancer region in the LTR U3 was fixed as the predominant sequence within the population. This change had no detectable effect on LTR transcription in our reporter gene studies. Almost no variation was detected in macP10 sequences, but variation slowly increased over the remaining passages in UVECs and ED cells. The slow increase in divergence during these passages suggested random genetic drift of a small population (19). As the viral stock was passaged through UVECs and fibroblasts that established a broader cell tropism, two more nucleotide changes were fixed within the LTR U3 population that created a new transcription factor binding motif that enhanced LTR activity in fibroblasts but not macrophages. The generation of a new motif within the adapting LTR has been previously demonstrated in HIV (54). A final mutation found in the last passage in fibroblast was a G-to-C change immediately upstream from a PU.1 site that has been reported earlier in fibroblast-tropic strains of virus (43). While the overall consensus changes observed in the LTR were modest, some of the genotypic changes that became predominant in the population resulted in phenotypic changes that correlated well with the observed cell tropism alterations.
EIAVwyo during the initial passage in MDMs replicated poorly, but by MDM passage 2 replication became robust. This finding suggests that EIAVwyo replication in MDMs in tissue culture is not analogous to replication in vivo in tissue macrophages and virus must overcome undefined replication barriers during early MDM passages that are not present in tissue macrophages in vivo. Since we did not observe changes in the LTR U3 during early macrophage passages, alterations of other regions such as SU may have been responsible for the initial in vitro adaptation. A study similar to this one investigating changes in those sequences and the role those changes play in cell tropism would be insightful. The SU has been implicated in the cell tropism of virtually all other retroviruses and is certainly the principal determinant of cell tropism in HIV (for recent reviews, see references 10 and 18). While Perry et al. have shown that SU sequences by themselves are not sufficient to control cell tropism of EIAV (46), these sequences are important virulence determinants (12, 44, 45) and would be predicted to control cell tropism in conjunction with the LTR and potentially other genes in the 3′ half of the genome.
Our studies do not directly address the source of the LTR variation. It is possible that the LTR sequences that became fixed as the predominant population at later time points were present in the inoculum but represented a minor fraction of the population that we did not identify during our PCR amplification and cloning of the inoculum sequences. Alternatively, the changes may have arisen de novo over the course of the passage as point mutations. If this latter possibility occurred, the mutations could have been generated during early passages or could have been generated over the passages in a stepwise manner. Our findings favor the stepwise development of the changes since we only see each of the mutations in the passage immediately prior to the mutation becoming the predominant variant in the population; however, the other alternatives cannot be excluded.
LTR evolution resulted in a new binding motif between an methylation-dependent binding protein site at the 5′ border of the enhancer and an Ets binding site that in macrophages is known to bind to PU.1 but is not bound in fibroblasts (17, 32, 34). While the presence of the new site in fibroblasts increased LTR activity, it did not enhance activity in DH82 cells, a macrophage cell line. From our EMSA studies, the shifting pattern in macrophages differed from that observed in fibroblasts or endothelial cells. This altered pattern indicates that a different set of proteins may be binding to the probe and it may be the binding of a different protein(s) that accounts for the altered LTR activity. Alternatively, in macrophages, a physical competition for binding may be occurring between the newly created site and the PU.1 binding site that resides immediately downstream and partially overlaps the new site within the enhancer (17, 32). Since PU.1 is not present in endothelial cells or fibroblasts, such competition would not exist in these cells.
The region of TM that was amplified in this study encodes the carboxy-terminal 78 amino acids of the cytoplasmic tail. Previous studies mapping the cell tropism of other retroviruses have not implicated this region in tropism determination. Thus, it is not surprising that this region of TM remained unchanged in our studies. Alternatively, we and others have reported that most of the cytoplasmic tail of TM can be lost during in vitro passage (37, 48), suggesting the possibility that large amounts of genetic variation might be tolerated in this region of the genome. In addition, we suspected that this portion of env might be variable because in vivo studies with EIAVwyo found that an adjacent upstream region of TM has significant genetic variation (2-4).
While a previous study found extensive LTR enhancer variation between different field isolates of EIAV (36), the EIAVwyo LTR was stable during a longitudinal study in an experimentally infected pony, consistent with other in vivo investigations of the EIAVwyo LTR (47). These findings suggest that rapid evolution of EIAVwyo LTR sequences does not occur within an infected horse. Furthermore, the observation that the EIAVwyo LTR is found in multiple infected horses and that the sequences do not significantly deviate from inoculum sequences indicate that the LTR U3wyo sequences are highly stable both within a horse and between infected horses. Thus, our current findings demonstrate the stability of the Wyoming-based LTR in vivo. This study does not account for the high level of LTR enhancer variation observed between field isolates that were observed in our previous study (36). Further exploration of field isolate LTR sequences may provide insights into both the level of enhancer variation with the field population and the mechanisms by which enhancer diversification occurs.
Supplementary Material
Acknowledgments
We kindly thank Charles Grose for helpful comments on the manuscript.
This work was supported in part by NIH grants CA72063 (W.J.M.) and AI144638 (J.L.O.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Anderson, J. P., A. G. Rodrigo, G. H. Learn, Y. Wang, H. Weinstock, M. L. Kalish, K. E. Robbins, L. Hood, and J. I. Mullins. 2001. Substitution model of sequence evolution for the human immunodeficiency virus type 1 subtype B gp120 gene over the C2-V5 region. J. Mol. Evol. 53:55-62. [DOI] [PubMed] [Google Scholar]
- 2.Baccam, P., R. J. Thompson, Y. Li, W. O. Sparks, M. Belshan, K. S. Dorman, Y. Wannemuehler, J. L. Oaks, J. L. Cornette, and S. Carpenter. 2003. Subpopulations of equine infectious anemia virus Rev coexist in vivo and differ in phenotype. J. Virol. 77:12122-12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belshan, M., P. Baccam, J. L. Oaks, B. A. Sponseller, S. C. Murphy, J. Cornette, and S. Carpenter. 2001. Genetic and biological variation in equine infectious anemia virus Rev correlates with variable stages of clinical disease in an experimentally infected pony. Virology 279:185-200. [DOI] [PubMed] [Google Scholar]
- 4.Belshan, M., M. E. Harris, A. E. Shoemaker, T. J. Hope, and S. Carpenter. 1998. Biological characterization of Rev variation in equine infectious anemia virus. J. Virol. 72:4421-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkhout, B. 1999. HIV-1 evolution under pressure of protease inhibitors: climbing the stairs of viral fitness. J. Biomed. Sci. 6:298-305. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, S., S. Alexandersen, M. J. Long, S. Perryman, and B. Chesebro. 1991. Identification of a hypervariable region in the long terminal repeat of equine infectious anemia virus. J. Virol. 65:1605-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter, S., and B. Chesebro. 1989. Change in host cell tropism associated with in vitro replication of equine infectious anemia virus. J. Virol. 63:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carvalho, M., and D. Derse. 1993. Physical and functional characterization of transcriptional control elements in the equine infectious anemia virus promoter. J. Virol. 67:2064-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho, M., M. Kirkland, and D. Derse. 1993. Protein interactions with DNA elements in variant equine infectious anemia virus enhancers and their impact on transcriptional activity. J. Virol. 67:6586-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clapham, P. R., J. D. Reeves, G. Simmons, N. Dejucq, S. Hibbitts, and A. McKnight. 1999. HIV coreceptors, cell tropism and inhibition by chemokine receptor ligands. Mol. Membr. Biol. 16:49-55. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, J. 2003. HIV. Escape artist par excellence. Science 299:1505-1508. [DOI] [PubMed] [Google Scholar]
- 12.Cook, R. F., C. Leroux, S. J. Cook, S. L. Berger, D. L. Lichtenstein, N. N. Ghabrial, R. C. Montelaro, and C. J. Issel. 1998. Development and characterization of an in vivo pathogenic molecular clone of equine infectious anemia virus. J. Virol. 72:1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derse, D., P. L. Dorn, L. Levy, R. M. Stephens, N. R. Rice, and J. W. Casey. 1987. Characterization of equine infectious anemia virus long terminal repeat. J. Virol. 61:743-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldman, N., and Yang, Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11:725-736. [DOI] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Hines, R., B. R. Sorensen, M. A. Shea, and W. Maury. 2004. PU.1 binding to Ets motifs within the equine infectious anemia virus long terminal repeat (LTR) enhancer: regulation of LTR activity and virus replication in macrophages. J. Virol. 78:3407-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman, T. L., and R. W. Doms. 1999. HIV-1 envelope determinants for cell tropism and chemokine receptor use. Mol. Membr. Biol. 16:57-65. [DOI] [PubMed] [Google Scholar]
- 19.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, United Kingdom.
- 20.Klevjer-Anderson, P., W. P. Cheevers, and T. B. Crawford. 1979. Characterization of the infection of equine fibroblasts by equine infectious anemia virus. Arch. Virol. 60:279-289. [DOI] [PubMed] [Google Scholar]
- 21.Kono, Y., and Y. Yokomizo. 1968. Attempts to cultivate the equine infectious anemia virus in various types of cells. Natl. Inst. Anim. Health Q. (Tokyo) 8:182-186. [PubMed] [Google Scholar]
- 22.Kuhner, M. K., J. Yamato, and J. Felsenstein. 1995. Estimating effective population size and mutation rate from sequence data using Metropolis-Hastings sampling. Genetics 140:1421-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 24.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lichtenstein, D. L., J. K. Craigo, C. Leroux, K. E. Rushlow, R. F. Cook, S. J. Cook, C. J. Issel, and R. C. Montelaro. 1999. Effects of long terminal repeat sequence variation on equine infectious anemia virus replication in vitro and in vivo. Virology 263:408-417. [DOI] [PubMed] [Google Scholar]
- 26.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madden, C. R., and D. S. Shih. 1996. Analysis of the long terminal repeat from a cytopathic strain of equine infectious anemia virus. Virology 225:395-399. [DOI] [PubMed] [Google Scholar]
- 28.Malmquist, W. A., D. Barnett, and C. S. Becvar. 1973. Production of equine infectious anemia antigen in a persistently infected cell line. Arch. Gesamte Virusforsch. 42:361-370. [DOI] [PubMed] [Google Scholar]
- 29.Mansky, L. M. 1996. Forward mutation rate of human immunodeficiency virus type 1 in a T lymphoid cell line. AIDS Res. Hum. Retroviruses 12:307-314. [DOI] [PubMed] [Google Scholar]
- 30.Mansky, L. M. 2002. HIV mutagenesis and the evolution of antiretroviral drug resistance. Drug Resist. Updates 5:219-223. [DOI] [PubMed] [Google Scholar]
- 31.Mansky, L. M., and H. M. Temin. 1995. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J. Virol. 69:5087-5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maury, W. 1994. Monocyte maturation controls expression of equine infectious anemia virus. J. Virol. 68:6270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maury, W. 1998. Regulation of equine infectious anemia virus expression. J. Biomed. Sci. 5:11-23. [DOI] [PubMed] [Google Scholar]
- 34.Maury, W., S. Bradley, B. Wright, and R. Hines. 2000. Cell specificity of the transcription-factor repertoire used by a lentivirus: motifs important for expression of equine infectious anemia virus in nonmonocytic cells. Virology 267:267-278. [DOI] [PubMed] [Google Scholar]
- 35.Maury, W., J. L. Oaks, and S. Bradley. 1998. Equine endothelial cells support productive infection of equine infectious anemia virus. J. Virol. 72:9291-9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maury, W., S. Perryman, J. L. Oaks, B. K. Seid, T. Crawford, T. McGuire, and S. Carpenter. 1997. Localized sequence heterogeneity in the long terminal repeats of in vivo isolates of equine infectious anemia virus. J. Virol. 71:4929-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maury, W., P. J. Wright, and S. Bradley. 2003. Characterization of a cytolytic strain of equine infectious anemia virus. J. Virol. 77:2385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maury, W. J., S. Carpenter, K. Graves, and B. Chesebro. 1994. Cellular and viral specificity of equine infectious anemia virus Tat transactivation. Virology 200:632-642. [DOI] [PubMed] [Google Scholar]
- 39.Oaks, J. L., T. C. McGuire, C. Ulibarri, and T. B. Crawford. 1998. Equine infectious anemia virus is found in tissue macrophages during subclinical infection. J. Virol. 72:7263-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Rourke, K. I., L. E. Perryman, and T. C. McGuire. 1989. Cross-neutralizing and subclass characteristics of antibody from horses with equine infectious anemia virus. Vet. Immunol. Immunopathol. 23:41-49. [DOI] [PubMed] [Google Scholar]
- 41.Orrego, A., C. J. Issel, R. C. Montelaro, and W. V. Adams, Jr. 1982. Virulence and in vitro growth of a cell-adapted strain of equine infectious anemia virus after serial passage in ponies. Am. J. Vet. Res. 43:1556-1560. [PubMed] [Google Scholar]
- 42.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 43.Payne, S. L., K. La Celle, X. F. Pei, X. M. Qi, H. Shao, W. K. Steagall, S. Perry, and F. Fuller. 1999. Long terminal repeat sequences of equine infectious anaemia virus are a major determinant of cell tropism. J. Gen. Virol. 80(Pt. 3):755-759. [DOI] [PubMed] [Google Scholar]
- 44.Payne, S. L., X. F. Pei, B. Jia, A. Fagerness, and F. J. Fuller. 2004. Influence of long terminal repeat and env on the virulence phenotype of equine infectious anemia virus. J. Virol. 78:2478-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Payne, S. L., J. Rausch, K. Rushlow, R. C. Montelaro, C. Issel, M. Flaherty, S. Perry, D. Sellon, and F. Fuller. 1994. Characterization of infectious molecular clones of equine infectious anaemia virus. J. Gen. Virol. 75(Pt. 3):425-429. [DOI] [PubMed] [Google Scholar]
- 46.Perry, S. T., M. T. Flaherty, M. J. Kelley, D. L. Clabough, S. R. Tronick, L. Coggins, L. Whetter, C. R. Lengel, and F. Fuller. 1992. The surface envelope protein gene region of equine infectious anemia virus is not an important determinant of tropism in vitro. J. Virol. 66:4085-4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reis, J. K., J. K. Craigo, S. J. Cook, C. J. Issel, and R. C. Montelaro. 2003. Characterization of EIAV LTR variability and compartmentalization in various reservoir tissues of long-term inapparent carrier ponies. Virology 311:169-180. [DOI] [PubMed] [Google Scholar]
- 48.Rice, N. R., L. E. Henderson, R. C. Sowder, T. D. Copeland, S. Oroszlan, and J. F. Edwards. 1990. Synthesis and processing of the transmembrane envelope protein of equine infectious anemia virus. J. Virol. 64:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 50.Seo, T. K., J. L. Thorne, M. Hasegawa, and H. Kishino. 2002. Estimation of effective population size of HIV-1 within a host: a pseudomaximum-likelihood approach. Genetics 160:1283-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simm, M., W. Chao, O. Pekarskaya, P. Sova, P. Gupta, R. Balachandran, and D. J. Volsky. 1996. Genetic variability and function of the long terminal repeat from syncytium-inducing and non-syncytium-inducing human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 12:801-809. [DOI] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verhoef, K., R. W. Sanders, V. Fontaine, S. Kitajima, and B. Berkhout. 1999. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-κB enhancer element into a GABP binding site. J. Virol. 73:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whetter, L., D. Archambault, S. Perry, A. Gazit, L. Coggins, A. Yaniv, D. Clabough, J. Dahlberg, F. Fuller, and S. Tronick. 1990. Equine infectious anemia virus derived from a molecular clone persistently infects horses. J. Virol. 64:5750-5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y. H., H. Sentsui, Y. Kono, and K. Ikuta. 2000. Mutations occurring during serial passage of Japanese equine infectious anemia virus in primary horse macrophages. Virus Res. 68:93-98. [DOI] [PubMed] [Google Scholar]
- 57.Zheng, Y. H., H. Sentsui, M. Sugita, T. Nakaya, M. Kishi, K. Hagiwara, Y. Inoshima, C. Ishihara, Y. Kono, J. L. Lu, and K. Ikuta. 2000. Replication ability in vitro and in vivo of equine infectious anemia virus avirulent Japanese strain. Virology 266:129-139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.