Abstract
The COVID-19 pandemic has presented a significant challenge to the world’s public health and led to over 6.9 million deaths reported to date. A rapid, sensitive, and cost-effective point-of-care virus detection device is essential for the control and surveillance of the contagious severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic. The study presented here aimed to demonstrate a solid-phase isothermal recombinase polymerase amplification coupled CRISPR-based (spRPA-CRISPR) assay for on-chip multiplexed, sensitive and visual COVID-19 DNA detection. The assay targets the SARS-CoV-2 structure protein encoded genomes and can simultaneously detect two specific genes without cross-interaction. The amplified target sequences were immobilized on the one-pot device surface and detected using the mixed Cas12a-crRNA collateral cleavage of reporter-released fluorescent signal when specific genes were recognized. The endpoint signal can be directly visualized for rapid detection of COVID-19. The system was tested with samples of a broad range of concentrations (20 to 2 × 104 copies) and showed analytical sensitivity down to 20 copies per microliter. Furthermore, a low-cost blue LED flashlight (~$12) was used to provide a visible SARS-CoV-2 detection signal of the spRPA-CRISPR assay which could be purchased online easily. Thus, our platform provides a sensitive and easy-to-read multiplexed gene detection method that can specifically identify low concentration genes.
Keywords: COVID-19, CRISPR-Based assay, Multiplexed spRPA-CRISPR assay, DNA sensor, Point-of-care, Visual detection
1. Introduction
The outbreak of the SARS-CoV-2, also known as COVID-19, has rapidly spread to produce a global pandemic and caused a huge number of deaths worldwide. The lack of rapid accessible and accurate molecular diagnostic testing methods has hindered the COVID-19 testing response as the pandemic quickly spread. Currently, the global standard approach approved for COVID-19 diagnosis is the quantitative reverse transcription polymerase chain reaction (RT-qPCR) (Corman et al., 2020), (Chuang, 2020). Although it is highly sensitive and specific, it requires specialized equipment and trained personnel, which limits its use in resource-limited settings. Due to the rapid growth of the infected population, there is a global shortage of RT-qPCR assay kits as well as their associated reagents (Lee et al., 2020). The detection of the virus using RT-qPCR usually takes 4–6 h, and the shipping of samples to the local laboratories has delayed the turnaround diagnosing time to >24 h (Broughton et al., 2020). Therefore, there is a pressing need for the development of rapid, high sensitive, multiplexed (Yang et al., 2015), (Shafiee et al., 2015) and precision point-of-care tools (Lee et al., 2021), (Kahng et al., 2020), (Zhou et al., 2014) for gene diagnostic technologies that can be used in a range of settings, including low-resource and remote areas.
The emerging isothermal nucleic acid amplification technologies, including recombinase polymerase amplification (RPA) and loop-mediated isothermal amplification (LAMP), eliminate the need for thermal cycling, that is required by traditional PCR method. Both RPA and LAMP amplification reactions use a specific set of primers that bind to the target DNA or RNA, initiating the amplification process. The LAMP technology typically requires six primers with the reaction temperature at 50–72 °C, while the RPA appears to have some attractive features as it requires only two primers with a faster reaction time and optimally works under lower working temperatures ranging from 37 °C to 42 °C (Iwamoto et al., 2003), (Park et al., 2020). However, in the RPA reaction, the formation of primer-dimers could cause the production of the nonspecifically amplified by-products with the presence of low concentration target DNA (del Río et al., 2014). Solid-phase amplification combines the inherent advantages of RPA and addresses its drawbacks. It is a detection technique developed based on RPA, where the primers are pre-immobilized onto the substrate (del Río et al., 2017). During amplification, a specific surface area is used to covalently immobilize one of the primers, allowing for the amplification of the target DNA through the elongation of the primers in both liquid and solid phases simultaneously. This solid-phase amplification approach minimizes the formation of primer-dimers and reduces the production of randomly sequenced by-products (Kersting et al., 2014). Previously reported multiplex testing platform (Sun et al., 2020) has pre-immobilized the LAMP primers by drying down on the microfluidic channel surface, which enabled a one-step amplification reaction by reconstituting the primers into the solution. Solid-phase RPA could easily be adaptable to multiplexed capacity by immobilizing different surface primers in an array format on-chip (del Río et al., 2017) and also allow further detection steps (e.g. CRISPR assay) on the immobilized amplified DNA targets. The solid-phase RPA technology was previously developed for bacterial pathogen DNA detection as point-of-care testing and to enable simplified and miniaturized nucleic acid-based diagnostics with a sensitivity of 10–100 colony forming units (Kersting et al., 2014). Also, it was reported that the Francisella tularensis detection has a LOD of 1.3 × 10−13 M (4 × 106 copies in 50 μL) in Ciara K. O’Sullivan’s work and have improved with one magnitude higher in their more recent work published in 2017 (del Río et al., 2014), (del Río et al., 2017).
RT-RPA or RT-LAMP have emerged as potential detection methods for COVID-19, with various studies demonstrating their efficacy (Iwamoto et al., 2003), (Sun et al., 2020)– (Zhang et al., 2020). To further improve the sensitivity of these isothermal DNA amplification assays, they can be coupled with the clustered regularly interspaced short palindromic repeats (CRISPR) system (Broughton et al., 2020), (Dara and Talebzadeh, 2020), (Ramachandran et al., 2020). Cas proteins in the CRISPR system can activate upon binding with target double-stranded DNA, unleashing nonspecific single-stranded DNA cleavage activity by AsCas12a (Cpf1) proteins (Kersting et al., 2014), (Ruff et al., 2016)– (Sin et al., 2009). When combined with DNA amplification, the CRISPR-based assay typically visualized the results on the lateral flow strips or fluorescent readers with good sensitivity of a few copies per microliter (Broughton et al., 2020), (Ning et al., 2021). However, these CRISPR-based DNA detection methods typically require multiple manual operations which potentially increase the risk of contaminations while transferring products between wells.
In this study, we presented the development of a novel one-pot solid-phase RPA coupled with the CRISPR-based (spRPA-CRISPR) multiplex detection system for the sensitive detection of SARS-CoV-2 related genes. The spRPA-CRISPR system utilizes single cartridge mixture of CRISPR RNA probes for E or N genes to specifically detect thiol-group immobilized primers and amplified gene targets without signal cross-walk. The endpoint assay results can be directly visualized under a commercially available blue LED flashlight and the system exhibits a lower limit of detection of 20 copies per microliter with a broad testing range at a constant temperature of 37 °C. The use of spRPA simplifies the chip design and handling procedures, making it easily adaptable for sensitive point-of-care multiplex virus detection.
2. Materials and methods
2.1. Materials and primer design
The TwistAmp™ basic kit was purchased from TwistDx (Babraham, UK, www.twistdx.co.uk) for RPA reactions. Primers and crRNA were designed by following the TwistDx kit instruction and IDT protocol, as shown in Table S1. Oligonucleotides (primers and crRNAs) and ssDNA-FQ reporters were purchased from IDT (Integrated DNA Technologies, US). The SARS-CoV-2 plasmid purchased from IDT is specially designed for research use only. The DNA template using COVID-19 N gene plasmid (2019-nCoV_N_Positive Control, IDT) and E gene (2019-nCoV_E_Positive Control, IDT). The RPA amplified DNA products were purified using QIAquick PCR Purification Kit (Qiagen) and were confirmed by agarose gel electrophoresis. The Sylgard 184 PDMS elastomer was mixed with the curing agent at a ratio of 10 and poured into a plastic Petri dish at 3 mm height. After curing, the PDMS was punched with 0.5 cm diameter holes and attached to the gold-coated substrates (EMS, US).
2.2. Probe verification by RPA assay
The materials for the RPA reaction including buffers and reagents were supplied in the TwistDx kit. The primers were first evaluated with RPA reaction for further solid-phase reaction. The RPA assay was tested in a 50 μL reaction volume containing 480 nM of forward and reverse primers, 1x rehydration buffer, and 2000 copies of genomic DNA strand. This reaction mixture was added to one tube of freeze-dried TwistAmp Basic reaction pellet and 14 mM Magnesium acetate was then added to the mix to start the reaction. The samples were incubated at 37 °C for 20 min. Amplicons generated were then cleaned of DNA-binding proteins using PCR purification columns to be visualized using 2% agarose gel electrophoresis. The amplified products were detected using ethidium bromide (Thermo Fisher).
2.3. CRISPR Cas12 cleavage reaction and LED readout
For the CRISPR assay, the crRNA of E gene or N gene were first assembled into Cas12a-crRNA complex with a total of 200 nM of AsCas12a (Cpf1) Nuclease (Cat. 1081068, IDT) was preincubated with 250 nM g-RNA in 1x binding buffer (20 mM Tris-HCl, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol) at 37 °C for 30 min. The reactions were performed separately for the N gene and E gene. After forming the Cas12a-crRNA complex, the FQ reporter was added to the reaction product at a final concentration of 500 nM. The complexes were placed on ice for immediate use or stored at 4 °C for up to 24 h. After the RPA amplification step, 2 μL of amplicons and 20 μL of CRISPR reaction complexes were mixed with 28 μL of 1x binding buffer to allow the Cas12a trans-cleavage assay to proceed. After 10 min incubation at 37 °C, the samples were ready to read by plate reader using blue excitation light or visualized under an LED flashlight (Vansky).
2.4. Solid-phase RPA reaction
2.4.1. Immobilization of forward primer
The forward primer was designed with a poly-15T at the 5′ end of the probe as a vertical spacer. 6-mercapto-1-hexanol (MCH, Sigma-Aldrich) is a commonly used, stable, and cost-effective lateral spacer. The MCH was optimized to be mixed with forward primers with a 1:10 ratio for surface immobilization (del Río et al., 2017). Reduction for oligos with thiol modification has followed the protocol provided by IDT. 1 μL of 10 μM thiolated forward primer for E gene or N gene with 100 μM MCH were prepared in 1M KH2PO4 (Sigma-Aldrich), and the mixtures were added into separate PDMS chambers for immobilization on the gold surface. The coating was carried out in a humid chamber at room temperature for 20 h. The gold surface was then washed to remove nonspecific immobilization of DNA by (1) rinsing in Milli-Q water for 5 min (2) drying in the stream of N2. (3) washing for 5 min in PBS, Tween 20, and Milli-Q. The surface was then blocked with Denhardt’s solution (ThermoFisher) at room temperature for 20 min to prevent nonspecific binding during subsequent steps. As a final step, the chamber was washed with PBS and Milli-Q water for 5 min and dried with the nitrogen stream.
2.4.2. Solid-phase RPA reaction on gold surface
Solid-phase RPA was performed by mixing 2.4 μL of 10 μM reverse primer, 13.2 μL of 10 nM DNA template, 1x rehydration buffer, DNase free water mixed with lyophilized pellet from TwistAmp Basic kit. The master mix of all reagents except for magnesium acetate was prepared and pipetted to transfer it into PDMS wells. Finally, add 2.5 μL of 280 mM magnesium acetate gently into each PDMS well as the last step to initiate the amplification. The samples were left to incubate at 37 °C for 30 min.
2.4.3. One-well spRPA-CRISPR detection with LED readout
The CRISPR reagent preparation was described in the “CRISPR reaction and LED readout” section. The spRPA amplified surface was rinsed twice with PBS. The crRNA of the N gene or E gene were first prepared with CRISPR-Cas12a protein and formed crRNA-CRISPR complex and mix with the fluorophore-quencher (FQ) reporter, and 1x binding buffer to bring the final reaction volume to 50 μL. After incubating for 10 min at 37 °C, the samples were ready to detect under an LED flashlight or plate reader. The visible colorimetric detection result was analyzed using ImageJ software by splitting the green channel and measuring the fluorescent intensity (Ding et al., 2020).
3. Results
3.1. The solid-phase diagnosis system design and validation
The solid-phase diagnosis system, as depicted in Fig. 1, was designed, and validated for the rapid detection of SARS-CoV-2. The system consists of PDMS wells, a gold-coated glass slide, a hot plate, and a blue LED flashlight. The utilization of DNA-based biosensors on gold-coated surfaces has demonstrated favorable properties for commercial applications. Extensive researches have confirmed the stability and consistent response of the robust covalent thiol-Au bond, as reported in previous studies (del Río et al., 2014), (Love et al., 2005)– (Su et al., 2016). To enhance the long-term stability of these biosensors during storage, investigations have revealed that the DNA-modified thiol-Au bond remains stable under most conditions including low temperatures and low salt concentrations (Li et al., 2013), (Herdt et al., 2006). Therefore, based on established protocols, we recommend a rinsing step of the primer-immobilized gold surface using Milli-Q water to eliminate ions, followed by drying the surface with a nitrogen beam. For long-term storage, the surface immobilization of the primers was performed in advance and could be stored under vacuum (del Río et al., 2017) or kept at 4 °C for at least 2 months (del Río et al., 2014) (Oberhaus et al., 2020). By adopting these recommended procedures, the stability of the DNA-modified thiol-Au bond can be preserved, ensuring the reliability and integrity of the biosensor during storage (Sun et al., 2007). These findings contribute to the optimization of long-term storage conditions for DNA-based biosensors, facilitating their practical use in various applications (Sun et al., 2007), (Poon et al., 2010), (Rasheed et al., 2022). Furthermore, in our study, Cas12a-crRNA complexes are prepared and added to the spRPA well with the ssDNA FQ reporter. The one-pot CRISPR-based reaction was conducted on a hot plate set at 37 °C, and was visualized after 10 min.
Fig. 1. The schematic of spRPA-CRISPR platform.
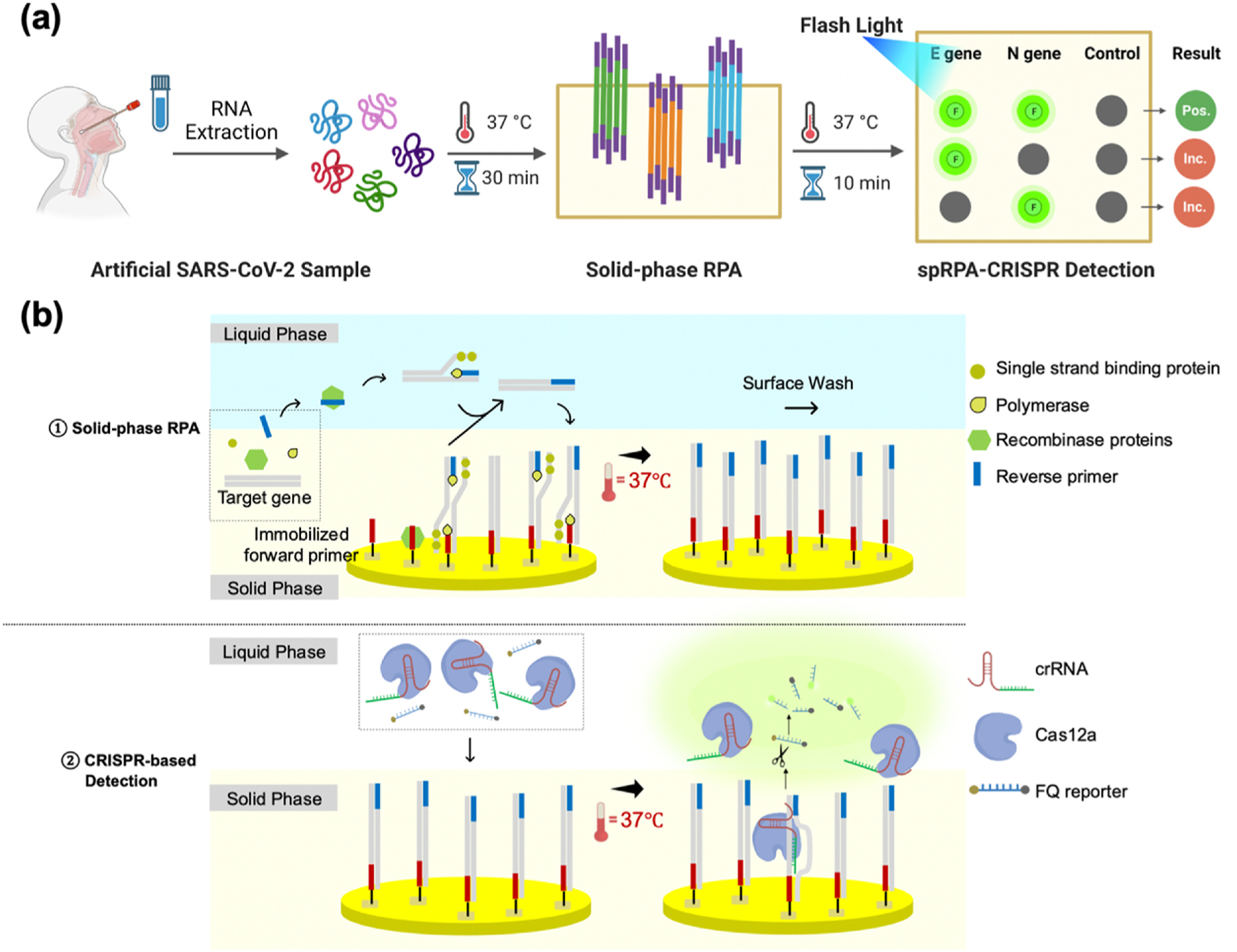
(a) The workflow of the detection of the artificial SARS-CoV-2 sample using spRPA-CRISPR platform. The “Pos.” stands for the positive results and the “Inc.” represents the inconclusive results. Illustration created with BioRender.com (b) spRPA (top) and CRISPR-based SARS-CoV-2 colorimetric detection (bottom).
To assess the ability of amplifying the SARS-CoV-2 target genes in the solid-phase RPA, liquid-phase RPA reactions were first performed using the designed primers (Table S1). The results showed that the liquid-phase RPA reaction was effective in binding the primers to the genomic DNA template and amplifying the target sequences. The successful amplification of the target N gene and E gene was confirmed by running an agarose gel, as shown in Fig. 2 (a). The performance of the CRISPR-Cas12a assay on the RPA products was then evaluated. The assay is considered positive if the fluorescent signal is detected for both E and N genes, and potentially positive if either the E or N gene is detected. This interpretation is in accordance with the Emergency Use Authorization (EUA) (Cepheid, 2022). The crRNAs were specifically designed for E and N genes separately. The CRISPR-Cas12a enzyme complexed crRNA could specifically detect the genes that carry complementary sequences from the RPA product. The fluorescent signal was generated when the Cas12a-crRNA complex recognized the predefined target sequences and collaterally cleaved the surrounding FQ reporter molecules. The flashlight allowed sensitive and quantitative readout of the CRISPR-Cas12-based assay. In Fig. 2 (b), the fluorescent signal for both E and N genes was detected under the excitation of a blue LED flashlight or fluorescent plate scanner. The visible signal was directly observed under the flashlight and imaged using a regular cellphone camera. The resulting images were then split into the red, blue, and green color channels using ImageJ. The fluorescent intensity measurements were analyzed from both the green color channel of the visual detection result and the measurement from the plate scanner. As shown in Fig. 2 (b), fluorescent intensity measurements for both blue LED flashlight visual detection and plate reader agreed with each other.
Fig. 2. Primers and probe verification with RPA-based CRISPR detection assay.
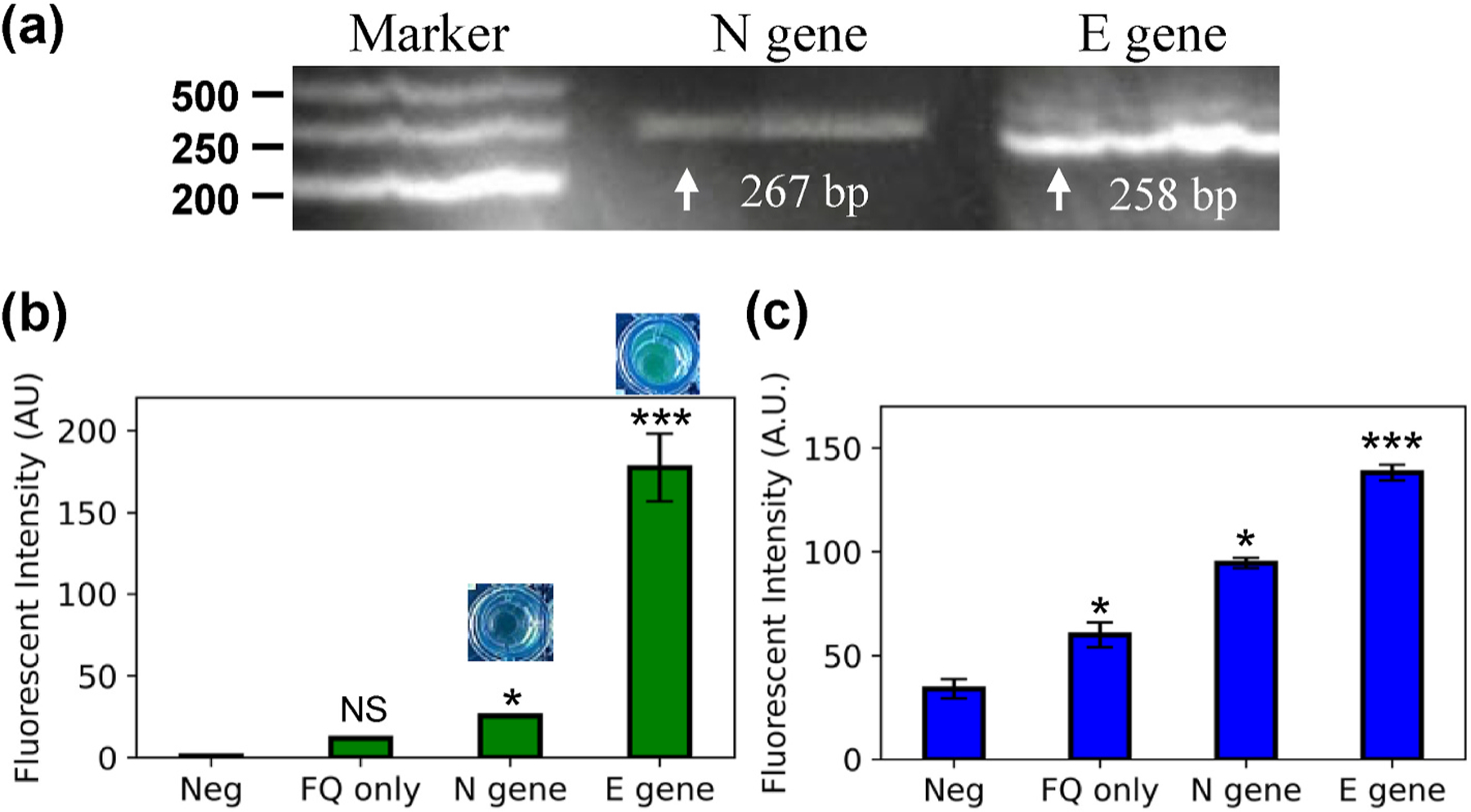
(a) Agarose gel (2%) illustration of liquid-phase RPA product of N gene and E gene. (b) CRISPR-based detection result read on plate reader. (c) CRISPR-based detection read directly under LED flashlight.
3.2. Evaluation of solid-phase RPA-CRISPR assay for COVID-19 diagnosis
The purchased SARS-CoV-2 plasmid containing the complete nucleocapsid gene from 2019-nCoV was used to demonstrate the feasibility of our multiplexed isothermal spRPA-CRISPR mechanism. The spRPA reaction was performed using a gold surface as the solid phase and bulk solution as liquid phase, with a thiolated forward primer covalently bonded to the gold layer supported by a glass substrate. The immobilized primers facilitated elongation by the polymerase reaction, with a vertical poly-15T spacer projecting the primer from the surface and a lateral MCH spacer avoiding undesirable interactions between neighboring primers (del Río et al., 2017). As shown in Fig. 1, spRPA was performed on both solid and liquid phases, with the immobilized forward primers serving as a starting point and elongating the strand following the polymerase. In the liquid phase, the DNA first elongates along the reverse primer and then releases it to hybridize specifically to the immobilized forward primers (Kersting et al., 2014). To perform the CRISPR assay with the DNA amplified only from the immobilized primers, the surface was rinsed twice with PBS before introducing the Cas12a-crRNA complex and FQ reporter to the system.
The system was then tested on the solid-phase multiplexing platform, where gene-specific amplification along the surface-immobilized primers was achieved within 30 min. The CRISPR-Cas-based assay was performed as an extended step to enhance the detection target viral genome amplicon signal and showed a visible signal within 10 min incubation. To identify the linear range and LOD of the optimized assay, the target plasmid templates were tested in the concentration range of 0 copies to 20000 copies per μL. The results were shown in Fig. 3. (a), where distinct fluorescent signals were shown between the negative control group where primers were not amplified with un-cleaved FQ reporter and the sample groups for the LOD test. A strong fluorescent signal was detected for both the blue LED flashlight excited visualized signal and the fluorescent imaging scanner.
Fig. 3. Sensitivity of spRPA-based CRISPR assay.
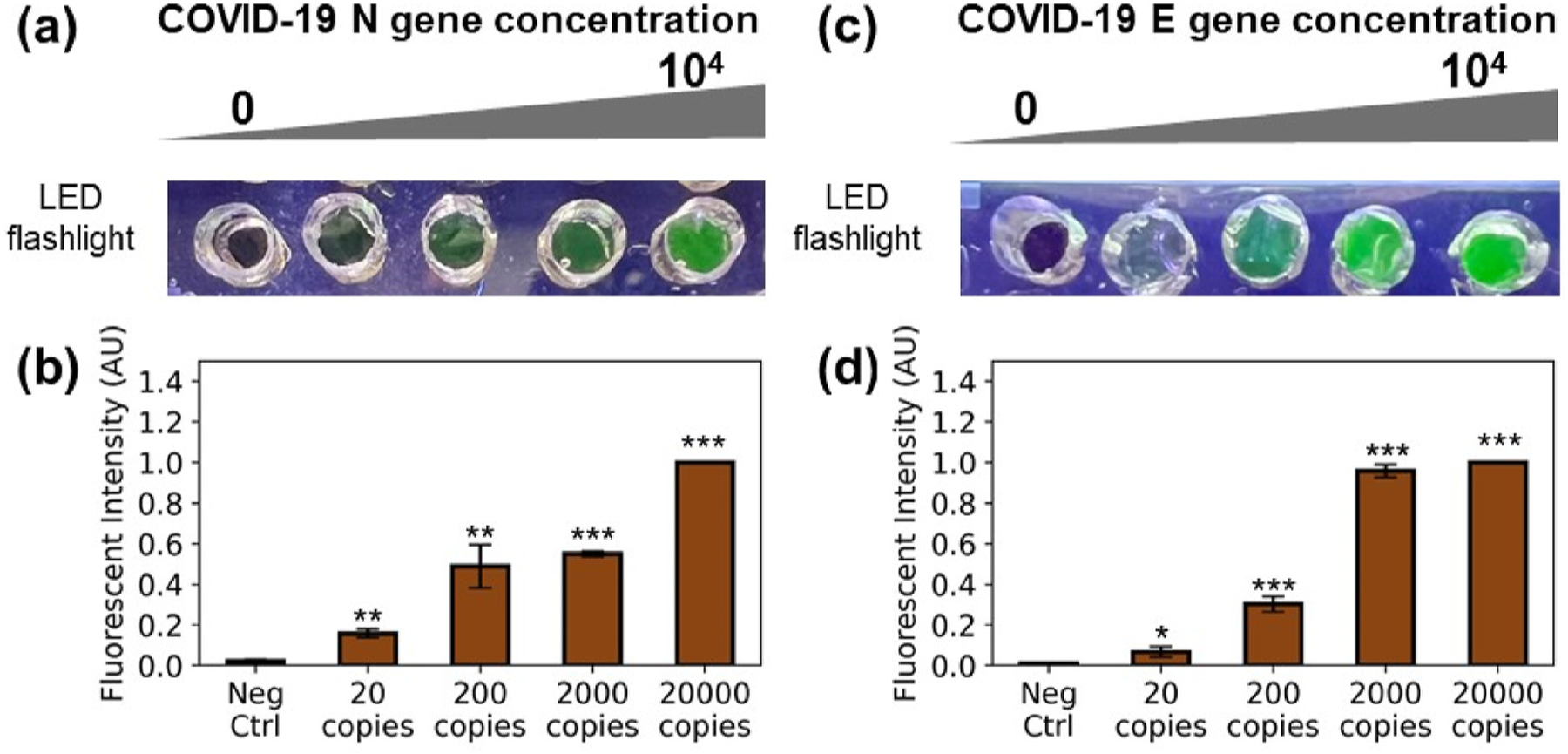
(a) Fluorescence signal excited with LED flashlight of N gene (0, 20, 200, 2000, 20000 copies per microliter). (b) Fluorescence signal measurement of N gene on the imaging scanner. (c) Fluorescence signal excited with LED flashlight of E gene (0, 20, 200, 2000, 20000 copies per microliter). (d) Fluorescence signal measurement of E gene on the imaging scanner.
To evaluate the specificity of our sp-RPA-based CRISPR system for the multiplexed detection, we simultaneously detect the N gene and E gene using Cas12a-crRNA complex of E gene or N gene separately into E gene or N gene amplified wells. The negative wells exhibited a low level of background fluorescent signal presumably from the weak auto-fluorescence from the FQ reporter and a low level of nonspecific primer-dimer that existed in the DNA amplification (Wang et al., 2020). As shown in Fig. 4 (a), the Cas12a-crRNA complexes of the E gene and N gene were added separately into the spRPA product amplified from the E gene primer. When the Cas12a-crRNA complex finds its complementary genes, the wells display strong fluorescent signals which indicate successful detection, and while the Cas12a-crRNA complex was not able to find its target sequences, the wells remain dark. In Fig. 4 (d), both N gene and E gene crRNA complexed Cas12a-crRNA were mixed and added separately into the solid phase N gene or E gene amplification well. Therefore, the COVID-19 E gene and N gene were accurately identified and distinguished from each other using the gene-specific spRPA-CRISPR assay, with direct visualization of the detection of both targets. The results highlight the effectiveness and versatility of our on-chip spRPA-CRISPR-based detection system for the simultaneous detection of multiple targets in a single reaction.
Fig. 4. Multiplex detection of COVID-19 genes.
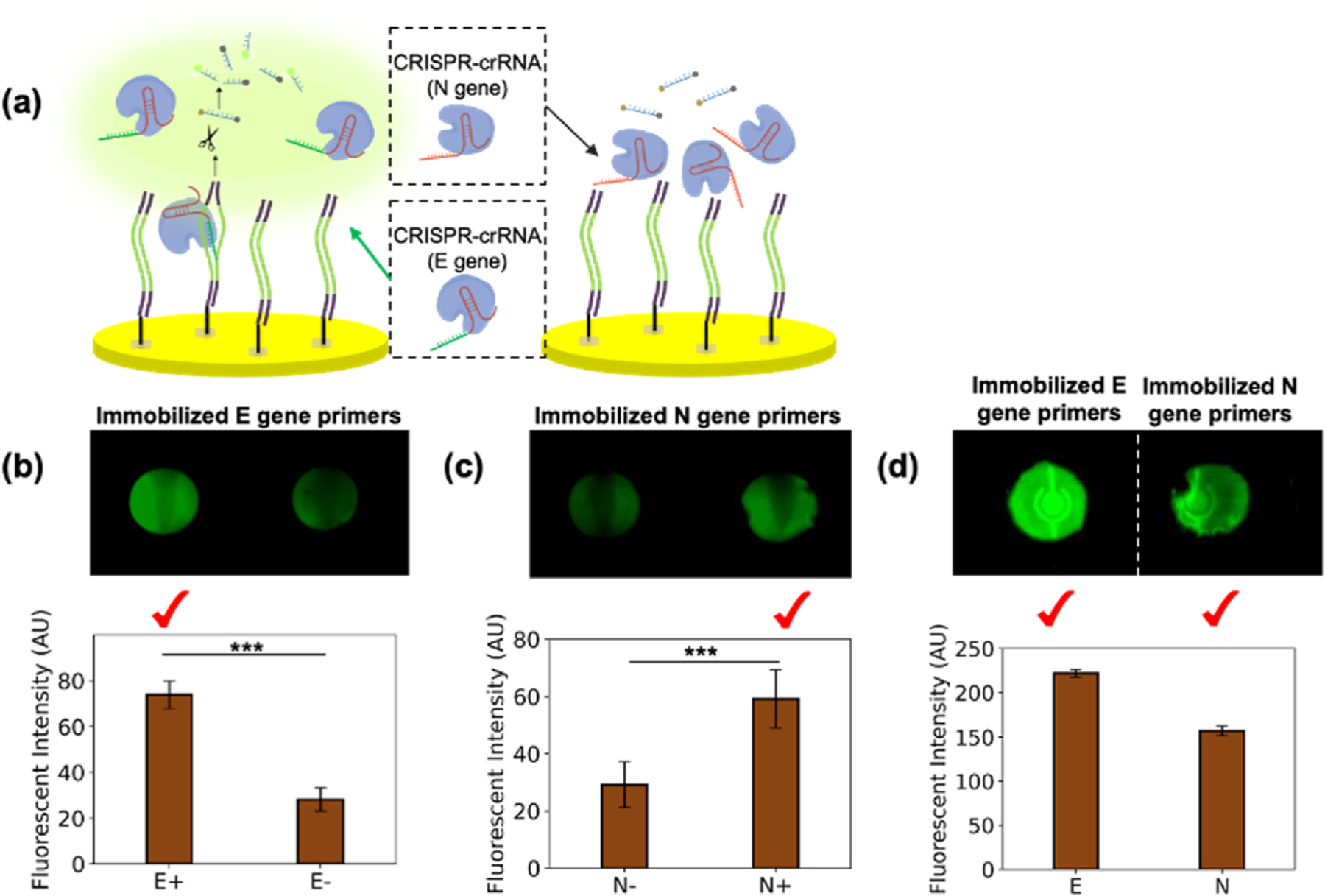
(a) Illustration of the gene specificity of CRISPR detection on the spRPA product and demonstrated the wells in (b) where the Cas12a-crRNA of E gene bind with amplified E gene thus triggered the trans-cleavage of FQ reporter, while the Cas12a-crRNA of N gene were not able to activate the amplified E gene. (b) CRISPR detection fluorescent image and the fluorescent intensity scan for E gene specificity test: spRPA amplified E gene on the well bottom; Left well: detected using Cas12a-crRNA of E gene; Right well: detected using Cas12a-crRNA of N gene. (c) The CRISPR detection fluorescent image and the fluorescent intensity scan for N gene specificity test: spRPA amplified N gene in both wells. Left well: detected using Cas12a-crRNA of E gene; Right well: detected using Cas12a-crRNA of N gene. (d) The single cartridge mixed Cas12a-crRNA of both E gene and N gene detected immobilized E gene (left) and immobilized N gene (right).
To further demonstrate the point-of-care diagnostic application, we analyzed the visual detection signal under a blue LED flashlight excitation. The on-chip spRPA-CRISPR assay was performed by preparing N gene, E gene primers, and a no-template negative control separately within each well. The CRISPR assay was performed using a single cartridge that mixed crRNA of both N gene and E gene with the spRPA product that amplified from 200 copies/μL for each of the target genes. The fluorescent signals were detectable after 10 min of incubation and visualized under the LED flashlight. The workflow of the spRPA-CRISPR assay, including signal visualization and fluorescent signal analysis, was shown in Fig. 5. The results indicate that the on-chip spRPA-CRISPR assay was able to detect both N and E genes of SARS-CoV-2 within a total detection time of ~40min. The fluorescent signals were distinguishable from the no-template negative control.
Fig. 5. Workflow of the spRPA-CRISPR chip for DNA detection.
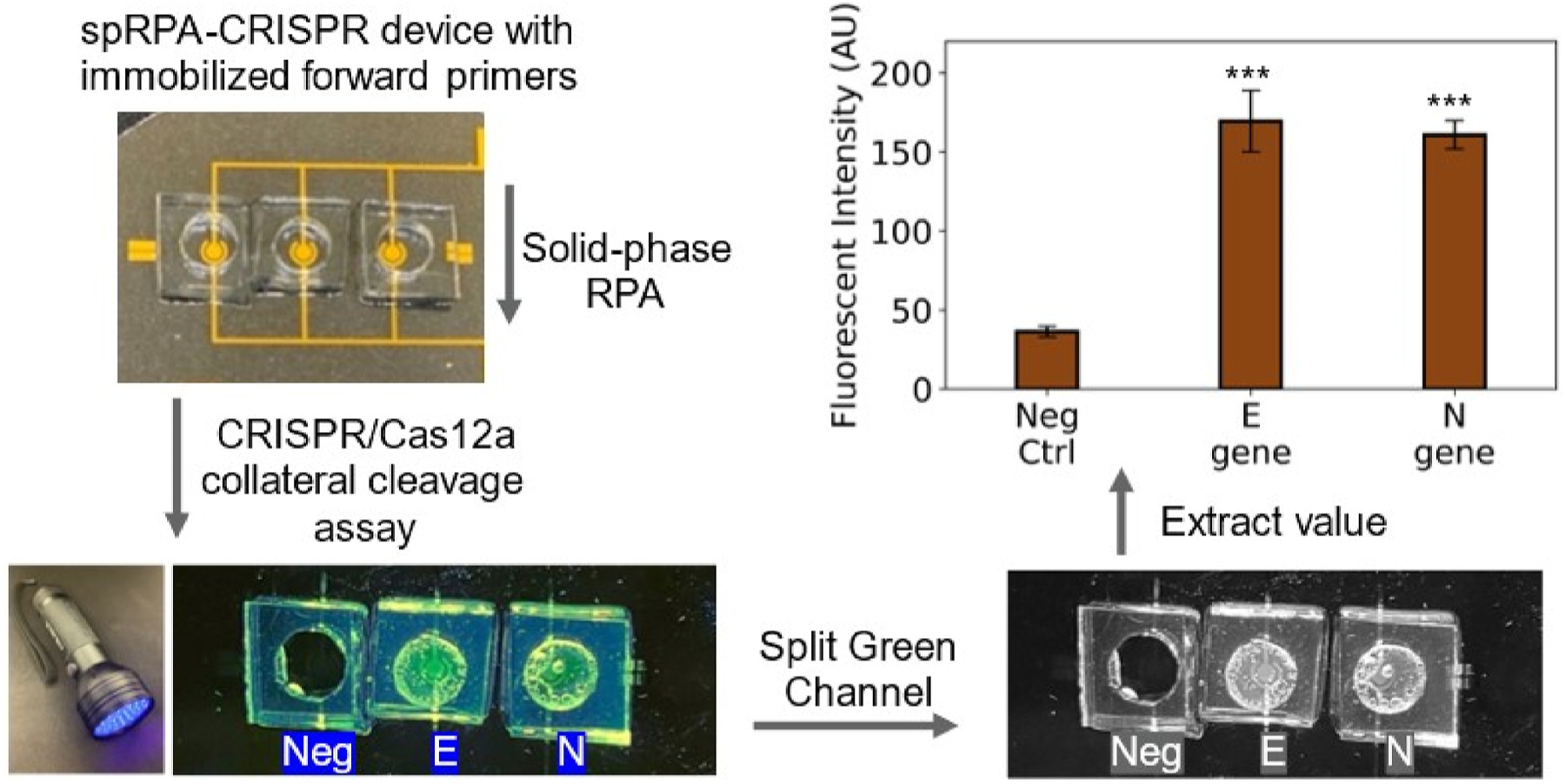
Fabricated prototype device bound with PDMS chamber and use LED flashlight to observe the endpoint fluorescent signal. Images were taken using a smartphone camera and analyzed the intensity by splitting green channel using ImageJ. Single cartridge reaction containing Cas12a-crRNA complex with both E gene and N gene test on the spRPA amplified target DNA templates with negative control with no amplified gene, E gene, and N gene (left to right). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusions and discussions
In this study, we have developed a one-pot solid-phase RPA coupled with a CRISPR-based (spRPA-CRISPR) multiplex detection system for the sensitive and visual detection of SARS-CoV-2. By integrating spRPA and CRISPR technologies, we achieve rapid and specific amplification of target genes, followed by detection through the cleavage of reporter molecules. The utilization of spRPA offers the advantage of simplified chip design and streamlined handling procedures, facilitating its seamless integration into sensitive point-of-care multiplex virus detection. Moreover, this innovative approach enhances the sensitivity of traditional spRPA through its coupling with CRISPR technologies, allowing for the detection of target genes at an unprecedented low concentration of 20 copies/μL. Additionally, it offers a reliable and user-friendly diagnostic platform, providing convenience and accuracy in virus detection. Furthermore, we have successfully demonstrated the direct visualization of detection results using a commercially available blue LED flashlight. This cost-effective and accessible visualization method eliminates the need for complex equipment, enhancing the practicality and broad applicability of this diagnostic approach.
Our spRPA-CRISPR assay shows great potential of our spRPA-CRISPR assay as a reliable diagnostic tool for the detection of SARS-CoV-2. It exhibits high sensitivity, specificity, and cost-effectiveness, which are crucial in the management of the ongoing COVID-19 pandemic. The utilization of solid-phase RPA used in our assay as a starting point for the amplification of target genes, combined with CRISPR-based assay, allows for the specific detection of the “pre-programmed” N gene, E gene, and control panels. The direct visualization of the endpoint results using a low-cost LED flashlight eliminates the need for extra lateral flow assay steps and simplifies the detection process, making it easier to use in point-of-care settings. Furthermore, our system has the potential to be integrated into a disposable microfluidic system that can be stored at room temperature without the need for cold chains, making it a more convenient and accessible diagnostic tool (Broughton et al., 2020).
Moving forward, we plan to conduct clinical evaluations and validations of our spRPA-CRISPR system. We also aim to integrate the system into a microfluidic device and take advantage of smartphone cameras to record and analyze the endpoint signals. This will enable quantitative test results to be quickly transmitted to healthcare professionals and public health organizations, allowing for timely interventions to be made.
In conclusion, our spRPA-CRISPR system has the potential to significantly improve the diagnosis of COVID-19 and other infectious diseases. Similar CRISPR biosensor chips can be used for the detection of HPV DNA, influenza, and other viral transfection diseases. We believe that the integration of our system into a disposable microfluidic device and the use of smartphone cameras for recording and analyzing results will greatly enhance the accessibility and convenience of diagnostic testing.
Supplementary Material
Acknowledgement
This work was supported by U.S. Department of Health & Human Services | National Institutes of Health (NIH) grant R21EB033102, National Science Foundation grant CBET 2039310, OAC 2215789, BCS 2200066, Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program (CURE), and Pennsylvania Infrastructure Technology Alliance (PITA). Discussion with Himanshu Jain and support of Institute for Functional Materials and Devices (I-FMD) are appreciated.
Footnotes
Credit author statement
Xiaochen Qin designed and constructed the solid-phase RPA based CRISPR detection system, performed the lithography and data analysis, designed primers and CRISPR probes, designed the surface gold patterns. interpreted the data and wrote the manuscript. Ratul Paul performed the lithography and data analysis, revised the manuscript. Yuyuan Zhou designed primers and CRISPR probes, revised the manuscript. Yue Wu designed the surface gold patterns, revised the manuscript. Xuanhong Cheng revised the manuscript. Yaling Liu interpreted the data, wrote the manuscript, developed the idea, and revised the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biosx.2023.100381.
Data availability
Data will be made available on request.
References
- Broughton JP, et al. , 2020. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol 38 (7), 870–874. 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepheid, 2022. Package Insert, “Xpert® Xpress SARS-CoV-2. https://www.fda.gov/media/136315/download.
- Chuang L, 2020. Labs in the time of COVID: an early-career scientist’s view. Dis. Model. Mech 13 (6) 10.1242/DMM.046151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, et al. , 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25 (3) 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dara M, Talebzadeh M, 2020. CRISPR/Cas as a potential diagnosis technique for COVID-19. Avicenna J. Med. Biotechnol. (AJMB) 12 (3), 201–202.: Aug. 03, 2020. [Online]. Available: [PMC free article] [PubMed] [Google Scholar]
- del Río JS, Yehia Adly N, Acero-Sanchez JL, Henry OYF, O’Sullivan CK, 2014. Electrochemical detection of francisella tularensis genomic DNA using solid-phase recombinase polymerase amplification. Biosens. Bioelectron 54, 674–678. 10.1016/j.bios.2013.11.035. [DOI] [PubMed] [Google Scholar]
- del Río JS, Lobato IM, Mayboroda O, Katakis I, O’Sullivan CK, 2017. Enhanced solid-phase recombinase polymerase amplification and electrochemical detection. Anal. Bioanal. Chem 409 (12), 3261–3269. 10.1007/s00216-017-0269-y. [DOI] [PubMed] [Google Scholar]
- Ding X, et al. , 2020. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun 111 10.1038/s41467-020-18575-6, 11, no. 1, pp. 1–10, Sep. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdt AR, Drawz SM, Kang Y, Taton TA, 2006. DNA dissociation and degradation at gold nanoparticle surfaces. Colloids Surf. B Biointerfaces 51 (2), 130–139. 10.1016/j.colsurfb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Sonobe T, Hayashi K, 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol 41 (6), 2616–2622. 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahng S-J, Soelberg SD, Fondjo F, Kim J-H, Furlong CE, Chung J-H, 2020. Carbon nanotube-based thin-film resistive sensor for point-of-care screening of tuberculosis. Biomed. Microdevices 22 (3), 1–10. 10.1007/S10544-020-00506-3, 2020 223. [DOI] [PubMed] [Google Scholar]
- Kersting S, Rausch V, Bier FF, von Nickisch-Rosenegk M, 2014. Multiplex isothermal solid-phase recombinase polymerase amplification for the specific and fast DNA-based detection of three bacterial pathogens. Microchim. Acta 181 (13–14), 1715–1723. 10.1007/s00604-014-1198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JYH, et al. , 2020. Validation of a single-step, single-tube reverse transcription-loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. bioRxiv 2020. 10.1101/2020.04.28.067363, 04.28.067363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kim Y-S, Yeo W-H, 2021. Advances in microsensors and wearable bioelectronics for digital stethoscopes in health monitoring and disease diagnosis. Adv. Healthc. Mater, 2101400 10.1002/ADHM.202101400. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang H, Dever B, Li XF, Le XC, 2013. Thermal stability of DNA functionalized gold nanoparticles. Bioconjugate Chem 24 (11), 1790–1797. 10.1021/bc300687z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM, 2005. Self-assembled monolayers thiolates Met. form of Nanotechnol 105 (4) 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- Ning B, et al. , 2021. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv 7 (2) 10.1126/SCIADV.ABE3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhaus FV, Frense D, Beckmann D, 2020. Immobilization techniques for aptamers on gold electrodes for the electrochemical detection of proteins: a review. Biosensors 10 (5). 10.3390/BIOS10050045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GS, et al. , 2020. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Mol. Diagnostics 22 (6), 729–735. 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon L, et al. , 2010. Photothermal release of single-stranded DNA from the surface of gold nanoparticles through controlled denaturating and Au-S bond breaking. ACS Nano 4 (11), 6395–6403. 10.1021/nn1016346. [DOI] [PubMed] [Google Scholar]
- Ramachandran A, et al. , 2020. “Electric-field-driven Microfluidics for Rapid CRISPR-Based Diagnostics and its Application to Detection of SARS-CoV-2,” 10.1101/2020.05.21.109637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed PA, Pandey RP, Jabbar KA, Mahmoud KA, 2022. Nb4C3Tx (MXene)/Au/DNA aptasensor for the ultraselective electrochemical detection of lead in water samples. Electroanalysis 34 (10), 1540–1546. 10.1002/elan.202100685. [DOI] [Google Scholar]
- Ruff P, Donnianni RA, Glancy E, Oh J, Symington LS, 2016. RPA stabilization of single-stranded DNA is critical for break-induced replication. Cell Rep 17 (12), 3359–3368. 10.1016/j.celrep.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafiee H, et al. , 2015. Paper and flexible substrates as materials for biosensing platforms to detect multiple biotargets. Sci. Rep 5 (1), 1–9. 10.1038/srep08719, 2015 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin MLY, Gau V, Liao JC, Haake DA, Wong PK, 2009. Active manipulation of quantum dots using ac electrokinetics. J. Phys. Chem. C 113 (16), 6561–6565. 10.1021/jp9004423. [DOI] [Google Scholar]
- Su S, et al. , 2016. Dual-target electrochemical biosensing based on DNA structural switching on gold nanoparticle-decorated MoS2 nanosheets. ACS Appl. Mater. Interfaces 8 (11), 6826–6833. 10.1021/acsami.5b12833. [DOI] [PubMed] [Google Scholar]
- Sun L, Yu C, Irudayaraj J, 2007. Surface-enhanced Raman scattering based nonfluorescent probe for multiplex DNA detection. Anal. Chem 79 (11), 3981–3988. 10.1021/ac070078z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, et al. , 2020. Smartphone-based multiplex 30-minute nucleic acid test of live virus from nasal swab extract. Lab Chip 20 (9), 1621–1627. 10.1039/d0lc00304b. [DOI] [PubMed] [Google Scholar]
- Wang L, et al. , 2020. Rapid and specific detection of Listeria monocytogenes with an isothermal amplification and lateral flow strip combined method that eliminates false-positive signals from primer–dimers. Front. Microbiol 10 10.3389/fmicb.2019.02959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, et al. , 2015. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS). BMC Genom 16 (1), 1–11. 10.1186/S12864-015-1572-7, 2015 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. , 2020. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv 2020 10.1101/2020.02.26.20028373, 02.26.20028373, Feb. [DOI] [Google Scholar]
- Zhou Y, Hu W, Peng B, Liu Y, 2014. Biomarker Binding on an Antibody-Functionalized Biosensor Surface: The Influence of Surface Properties, Electric Field, and Coating Density. Journal of Physical Chemistry C 118 (26), 14586–14594. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.


