Abstract
Plants produce a burst of reactive oxygen species (ROS) after pathogen infection to successfully activate immune responses. During pattern-triggered immunity (PTI), ROS are primarily generated by the NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD). RBOHD is degraded in the resting state to avoid inappropriate ROS production; however, the enzyme mediating RBOHD degradation and how to prevent RBOHD degradation after pathogen infection is unclear. In this study, we identified an Arabidopsis (Arabidopsis thaliana) vacuole-localized papain-like cysteine protease, XYLEM CYSTEINE PEPTIDASE 1 (XCP1), and its inhibitor CYSTATIN 6 (CYS6). Pathogen-associated molecular pattern-induced ROS burst and resistance were enhanced in the xcp1 mutant but were compromised in the cys6 mutant, indicating that XCP1 and CYS6 oppositely regulate PTI responses. Genetic and biochemical analyses revealed that CYS6 interacts with XCP1 and depends on XCP1 to enhance PTI. Further experiments showed that XCP1 interacts with RBOHD and accelerates RBOHD degradation in a vacuole-mediated manner. CYS6 inhibited the protease activity of XCP1 toward RBOHD, which is critical for RBOHD accumulation upon pathogen infection. As CYS6, XCP1, and RBOHD are conserved in all plant species tested, our findings suggest the existence of a conserved strategy to precisely regulate ROS production under different conditions by modulating the stability of RBOHD.
RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD) is degraded in the vacuole to limit reactive oxygen species (ROS) production in the resting state; RBOHD degradation is inhibited upon pathogen infection to promote the ROS burst and disease resistance.
Introduction
Plants have developed 2-tiered innate immune responses to fight pathogens (Ngou et al. 2022). The first tier is pattern-triggered immunity (PTI), which is initiated by membrane-associated pattern recognition receptors (PRRs) via recognition of pathogen-associated molecular patterns (PAMPs), including bacterial flagellin (represented by flg22) and elongation factor Tu (represented by elf18) (Chinchilla et al. 2006; Zipfel et al. 2006a; Bigeard et al. 2015). PTI responses include transient production of reactive oxygen species (ROS) and a transient spike in calcium levels, phosphorylation of MITOGEN-ACTIVATED PROTEIN KINASEs (MAPKs or MPKs), accumulation of defense-related phytohormones, and induction of defense-related genes (Bigeard et al. 2015). Some pathogen effectors can repress PTI (Khan et al. 2018), and plant intracellular NUCLEOTIDE-BINDING LEUCINE-RICH REPEAT (NB-LRR) proteins can recognize pathogen effectors, which leads to the second layer of plant immune responses, called effector-triggered immunity (ETI) (Martel et al. 2021). A recent study suggested the existence of an intricate interaction between PTI and ETI (Yuan et al. 2021).
The ROS burst is a conserved signaling output in response to biotic and abiotic stresses that is precisely controlled to activate defense responses without causing detrimental effects to plants (Kadota et al. 2015; Mittler 2017; Waszczak et al. 2018; Lee et al. 2020). During PTI, the ROS burst is primarily produced by the NADPH oxidase RESPIRATORY BURST OXIDASE HOMOLOG D (RBOHD), a membrane-localized protein containing 6 conserved transmembrane domains in the middle with its N and C termini in the intracellular cytosol (Miller et al. 2009). Phosphorylation plays a critical role in regulating RBOHD activity and stability. Upon PAMP perception, the plasma membrane-associated kinase BOTRYTIS-INDUCED KINASE 1 (BIK1) is activated by the PRR complex to phosphorylate RBOHD (Kadota et al. 2014). Moreover, calcium burst activates CALCIUM-DEPENDENT PROTEIN KINASEs (CPKs), which also play a role in RBOHD phosphorylation (Kimura et al. 2020). These enzymes phosphorylate RBOHD at different sites to enhance ROS production (Kadota et al. 2014; Kimura et al. 2020). The receptor-like cytoplasmic kinase AvrPphB SUSCEPTIBLE1-LIKE 13 (PBL13) also phosphorylates RBOHD, resulting in enhanced RBOHD ubiquitination by PBL13-INTERACTING RING DOMAIN E3 LIGASE (PIRE); RBOHD is then degraded in a vacuole-mediated manner to ensure that only an appropriate amount of RBOHD is located at the plasma membrane in the resting state (Lee et al. 2020). However, the enzyme in the vacuole responsible for RBOHD degradation and the mechanism through which plants suppress RBOHD degradation upon pathogen infection remains unknown.
Numerous proteases localize in the vacuole to promote protein degradation, among which PAPAIN-LIKE CYSTEINE PROTEASES (PLCPs) are prominent (Van der Hoorn 2008). PLCPs cleave specific proteins using their catalytic cysteine residues and play crucial roles in plant–pathogen interactions (Shindo and Van der Hoorn 2008). For example, PHYTOPHTHORA-INHIBITED PROTEASE 1 (PIP1) and REQUIRED FOR CLADOSPORIUM FULVUM RESISTANCE 3 (RCR3) in tomato (Solanum lycopersicum) activate the salicylic acid–mediated defense pathway (Ilyas et al. 2015); RESPONSIVE TO DEHYDRATION 21A (RD21A) is required for drought-induced resistance to Pseudomonas syringae in Arabidopsis (Arabidopsis thaliana) (Liu et al. 2020). XYLEM CYSTEINE PEPTIDASE 2 (XCP2) enhances susceptibility to Ralstonia solanacearum in Arabidopsis (Zhang et al. 2014). Owing to their important roles in plant defense, PLCPs have been identified as targets of pathogen effectors (Clark et al. 2018; Misas Villamil et al. 2019).
Cystatins are naturally occurring PLCP inhibitors that also regulate plant immune responses (Van Wyk et al. 2016). For example, CYSTATIN GENE FROM FRAGARIA (Cyf1) exhibits antifungal activity in strawberry (Fragaria × ananassa) (Martinez et al. 2005b), and Arabidopsis CYSTATIN 1 (CYS1) inhibits cell death during ETI (Belenghi et al. 2003). Typically, cystatins inhibit the activities of specific PLCPs to modulate immunity. For example, TcCYS4 in cocoa (Theobroma cacao) plays a role in Moniliophthora perniciosa infection via interaction with the cysteine protease TcCYPR04 (Cardoso et al. 2015). In addition, the cystatin CORN CYSTATIN 9 (CC9) in maize (Zea mays) suppresses defense against Ustilago maydis by inhibiting apoplastic PLCPs (Van der Linde et al. 2012). However, to date, target PLCPs of most cystatins remain unidentified.
Arabidopsis contains 7 cystatins and 31 PLCPs (Martinez et al. 2005a; Liu et al. 2018). In this study, we demonstrate that CYS6 promotes plant pathogen resistance by regulating PTI. Through protein interaction assays and genetic complementation assays, we identified the vacuole-localized protein XCP1 as the PLCP functioning downstream of CYS6. Furthermore, we show that XCP1 is the enzyme that destabilizes RBOHD in a vacuole-dependent manner and is inhibited by CYS6. Abolishing the function of CYS6 prevented PAMP-triggered RBOHD accumulation, indicating that CYS6 plays a critical role in stabilizing RBOHD upon pathogen infection. These results suggest that XCP1 is the enzyme in the vacuole that promotes RBOHD degradation in the resting state and is inhibited by CYS6 to enhance ROS production upon pathogen infection. These findings fill the gaps in our knowledge about the regulation of RBOHD stability.
Results
CYS6 mediates plant defenses against both the hemibiotroph Psm ES4326 and the necrotroph Botrytis cinerea
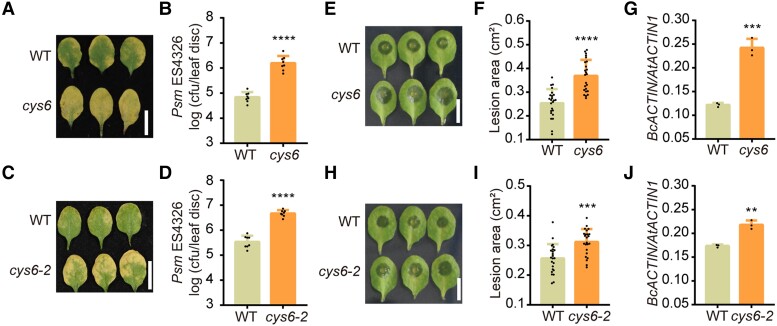
Cystatins are important regulators of plant defenses against pathogens (Lima et al. 2015). To investigate the key basal defense-related cystatin(s) in Arabidopsis, we compared the resistance of wild type (WT) and knockout or knockdown mutants of all Arabidopsis CYS genes to P. syringae pv. maculicola ES4326 (Psm ES4326). Among them, the cys6 mutant displayed a hypersensitive phenotype to Psm ES4326 (Supplemental Fig. S1). Compared with WT, the cys6 mutant exhibited extensive chlorosis and allowed 22.4-fold greater growth of Psm ES4326 and 7.2-fold higher growth of Pst DC3000 at 3 d after pathogen inoculation (Fig. 1, A and B; Supplemental Fig. S2). To demonstrate that CYS6 positively regulates plant defense against Psm ES4326, we generated another cys6 mutant using clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing. We identified a successfully edited cys6 mutant, named cys6-2, with a 1 bp insertion leading to the introduction of a premature stop codon and early termination of translation (Supplemental Fig. S2). As with the first cys6 mutant tested, the cys6-2 mutant was hypersensitive to Psm ES4326 and Pst DC3000 infection (Fig. 1, C and D; Supplemental Fig. S2).
Figure 1.
CYS6 enhances plant resistance to broad-spectrum pathogens. A) and C) Representative leaves of 3-wk-old WT, cys6A), and cys6-2C) after Psm ES4326 (OD600 nm = 0.0001) infection at 3 d postinoculation (dpi). B) and D) Bacterial titer after Psm ES4326 infection in WT, cys6B), and cys6-2D). cfu, colony forming unit. E) Representative leaves of 3-wk-old WT and cys6 mutant after B. cinerea (1 × 105 spores/mL) infection at 40 h postinoculation (hpi). F) and G) Lesion sizes caused by B. cinerea on leaves of WT and the cys6 mutant as measured with ImageJ F) and quantification of B. cinerea biomass, based on RT-qPCR analysis of genomic DNA from diseased leaves for BcACTIN with AtACTIN1 as a reference G). H) Representative leaves of 3-wk-old WT and the CRISPR/Cas9-generated cys6-2 mutant after B. cinerea (1 × 105 spores/mL) infection at 40 hpi. I) and J) Lesion sizes caused by B. cinerea on leaves of WT and the cys6-2 mutant as measured with ImageJ I) and quantification of B. cinerea biomass, based on RT-qPCR analysis of genomic DNA from diseased leaves for BcACTIN with AtACTIN1 as a reference J). Significant differences relative to WT were detected using Student’s t test. Data are shown as means ± Sd (n = 8 for B) and D); n = 24 for F) and I); n = 3 for G) and J)). *P < 0.01; ***P < 0.001; ****P < 0.0001.
We wondered whether CYS6-mediated defense is specific to Psm ES4236 or acts against multiple pathogens. As Psm ES4326 is a hemibiotroph, we chose a necrotroph, B. cinerea, to test whether CYS6 also enhances plant defense against other pathogens. Indeed, B. cinerea caused larger lesions on the leaves of cys6 than on those of WT (Fig. 1E). The mean lesion areas of WT and cys6 were 0.25 and 0.37 cm2, respectively (Fig. 1F). We also detected pathogen accumulation by quantifying the number of copies of BcACTIN in leaves, using Arabidopsis ACTIN1 for normalization; compared with WT, the cys6 mutant had a higher amount of pathogen genomic DNA (Fig. 1G). We obtained similar results with the cys6-2 mutant, confirming that CYS6 positively regulates plant defense against B. cinerea (Fig. 1, H to J). As cys6 and cys6-2 mutants showed similar phenotypes, we only used cys6 in subsequent experiments.
To further confirm that CYS6 positively regulates plant defense, we created a construct that expresses CYS6 under the control of the cauliflower mosaic virus (CaMV) 35S promoter and transformed it into WT. We chose 2 35S:CYS6 transgenic lines to determine whether the overexpression of CYS6 enhanced plant defenses against pathogens. The CYS6-overexpressing lines indeed showed higher resistance to Psm ES4326 and B. cinerea than WT (Supplemental Fig. S3). These results indicated that CYS6 mediates broad-spectrum defenses against plant pathogens.
CYS6 depends on its PLCP inhibitor activity to positively regulate PTI
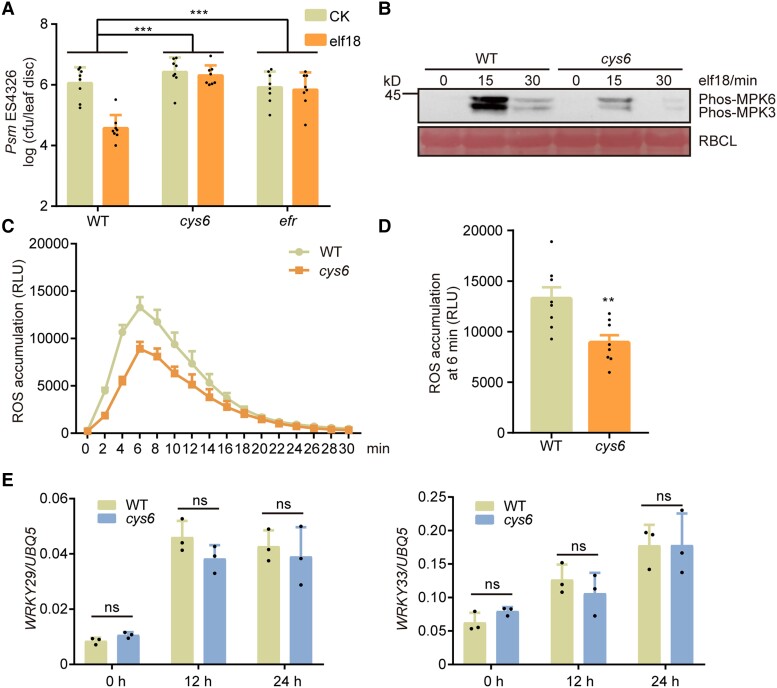
Based on the above findings, we hypothesized that CYS6 may regulate PTI, which mediates plant defenses against both biotrophs and necrotrophs. elf18 is a PAMP that induces typical PTI responses in plants, including enhanced pathogen resistance, phosphorylation of MPK3 and MPK6, ROS burst, and the induction of PTI-related genes (Bigeard et al. 2015). To test our hypothesis, we infiltrated elf18 into the leaves of 3-wk-old WT and cys6 plants and measured their PTI responses. As shown in Fig. 2A, elf18 treatment induced dramatic resistance against Psm ES4326 in WT, whereas resistance was significantly attenuated in the cys6 mutant. Moreover, we determined that PAMP-triggered phosphorylation of MPK3 and MPK6 and ROS burst are also compromised in the cys6 mutant (Fig. 2, B to D). Interestingly, the induction of PAMP-responsive genes, such as WRKY29 and WRKY33, was comparable between WT and cys6, indicating that CYS6 mainly functions at the protein level to regulate PTI (Fig. 2E; Supplemental Fig. S4). To further confirm the PTI-deficient phenotypes of the cys6 mutant, we used a nonvirulent pathogen, P. syringae pv tomato (Pst) DC3000 hrcC (D'Ambrosio et al. 2017), and another PAMP, flg22. We detected a higher amount of pathogen genomic DNA in the cys6 mutant than in WT after hrcC infection, confirming the negative role of CYS6 in PTI (Supplemental Fig. S5). Although flg22-induced resistance against Psm ES4326 was not inhibited in the cys6 mutant, flg22-triggered phosphorylation of MPK3 and MPK6 and ROS burst were partially dependent on CYS6 (Supplemental Fig. S6). Overall, these findings indicate that CYS6 is required for the complete activation of PTI responses.
Figure 2.
elf18-triggered defenses are compromised in the cys6 mutant. A) Three-wk-old WT, cys6, and efr mutants were preinoculated with 10 mM MgSO4 (CK) or 1 µM elf18 (elf18) for 1 d before infiltration with Pm ES4326 (OD600 nm = 0.001). Bacterial titer was determined at 3 dpi. Significant differences were detected by 2-way ANOVA. Data are shown as means ± Sd (n = 8). B) Twelve-d-old seedlings were treated with 1 µM elf18 for the indicated times, and samples were collected for immunoblots to detect the activation of MPK3 and MPK6 using a phospho-p44/42 MAPK antibody. The large subunit of Rubisco (RBCL) was stained with Ponceau S stain to serve as a loading control. C) and D) Leaf discs from 3-wk-old plants were immersed in elicitation solution containing 1 µM elf18, and ROS production was measured using a luminometer at the indicated time points C). ROS production at 6 min after elf18 treatment; a significant difference was detected by Student’s t test D). Data are shown as means ± Sem (n = 8). RLU, relative light units. E) Expression levels of PTI marker genes in 12-d-old WT and cys6 at the indicated time points after 1 µM elf18 treatment. Significant differences were detected by Student’s t test. Data are shown as means ± Sd (n = 3). **P < 0.01; ***P < 0.001; ns, no significant difference.
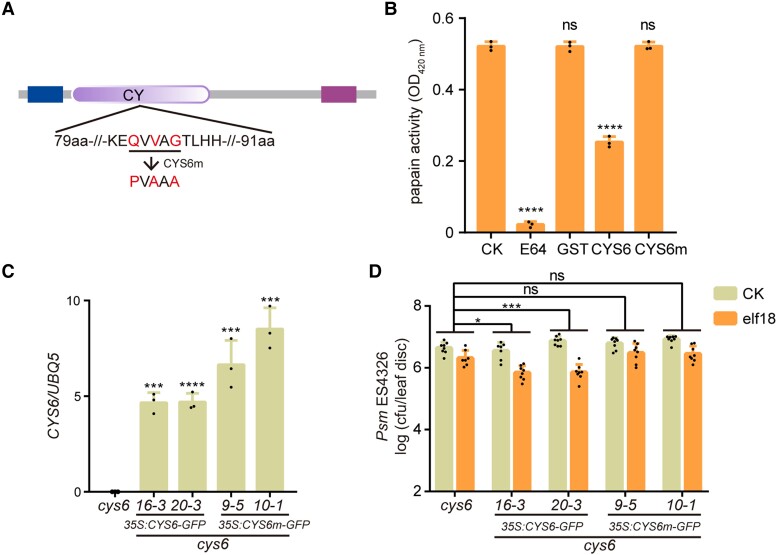
CYS6 exhibits PLCP inhibitor activity (Zhang et al. 2008); accordingly, we asked whether the regulation of PTI by CYS6 is dependent on its protease inhibitor activity. To answer this question, we created CYS6m, a point mutant variant of CYS6 that may affect its protease inhibitor activity according to a previous report (Fig. 3A) (Tremblay et al. 2019). As anticipated, the addition of the commercial PLCP inhibitor E64 or in vitro-produced glutathione S-transferase (GST)-CYS6 inhibited the protease activity of papain (Fig. 3B; Supplemental Fig. S7). However, the same amount of GST-CYS6m or GST exerted no effect on the protease activity of papain (Fig. 3B). To explore the requirement of CYS6 protease inhibitor activity for PTI in planta, we generated transgenic lines overexpressing CYS6 or CYS6m from the 35S promoter in the cys6 mutant background. We chose 2 independent lines for each construct and determined the gene expression levels of CYS6 and CYS6m (Fig. 3C). We observed that CYS6 overexpression largely restores the elf18-induced plant resistance to Psm ES4326 that is defective in cys6; however, the levels of induced resistance observed in the CYS6m transgenic lines were comparable with those in the cys6 mutant (Fig. 3D). Collectively, these data demonstrate that CYS6 acts as a PLCP inhibitor to promote PTI responses.
Figure 3.
The protease inhibitor activity of CYS6 is required for elf18-triggered resistance. A) Structure of CYS6 as determined by SMART (http://smart.embl-Heidelberg.de/). The N-terminal box indicates the predicted signal peptide, the middle box represents the cystatin (CY) domain, and the C-terminal bar is a low-complexity region. The amino acids mutated in CYS6m are marked in red. B)E. coli–produced GST, GST-CYS6 (CYS6), and GST-CYS6m (CYS6m) were used to test their inhibition on papain with E64 as a positive control. The products of papain digestion were detected by measuring the optical density at 420 nm. Significant differences compared with the CK samples were detected by Student’s t test. Data are shown as means ± Sd (n = 3). C) Relative expression levels of CYS6 and CYS6m in their transgenic lines in the cys6 mutant background. Significant differences compared with the cys6 mutant were detected by Student’s t test. Data are shown as means ± Sd (n = 3). D) Overexpressing CYS6 but not CYS6m rescues the deficiency of elf18-triggered resistance in the cys6 mutant. Experiments were performed as described in Fig. 2A. Significant differences were detected by 2-way ANOVA. Data are shown as means ± Sd (n = 8). *P < 0.05; ***P < 0.001; ****P < 0.0001; ns, no significant difference.
CYS6 interacts with XCP1 in the vacuole
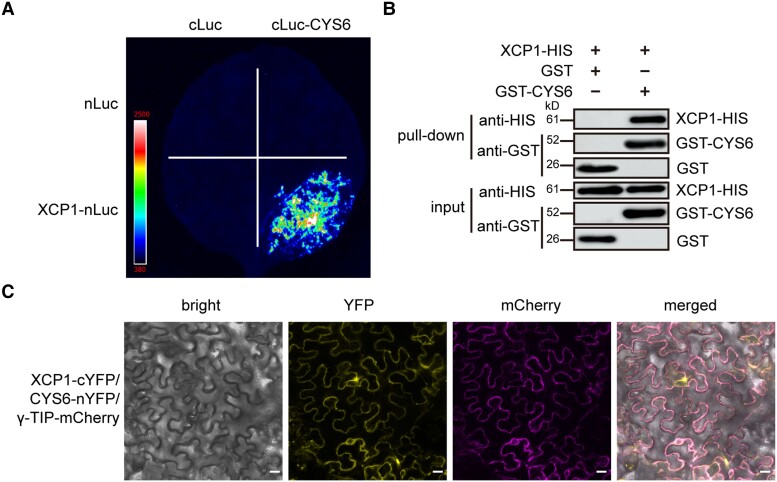
PLCPs are known target proteases of cystatins (Van Wyk et al. 2016). To investigate the molecular mechanism underlying the CYS6-mediated regulation of PTI, we employed a split-luciferase complementation assay to screen for PLCPs that can interact with CYS6 by selecting 1 PLCP from each subfamily (Richau et al. 2012). Among these selected PLCPs, we observed that XCP1 and PAP5 can interact with CYS6 (Fig. 4A; Supplemental Fig. S8). As the loss of PAP5 function did not show a clear PTI phenotype and because XCP2, the other PLCP in the same subfamily as XCP1, did not interact with CYS6 (Supplemental Fig. S9), we only focused on XCP1 in this study. To clarify the specific interaction between CYS6 and XCP1, which is a vacuole-localized protein, we tested if other vacuole-localized proteins, such as VACUOLAR MORPHOLOGY 3 (VAM3) (Radin et al. 2021) and VACUOLAR H+-ATPASE SUBUNIT a2 (VHA-a2) (Bak et al. 2013), might also interact with CYS6. As shown in Supplemental Fig. S10, we observed no fluorescence signal when the encoding constructs were coexpressed with CYS6 in split-luciferase assays. We next verified the interaction between CYS6 and XCP1 using pull-down assays. To this end, we purified recombinant XCP1-HIS and GST-CYS6 from Escherichia coli. As shown in Fig. 4B, XCP1-HIS could bind to GST-CYS6 but not to GST in GST pull-down assays, indicating that XCP1 can directly interact with CYS6. We further confirmed the interaction between CYS6 and XCP1 using bimolecular fluorescence complementation (BiFC) assays, which showed that only the combination of XCP1-cYFP (a fusion between XCP1 and the C-terminal half of the yellow fluorescent protein) and CYS6-nYFP (a fusion between CYS6 and the N-terminal half of YFP) reconstitutes fluorescence signals (Fig. 4C; Supplemental Fig. S11).
Figure 4.
CYS6 interacts with XCP1. A) Split-luciferase assays were performed on XCP1 and CYS6. Different combinations of Agrobacteria carrying nLuc, cLuc, XCP1-nLuc, or cLuc-CYS6 as indicated were infiltrated into N. benthamiana leaves, and luciferin was sprayed onto the infiltrated leaves 3 d later to detect luminescence signals with a CCD camera. B) Purified recombinant GST and GST-CYS6 were used to pull down XCP1-HIS from cell lysates. The protein levels of XCP1-HIS, GST, and GST-CYS6 were detected by immunoblots using anti-HIS and anti-GST antibodies. C)XCP1-cYFP and CYS6-nYFP were coexpressed in N. benthamiana leaves together with the vacuolar marker γ-TIP-mCherry for 3 d. The YFP and mCherry signals were visualized using a confocal microscope. Scale bar, 20 µm.
Because XCP1 is a vacuole-localized PLCP (Supplemental Fig. S12A) (Funk et al. 2002), we speculated that CYS6 is also localized in the vacuole. To test this possibility, we coexpressed CYS6-GFP with the tonoplast localization marker gene γ-TIP-mCherry, encoding a fusion between mCherry and a vacuolar membrane-localized aquaporin (γ-TONOPLAST INTRINSIC PROTEIN) (Nelson et al. 2007). As shown in Supplemental Fig. S12B, CYS6-GFP colocalized well with γ-TIP-mCherry, indicating that CYS6 is a vacuole-localized protein. In addition, the YFP signals from CYS6-nYFP and XCP1-cYFP overlapped well with mCherry signals from the vacuolar marker, indicating that CYS6 and XCP1 interact in vacuoles (Fig. 4C).
CYS6 depends on XCP1 to regulate PTI
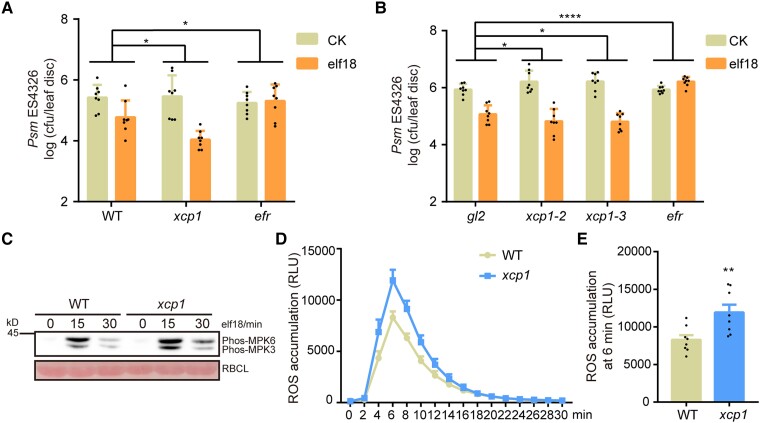
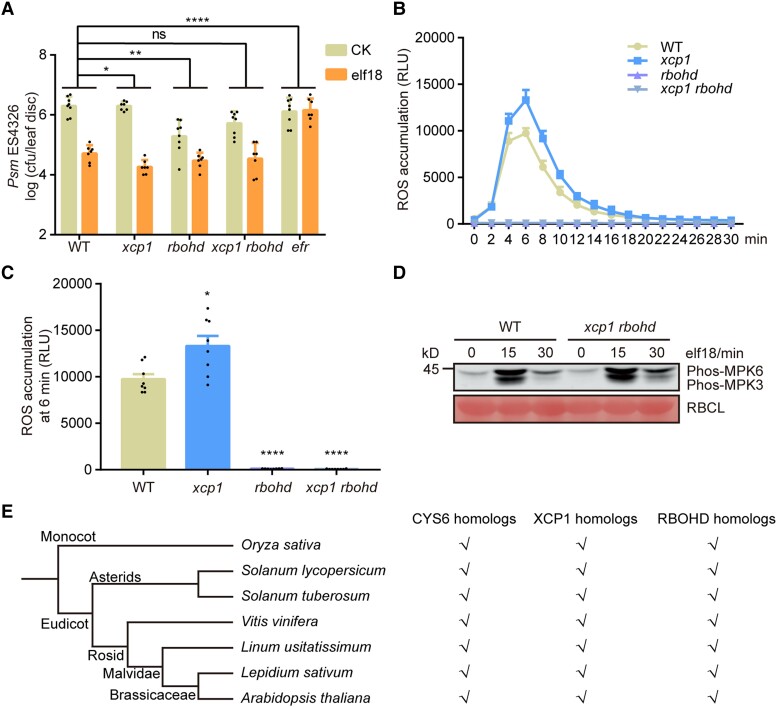
XCP1 regulates plant defense against pathogens (Pérez-López et al. 2021; Chen et al. 2023); however, its role in PTI has not been explored. To detect whether mutation of XCP1 alters plant responses to PAMPs, we infiltrated elf18 into the leaves of 3-wk-old WT and xcp1 mutant plants. We observed that elf18 triggers a stronger resistance to Psm ES4326 in the xcp1 mutant than in WT (Fig. 5A). To confirm the function of XCP1 in PAMP-triggered defense, we created 2 more xcp1 mutants, xcp1-2 and xcp1-3, using CRISPR/Cas9 technology (Supplemental Fig. S13). The xcp1-2 and xcp1-3 mutants also showed an enhanced response to elf18 compared with WT (Fig. 5B). Furthermore, elf18 induced higher phosphorylation of MPK3 and MPK6 and ROS burst in the xcp1 mutant than in WT (Fig. 5, C to E). These results demonstrate that, in contrast to CYS6, XCP1 is a negative regulator of PTI.
Figure 5.
XCP1 negatively regulates elf18-triggered defenses. A) and B) elf18-induced plant resistance to Psm ES4326 is enhanced in the xcp1 mutant. Experiments were performed as described in Fig. 2A. Significant differences were detected by 2-way ANOVA. Data are shown as means ± Sd (n = 8). The gl2 mutant was used as a control because the xcp1-2 and xcp1-3 mutants were in the g12 mutant background. C) Elf18-triggered phosphorylation of MPK3 and MPK6 is enhanced in the xcp1 mutant. Experiments were performed as described in Fig. 2B. D) and E) elf18-triggered ROS production is enhanced in the xcp1 mutant. Experiments were performed as shown in Fig. 2C. Significant difference were detected by Student’s t test. Data are shown as means ± Sem (n = 8). RLU, relative light units. *P < 0.05; **P < 0.01; ****P < 0.0001.
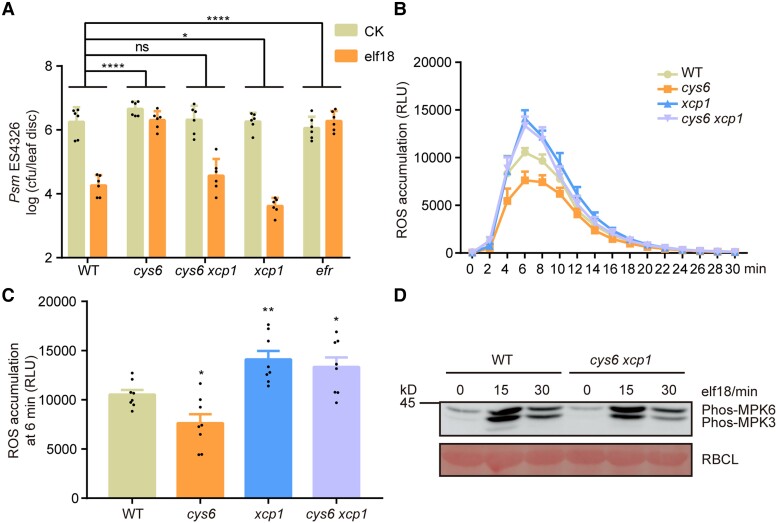
To determine whether CYS6 depends on XCP1 to enhance PTI, we created the cys6 xcp1 double mutant by crossing the cys6 and xcp1 single mutants. As shown in Fig. 6A, the compromised elf18-triggered resistance phenotype of cys6 was rescued in the cys6 xcp1 double mutant. The mutation of CYS6 decreased the elf18-induced ROS burst, whereas mutation of XCP1 enhanced the elf18-induced ROS burst, and ROS production in the cys6 xcp1 double mutant resembled that in the xcp1 single mutant (Fig. 6, B and C). The elf18-triggered phosphorylation of MPK3 and MPK6 was comparable in WT and the cys6 xcp1 double mutant, indicating that mutation of XCP1 rescues the compromised phosphorylation of MPK3 and MPK6 seen in the cys6 mutant (Fig. 6D). Collectively, these data suggest that CYS6 promotes PTI by inhibiting XCP1.
Figure 6.
CYS6 depends on XCP1 to regulate elf18-triggered defenses. A) Mutation of XCP1 rescues the compromised elf18-triggered resistance of cys6. Experiments were performed as described in Fig. 2A. Significant differences were detected by 2-way ANOVA. Data are shown as means ± Sd (n = 6). B) and C) Mutation of XCP1 rescues the compromised elf18-triggered ROS burst of cys6. Experiments were performed as shown in Fig. 2C. Significant differences were detected by Student’s t test. Data are shown as means ± Sem (n = 8). RLU, relative light units. D) Mutation of XCP1 rescues the compromised elf18-triggered phosphorylation of MPK3 and MPK6 of cys6. Experiments were performed as described in Fig. 2B. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, no significant difference.
The NADPH oxidase RBOHD is destabilized by XCP1 in a vacuole-mediated manner
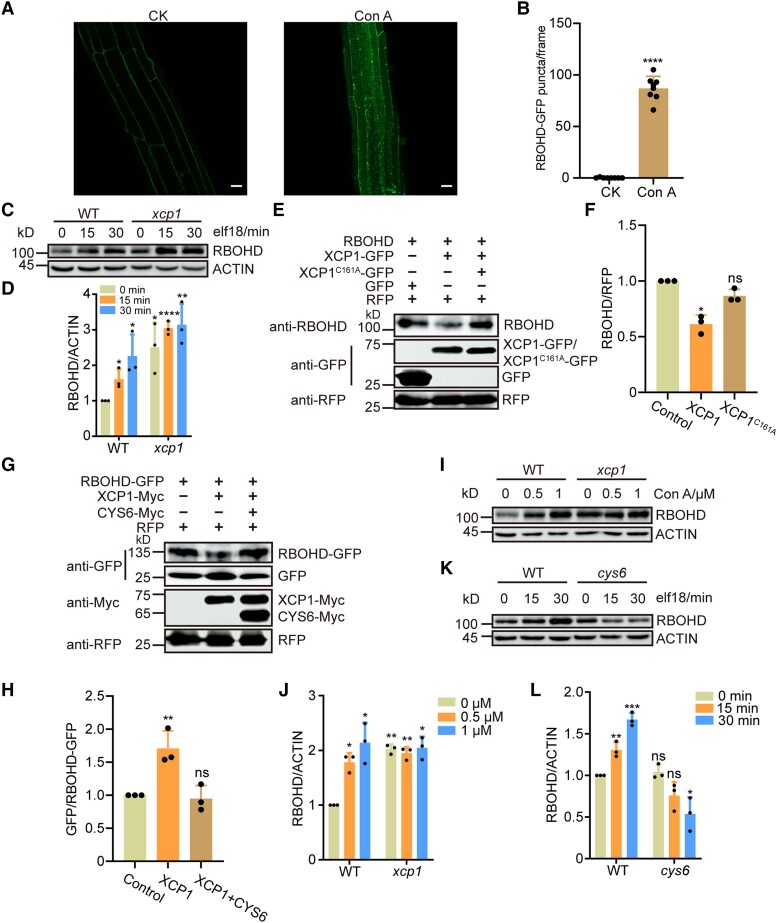
RBOHD-mediated ROS burst is critical for the establishment of PTI. RBOHD was reported to be degraded in a vacuole-dependent manner (Lee et al. 2020). In this study, we treated RBOHD-GFP (a construct encoding a fusion between RBOHD and the green fluorescent protein [GFP]) seedlings with concanamycin A (Con A), a well-established inhibitor of vacuole-dependent degradation (Drose and Altendorf 1997). After the treatment, we detected clear RBOHD-GFP puncta within the vacuole, thereby reinforcing the notion that RBOHD degradation is mediated by the vacuole (Fig. 7, A and B). As XCP1 is a vacuole-localized PLCP and regulates ROS burst after PAMP treatment (Fig. 5, D and E), we speculated that XCP1 represses PTI by destabilizing RBOHD. To test this hypothesis, we examined the abundance of RBOHD transcripts and RBOHD protein in the xcp1 mutant. We detected similar RBOHD mRNA levels in WT and the xcp1 mutant (Supplemental Fig. S14); however, RBOHD protein levels were higher in the xcp1 mutant than in WT both before and after elf18 or flg22 treatment, indicating that XCP1 destabilizes RBOHD (Fig. 7, C and D; Supplemental Fig. S15). Moreover, we examined the protein levels of MPK3 and MPK6 because their phosphorylation forms were enhanced in the xcp1 mutant. However, MPK3 and MPK6 levels were comparable in the WT and xcp1 mutant, indicating that XCP1 does not directly regulate the stability of MPK3 or MPK6 (Supplemental Fig. S16). To further confirm that XCP1 decreases RBOHD abundance, we fused XCP1 to a GFP tag and then expressed RBOHD together with empty vector or XCP1-GFP in the leaves of Nicotiana benthamiana plants. We examined RBOHD levels using an anti-RBOHD antibody, which could not detect endogenous RBOHD from N. benthamiana (Supplemental Fig. S17). Coexpression with XCP1, but not PAP5, resulted in lower levels of RBOHD compared with the control, confirming that XCP1 destabilizes RBOHD in planta (Supplemental Fig. S17). To investigate whether the enzymatic activity of XCP1 is required for destabilizing RBOHD, we mutated the enzymatically critical residue cysteine 161 to alanine (Pogorelko et al. 2019). The mutant form, XCP1C161A, was unable to promote the degradation of RBOHD (Fig. 7, E and F). To substantiate the role of vacuolar degradation in the processing of RBOHD, we conducted a free GFP cleavage assay (Huang et al. 2019a). Consistent with previous results, coexpression of XCP1 with RBOHD-GFP resulted in the destabilization of full-length RBOHD-GFP, with an increased level of cleaved free GFP compared with control samples (Fig. 7, G and H). These data support the notion that XCP1-modulated RBOHD degradation occurs within the vacuole. We further treated Arabidopsis seedlings with Con A and established that Con A increases RBOHD abundance in a dose-dependent manner (Fig. 7, I and J). However, the stability of RBOHD remained unaffected by Con A in the xcp1 mutant, which also demonstrated that XCP1 is the enzyme in the vacuole that destabilizes RBOHD (Fig. 7, I and J).
Figure 7.
The stability of RBOHD is modulated by XCP1 and CYS6 in a vacuole-dependent manner. A) and B) Con A treatment results in the accumulation of RBOHD-GFP in puncta within the vacuole. Confocal microscopy images of the roots of 7-d-old seedlings expressing RBOHD-GFP treated with DMSO (CK) or 1 µM Con A for 20 h A) and number of RBOHD-GFP puncta B). Data are shown as means ± Sd (n = 8). Scale bar, 20 µm. C) and D) RBOHD levels in 12-d-old WT and xcp1 seedlings after 1 µM elf18 treatment; samples were collected at the indicated time points after treatment for immunoblots C). Quantification of relative protein levels using ImageJ D). Data are shown as means ± Sd (n = 3). E) and F) Agrobacterium cultures expressing RBOHD were mixed with Agrobacteria harboring XCP1-GFP or XCP1C161A-GFP and infiltrated into N. benthamiana leaves. Samples were collected 3 d later for immunoblots E). Quantification of relative protein levels using ImageJ F). Data are shown as means ± Sd (n = 3). G) and H)RBOHD-GFP was coexpressed with XCP1-Myc or XCP1-Myc and CYS6-Myc in N. benthamiana leaves. Samples were collected 3 d after infiltration for immunoblots G). Quantification of relative protein levels by ImageJ H). Data are shown as means ± Sd (n = 3). I) and J) Twelve-d-old WT and xcp1 seedlings were treated with different concentrations of Con A for 18 h before sample collection; RBOHD levels were measured by immunoblots I). Quantification of relative protein levels using ImageJ J). Data are shown as means ± Sd (n = 3). K) and L) RBOHD levels measured in 12-d-old WT and cys6 seedlings after 1 µM elf18 treatment; samples were collected at the indicated time points after treatment K). Quantification of relative protein levels by ImageJ L). Data are shown as means ± Sd (n = 3). Significant differences were detected using Student’s t test. *P < 0.05: **P < 0.01: ****P < 0.0001; ns, no significant difference.
Furthermore, we fused CYS6 to a Myc tag and coexpressed the encoding construct with XCP1-Myc and RBOHD-GFP in N. benthamiana leaves to determine whether CYS6 is a functional cystatin of XCP1. The addition of CYS6 prevented the typical drop of RBOHD abundance and the change of free GFP/RBOHD-GFP ratio by XCP1, indicating that CYS6 does inhibit the protease activity of XCP1 toward RBOHD (Fig. 7, G and H; Supplemental Fig. S17B). We also examined RBOHD levels in WT and the cys6 mutant by immunoblotting, revealing that RBOHD levels are comparable at 0 min of elf18 or flg22 treatment between WT and cys6, suggesting that the basal activity of XCP1 is sufficient to destabilize RBOHD in healthy tissues (Fig. 7, K and L; Supplemental Fig. S15). The overexpression of CYS6 suppressed the basal activity of XCP1, resulting in RBOHD accumulation (Supplemental Fig. S18). After elf18 treatment, the cys6 mutant, but not the mutants of other positive PTI regulators, exhibited a lower abundance of RBOHD than WT, suggesting that the activity of CYS6 is essential for RBOHD accumulation upon pathogen infection (Fig. 7, K and L; Supplemental Fig. S19). Remarkably, the PAMP-induced accumulation of RBOHD was completely eliminated in the cys6 mutant, indicating that PAMP may regulate the stability of RBOHD by controlling CYS6 (Fig. 7, K and L; Supplemental Fig. S19). Although mutating BAK1-INTERACTING RECEPTOR-LIKE KINASE 2 (BIR2), another negative regulator of ROS production during PTI, also rescued the PTI-deficient immune phenotype of the cys6 mutant, we observed no significant difference in RBOHD abundance between WT and the bir2 mutant, indicating that CYS6 and XCP1 specifically regulate RBOHD stability to modulate PAMP-triggered ROS burst (Supplemental Figs. S20 and S21).
XCP1 regulates PTI in an RBOHD-dependent manner
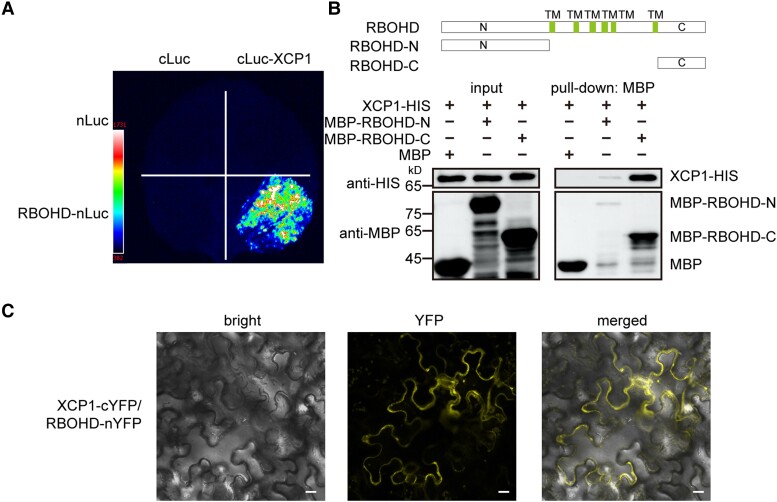
As XCP1 destabilizes RBOHD, we wondered whether XCP1 directly interacted with RBOHD to promote its degradation. To test this possibility, we separately fused XCP1 to cLuc (the C-terminal half of firefly luciferase [Luc]) and RBOHD to nLuc (the N-terminal half of Luc) to perform split-luciferase complementation assays in N. benthamiana leaves. We only detected reconstituted luciferase in the leaf regions coexpressing cLuc-XCP1 and RBOHD-nLuc (Fig. 8). To verify the direct interaction between XCP1 and RBOHD, we carried out a pull-down assay. Since RBOHD is a membrane protein, we separately produced its soluble N terminus (RBOHD-N) and C terminus (RBOHD-C) fused to maltose-binding protein (MBP) in E. coli (Fig. 8). As shown in Fig. 8B, both MBP-RBOHD-N and MBP-RBOHD-C could pull down XCP1, although only a small amount of RBOHD-N was pulled down by the beads, indicating that RBOHD possesses multiple domains that interact with XCP1. We further confirmed their interaction using BIFC assay: only the combination of XCP1-cYFP and RBOHD-nYFP emitted fluorescence signals (Fig. 8C; Supplemental Fig. S22).
Figure 8.
XCP1 interacts with RBOHD. A) Split-luciferase assays were performed to test the interaction of XCP1 and RBOHD. Different combinations of Agrobacteria containing nLuc, cLuc, RBOHD-nLuc, or cLuc-XCP1 constructs as indicated were infiltrated into N. benthamiana leaves, and luciferin was sprayed onto the infiltrated leaves 3 d later to detect luminescence signals with a CCD camera. B) Purified recombinant MBP, MBP-RBOHD-N, and MBP-RBOHD-C were used to pull down XCP1-HIS from cell lysates. The protein levels of XCP1-HIS, MBP, MBP-RBOHD-N, and MBP-RBOHD-C were detected by immunoblots using anti-HIS and anti-MBP antibodies. TM, transmembrane region. C) Different combinations of Agrobacteria carrying nYFP, cYFP, XCP1-cYFP, or RBOHD-nYFP constructs as indicated were infiltrated into N. benthamiana leaves. YFP signals were visualized 3 d later using a confocal microscope. Scale bar, 20 µm.
To investigate whether XCP1 mediates PTI by destabilizing RBOHD, we crossed xcp1 to the rbohd mutant and generated the xcp1 rbohd double mutant. Plants were treated with elf18 to examine PTI responses in the xcp1 rbohd double mutant. Mutation of RBOHD compromised elf18-triggered plant resistance to Psm ES4326, which is consistent with the critical role of ROS in PTI responses (Fig. 9A). Furthermore, we observed that the loss of RBOHD function returns the enhanced elf18-triggered plant resistance phenotype of xcp1 to a level similar to that in WT (Fig. 9A). In addition, ROS accumulation in the xcp1 mutant was completely dependent on RBOHD (Fig. 9, B and C). However, compared with WT, the xcp1 rbohd mutant showed stronger elf18-triggered phosphorylation of MPK3 and MPK6, indicating that XCP1 may have other targets during PTI (Fig. 9D). Collectively, these results demonstrate that XCP1 depends on RBOHD to modulate PAMP-induced resistance.
Figure 9.
XCP1 depends on RBOHD to repress elf18-triggered defenses. A) Mutating RBOHD suppresses the enhanced elf18-triggered resistance phenotype of xcp1. Experiments were performed as described in Fig. 2A. Significant differences were detected by 2-way ANOVA. Data are shown as means ± Sd (n = 8). B) and C) Mutating XCP1 abolishes the enhanced ROS production of the xcp1 mutant. Experiments were performed as shown in Fig. 2C. Significant differences were detected by Student’s t test. Data are shown as means ± Sem (n = 8). D) Mutating RBOHD does not affect the elf18-triggered phosphorylation of MPK3 and MPK6. Experiments were performed as described in Fig. 2B. E) Presence of putative orthologs for CYS6, XCP1, and RBOHD in different species. Tick marks suggest that the species possess the indicated related proteins. The phylogenetic tree was generated by Phytozome. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, no significant difference.
As RBOHD homologs are also in charge of ROS production upon pathogen infection in other plant species, such as rice (Oryza sativa), tobacco (Nicotiana tabacum), and potato (Solanum tuberosum) (Simon-Plas et al. 2002; Wong et al. 2007; Lukan et al. 2020), we wondered whether the CYS6–XCP1 regulatory module on RBOHD stability is conserved in plants. To evaluate this idea, we searched for their homologs in other plant species by BLAST analysis (Supplemental Data Set 1). As shown in Fig. 9E, homologs of CYS6, XCP1, and RBOHD were ubiquitous in all tested plants, indicating that the CYS6–XCP1–RBOHD cascade may be conserved in plants.
Discussion
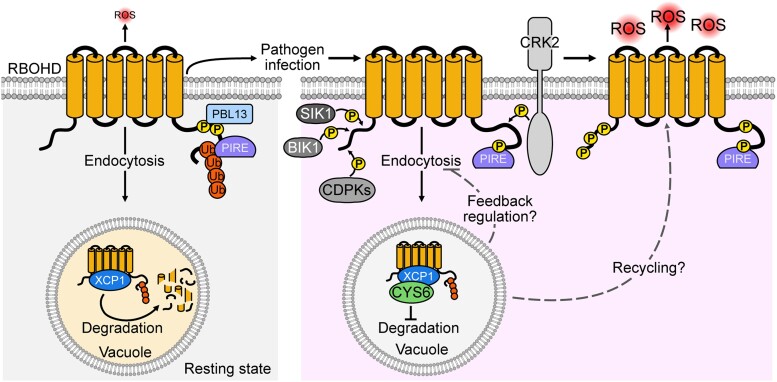
Plants grow in a dynamic environment that requires them to rapidly respond to challenges via multiple signals, including ROS. After sensing PAMPs from pathogens, ROS are rapidly produced by RBOHD, which is positively and negatively regulated via posttranslational modifications such as phosphorylation and ubiquitination (Kadota et al. 2015; Kimura et al. 2020; Lee et al. 2020; Li et al. 2021). In this study, we reported that the vacuolar cystatin, CYS6, and its target PLCP, XCP1, modulate PTI by regulating the stability of RBOHD. Protein ubiquitination is typically the signal for removing membrane proteins via endocytosis and eventual vacuole-mediated degradation, which is the only known RBOHD degradation pathway in Arabidopsis (Foot et al. 2017). We assume that RBOHD undergoes phosphorylation and ubiquitination by PBL3 and PIRE, respectively, at its C terminus, and is then sorted to the vacuole for XCP1-mediated degradation to ensure that only the required amount of RBOHD is located at the plasma membrane in the resting state (Fig. 10) (Lee et al. 2020). Upon pathogen infection, CYS6 inhibits the protease activity of XCP1, resulting in RBOHD accumulation. At the same time, RBOHD is phosphorylated at several residues to activate RBOHD for ROS production (Fig. 10) (Li et al. 2014; Kadota et al. 2015; Kimura et al. 2020). Because CYS6 and XCP1 are vacuole-localized proteins, we speculate that feedback regulation may exist or that nondegraded RBOHD in the vacuole may be recycled to the plasma membrane (Lee et al. 2022). However, the mechanisms through which PAMPs regulate CYS6 to inhibit the protease activity of XCP1 and then stabilize RBOHD require further investigation. Further study is also required to confirm the change in protease activity of XCP1 after pathogen infection in the presence and absence of CYS6.
Figure 10.
Working model of XCP1–CYS6-mediated regulation of RBOHD. In the resting state, RBOHD is phosphorylated and ubiquitinated by PBL13 and PIRE, respectively. Ubiquitinated RBOHD relocates to the vacuole through endocytosis, where it can be degraded by XCP1. After pathogen infection, CYS6 inhibits the activity of XCP1 and results in RBOHD accumulation. At the same time, RBOHD is activated by phosphorylation mediated by CDPKs, BIK1, CRK2, and SIK1 to enhance ROS production.
In addition to biotic stresses, abiotic stresses such as high light and drought also induce ROS accumulation in an RBOHD-dependent manner (Liu and He 2016; Yao et al. 2017; Zandalinas et al. 2020; Luo et al. 2021). For example, ETHYLENE RESPONSE FACTOR 74 (ERF74) induces the expression of RBOHD to improve plant defenses against various abiotic stresses including drought, high light, and heat (Yao et al. 2017); mutation of EPSIN-LIKE CLATHRIN ADAPTOR 4 (ECA4) compromises plant responses to salinity owing to the lower recycling of RBOHD to the plasma membrane in this mutant (Lee et al. 2022). The regulation of RBOHD activity through endocytosis, which shuttles membrane proteins to the vacuole for degradation, is also involved in plant tolerance to salt stress (Hao et al. 2014); moreover, CYS6 overexpression can promote plant tolerance to multiple abiotic stresses (Zhang et al. 2008). Based on these reports, we propose that the CYS6–XCP1 module that affects RBOHD stability also regulates ROS production under abiotic stress conditions.
Owing to the important role of PLCPs in plant–pathogen interactions, multiple PLCPs have been identified as targets of pathogen effectors (Shindo and Van der Hoorn 2008). XCP1 is also an effector target that can be inhibited by PLASMODIOPHORA BRASSICAE PROTEIN 53 (SSPbP53), an effector of the clubroot pathogen Plasmodiophora brassicae, to diminish the formation of root galls (Pérez-López et al. 2021). Moreover, a recently published article claims that XCP1 can act as a caspase to regulate plant immune responses by proteolyzing PATHOGENESIS-RELATED 1 (PR1) for systemic immunity (Chen et al. 2023). Combined with our finding that XCP1 mediates RBOHD degradation, XCP1 appears to be a multifunctional protein in plant–pathogen interactions.
CYS6 may also regulate other proteins in PTI, as mutating XCP1 did not completely rescue the PTI-deficient phenotype of the cys6 mutant. In addition, XCP1 may have other substrates as well. We observed that the elf18-triggered phosphorylation of MPK3 and MPK6 was enhanced in the xcp1 mutant and lower in the cys6 mutant (Fig. 2, B and 5C). However, the elf18-triggered phosphorylation of MPK3 and MPK6 remained higher in the xcp1 rbohd double mutant than in WT, suggesting that XCP1 targets other proteins to repress the MPK cascade (Fig. 9D). Because endocytosis regulates the signal duration and amplitude of some receptor complexes, such as the endocytosis of the flagellin receptor FLAGELLIN-SENSITIVE 2 (FLS2) induced by its ligand flg22, we speculate that the components of the PAMP receptor complex may be another target of XCP1 during PTI (Robatzek et al. 2006; Claus et al. 2018). Further investigation of XCP1 targets may uncover new mechanisms regulating PTI.
In summary, our study illustrates a strategy to regulate ROS burst upon pathogen infection. Because RBOHD and its homologs are the key NADPH oxidases involved in ROS production upon pathogen infection (Simon-Plas et al. 2002; Kobayashi et al. 2007; Wong et al. 2007; Huang et al. 2019b; Lukan et al. 2020) and as CYS6 and XCP1 are conserved among all the plant species tested (Fig. 9E), we speculate that other plant species may also use the CYS6–XCP1–RBOHD cascade to control ROS production under different conditions.
Materials and methods
Plant materials and growth conditions
Arabidopsis (A. thaliana) WT, mutants, and transgenic lines were all in the Col-0 accession in this study. The T-DNA insertion mutants cys1 (SALK_109268), cys2 (SALK_113078), cys3 (cs838011), cys4 (SALK_064249), cys5 (SALK_149928C), cys6 (SALK_027847C), cys7 (SALK_068510C), xcp1 (SALK_084789), and pap5 (SALK_131226C) were ordered from the Arabidopsis biological resource center or Arashare. Seeds of rbohd, efr, fls2, mpk3-1, and cpk5 cpk6 cpk11 were described previously (Zipfel et al. 2004; Zipfel et al. 2006a; Boudsocq et al. 2010; Tian et al. 2018; Yan et al. 2021). The cys6 xcp1 and xcp1 rbohd double mutants were generated by crossing the single mutants. The cys6-2, xcp1-2, xcp1-3, bir2, 35S:CYS6-GFP, 35S:CYS6m-GFP, 35S:RBOHD-GFP, and cys6 bir2 were created in this study using floral dip-mediated transformation (Clough and Bent 1998).
Seeds were surface sterilized with 2% (v/v) plant preservative mixture (Coolaber, PTC1000), stratified at 4°C for 2 d in the dark, and then sown on solidified half-strength MS medium (2.4 g MS medium, 15 g sucrose, 0.5 g MES-H, 8 g agar powder per liter, pH 5.8) or directly in soil. Plants were grown in growth chambers under conditions of 12 h light/12 h dark photoperiod (100 to 120 µmol m−2 s−1 provided by a combination of white and full spectrum LED lamps), 22°C, and 60% relative humidity.
Vector construction
The cys6-2, xcp1-2, xcp1-3, and bir2 mutants were produced by CRISPR/Cas9-mediated genome editing (Xing et al. 2014; Kong et al. 2021). To construct the pHEE401E-CYS6-GL2 vector, 2 simple guide RNAs (sgRNAs) were designed for CYS6, CYS6-T1 sgRNA, and CYS6-T2 sgRNA (Supplemental Data Set 2). The CYS6-T1 sgRNA was introduced into the primer CYS6-T1-F, the CYS6-T2 sgRNA was introduced into the primer CYS6-T2-F, and the GL2 sgRNA was introduced into the primer GL2-R. The primer pair CYS6-T1-F/CYS6-T2-R was used to amplify a PCR fragment from pCBC-DT1T2, and the primer pair CYS6-T2-F/GL2-R was used to amplify a PCR fragment from pCBC-DT2T3. Then, these 2 fragments were purified and ligated into the pHEE401E binary vector digested by BsaI. The pHEE401E-XCP1-GL2 vector was constructed as described above by replacing CYS6 sgRNAs with XCP1 sgRNAs. To construct the pHEE401E-BIR2 vector, the primer pair BIR2-T1-F/BIR2-T2-R was used to amplify a PCR fragment from pCBC-DT1T2. Then, the fragment was purified and ligated into the pHEE401E binary vector digested by BsaI.
The full-length coding sequences of CYS6, XCP1, VAM3, VHA-a2, CTB2, AALP, RD21A, CEP1, XBCP3, PAP5, THI1, RD19A, and RBOHD were individually amplified by PCR using the cDNA of Col-0 seedlings as template and cloned into the pENTRY vector. To mutate CYS6 to CYS6m, the primer pair CYS6m-F/R was designed (Supplemental Data Set 2). The primer pairs CYS6-CDS-F/CYS6m-R and CYS6m-F/CYS6-CDS-R were used to amplify 2 PCR fragments from the pENTRY-CYS6 plasmid. Then, CYS6m was amplified by overlap PCR using these 2 fragments and cloned into the pENTRY vector. Fragments encoding the N terminus (amino acids [aa] 1 to 376) and the C terminus (aa 756 to 921) of RBOHD were PCR amplified from the pENTRY-RBOHD plasmid. Then, RBOHD-N and RBOHD-C were cloned into the pMALc2x vector (Tian et al. 2023). To build the corresponding constructs needed, the CYS6, CYS6m, XCP1, and RBOHD sequences cloned into pENTRY were recombined with the destination vectors pEG103 (35S:X-GFP), pDEST15 (GST-X), pET32a (X-HIS), pX-nYFP (35S:X-nYFP), pX-cYFP (35S:X-cYFP), p1390-MH (35S:X-Myc-HIS), pDESTGwnluc (35S:X-nLuc), and pDESTclucGw (35S:cLuc-X), accordingly (Moreland et al. 2005; Earley et al. 2006; Robertson et al. 2008; Liu et al. 2016; Tian et al. 2018).
Bacterial growth and infiltration
Psm ES4326, Pst DC3000, and Pst DC3000 hrcC were streaked onto King's B medium containing 50 µg/mL streptomycin or 50 μg/mL rifampicin and grown in a 28°C incubator for 36 h. Psm ES4326 and Pst DC3000 were resuspended in 10 mM MgSO4 to OD600 nm = 0.001 for testing the resistance of CYS6 overexpression lines or to OD600 nm = 0.0001 for measuring pathogen growth in cys6 mutants. Pst DC3000 hrcC was resuspended in 10 mM MgSO4 to OD600 nm = 0.01. For each assay, the bacterial suspension was infiltrated into the 3rd and 4th true leaves of 3-wk-old Arabidopsis plants with needleless syringes. For PTI experiments, elf18 or flg22 was infiltrated into the same leaves 1 d in advance of syringe infiltration of Psm ES4326 solution (OD600 nm = 0.001). Three days later (6 d later for Pst DC3000 hrcC), 16 leaf discs (5 mm in diameter) from 8 plants of each genotype were surface sterilized and then randomly divided into 8 sample pools for bacterial growth measurement according to a previously reported method (Sun et al. 2021).
B. cinerea inoculation
B. cinerea was cultivated in Petri plates containing V8 medium (36% [v/v] V8 original vegetable juice, 0.02 mM CaCO3, 2% [w/v] agar) at 22°C in the dark for ∼10 d. Spores were harvested from the B. cinerea plate and resuspended to 5 × 105 spores/mL for testing the resistance of CYS6 overexpression lines or 1 × 105 spores/mL for measuring the sensitivity of cys6 mutants in potato dextrose agar (PDA) liquid medium. The 3rd and 4th true leaves of Arabidopsis plants were detached and inoculated with 5 µL drops of spores. The inoculated leaves were kept on plates with 0.8% (w/v) agar at 22°C in the dark for 40 h (Sun et al. 2021). The area of diseased spots was measured using ImageJ. Leaves were further collected to extract genomic DNA for qPCR to detect the levels of BcACTIN and AtACTIN1.
Measurement of ROS production
One day before the ROS assays, leaf discs (5 mm in diameter) were cut from the 3rd or 4th true leaves of 3-wk-old plants. Each leaf disc was floated in individual wells of a 96-well microtiter plate containing 200 µL deionized water and then incubated overnight at 22°C. Prior to the assay, the elicitation solution (horseradish peroxidase [HRP] [20 µg/mL], luminol [0.2 µM], and elf18 [1 µM] or flg22 [100 nM]) was prepared (Smith and Heese 2014). Then, 100 µL of the elicitation solution was rapidly added to each well after the water was removed. ROS production was measured using a 96-well microplate luminometer (BERTHOLD, Centro XS LB 960). In the final result, n represents the number of leaf discs used for each genotype.
Protein extraction and immunoblot analysis
To detect protein levels after PAMP treatment, 12-d-old seedlings were inoculated with 1 µM elf18 or flg22 for the indicated times (Heese et al. 2007). For Con A (Abcam, ab144227) treatment, different concentrations of Con A as indicated were added to half-strength MS liquid and 12-d-old seedlings were treated for 18 h before sample collection. Total proteins were extracted using native buffer (50 mM Tris-HCl, PH 7.5, 150 mM NaCl, 0.1% [v/v] Triton X-100, 0.2% [v/v] NP-40, protease inhibitor cocktail, and phosphatase inhibitor cocktail). Immunoblots were performed with the following antibodies: anti-phospho-MAPK (Cell Signaling, 4370, 1:2,000), anti-MPK3 (Sigma, M8318, 1:2,000), anti-MPK6 (Sigma, A7140, 1:2,000), anti-RBOHD (Agrisera, AS152962, 1:2,000), and anti-ACTIN (Easybio, BE0027, 1:5,000).
To check XCP1-mediated RBOHD degradation in N. benthamiana, combinations of Agrobacterium (Agrobacterium tumefaciens) cell suspensions harboring the related constructs were infiltrated into the lower epidermis of N. benthamiana leaves, and samples were collected 3 d later. Total proteins were extracted, and protein levels were detected as mentioned above. The primary antibodies used in these assays were anti-Myc (Easybio, BE2010, 1:2,000), anti-GFP (Easybio, BE2002, 1:2,000), anti-RFP (Easybio, BE2023, 1:2,000), and anti-RBOHD antibodies.
BIFC assay
Combinations of Agrobacterium cell suspensions containing 35S:nYFP, 35S:cYFP, 35S:RBOHD-nYFP, 35S:CYS6-nYFP, 35S:XCP1-cYFP, or 35S:γ-TIP-mCherry (Nelson et al. 2007) as indicated were infiltrated into the lower epidermis of N. benthamiana leaves at an OD600 nm = 0.4 (Han et al. 2020). The tombusvirus silencing suppressor p19 was coinfiltrated to ensure efficient expression of the fusion proteins. Three days later, the fluorescence signal was visualized by using an LSM-900 laser scanning confocal microscope (Zeiss). The excitation was set at 488 nm, and the emission was recorded at 505 to 550 nm for YFP.
Split-luciferase assay
Combinations of Agrobacterium cell suspensions containing 35S:cLuc, 35S:nLuc, 35S:RBOHD-nluc, 35S:cluc-XCP1, 35S:VAM3-nluc, 35S:VHA-a2-nluc, 35S:CTB2-nluc, 35S:AALP-nluc, 35S:RD21A-nluc, 35S:CEP1-nluc, 35S:XBCP3-nluc, 35S:PAP5-nluc, 35S:THI1-nluc, 35S:RD19A-nluc, 35S:XCP1-nluc, or 35S:cluc-CYS6 as indicated were transiently coinfiltrated in N. benthamiana leaves at an OD600 nm = 0.4. The viral suppressor p19 was coexpressed to ensure efficient expression of the fusion proteins. Three days later, 1 mM luciferin was sprayed onto the infiltrated leaves, which were incubated for 10 min. Chemiluminescence images were captured using a charge-coupled device camera (Tanon, 5200Multi or Amersham, Imager 680).
Subcellular localization
To determine the localization of XCP1 and CYS6, N. benthamiana leaves were infiltrated with Agrobacterium suspensions carrying 35S:XCP1-GFP or 35S:CYS6-GFP, 35S:γ-TIP-mCherry, and 35S:p19. Fluorescence signals were monitored 3 d later. To confirm the vacuole-mediated degradation of RBOHD, Arabidopsis 35S:RBOHD-GFP seedlings were grown on half-strength MS medium for 7 d before being treated with 1 µM Con A for 20 h. Images were captured using an LSM-900 laser scanning confocal microscope (Zeiss). The excitation was set at 488 nm for GFP and 587 nm for mCherry. The emission was recorded at 505 to 550 nm for GFP and 600 to 650 nm for mCherry.
In vitro papain inhibition assay
For the papain inhibition assay, the sequence encoding the N-terminal predicted signal peptide of CYS6 was removed to improve protein solubility in E. coli. Bacterial culture and protein purification were performed as described previously with minor adjustments (Liu and Zhang 2004). Briefly, the production of GST, GST-CYS6, and GST-CYS6m proteins was induced by 0.5 mM IPTG at 30°C for 5 h. For purification of fusion proteins, the bacterial pellet was resuspended and sonicated (200 W, the treatment/interval was 10 s/10 s, 15 to 20 min for each pellet) in lysis buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 and 0.5% [v/v] Triton X-100, 1 mM DTT, 1 mM PMSF, 1 mg/mL lysozyme, pH 7.4). The supernatant was incubated with a GST-tag purification column (Beyotime, P2262) at 4°C for 2 h, after which the purified proteins were eluted with elution buffer (50 mM Tris-HCl and 10 mM GSH, pH 8).
For the in vitro test, 80 μg purified protein was mixed with 80 μL of papain solution (1 mg/mL in 0.1 M sodium phosphate pH 6.1, 10 mM cysteine, 10 mM EDTA) in a final volume of 400 μL and preincubated at 37°C for 15 min. Then, 2 mL of azocasein solution (1 mg/mL in 0.1 M sodium phosphate pH 6.1, 10 mM proteinase, 10 mM EDTA) was added to the mixture, and the samples were incubated at 37°C for 1 h. The reaction was terminated by adding 1 mL precooled 10% (w/v) trichloroacetic acid and keeping on ice for 30 min. Samples were centrifuged for 5 min at 13,000 × g, and supernatants were used to determine the inhibition of papain digestion of its substrate for CYS6 by measuring the optical density at 420 nm (Zhang et al. 2008).
Pull-down assay
The production of recombinant GST-CYS6 and XCP1-HIS was performed as described previously. The supernatants from GST and GST-CYS6 cell lysates were incubated with the GST-tag purification column at 4°C for 2 h and then washed twice using lysis buffer. Cell lysates containing XCP1-HIS soluble proteins were added to the column and incubated at 4°C for 2 h. Thereafter, the column was washed 3 times with lysis buffer to remove nonspecific binding proteins. The bound proteins were eluted with elution buffer and detected by immunoblotting with anti-GST antibody (Easybio, BE2013, 1:2,000) and anti-HIS antibody (Trans, HT501, 1:2,000).
The production of MBP, MBP-RBOHD-N, and MBP-RBOHD-C proteins was induced by IPTG (0.5 mM) at 30°C for 5 h. For protein purification, the cell pellet was resuspended and sonicated in lysis buffer (20 mM Tris-HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 0.1% [v/v] Triton X-100, 1 mM DTT, 1 mM PMSF, and 1 mg/mL lysozyme). The supernatant was incubated with amylose resin (NEB, E8021V) at 4°C for 2 h and then washed twice using lysis buffer. Cell lysates containing XCP1-HIS soluble protein were added to the column and incubated at 4°C for 2 h. The bound proteins were eluted using 10 mM maltose and detected by immunoblotting with anti-MBP antibody (Trans, HT701, 1:2,000) and anti-HIS antibody.
Reverse transcription quantitative PCR (RT-qPCR) analysis
To check the expression levels of CYS genes in their mutants, samples were collected from 12-d-old Arabidopsis seedlings of WT, cys1, cys2, cys3, cys4, cys5, cys6, and cys7. To measure CYS6 expression levels in the transgenic lines of 35S:CYS6-GFP and 35S:CYS6m-GFP, leaves were harvested from 3-wk-old plants. Twelve-d-old xcp1 mutant seedlings were used to measure RBOHD expression levels. To detect marker gene expression after elf18 treatment, 12-d-old seedlings were inoculated with 1 µM elf18 for different times as indicated. Total RNA was extracted using TRIzol reagent (Takara, 9109). First-strand cDNA was synthesized with a HiScript III RT SuperMix kit (Vazyme, R323-01) and used as RT-qPCR template.
Quantitative PCR (qPCR) analysis was performed employing a QuantStudio 5 real-time PCR system and 2×M5 SYBR Premix EsTaq (with Tli RNaseH) (Mei5Bio, MF787) with gene-specific primers (Supplemental Data Set 2). The expression levels of target genes were calculated using the delta Ct method.
Phylogenetic analysis
The amino acid sequences of Arabidopsis cystatins were retrieved from the TAIR database and then were aligned with Clustal software. The phylogenetic tree was produced using MEGA 7.0 with the neighbor-joining method with 500 bootstrap replications (Supplemental Files 1 and 2).
Accession numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under the following accession numbers: CYS1 (At5g12140), CYS2 (At2g31980), CYS3 (At2g40880), CYS4 (At4g16500), CYS5 (At5g47550), CYS6 (At3g12490), CYS7 (At5g05110), XCP1 (At4g35350), RBOHD (At5g47910), GL2 (At1g79840), BIR2 (At3g28450), VAM3 (At5g46860), VHA-a2 (At2g21410), RD21A (At1g47128), XBCP3 (At1g09850), CEP1 (At5g50260), THI1 (At1g06260), PAP5 (At3g49340), AALP (At5g60360), RD19A (At4g39090), CTB2 (At1g02305), AtACTIN1 (At2g37620), BcACTIN (BCIN_16g02020), and UBQ5 (At3g62250).
Supplementary Material
Acknowledgments
We thank Haiyan Yu and Xiaomin Zhao from the Analysis and Testing Center of State Key Laboratory of Microbial Technology (SKLMT, Shandong University) for assistance with the laser scanning confocal microscopy.
Contributor Information
Yang Liu, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Tingting Gong, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Xiangjiu Kong, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Jiaqi Sun, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Lijing Liu, The Key Laboratory of Plant Development and Environmental Adaptation Biology, Ministry of Education, School of Life Sciences, Shandong University, Qingdao 266237, China.
Author contributions
L.L. designed the research and supervised the experiments. Y.L. performed the experiments and analyzed the data with help from T.G., X.K., and J.S. L.L. and Y.L. wrote the manuscript, and all authors helped to improve it.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sensitivity of cystatin mutants to Psm ES4326.
Supplemental Figure S2. cys6 mutants are sensitive to Pst DC3000.
Supplemental Figure S3. CYS6 overexpression enhances plant defense against broad-spectrum pathogens.
Supplemental Figure S4. Expression of PAMP-responsive genes is comparably induced in WT and the cys6 mutant.
Supplemental Figure S5. The cys6 mutant is hypersensitive to Pst DC3000 hrcC.
Supplemental Figure S6. flg22-triggered defenses in the cys6 mutant.
Supplemental Figure S7. Reagents used for the papain inhibition assay.
Supplemental Figure S8. Split-luciferase complementation assays to screen the PLCPs that may interact with CYS6.
Supplemental Figure S9. PAP5 and XCP2 are not the PLCPs working downstream of CYS6 in PTI.
Supplemental Figure S10. The vacuole-localized proteins VAM3 and VHA-a2 do not interact with CYS6.
Supplemental Figure S11. BIFC assays to detect the interaction of CYS6 and XCP1.
Supplemental Figure S12. The subcellular localization of XCP1 and CYS6.
Supplemental Figure S13. Generation of the xcp1-2 and xcp1-3 mutants by CRISPR/Cas9-mediated genome editing.
Supplemental Figure S14. Relative expression levels of RBOHD in WT and the xcp1 mutant.
Supplemental Figure S15. Protein levels of RBOHD in xcp1 and cys6 mutants after flg22 treatment.
Supplemental Figure S16. Protein levels of MPK3 and MPK6 in the xcp1 mutant.
Supplemental Figure S17. XCP1, but not PAP5, promotes the degradation of RBOHD.
Supplemental Figure S18. RBOHD accumulates in CYS6 overexpression lines.
Supplemental Figure S19. Abundance of RBOHD in the mutants of PTI positive regulators.
Supplemental Figure S20. Generation of the bir2 mutant by the CRISPR/Cas9-mediated genome editing.
Supplemental Figure S21. elf18-triggered defense responses in the cys6 bir2 mutant.
Supplemental Figure S22. Controls for the BIFC assay between XCP1 and RBOHD.
Supplemental Data Set 1. CYS6, XCP1, and RBOHD homologs identified in the indicated species.
Supplemental Data Set 2. Primers used in this study.
Supplemental Data Set 3. Summary of statistical analyses from this study.
Supplemental File 1. Alignment of protein sequences used for the phylogenetic tree shown in Supplemental Fig. S1.
Supplemental File 2. Newick format of the phylogenetic tree.
Funding
This work was supported by the National Natural Science Foundation of China (32000224) awarded to L.L.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Bak G, Lee EJ, Lee Y, Kato M, Segami S, Sze H, Maeshima M, Hwang JU, Lee Y. Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell. 2013:25(6):2202–2216. 10.1105/tpc.113.110411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenghi B, Acconcia F, Trovato M, Perazzolli M, Bocedi A, Polticelli F, Ascenzi P, Delledonne M. AtCYS1, a cystatin from Arabidopsis thaliana, suppresses hypersensitive cell death. Eur J Biochem. 2003:270(12):2593–2604. 10.1046/j.1432-1033.2003.03630.x [DOI] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant. 2015:8(4):521–539. 10.1016/j.molp.2014.12.022 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan LB, He P, Bush J, Cheng SH, Sheen J. Differential innate immune signalling via Ca2 + sensor protein kinases. Nature 2010:464(7287):418–422. 10.1038/nature08794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso TH, Freitas AC, Andrade BS, Sousa AO, Santiago Ada S, Koop DM, Gramacho KP, Alvim FC, Micheli F, Pirovani CP. TcCYPR04, a cacao papain-like cysteine-protease detected in senescent and necrotic tissues interacts with a cystatin TcCYS4. PLoS One 2015:10(12):e0144440. 10.1371/journal.pone.0144440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Lin FW, Cheng KT, Chang CH, Hung SC, Efferth T, Chen YR. XCP1 Cleaves pathogenesis-related protein 1 into CAPE9 for systemic immunity in Arabidopsis. Nat Commun. 2023:14(1):4697. 10.1038/s41467-023-40406-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006:18(2):465–476. 10.1105/tpc.105.036574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Franco JY, Schwizer S, Pang Z, Hawara E, Liebrand TWH, Pagliaccia D, Zeng L, Gurung FB, Wang P, et al. . An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat Commun. 2018:9(1):1718. 10.1038/s41467-018-04140-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus LAN, Savatin DV, Russinova E. The crossroads of receptor-mediated signaling and endocytosis in plants. J Integr Plant Biol. 2018:60(9):827–840. 10.1111/jipb.12672 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998:16(6):735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- D'Ambrosio JM, Couto D, Fabro G, Scuffi D, Lamattina L, Munnik T, Andersson MX, Alvarez ME, Zipfel C, Laxalt AM. Phospholipase C2 affects MAMP-triggered immunity by modulating ROS production. Plant Physiol. 2017:175(2):970–981. 10.1104/pp.17.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drose S, Altendorf K. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J Exp Biol. 1997:200(1):1–8. 10.1242/jeb.200.1.1 [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song KM, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006:45(4):616–629. 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Foot N, Henshall T, Kumar S. Ubiquitination and the regulation of membrane proteins. Physiol Rev. 2017:97(1):253–281. 10.1152/physrev.00012.2016 [DOI] [PubMed] [Google Scholar]
- Funk V, Kositsup B, Zhao C, Beers EP. The Arabidopsis xylem peptidase XCP1 is a tracheary element vacuolar protein that may be a papain ortholog. Plant Physiol. 2002:128(1):84–94. 10.1104/pp.010514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Liu Y, Shi W, Qiao Y, Wang LY, Tian YC, Fan M, Deng ZP, Lau OS, De Jaeger G, et al. . KIN10 promotes stomatal development through stabilization of the SPEECHLESS transcription factor. Nat Commun. 2020:11(1):4214. 10.1038/s41467-020-18048-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H, Fan L, Chen T, Li R, Li X, He Q, Botella MA, Lin J. Clathrin and membrane microdomains cooperatively regulate RbohD dynamics and activity in Arabidopsis. Plant Cell. 2014:26(4):1729–1745. 10.1105/tpc.113.122358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AME, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci U S A. 2007:104(29):12217–12222. 10.1073/pnas.0705306104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yu LJ, Zhang X, Fan B, Wang FZ, Dai YS, Qi H, Zhou Y, Xie LJ, Xiao S. Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 2019a:15(3):407–422. 10.1080/15548627.2018.1520547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Cao H, Yang L, Chen C, Shabala L, Xiong M, Niu M, Liu J, Zheng Z, Zhou L, et al. . Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. J Exp Bot. 2019b:70(20):5879–5893. 10.1093/jxb/erz328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M, Horger AC, Bozkurt TO, van den Burg HA, Kaschani F, Kaiser M, Belhaj K, Smoker M, Joosten MH, Kamoun S, et al. . Functional divergence of two secreted immune proteases of tomato. Curr Biol. 2015:25(17):2300–2306. 10.1016/j.cub.2015.07.030 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015:56(8):1472–1480. 10.1093/pcp/pcv063 [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. . Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol Cell. 2014:54(1):43–55. 10.1016/j.molcel.2014.02.021 [DOI] [PubMed] [Google Scholar]
- Khan M, Seto D, Subramaniam R, Desveaux D. Oh, the places they’ll go! A survey of phytopathogen effectors and their host targets. Plant J. 2018:93(4):651–663. 10.1111/tpj.13780 [DOI] [PubMed] [Google Scholar]
- Kimura S, Hunter K, Vaahtera L, Tran HC, Citterico M, Vaattovaara A, Rokka A, Stolze SC, Harzen A, Meissner L, et al. . CRK2 and C-terminal phosphorylation of NADPH oxidase RBOHD regulate reactive oxygen species production in Arabidopsis. Plant Cell. 2020:32(4):1063–1080. 10.1105/tpc.19.00525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H. Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell. 2007:19(3):1065–1080. 10.1105/tpc.106.048884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Pan W, Sun N, Zhang T, Liu L, Zhang H. GLABRA2-based selection efficiently enriches Cas9-generated nonchimeric mutants in the T1 generation. Plant Physiol. 2021:187(2):758–768. 10.1093/plphys/kiab356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Lal NK, Lin ZD, Ma S, Liu J, Castro B, Toruno T, Dinesh-Kumar SP, Coaker G. Regulation of reactive oxygen species during plant immunity through phosphorylation and ubiquitination of RBOHD. Nat Commun. 2020:11(1):1838. 10.1038/s41467-020-15601-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hanh Nguyen H, Park Y, Lin J, Hwang I. Spatial regulation of RBOHD via AtECA4-mediated recycling and clathrin-mediated endocytosis contributes to ROS accumulation during salt stress response but not flg22-induced immune response. Plant J. 2022:109(4):816–830. 10.1111/tpj.15593 [DOI] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. . The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 2014:15(3):329–338. 10.1016/j.chom.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Li P, Zhao L, Qi F, Htwe N, Li Q, Zhang D, Lin F, Shang-Guan K, Liang Y. The receptor-like cytoplasmic kinase RIPK regulates broad-spectrum ROS signaling in multiple layers of plant immune system. Mol Plant. 2021:14(10):1652–1667. 10.1016/j.molp.2021.06.010 [DOI] [PubMed] [Google Scholar]
- Lima AM, dos Reis SP, de Souza CR. Phytocystatins and their potential to control plant diseases caused by fungi. Protein Pept Lett. 2015:22(2):104–111. 10.2174/0929866521666140418101711 [DOI] [PubMed] [Google Scholar]
- Liu H, Hu M, Wang Q, Cheng L, Zhang Z. Role of papain-like cysteine proteases in plant development. Front Plant Sci. 2018:9:1717. 10.3389/fpls.2018.01717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Sonbol FM, Huot B, Gu YN, Withers J, Mwimba M, Yao J, He SY, Dong XN. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat Commun. 2016:7(1):13099. 10.1038/ncomms13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SK, Zhang XX. Expression and purification of a novel rice (Oryza sativa L.) mitochondrial ATP synthase small subunit in Escherichia coli. Protein Expr Purif. 2004:37(2):306–310. 10.1016/j.pep.2004.06.010 [DOI] [PubMed] [Google Scholar]
- Liu Y, He C. Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 2016:35(5):995–1007. 10.1007/s00299-016-1950-x [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang K, Cheng Q, Kong D, Zhang X, Wang Z, Wang Q, Qi X, Yan J, Chu J, et al. . Cysteine protease RD21A regulated by E3 ligase SINAT4 is required for drought-induced resistance to Pseudomonas syringae in Arabidopsis. J Exp Bot. 2020:71(18):5562–5576. 10.1093/jxb/eraa255 [DOI] [PubMed] [Google Scholar]
- Lukan T, Pompe-Novak M, Baebler S, Tusek-Znidaric M, Kladnik A, Kriznik M, Blejec A, Zagorscak M, Stare K, Dusak B, et al. . Precision transcriptomics of viral foci reveals the spatial regulation of immune-signaling genes and identifies RBOHD as an important player in the incompatible interaction between potato virus Y and potato. Plant J. 2020:104(3):645–661. 10.1111/tpj.14953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Dai Y, Zheng C, Yang Y, Chen W, Wang Q, Chandrasekaran U, Du J, Liu W, Shu K. The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 2021:229(2):950–962. 10.1111/nph.16921 [DOI] [PubMed] [Google Scholar]
- Martel A, Ruiz-Bedoya T, Breit-McNally C, Laflamme B, Desveaux D, Guttman DS. The ETS-ETI cycle: evolutionary processes and metapopulation dynamics driving the diversification of pathogen effectors and host immune factors. Curr Opin Plant Biol. 2021:62:102011. 10.1016/j.pbi.2021.102011 [DOI] [PubMed] [Google Scholar]
- Martinez M, Abraham Z, Carbonero P, Diaz I. Comparative phylogenetic analysis of cystatin gene families from Arabidopsis, rice and barley. Mol Genet Genomics. 2005a:273(5):423–432. 10.1007/s00438-005-1147-4 [DOI] [PubMed] [Google Scholar]
- Martinez M, Abraham Z, Gambardella M, Echaide M, Carbonero P, Diaz I. The strawberry gene Cyf1 encodes a phytocystatin with antifungal properties. J Exp Bot. 2005b:56(417):1821–1829. 10.1093/jxb/eri172 [DOI] [PubMed] [Google Scholar]
- Miller G, Schlauch K, Tam R, Cortes D, Torres MA, Shulaev V, Dangl JL, Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci Signal. 2009:2(84):ra45. 10.1126/scisignal.2000448 [DOI] [PubMed] [Google Scholar]
- Misas Villamil JC, Mueller AN, Demir F, Meyer U, Okmen B, Schulze Huynck J, Breuer M, Dauben H, Win J, Huesgen PF, et al. . A fungal substrate mimicking molecule suppresses plant immunity via an inter-kingdom conserved motif. Nat Commun. 2019:10(1):1576. 10.1038/s41467-019-09472-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. ROS are good. Trends Plant Sci. 2017:22(1):11–19. 10.1016/j.tplants.2016.08.002 [DOI] [PubMed] [Google Scholar]
- Moreland N, Ashton R, Baker HM, Ivanovic I, Patterson S, Arcus VL, Baker EN, Lott JS. A flexible and economical medium-throughput strategy for protein production and crystallization. Acta Crystallogr D Biol Crystallogr. 2005:61(10):1378–1385. 10.1107/S0907444905023590 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007:51(6):1126–1136. 10.1111/j.1365-313X.2007.03212.x [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Ding PT, Jones JDG. Thirty years of resistance: zig-zag through the plant immune system. Plant Cell. 2022:34(5):1447–1478. 10.1093/plcell/koac041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-López E, Hossain MM, Wei Y, Todd CD, Bonham-Smith PC. A clubroot pathogen effector targets cruciferous cysteine proteases to suppress plant immunity. Virulence 2021:12(1):2327–2340. 10.1080/21505594.2021.1968684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko GV, Juvale PS, Rutter WB, Hutten M, Maier TR, Hewezi T, Paulus J, van der Hoorn RAL, Grundler FMW, Siddique S, et al. . Re-targeting of a plant defense protease by a cyst nematode effector. Plant J. 2019:98(6):1000–1014. 10.1111/tpj.14295 [DOI] [PubMed] [Google Scholar]
- Radin I, Richardson RA, Coomey JH, Weiner ER, Bascom CS, Li T, Bezanilla M, Haswell ES. Plant PIEZO homologs modulate vacuole morphology during tip growth. Science 2021:373(6554):586––590.. 10.1126/science.abe6310 [DOI] [PubMed] [Google Scholar]
- Richau KH, Kaschani F, Verdoes M, Pansuriya TC, Niessen S, Stuber K, Colby T, Overkleeft HS, Bogyo M, Van der Hoorn RAL. Subclassification and biochemical analysis of plant papain-like cysteine proteases displays subfamily-specific characteristics. Plant Physiol. 2012:158(4):1583–1599. 10.1104/pp.112.194001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev. 2006:20(5):537–542. 10.1101/gad.366506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson PD, Warren EM, Zhang H, Friedman DB, Lary JW, Cole JL, Tutter AV, Walter JC, Fanning E, Eichman BF. Domain architecture and biochemical characterization of vertebrate Mcm10. J Biol Chem. 2008:283(6):3338–3348. 10.1074/jbc.M706267200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T, Van der Hoorn RA. Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol Plant Pathol. 2008:9(1):119–125. 10.1111/j.1364-3703.2007.00439.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Plas F, Elmayan T, Blein JP. The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 2002:31(2):137–147. 10.1046/j.1365-313X.2002.01342.x [DOI] [PubMed] [Google Scholar]
- Smith JM, Heese A. Rapid bioassay to measure early reactive oxygen species production in Arabidopsis leave tissue in response to living Pseudomonas syringae. Plant Methods 2014:10(1):6. 10.1186/1746-4811-10-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Kong X, Liu Y, Gong T, Gu X, Liu L. The THO/TREX complex active in alternative splicing mediates plant responses to salicylic acid and jasmonic acid. Int J Mol Sci. 2021:22(22):12197. 10.3390/ijms222212197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Wang SB, Wu JH, Wang YX, Wang XT, Liu SW, Han DJ, Xia GM, Wang MC. Allelic variation of TaWD40–4B.1 contributes to drought tolerance by modulating catalase activity in wheat. Nat Commun. 2023:14(1):1200. 10.1038/s41467-023-36901-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian YC, Fan M, Qin ZX, Lv HJ, Wang MM, Zhang Z, Zhou WY, Zhao N, Li XH, Han C, et al. . Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat Commun. 2018:9(1):1063. 10.1038/s41467-018-03463-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay J, Goulet MC, Michaud D. Recombinant cystatins in plants. Biochimie 2019:166:184–193. 10.1016/j.biochi.2019.06.006 [DOI] [PubMed] [Google Scholar]
- Van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008:59(1):191–223. 10.1146/annurev.arplant.59.032607.092835 [DOI] [PubMed] [Google Scholar]
- Van der Linde K, Hemetsberger C, Kastner C, Kaschani F, van der Hoorn RA, Kumlehn J, Doehlemann G. A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell. 2012:24(3):1285–1300. 10.1105/tpc.111.093732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wyk SG, Kunert KJ, Cullis CA, Pillay P, Makgopa ME, Schluter U, Vorster BJ. Review: the future of cystatin engineering. Plant Sci. 2016:246:119–127. 10.1016/j.plantsci.2016.02.016 [DOI] [PubMed] [Google Scholar]
- Waszczak C, Carmody M, Kangasjarvi J. Reactive oxygen species in plant signaling. Annu Rev Plant Biol. 2018:69(1):209–236. 10.1146/annurev-arplant-042817-040322 [DOI] [PubMed] [Google Scholar]
- Wong HL, Pinontoan R, Hayashi K, Tabata R, Yaeno T, Hasegawa K, Kojima C, Yoshioka H, Iba K, Kawasaki T, et al. . Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell. 2007:19(12):4022–4034. 10.1105/tpc.107.055624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014:14(1):327. 10.1186/s12870-014-0327-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan ZW, Wang JX, Wang FX, Xie CAT, Lv BS, Yu ZP, Dai SJ, Liu X, Xia GM, Tian HY, et al. . MPK3/6-induced degradation of ARR1/10/12 promotes salt tolerance in Arabidopsis. Embo Rep. 2021:22(10):e52457. 10.15252/embr.202152457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, He RJ, Xie QL, Zhao XH, Deng XM, He JB, Song L, He J, Marchant A, Chen XY, et al. . ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 2017:213(4):1667–1681. 10.1111/nph.14278 [DOI] [PubMed] [Google Scholar]
- Yuan M, Ngou BPM, Ding P, Xin XF. PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol. 2021:62:102030. 10.1016/j.pbi.2021.102030 [DOI] [PubMed] [Google Scholar]
- Zandalinas SI, Fichman Y, Mittler R. Vascular bundles mediate systemic reactive oxygen signaling during light stress. Plant Cell. 2020:32(11):3425–3435. 10.1105/tpc.20.00453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Tremousaygue D, Denance N, van Esse HP, Horger AC, Dabos P, Goffner D, Thomma BP, van der Hoorn RA, Tuominen H. PIRIN2 stabilizes cysteine protease XCP2 and increases susceptibility to the vascular pathogen Ralstonia solanacearum in Arabidopsis. Plant J. 2014:79(6):1009–1019. 10.1111/tpj.12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu S, Takano T. Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol. 2008:68(1–2):131–143. 10.1007/s11103-008-9357-x. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006a:125(4):749–760. 10.1016/j.cell.2006.03.037 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 2004:428(6984):764–767. 10.1038/nature02485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.