Abstract
Recently it has been shown that kaposica, an immune evasion protein of Kaposi's sarcoma-associated herpesvirus, inactivates complement by acting on C3-convertases by accelerating their decay as well as by acting as a cofactor in factor I-mediated inactivation of their subunits C3b and C4b. Here, we have mapped the functional domains of kaposica. We show that SCRs 1 and 2 (SCRs 1-2) and 1-4 are essential for the classical and alternative pathway C3-convertase decay-accelerating activity (DAA), respectively, while the SCRs 2-3 are required for factor I cofactor activity (CFA) for C3b and C4b. SCR 3 and SCRs 1 and 4, however, contribute to optimal classical pathway DAA and C3b CFA, respectively. Binding data show that SCRs 1-4 and SCRs 1-2 are the smallest structural units required for measuring detectable binding to C3b and C4b, respectively. The heparin-binding site maps to SCR 1.
The complement system is an essential arm of innate immunity, providing the first line of defense against a repertoire of invading microorganisms, including viruses (6, 20). Invasion by viral predators leads to activation of the complement system in a cascade-dependent manner via three activation pathways, classical, alternative, and lectin, and results in efficient elimination of viruses as a consequence of opsonization, phagocytosis, and lysis (3, 13). Since viruses solely depend on their host for survival and propagation, coevolution of viruses along with their host has led to the development of various mechanisms to evade the host complement attack. Viruses known to subvert the complement system include the poxviruses, herpesviruses, retroviruses, paramyxoviruses, and picornaviruses (1, 7, 14, 27).
Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8 (HHV-8) is the most recently discovered human tumor virus belonging to the gammaherpesvirus family. KSHV DNA has been found in all the epidemiological forms of Kaposi's sarcoma and has thus been implicated in the pathogenesis of Kaposi's sarcoma. It is also associated with two lymphoproliferative disorders, body cavity-based B-cell lymphoma, or primary effusion lymphoma, and a subset of multicentric Castleman's disease (16, 18). The KSHV genome and its encoded gene products have been identified in vivo in human B cells, macrophages, endothelial cells, epithelial cells, and keratinocytes (26). Sequencing of the KSHV genome revealed the presence of 80 complete open reading frames (ORFs) encoded within it, with several of them having significant homology to human cellular genes captured during virus evolution (23). The sequence of ORF 4 was strikingly similar to the human complement control proteins belonging to the regulators of complement activation (RCA) family (16, 23). The members of this family are characterized by the presence of conserved elemental units termed the complement control protein domain (CCP) or short consensus repeat (SCR). Each SCR is composed of ∼60 amino acids with four invariant cysteines linked by two disulfide bonds, an invariant tryptophan, and highly conserved prolines, glycines, and other hydrophobic residues, which together fold into a bead-like structure. Multiple SCRs are separated by linkers of 2 to 7 residues, giving the proteins a “beads-on-a-string”-like appearance. The RCA proteins regulate complement by two different mechanisms: (i) by accelerating the irreversible dissociation of the C3-convertases and (ii) by serving as cofactors for the serine protease factor I-mediated cleavage of C3b and C4b (the subunits of C3-convertases) (9, 21, 25).
The KSHV ORF 4 (1,650 bp) encodes a protein that contains four extracellular SCRs followed by a dicysteine motif, a serine/threonine (S/T)-rich region, and a transmembrane region. The molecule contains three potential N-linked and several O-linked carbohydrate sites (Fig. 1). Analysis of posttranscriptional processing of ORF 4 suggested that, in addition to the unspliced mRNA, two spliced transcripts are produced. Both of them contain the transmembrane region; however, they lack either the S/T region or the dicysteine motif and the S/T region (31). In our previous study, we have expressed the soluble form of this protein (SCRs 1-4 without other regions) using the Pichia expression system and assigned a function to this protein (19). The purified KSHV ORF 4 protein (molecular mass, 56,000 Da) inhibited human complement-mediated lysis of erythrocytes, blocked cell surface deposition of C3b, and served as a cofactor for factor I-mediated inactivation of C3b and C4b. Based on its function, the protein was named as kaposica (the Kaposi's sarcoma-associated herpesvirus inhibitor of complement activation) (19). In a parallel study, another group has shown that apart from factor I cofactor activity (CFA), this protein also possesses considerable decay-accelerating activity (DAA) for the classical C3-convertase (C4b,2a), but poor DAA for the alternative pathway C3-convertase (C3b,Bb) (30).
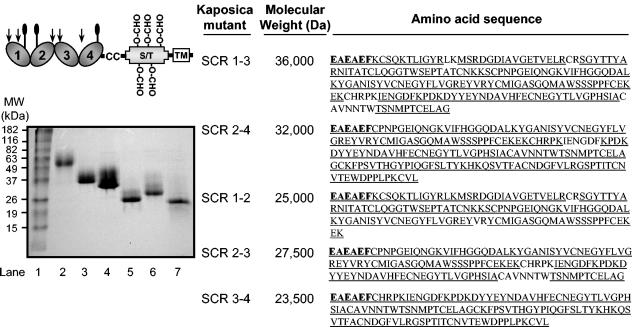
FIG. 1.
SDS-PAGE and sequence analysis of the purified kaposica mutants. (Left) Schematic representation of the structure of kaposica. It contains four N-terminal SCRs followed by a dicysteine motif, a serine/threonine (S/T) region, and a transmembrane (TM) region. The predicted N-linked and O-linked carbohydrate sites are indicated with balloons and O-CHO, respectively. The predicted O-linked carbohydrate sites within SCRs 1-4 are not marked. The putative heparin-binding sites with a characteristic motif of K/RXK/R (28) are indicated with arrows. Our data shows that the heparin-binding site is located in SCR 1. Kaposica mutants were cloned in P. pastoris and purified as described. The purified mutants were run on an 11% SDS-PAGE gel under reducing conditions and stained with Coomassie blue. Lane 1, molecular mass (MW) markers; lane 2, kaposica; lane 3, SCR 1-3; lane 4, SCR 2-4; lane 5, SCR 1-2; lane 6, SCR 2-3; and lane 7, SCR 3-4. For amplification of the respective deletion mutants, the following sequence-specific primers were used: SCR 1-3, AS-9 (5′ GGAATTCAAGTGTTCCCAAAAAACCTTAATTGG 3′) and AS-15 (5′ GCTCTAGATTAGCCTGCGAGTTCACAGGTTGG 3′); SCR 2-4, AS-17 (5′ GGAATTCTGTCCAAACCCAGGTGAAATAC 3′) and AS-10 (5′ GCTCTAGATTACAAAACACACTTAGGAAGTGG 3′); SCR 1-2, AS-9 and AS-16 (5′ GCTCTAGATTACTTTTCTTTTTCACAAAAAGGAGG 3′); and SCR 2-3, AS-17 and AS-15, and SCR 3-4, AS-18 (5′GGAATTCTGTCACAGACCGAAAATCGAAAATG 3′) and AS-10. The primers incorporated restriction sites EcoRI and XbaI (italicized) in the forward and the reverse orientations, respectively, and a stop codon (indicated in bold) in the reverse orientation primers. (Right) Amino acid sequences of kaposica mutants were determined by mass spectrometry. Bold amino acid residues indicate the residues that are added at the N terminus due to cloning. The underlined sequences on the figure indicate the sequences determined by mass spectrometry. The apparent molecular masses were determined by running the mutants on SDS-PAGE.
Since kaposica inhibits complement and provides a mechanism by which KSHV can subvert complement attack by the host, identification of its structural determinants important in its function would not only help in elucidating the complement regulatory mechanisms of kaposica, but would also lead to the identification of the target regions for future drug development. To identify the functional domains of kaposica, we have generated a series of deletion mutants and expressed them using the Pichia expression system. Importantly, we did not add the His tag which is normally added to aid purification, since addition of a His stretch might add a charge on the protein if the pKa of His is altered upon binding and may influence its activity. In brief, the recombinant kaposica constructs comprising SCRs 1-3, 2-4, 1-2, 2-3, and 3-4 were constructed from the full-length kaposica clone (19) by PCR amplification (primer combinations shown in the legend) and cloned into the yeast expression vector pPICZα (Invitrogen, Carlsbad, Calif.) at the EcoRI and XbaI sites downstream of the AOX1 methanol-inducible promoter. After sequencing the clones for their authenticity, each of the mutants was integrated into the Pichia genome as per the manufacturer's instruction (Pichia expression kit, version F; Invitrogen).
The mutant proteins were expressed and concentrated from the culture supernatants by ultrafiltration followed by ammonium sulfate precipitation (80%) and purified as given below. Kaposica mutants 1-2 and 1-3 were allowed to bind to heparin-agarose column (Sigma, St. Louis, Mo.) in 10 mM sodium phosphate buffer pH 7.4 and eluted using 500 mM NaCl in the above buffer. Mutants 2-3, 3-4, and 2-4, however, failed to bind to the heparin-agarose column and thus were purified using DEAE Sephacel (Sigma) in 10 mM sodium phosphate (pH 7.4). The proteins were eluted using a linear salt gradient from 0 to 250 mM. Based on the pI of the mutant proteins, they were subjected to further purification either on a Mono S or a Mono Q column (Pharmacia, Uppsala, Sweden). Kaposica 1-2 was purified using a Mono S column in 5 mM sodium acetate buffer (pH 4.0), and the protein was eluted by a linear salt gradient of 0 to 1 M. The other mutants (kaposica 1-3, 2-4, 2-3, and 3-4) were purified on a Mono Q column in 20 mM Tris (pH 8.0) and eluted using a linear salt gradient of 0 to 500 mM. The purified proteins ran as single bands on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1). To confirm the identity of the mutants, they were subjected to sequencing by mass spectrometry as previously described (11) at the Biomolecular Research Facility, University of Virginia, Charlottesville. The amino acid sequences of the expressed mutants were consistent with the predicted sequences confirming the identity of mutants (Fig. 1). The purified mutants were also verified for proper folding using size exclusion chromatography (Superose 12; Pharmacia). All the mutants ran as a monodisperse population (data not shown). These data along with preservation of various functions in mutants, which require different SCRs (described below), suggest that mutants have maintained proper conformation. The molecular masses of kaposica and the mutants 1-3, 2-4, 1-2, 2-3, and 3-4 on size exclusion chromatography were 120,000 Da, 71,000 Da, 56,000 Da, 46,000 Da, 42,000 Da, and 23,000 Da, suggesting that kaposica as well as its mutants except SCR 3-4, exist as dimers. The physiologic importance of dimerization in kaposica is not clear at present.
Previously, we have shown that like vaccinia virus complement control protein (VCP), kaposica also binds to heparin (19). From our purification data, it is clear that only SCR 1-3 and SCR 1-2 mutants retained the heparin-binding activity. Since the SCR 2-4 mutant did not bind to heparin, we conclude that the heparin-binding site is located in SCR 1 of kaposica. We have also verified these data by performing binding of purified kaposica mutants to heparin-agarose (data not shown). It is well known that interaction of human complement regulator factor H with heparin is important for regulation of the alternative pathway (17). Whether the heparin-binding site present on SCR 1 of kaposica plays any role in complement regulation is not known at present.
It is now clear that kaposica inactivates C3-convertases by acting as a cofactor in factor I-mediated inactivation of C3b and C4b, as well as by accelerating decay of the classical pathway (C4b,2a) and, to a limited extent, the alternative pathway (C3b,Bb) C3-convertases into their subunits (19, 30). The data presented in Fig. 2 and 3 show that SCRs 2-3 is the minimum region of kaposica required for factor I cofactor activity for C3b and C4b. Further, these data indicate that the presence of SCRs 1 and 4 are essential for providing optimal cofactor activity against C3b (Fig. 2). Previous data on mapping of the functional domains of human RCA proteins (factor H, membrane cofactor protein, complement receptor 1 and C4b-binding protein) suggested that a minimum of three SCRs are necessary for factor I cofactor activities for C3b and/or C4b (4, 8, 9). Hence kaposica is the first example where factor I cofactor activity is conferred by only two SCRs.
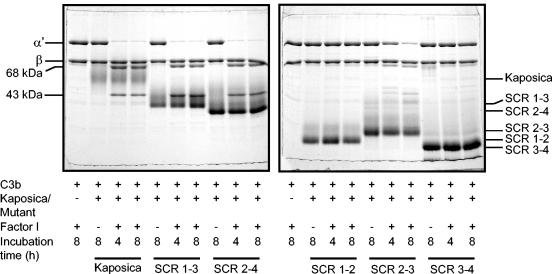
FIG. 2.
Analysis of factor I cofactor activity of kaposica and its mutants for complement protein C3b. Cofactor activity was determined by incubating C3b (2.5 μg) with kaposica (2 μg) or mutants (2 μg) and factor I (150 ng) in 15 μl of 10 mM sodium phosphate, pH 7.4, containing 145 mM NaCl at 37°C for the indicated time period. Cleavage products were visualized by running the samples on 10% SDS-PAGE gels and staining with Coomassie blue. During C3b cleavage, the α′-chain is cleaved into N-terminal 68-kDa and C-terminal 43-kDa fragments; appearance of these fragments indicates the generation of iC3b (inactive C3b).
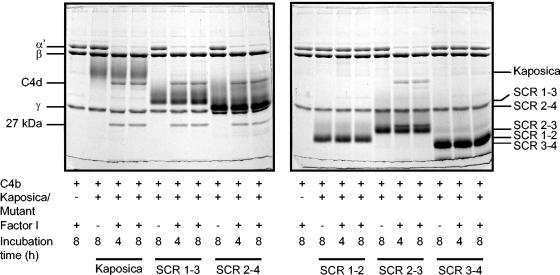
FIG. 3.
Analysis of factor I cofactor activity of kaposica and its mutants for complement protein C4b. Cofactor activity was determined by incubating C4b (3.2 μg) with kaposica (2 μg) or mutants (2 μg) and factor I (150 ng) in 15 μl of 10 mM sodium phosphate, pH 7.4, containing 145 mM NaCl at 37°C for the indicated time period. Cleavage products were visualized by running the samples on 10% SDS-PAGE and staining with Coomassie blue. During C4b cleavage, the α′-chain is cleaved into N-terminal 27-kDa, C-terminal 16-kDa (not visualized on the gel), and central C4d fragments; these cleavages result in inactivation of C4b and generation of C4c and C4d.
The effect of kaposica and its mutants on decay of the classical pathway (CP) C3-convertase was assessed by forming C4b,2a on sheep erythrocytes (15, 22). The data depicted in Fig. 4 shows that the CP DAA resides in SCRs 1-2 but SCR 3 also contributes to the optimal function. This data is consistent with the previous data on the requirement of multiple SCRs for CP DAA of human RCA proteins; in the case of decay-accelerating factor, it has been shown that SCRs 2-3 are required for CP DAA (5) while in complement receptor 1, SCRs 1-3 contribute to CP DAA (12).
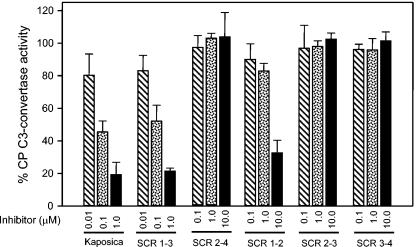
FIG. 4.
Classical pathway decay-accelerating activity of kaposica and its mutants. The classical pathway (CP) C3-convertase C4b,2a was formed on antibody-coated sheep erythrocytes, using purified C1, C4, and C2 (Calbiochem) (15, 22). The cells coated with C4b,2a were incubated with increasing concentrations of kaposica or its mutants at 22°C for 5 min, and the remaining C3-convertase activity was assayed by incubating the cells with guinea pig sera diluted 1:100 in dextrose gelatin Veronal buffer, pH 7.4, containing 40 mM EDTA. The data were normalized by setting 100% C3-convertase activity to be equal to the average activity in the absence of inhibitor (kaposica or its mutants).
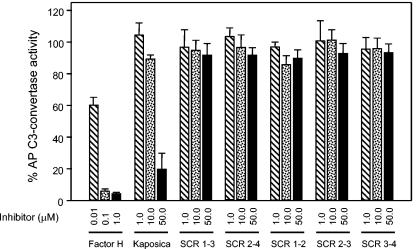
In comparison to human complement regulators, kaposica is a poor regulator of the alternative pathway C3-convertase C3b,Bb. Earlier, it was shown that it is at least 1,000-fold less active compared to human factor H and decay-accelerating factor (30). In the present study, we have compared alternative pathway DAA of kaposica with human factor H by forming C3-convertase on rabbit erythrocytes (15, 24). At 10 μM and 50 μM concentrations, kaposica caused 10.5% ± 2.5% and 80.1% ± 9.9% decay of the AP C3 convertase relative to the spontaneous decay in buffer (Fig. 5). The decay by factor H was 39.7% ± 4.9% at 0.01 μM and 94.7% ± 2.0% at a 0.1 μM concentration, indicating that kaposica is about 1,500-fold less active compared to factor H. Further, when kaposica mutants were tested for DAA of AP convertase, none showed any activity even at 50 μM (Fig. 5), indicating thereby that all four SCRs contribute to alternative pathway DAA.
FIG. 5.
Alternative pathway decay-accelerating activity of kaposica and its mutants. The alternative pathway (AP) C3-convertase C3b,Bb was formed on rabbit erythrocytes, using purified C3, factor B, and factor D in the presence of NiCl2 (15, 22). The cells coated with C3b,Bb were incubated with increasing concentrations of kaposica or its mutants at 37°C for 10 min, and the remaining C3-convertase activity was assayed by incubating the cells with human sera diluted 1:5 in gelatin Veronal buffer, pH 7.4, containing 20 mM EDTA. Factor H was used as a positive control. The data were normalized by setting 100% C3-convertase activity to be equal to the average activity in the absence of inhibitor (kaposica or its mutants).
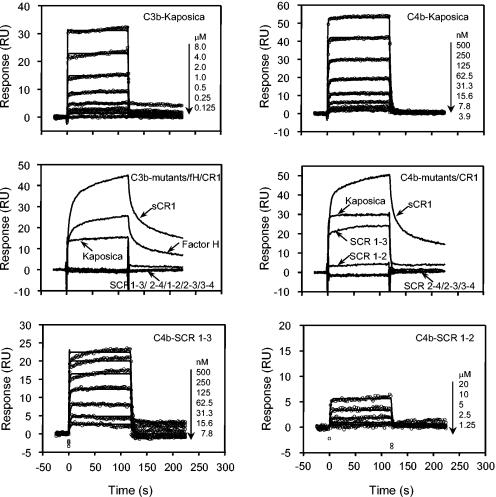
Even though ligand binding is a prerequisite for CFA and DAA, a large body of evidence indicates that ligand binding does not always correlate well with cofactor and decay activities (9). From the data presented above, it is clear that SCRs 1-2 and 1-4 are the minimum regions required for the CP DAA and AP DAA, respectively, while SCRs 2-3 are enough to confer CFA for C3b and C4b. In order to determine whether DAA and CFA of various mutants correlate with binding to C3b and C4b, we performed a quantitative analysis of binding of kaposica and its mutants to C3b and C4b using the surface plasmon resonance assay (Fig. 6). In this assay, C3b and C4b were oriented on a streptavidin chip by labeling their free -SH groups with biotin (2). This setup mimicked the physiological orientation of these proteins and also allowed accurate determination of the binding constants (2). Our data showed that like VCP-C3b/C4b interactions (2), interactions of kaposica with C3b and C4b also followed a simple 1:1 binding model (Fig. 6). It is important to point out here that, though kaposica ran as a dimer on size exclusion chromatography, it showed 1:1 interaction. It is possible that a binding site for C3b/C4b on one of the molecules is unavailable for binding due to sterically hindered access to the site. Interestingly, when deletion mutants were allowed to flow over the sensor chip, none of the deletion mutants showed binding to C3b and only SCR 1-2 and 1-3 mutants showed binding to C4b (Fig. 6). Thus, the requirement of all the four SCRs for AP DAA could be explained on the basis of binding data to C3b. Further analysis of binding of mutants to C4b showed that the SCR 1-3 mutant which showed comparable CP DAA to kaposica displayed similar affinity for C4b (Table 1), and the SCR 1-2 mutant which showed a 61-fold decrease in CP DAA showed a dramatic (263-fold) decrease in affinity for C4b. The data on CFA, however, did not correlate with binding data; the SCR 2-3 mutant which displayed CFA against both C3b and C4b showed no detectable binding either to C3b or to C4b. A prevailing model of factor I-mediated inactivation of C3b and C4b suggests that this process involves two steps: (i) recognition of C3b/C4b by the cofactor and (ii) interaction of factor I with C3b/C4b and the cofactor (29). Since the SCR 2-3 mutant did not show detectable binding either to C3b or to C4b, it is likely that the interaction of SCRs 2-3 with C3b/C4b is very weak and binding of factor I to C3b/C4b and SCRs 2-3 stabilizes this interaction.
FIG. 6.
Analysis of binding of kaposica and its mutants to immobilized C3b and C4b. C3b and C4b were oriented in their physiological orientation on a streptavidin chip (Sensor Chip SA; Biacore AB) by labeling their free -SH groups with biotin (2). Various concentrations of kaposica or the mutant proteins in PBS-T (10 mM sodium phosphate and 145 mM NaCl, pH 7.4, containing 0.05% Tween 20) were injected over a streptavidin chip, wherein Fc-1 served as a control flow cell and Fc-2 was immobilized with C3b (2,300 relative units [RU]) or C4b (1,130 RU). (Top panels) Sensogram overlays for binding of kaposica to immobilized C3b and C4b. (Middle panels) Binding of kaposica and the mutant proteins to C3b and C4b. Sensograms for binding to C3b were generated by injecting 2.0 μM kaposica or its mutants (20 μM each). Factor H (200 nM) and soluble complement receptor 1 (sCR1; 50 nM) were included as controls. Sensograms for binding to C4b were generated by injecting 125 nM kaposica, 500 nM SCR 1-3, 10 μM SCR 1-2, or 20 μM of SCR 2-4, 2-3, and 3-4 mutants. sCR1 was injected at 125 nM concentration. (Bottom panels) Sensogram overlays for binding of SCR 1-3 and SCR 1-2 mutants to immobilized C3b and C4b. Solid lines in the top and bottom panels represent the global fitting of the data to a 1:1 Langmuir binding model (A+B ↔ AB; BIAevaluation 4.1). The concentration of analytes injected is indicated to the right of each sensogram overlay.
TABLE 1.
Kinetic and affinity data for the interactions of kaposica with C3b and C4b and SCR 1-3 and SCR 1-2 mutants with C4bd
| Ligand | Analyte | kd(1/s)/ka(1/Ms) | SE (kd/ka) | KDa (μM) | χ2 |
|---|---|---|---|---|---|
| C3b | Kaposica | 0.679/(1.49 × 105) | 0.0355/(8.27 × 103) | 4.56a | 2.01 |
| C3b | Kaposica | NAc | NA | 4.43b | 0.0747 |
| C4b | Kaposica | 0.371/(2.11 × 106) | (4.40 × 10−3)/(2.71 × 104) | 0.176a | 0.773 |
| C4b | Kaposica | NA | NA | 0.174b | 0.269 |
| C4b | SCR 1-3 | 0.334/(2.47 × 106) | 0.0139/(1.11 × 105) | 0.135a | 2.26 |
| C4b | SCR 1-3 | NA | NA | 0.147b | 0.197 |
| C4b | SCR 1-2 | 0.413/(8.93 × 103) | 0.0355/(1.04 × 103) | 46.3a | 0.251 |
| C4b | SCR 1-2 | NA | NA | 46.6b | 0.0585 |
Data were calculated by global fitting to a 1:1 Langmuir binding model (BIAevaluation 4.1).
Data were calculated by steady-state analysis (BIAevaluation 4.1).
NA, not applicable.
ka, association rate consistant; kd, dissociation rate constant; KD, equilibrium rate constant; SE, standard error.
In summary, our data clearly show that different SCRs of kaposica are important for its various functional activities. It is pertinent to point out here that although KSHV encodes a small complement regulatory protein (composed of four SCRs) compared to human regulators (composed of 4 to 59 SCRs), it possesses at least three out of four C3-convertase regulatory activities of the host RCA proteins. The human RCA proteins that have all these activities are complement receptor 1 and C4b-binding protein, which are 30 and 59 SCR-containing proteins, respectively. Thus, the small size of kaposica reflects the limited size of the KSHV genome, but this does not limit its activities. Gammaherpesvirus 68 (γHV-68), a murine gammaherpesvirus, also encodes a four-SCR-containing protein similar to kaposica and regulates complement at the C3-convertase level. In an elegant in vivo study, it has been shown that γHV-68 RCA-C3 interactions are important for acute as well as persistent viral infection (10). Given the fact that kaposica inactivates complement by acting on the C3-convertases and the in vivo data on γHV-68 (10), it is likely that kaposica may play a similar role during KSHV infections. Further delineation of the control points in kaposica and targeting them may provide an alternative way to control KSHV infections.
Acknowledgments
We thank John D. Lambris (Department of Pathology and Laboratory Medicine, University of Pennsylvania, Philadelphia, PA) for his continuous support; Michael K. Pangburn (Department of Biochemistry, University of Texas Health Science Center, Tyler, Texas) for his support and the generous gift of complement proteins C3 and factors B, D, H and I; Henry March (AVANT Immunotherapeutics, Inc., Needham, Mass.) for providing sCR1; and Nicholas E. Sherman (Biomolecular Research Facility, University of Virginia, Charlottesville) for sequencing kaposica mutants. We also express appreciation to Sharanabasava Hallihosur for his excellent technical assistance.
This work was supported by the Wellcome Trust Overseas Senior Research Fellowship in Biomedical Science in India to A.S. The authors also acknowledge financial assistance to A.K.S. by the Council of Scientific and Industrial Research, India.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernet, J., J. Mullick, Y. Panse, P. B. Parab, and A. Sahu. 2004. Kinetic analysis of the interactions between vaccinia virus complement control protein and human complement proteins C3b and C4b. J. Virol. 78:9446-9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernet, J., J. Mullick, A. K. Singh, and A. Sahu. 2003. Viral mimicry of the complement system. J. Biosci. 28:249-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blom, A. M., L. Kask, and B. Dahlback. 2001. Structural requirements for the complement regulatory activities of C4BP. J. Biol. Chem. 276:27136-27144. [DOI] [PubMed] [Google Scholar]
- 5.Brodbeck, W. G., D. Liu, J. Sperry, C. Mold, and M. E. Medof. 1996. Localization of classical and alternative pathway regulatory activity within the decay-accelerating factor. J. Immunol. 156:2528-2533. [PubMed] [Google Scholar]
- 6.Cooper, N. R. 1998. Complement and viruses, p. 393-407. In J. E. Volanakis and M. M. Frank (ed.), The human complement system in health and disease. Marcel Dekker, Inc., New York, N.Y.
- 7.Gewurz, B. E., R. Gaudet, D. Tortorella, E. W. Wang, and H. L. Ploegh. 2001. Virus subversion of immunity: a structural perspective. Curr. Opin. Immunol. 13:442-450. [DOI] [PubMed] [Google Scholar]
- 8.Gordon, D. L., R. M. Kaufman, T. K. Blackmore, J. Kwong, and D. M. Lublin. 1995. Identification of complement regulatory domains in human factor H. J. Immunol. 155:348-356. [PubMed] [Google Scholar]
- 9.Hourcade, D., M. K. Liszewski, M. Krych-Goldberg, and J. P. Atkinson. 2000. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology 49:103-116. [DOI] [PubMed] [Google Scholar]
- 10.Kapadia, S. B., B. Levine, S. H. Speck, and H. W. Virgin. 2002. Critical role of complement and viral evasion of complement in acute, persistent, and latent gamma-herpesvirus infection. Immunity 17:143-155. [DOI] [PubMed] [Google Scholar]
- 11.Kinter, M., and N. E. Sherman. 2000. Protein sequencing and identification using tandem mass spectrometry. Wiley-Interscience, New York, N.Y.
- 12.Krych-Goldberg, M., R. E. Hauhart, V. B. Subramanian, B. M. Yurcisin, D. L. Crimmins, D. E. Hourcade, and J. P. Atkinson. 1999. Decay accelerating activity of complement receptor type 1 (CD35). Two active sites are required for dissociating C5 convertases. J. Biol. Chem. 274:31160-31168. [DOI] [PubMed] [Google Scholar]
- 13.Lachmann, P. J. 2002. Microbial subversion of the immune response. Proc. Natl. Acad. Sci. USA 99:8461-8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lubinski, J., T. Nagashunmugam, and H. M. Friedman. 1998. Viral interference with antibody and complement. Semin. Cell Dev. Biol. 9:329-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenzie, R., G. J. Kotwal, B. Moss, C. H. Hammer, and M. M. Frank. 1992. Regulation of complement activity by vaccinia virus complement-control protein. J. Infect. Dis. 166:1245-1250. [DOI] [PubMed] [Google Scholar]
- 16.Means, R. E., J. K. Choi, H. Nakamura, Y. H. Chung, S. Ishido, and J. U. Jung. 2002. Immune evasion strategies of Kaposi′s sarcoma-associated herpesvirus. Curr. Top. Microbiol. Immunol. 269:187-201. [DOI] [PubMed] [Google Scholar]
- 17.Meri, S., and M. K. Pangburn. 1990. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc. Natl. Acad. Sci. USA 87:3982-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, P. S., and Y. Chang. 2003. Kaposi's sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu. Rev. Microbiol. 57:609-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullick, J., J. Bernet, A. K. Singh, J. D. Lambris, and A. Sahu. 2003. Kaposi′s sarcoma-associated herpesvirus (human herpesvirus-8) open reading frame 4 protein (kaposica) is a functional homolog of complement control proteins. J. Virol. 77:3878-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mullick, J., A. Kadam, and A. Sahu. 2003. Herpes and pox viral complement control proteins: “the mask of self.” Trends Immunol. 24:500-507. [DOI] [PubMed] [Google Scholar]
- 21.Oran, A. E., and D. E. Isenman. 1999. Identification of residues within the 727-767 segment of human complement component C3 important for its interaction with factor H and with complement receptor 1 (CR1, CD35). J. Biol. Chem. 274:5120-5130. [DOI] [PubMed] [Google Scholar]
- 22.Pan, Q., R. O. Ebanks, and D. E. Isenman. 2000. Two clusters of acidic amino acids near the NH2 terminus of complement component C4 alpha′-chain are important for C2 binding. J. Immunol. 165:2518-2527. [DOI] [PubMed] [Google Scholar]
- 23.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sahu, A., T. R. Kozel, and M. K. Pangburn. 1994. Specificity of the thioester-containing reactive site of human C3 and its significance to complement activation. Biochem. J. 302:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahu, A., D. Morikis, and J. D. Lambris. 2000. Complement inhibitors targeting C3, C4, and C5, p. 75-112. In J. D. Lambris and V. M. Holers (ed.), Therapeutic interventions in the complement system. Humana Press Inc., Totowa, N.J.
- 26.Schulz, T. F., Y. Chang, and P. S. Moore. 1998. Kaposi′s sarcoma-associated herpesvirus (human herpesvirus 8), p. 87-134. In D. J. McCance (ed.), Human tumor viruses. ASM Press, Washington, D.C.
- 27.Seet, B. T., J. B. Johnston, C. R. Brunetti, J. W. Barrett, H. Everett, C. Cameron, J. Sypula, S. H. Nazarian, A. Lucas, and G. McFadden. 2003. Poxviruses and immune evasion. Annu. Rev. Immunol. 21:377-423. [DOI] [PubMed] [Google Scholar]
- 28.Smith, S. A., N. P. Mullin, J. Parkinson, S. N. Shchelkunov, A. V. Totmenin, V. N. Loparev, R. Srisatjaluk, D. N. Reynolds, K. L. Keeling, D. E. Justus, P. N. Barlow, and G. J. Kotwal. 2000. Conserved surface-exposed K/R-X-K/R motifs and net positive charge on poxvirus complement control proteins serve as putative heparin binding sites and contribute to inhibition of molecular interactions with human endothelial cells: a novel mechanism for evasion of host defense. J. Virol. 74:5659-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soames, C. J., and R. B. Sim. 1997. Interactions between human complement components factor H, factor I and C3b. Biochem. J. 326:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiller, O. B., D. J. Blackbourn, L. Mark, D. G. Proctor, and A. M. Blom. 2003. Functional activity of the complement regulator encoded by Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 278:9283-9289. [DOI] [PubMed] [Google Scholar]
- 31.Spiller, O. B., M. Robinson, E. O'Donnell, S. Milligan, B. P. Morgan, A. J. Davison, and D. J. Blackbourn. 2003. Complement regulation by Kaposi's sarcoma-associated herpesvirus ORF4 protein. J. Virol. 77:592-599. [DOI] [PMC free article] [PubMed] [Google Scholar]