Abstract
Kaposi's sarcoma-associated herpesvirus (also called human herpesvirus 8 [HHV-8]) replication and transcription activator (RTA) is apparently necessary and sufficient for the switch from viral latency to lytic replication. RTA may regulate open reading frame (ORF) K14 (viral OX-2 homologue) and ORF74 (viral G-protein-coupled receptor homologue) genes through an interferon-stimulated response element (ISRE)-like sequence (K14 ISRE) in the promoter region. RTA strongly activated a K14 ISRE-containing K14-ORF74 promoter reporter construct and a heterologous promoter reporter construct containing K14 ISRE. RTA could bind to K14 ISRE and other ISREs, activate promoter reporter constructs from interferon-simulated genes (ISGs), and selectively induce three endogenous ISGs in primary endothelial cells: ISG-54, myxovirus resistance protein 1 (MxA), and stimulated trans-acting factor of 50 kDa. In addition, a region in the RTA DNA-binding domain has been identified with certain sequence similarity to the DNA-binding domains of the interferon regulatory factor (IRF) family. Mutation in one conserved amino acid within this region reduced the ability of RTA to bind to ISRE as well as other RTA response elements. Furthermore, the mutant failed to activate RTA-responsive promoters and to induce viral lytic gene expression. The mutation at the same conserved amino acid residue in IRF-7 drastically reduced its ability to bind to DNA and to activate the beta interferon promoter. The sequence and functional similarities between RTA and IRFs suggest that the HHV-8 RTA may usurp the cellular IRF pathway.
Human herpesvirus 8 (HHV-8), also known as Kaposi's sarcoma (KS)-associated herpesvirus, is a gammaherpesvirus. It was first discovered in KS specimens and is believed to be the etiological agent of KS (7, 8, 39). In addition, HHV-8 is implicated in the pathogenesis of other human cancers, including AIDS-associated body cavity-based lymphoma (BCBL), also called primary effusion lymphoma (PEL), and a lymphoproliferative disorder known as multicentric Castleman's disease (16, 39, 68).
HHV-8 goes through both latency and lytic replication cycles. The switch from latency to lytic replication in HHV-8 resembles that of other gammaherpesviruses, such as Epstein-Barr virus (EBV). The expression of the HHV-8 replication and transcription activator (RTA) is apparently necessary and sufficient for the switch from latency to lytic replication (16, 68). RTA is an immediate-early gene (51, 61, 77) and a sequence-specific DNA-binding protein. The N-terminal amino acids 1 to 272 are sufficient for DNA binding in vitro (35). A number of RTA-responsive elements (RREs) were identified in the transcriptional regulatory regions of different subsequently expressed viral genes (21, 36, 49, 52, 60, 61, 77) and were found to consist of long and poorly defined sequences that could be tandem arrays of phased A/T trinucleotide motifs (Table 1) (14, 17, 30, 47, 49, 57, 59).
TABLE 1.
Sequence comparison of RRE and ISREsa
| Consensus RREb | (A/T)3N7(A/T)3N7(A/T)3 or (A/T)3N17(A/T)3 |
| K14 ISRE | GAGAAACAGAAACG |
| Tap2 ISRE | GCGAAAGCGAAAGC |
| vIL6 ISRE1 | GGGAAT-CGAAAGC |
| ISG15 ISRE | GGGAAAGGGAAACCGAAACT |
| ISG54 ISRE | GGGAAAGTGAAACT |
| IFI6-16 ISRE1/2c | GGGAAAATGAAACT |
| MxA ISRE2d | CAGAAA-CGAAACC |
| MxA ISRE3 | AAGAAA-TGAAACA |
| Consensus ISREe | (A/G)NGAAANNGAAACT |
RTA interacts with other factors to modulate its transcription potential (22, 24, 31, 65). A cellular zinc finger protein, K-RBP (KS-associated herpesvirus RTA-binding protein), interacts with RTA and functions as a cellular cofactor to modulate viral gene expression (65). RTA also interacts physically with recombination signal sequence-binding protein J kappa (RBP-Jκ) (also known as CBF-1 and CSL) in the transactivation of viral promoters (28). The RTA-RBP-Jκ complex appears to be essential for the RTA-mediated switch from latency to lytic replication (29). Beyond functioning in initiating viral lytic replication, RTA is involved in the induction of cellular interleukin-6 (IL-6) (13). RTA also blocks p53-mediated apoptosis by competing for binding to CBP (23).
HHV-8 ORF K14 (viral OX-2 homologue [vOX-2]) and ORF74 (viral G-protein-coupled receptor homologue [vGPCR]) genes are expressed from a bicistronic mRNA in the lytic replication cycle (26). K14 (vOX2) provides an activating signal for the production of inflammatory cytokines that potentially promote cytokine-mediated angiogenic proliferation of HHV-8-infected cells (11). ORF74 (vGPCR) can induce immortalization and recapitulate certain aspects of KS angiogenesis and oncogenesis in human primary endothelial cells (4, 38), and it is thus thought to be one of the pathogenic determinants in the development of KS.
Interferon (IFN) regulatory factors (IRFs) are a small family of transcription factors with multiple functions, including virus-induced IFN production, modulation of apoptosis, latency, transformation, and immune response pathways (43, 55, 62, 75). The hallmark of the family is a conserved N-terminal DNA-binding domain which mediates binding to the core region of IFN-stimulated response elements (ISREs) and thereby regulates IFN-responsive promoters. The C-terminal portion of the IRF family is variable and defines its biological functions (43, 55, 62, 75).
HHV-8 and IRFs have an intriguing relationship. The HHV-8 genome contains four viral IRF homologues (vIRFs) (45). Although none of them have been shown to bind to ISRE, both vIRF-1 and -3 are capable of blocking the production of IFN upon virus infection (19, 32, 34). In addition, the ORF45 protein interacts with IRF-7 and blocks IRF-7-mediated IFN production (78). Other gammaherpesviruses also have been shown to interact with members of the IRF family. Notably, EBV induces the expression of IRF-7 and activates IRF-7 protein by phosphorylation and nuclear translocation (71-75), and the rhesus rhadinovirus genome has been shown to contain eight IRF homologues (1).
In this report, we show that RTA has a region with similarity to DNA-binding domains of IRF family proteins. RTA binds to ISRE specifically and regulates HHV-8 K14-ORF74 genes through an ISRE-like sequence in the promoter region. RTA also selectively induces cellular IFN-stimulated genes (ISGs). Since ISGs are induced during HHV-8 lytic replication, our findings suggest a strong interplay between HHV-8 RTA and the IFN signaling pathway, which may play an important role in HHV-8 biology.
MATERIALS AND METHODS
Plasmids and antibodies.
Expression plasmids of RTA (pCMV-Tag50) and mutants (pCMV-RTA-AD1, AD2, AD3, and AD4) were described elsewhere (64). Another RTA expression plasmid, pCMV50, was a gift from Byrd Quinlivan (54). Human ISG54-luc and IFI6-16-luc were gifts from Ahmet Civas (20). The IFN-β-promoter reporter was a gift from Rongtuan Lin and John Hiscott (33). The IRF-7 expression plasmid has been described previously (74). The mutants RTA-P102T, RTA-F120D, RTA-K158E, RTA-A158P, and IRF7-K92E were obtained with the use of a PCR mutagenesis kit (Invitrogen). The complete coding sequence of Tag-RTA or Tag-RTA K152E was inserted into pET28b (Novagen) at the EcoRI and XhoI sites to generate pET28b-RTA or pET28b-RTA K152E, respectively, for bacterial expression. The recombinant adenovirus for RTA (AdRTA) and for green fluorescence protein (AdGFP) were gifts from Byrd Quinlivan (54). The β-galactosidase expression plasmid pCMVβ was purchased from Clontech (BD Biosciences, Palo Alto, CA). The K14-ORF74 promoter reporter constructs were generated by inserting the PCR products into the pGL3-basic vector (Promega, Madison, WI). Two oligos, 5′-GATCGTGAGAAACAGAAACGGCGATCGTGAGAAACAGAAACGGCGATCGTGAGAAACAGAAACGGC-3′ and its complementary strand, with GATC at the 5′ end, were used for cloning 3xK14 ISRE into the pGL3-promoter vector (K14ISRE-Luc). mK14 ISRE-Luc was made in the same way except for mutations at the desired nucleotides, as shown below in Fig. 1A. All mutations were verified by sequence analysis by the UNL Sequencing Facility. RTA antibodies were from David Lukac (see Fig. 6 and 7 below) and Gary Hayward (see Fig. 3, 4, and 5, below); K8 and K8.1 antibodies were obtained from Jae Jung; vIRF-1 antibody was from Yuan Chang; vIL-6 antibody was from John Nicholas; ISG-15 was from Ernest Bordin; and MxA antibody was from Mark A. McNiven. Tubulin and Flag-antibodies were purchased from Sigma. IRF-1 antibody was from Santa Cruz. IRF-7 antibody was described elsewhere (74). Sendai virus stock was purchased from Spafas, Inc.
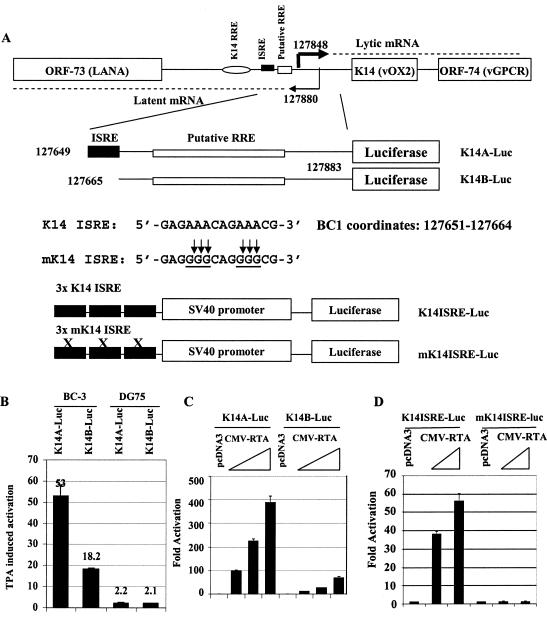
FIG. 1.
RTA activates the K14-ORF74 promoter though an ISRE-like element. A. Schematic representation of K14-ORF74 promoter region and reporter constructs. RNA start sites for K14-ORF74 and for LANA are shown and are based on the report of Jeong et al. (26). The K14 ISRE and its sequence, its mutations, the reporter constructs, and BC-1 genomic coordinates are shown. The mutated nucleotides are underlined. A previously identified K14 RRE and a putative RRE are also shown. B. K14-ORF74 promoter reporter constructs are inducible in HHV-8-positive cells. DG 75 is HHV-8 negative, while BC3 is an HHV-8-positive cell line. The reporter constructs were transfected into cells by electroporation. The fold activation after TPA treatment (24 to 36 h posttreatments) is shown. C. RTA activates K14-ORF74 reporter constructs. 293T cells were transfected with reporter construct, K14A-luc, or K14B-luc, along with CMV-β-Gal and various amounts of RTA expression plasmids (10, 25, and 50 ng). The reporter activity is expressed relative to the vector control. D. RTA activates K14 ISRE. K14 ISRE-luc or mK14ISRE-luc was transfected with the RTA expression plasmid (0.1 and 0.3 μg). The reporter activity is expressed relative to vector control. In these assays, luciferase activity was normalized by β-galactosidase activity. Standard deviations are shown.
FIG. 6.
RTA K152 E failed to induce lytic gene expression efficiently. A. RTA K152E failed to activate RTA-responsive promoter reporter constructs. The promoter constructs were transfected into 293T cells along with the same amounts (0.3 μg) of RTA or RTA K152E expression plasmid. Luciferase activity was normalized by β-galactosidase activity. The relative reporter activities are shown with standard deviations. B. RTA K152E failed to induce lytic gene expression. RTA or RTA K152E plasmids were transfected with CD4 expression plasmid into BCBL-1 cells, and the transfected cells were selected by using CD4 magnetic beads. Lysates from transfected and selected cells were used for Western blot analysis. The identities of proteins are shown.
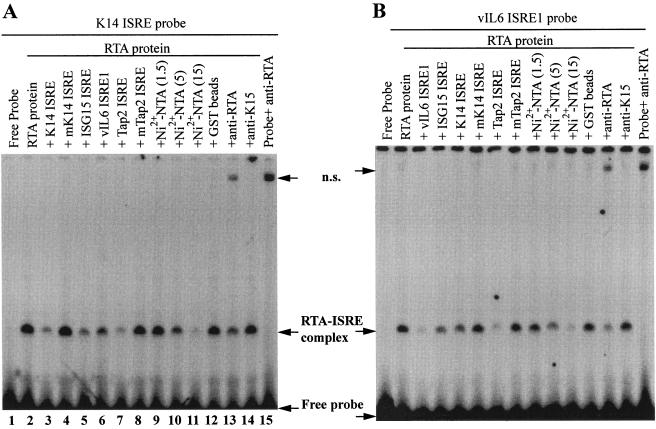
FIG. 3.
RTA binds to ISRE. A. The purified RTA protein binds to K14 ISRE. The probe was labeled with [α-32P]dATP. K14 ISRE, mK14 ISRE, ISG15 ISRE, Tap-2 ISRE, and mTap2-ISRE were used as cold competitors. Cold competitors were added at a 50-fold molar excess over hot probe. Various amounts of Ni2+-NTA agarose beads (1.5, 5, and 15 μl) were used to remove the histidine-tagged RTA proteins in an EMSA. Fifteen microliters of GST beads was used as a control. Rabbit polyclonal anti-RTA or K15 serum was also used. ns, nonspecific. B. RTA binds to known ISREs. The probe was ISRE-1 from the vIL-6 promoter. The cold competitors, Ni2+-NTA agarose beads, GST beads, and antisera used are shown. Specific protein-DNA complexes are shown.
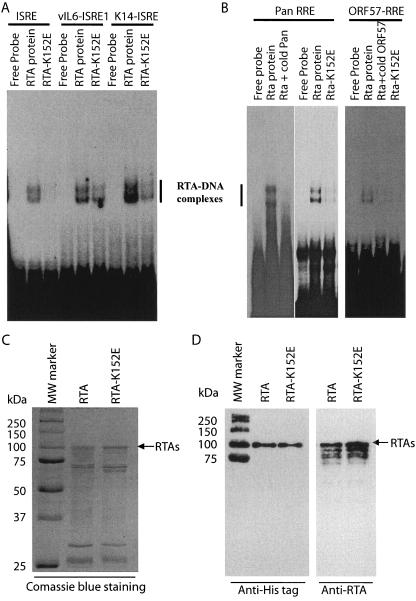
FIG. 5.
RTA K152 E failed to efficiently bind to ISREs or RREs. A. RTA K152 E binds to ISRE with reduced affinities. Equal amounts of partially purified recombinant RTA and RTA K152E proteins from bacteria were used for EMSA with various probes. The probes were ISRE from the ISG-15 gene, vIL6 ISRE1, and K14 ISRE. B. RTA K152 E binds to RREs with reduced affinities. Two known RREs, PAN and ORF57 RREs, were used for EMSA. Cold competitors and protein used are indicated on top. The RTA-DNA complexes are shown. C. Coomassie blue staining of the partially purified E. coli-derived RTA and its mutant RTA K152E proteins. Molecular mass (MW) markers and their sizes in kilodaltons are also shown. D. Western blot analysis of E. coli-derived RTA and its mutant RTA K152E. His tag and RTA antibodies were used for detection of partially purified proteins as shown in panel C. Some MW markers were also reactive to His tag antibody as shown.
Western blot analysis, RNA extraction, and RPA.
Standard Western blot analysis was performed as described previously (70-72). Total RNA was isolated from cells by using the RNeasy total RNA isolation kit (QIAGEN, Valencia, CA). RNase protection assays (RPA) were performed with total RNA using the RNase protection assay kit II (Ambion, Houston, TX). The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe was from U.S. Biochemicals, Inc. The probe for stimulated trans-acting factor of 50 kDa (STAF-50) was made from the PCR product amplified using the two primers from STAF-50: 5′-GGGGTACCGACGTCATGAAAAGGAGTG-3′ and 5′-GGATTTGAATTCTTAAATGTG-3′. The resultant PCR products were cloned into a pcDNA3.1 vector. The probe for IRF-7 was described before (69, 71).
Expression and purification of HHV-8 RTA.
Escherichia coli BL21(DE3) cells harboring the pET28b-RTA or pET28b-RTA K152E plasmid were induced for 4 h with 0.5 mM isopropylthio-β-d-galactoside at 28°C. After the induction, proteins were purified with Ni2+-nitrilotriacetic acid (NTA) agarose (QIAGEN, Valencia, CA) under the nondenaturing conditions suggested in the QIAexpressionist manual. Fractions containing target proteins were dialyzed against phosphate-buffered saline buffer (pH 7.4), separated into aliquots, and stored at −70°C.
EMSA.
The probes were obtained by first annealing complementary oligonucleotides and then labeling them with [α-32P]dATP (Amersham) using DNA polymerase Klenow fragment (Fermentas). The K14 ISRE probe was obtained by annealing two oligonucleotides, 5′-ACAGGTACCGTGAGAAACAGAAACGGC-3′ and its complementary strand, with ACAGGTACC at the 5′ end. The mK14ISRE probe was made exactly the same way as the K14 ISRE, with the mutations in the proper places as shown below in Fig. 1A. The oligos, 5′-GATCAGCCCGCGGGAATCGAAAGCC-3′ and its complementary strand, with GATC at the 5′ end, were used for the vIL6 ISRE-1 probe. The consensus ISRE probe from ISG-15 was synthesized according to the methods described in a previous report (27). Tap-2 ISRE and mTap2 ISRE probes were described earlier (72). The 5′ overhangs of the probes were filled in by DNA polymerase I Klenow fragment and purified with the QIAquick nucleotide removal kit (QIAGEN). An electrophoretic mobility shift assay (EMSA) was performed essentially as described previously (59). Basically, labeled probes were purified with the QIAquick nucleotide removal kit (QIAGEN). The purified RTA protein was incubated with 20,000 to 50,000 cpm of labeled probe in a volume of 12.5 μl containing 20 mM HEPES-KOH (pH 7.9), 1 mM MgCl2, 0.1 mM EGTA, 2 μg of poly(dI-dC), 4% Ficoll for 20 min at room temperature. When antibody was used, 0.1 μl of antibody was incubated with the reaction mixture for 15 min prior to the addition of probe. Various amounts of Ni2+-NTA agarose beads were incubated with RTA proteins, the beads were collected by centrifuging, and the supernatants were used for EMSA. Glutathione-Sepharose 4B beads (Amersham) (GST beads) were used as a control for Ni2+-NTA beads. The binding mixtures were separated on a 4.5% polyacrylamide gel in 0.25× Tris-borate-EDTA buffer.
Cell culture, transient transfection, and reporter assays.
BCBL-1 and BC-3 are HHV-8-positive PEL cell lines (2). DG75 is an HHV-8-negative Burkitt's lymphoma cell line. These cells were maintained in RPMI 1640 plus 10% fetal bovine serum. Electroporation was used for transfection of these B cells as described previously (71, 72, 76). 293T cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum. Effectene (QIAGEN) was used for the transfection of 293T cells. HUVEC are human primary umbilical vascular endothelial cells (gift from Lingjun Zhao) and were maintained in the EBM-2 plus medium (Cambrex). The luciferase assays were performed using the assay kit from Promega according to the manufacturer's recommendations.
Isolation of transfected cells and in vitro translation.
Enrichment for CD4-positive cells was performed with anti-CD4 antibody conjugated to magnetic beads according to the manufacturer's recommendations (Dynal, Inc.). The isolated cells were lysed immediately and used for Western blot analysis. For in vitro transcription and translation, the TNT-coupled transcription and translation kit (Promega) was used essentially as specified by the manufacturer.
Induction of viral lytic replication.
BC-3 was transfected with various reporters. After overnight recovery, these cells were treated with 20 ng/ml 12-O-tetradecanoylphorbol-13-acetate (TPA) for 24 h to induce lytic viral replication. After induction, the cells were harvested for reporter assays. For BCBL-1 cells, 20 ng/ml of TPA for 48 h was used for induction of lytic replication.
RT-coupled PCR.
Total RNA was isolated from samples by using an RNeasy kit (QIAGEN). The RNA samples were treated with DNase I at 37°C for 30 min. The primers used in this experiment were as follows: for the actin gene, Actin 1 (5′-TTC TAC AAT GAG CTG CGT GT-3′) and Actin 2 (5′-GCC AGA CAG CAC TGT GTT GG-3′); for the ISG-54 gene, ISG545 (5′-AGAAATCAAGGGAGAAAG-3′) and ISG5431 (5′-TCATTCCCCATTCCAGC-3′). These primers were mixed with 0.1 μg of RNA, reverse transcription (RT) reactions were done in 50°C for 30 min, and PCRs were followed with standard protocols, with 30 cycles for the actin gene while 35 cycles for ISG-54 were used for PCR amplification.
RESULTS
RTA may regulate ORF-K14 and ORF74 genes through an ISRE-like sequence.
K14 and ORF74 are transcribed together as a bicistronic mRNA in the lytic replication cycle (10, 26). By sequence analysis, an ISRE-like element, hereafter named K14 ISRE, was identified in the promoter region of K14-ORF74 (Fig. 1A). The K14 ISRE is almost identical to the consensus ISRE sequence (12). The possible role of K14 ISRE in the regulation of K14-ORF74 expression in lytic replication was examined by using two promoter reporter constructs, K14A-Luc and K14B-Luc, with or without the K14 ISRE sequence (Fig. 1A). Since TPA can induce HHV-8-positive PEL cells into lytic replication (60, 61, 77), whether the K14-ORF74 promoter constructs are responsive to TPA induction in HHV-8-positive cells was examined. In HHV-8-positive BC-3 cells, induction by TPA was threefold higher in cells transfected with ISRE-containing K14A-luc compared to those with ISRE-lacking K14B-luc (Fig. 1B). However, in HHV-8-negative DG75 cells, there was very little induction for either promoter reporter construct (Fig. 1B). These data suggest that a viral gene(s) in the lytic replication cycle may be responsible for activation of the K14-ORF74 promoter via K14 ISRE. RTA is among the earliest gene products expressed upon TPA treatment and has been shown to activate the K14-ORF74 promoter (10, 26, 77); therefore, the putative role of RTA on K14 ISRE was examined. As shown in Fig. 1C, the K14-ORF74 promoter reporter construct with the ISRE (K14A-luc) responded to RTA extremely well, while the construct lacking the element (K14B-luc) was only weakly activated by RTA. To further test if K14 ISRE could mediate RTA activation, K14 ISRE was multimerized and placed upstream of a simian virus 40 minimum promoter reporter construct (Fig. 1A), and the resulting reporter construct (K14LSRE-luc) was transfected into cells with RTA. RTA was found to activate the K14 ISRE-containing heterologous promoter reporter construct but not the corresponding mK14ISRE-containing reporter construct (Fig. 1D). In addition, RTA failed to activate the pGL3-promoter vector (data not shown). Thus, these results suggest that K14 ISRE may be an important element used by RTA for the activation of the K14-ORF74 promoter. Other putative RREs, which are not K14 ISRE, have been identified or predicted in the K14-ORF74 promoter region (Fig. 1A) (26, 30). Obviously, without the K14 RRE identified by Jeong et al. (26), the K14 promoter is still activated by RTA (Fig. 1B and C). However, additional putative RRE identified by Liao et al. (30) could be responsible for the activation of the ISRE-lacking K14-ORF74 promoter reporter construct (K14B-luc).
The RTA activation domain is required for activation of K14 ISRE.
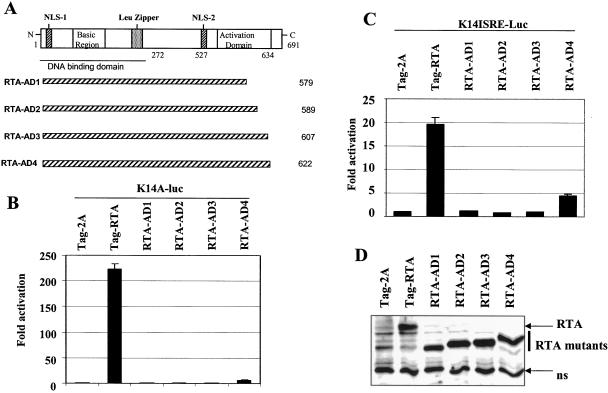
RTA has been shown to activate K14-ORF74 promoter reporter constructs (10, 26), but the domain(s) that is involved in the activation is not known. Therefore, different deletion mutants of RTA were tested in order to identify the domains of RTA that are necessary for K14-ORF74 activation. The RTA AD1 to -4 mutants have various deletions in the activation domain, as shown in Fig. 2A, and none of them were able to activate the K14-ORF74 promoter to the same extent as the wild-type RTA (Fig. 2B). Similarly, the activation domain was also required for the activation of K14 ISRE (Fig. 2C). Since the expression levels of all RNA mutants were similar (Fig. 2D), this suggested that the activation domain of RTA is required for K14 ISRE-mediated activation of the K14-ORF74 promoter.
FIG. 2.
The activation domain of RTA is required for activation of the K14-ORF74 promoter. A. Schematic diagram of RTA domains and mutants. The number of amino acid residues is shown. The basic region of the RTA and leucine zipper regions, activation domain, DNA-binding domain, and nuclear localization signal (NLS) are shown. B. The activation domain of RTA is required for activation of the K14-ORF74 promoter. 293T cells were transfected with reporter construct, K14A-luc, along with CMV-β-Gal and equal amounts (50 ng) of RTA or various mutant expression plasmids. The relative reporter activity is shown. C. The activation domain of RTA is required for activation of the K14 ISRE. 293T cells were transfected with reporter construct, K14ISRE-luc along with CMV-β-Gal, and equal amounts (0.3 μg) of RTA or various mutant expression plasmids. The reporter activity is expressed relative to the vector control. Luciferase activity was normalized by β-galactosidase activity. Standard deviations are shown. D. Expression status of RTA and its deletion mutants. Western blotting with Flag monoclonal antibody was performed. The cell lysates from transfected cells are indicated. The identity of proteins is as shown. ns, nonspecific.
RTA binds to ISRE directly.
The involvement of ISRE in the activation of K14-ORF74 and the results in Fig. 2 suggest that RTA might bind to ISRE directly. Partially purified RTA protein, which has been used widely to study RTA binding (14, 35, 58, 59, 66), was used for EMSA with the K14 ISRE probe. Poly-histidine-tagged full-length RTA was expressed in bacteria and partially purified using Ni2+-NTA agarose beads (see Materials and Methods for details). It was noted that the purified RTA had the expected properties, as it binds to other known RREs (see Fig. 5, below, and data not shown). As shown in Fig. 3A, specific binding of RTA to the K14 ISRE was observed in the EMSA. The band was specific, because it disappeared in the presence of cold competitors such as K14 ISRE itself, the consensus ISRE for the ISG15 promoter, or the ISRE from the transporter associated with antigen processing 2 promoter (Tap2 ISRE) (lanes 3 and 5 to 7). However, the mutated K14 ISRE and Tap2 ISRE had little effect on the RTA-DNA complex formation (lanes 4 and 8). When Ni2+-NTA beads were incubated with purified RTA, the intensity of the major protein-DNA complex decreased; however, the GST beads had no effect (lanes 9 to 12). When RTA antibody was added to the reaction mixture, a decrease in the intensity of the major specific band was observed; however, an irrelevant K15 antibody had no effect (lanes 13 and 14). There was also a nonspecific band. The nonspecific band was due to the reactivity of the RTA antibody to DNA probe itself, as the probe plus antibody also generated the same complex (lane 15). Thus, from specific competition by cold probes, the effect of Ni2+-NTA beads, as well as reactivity to RTA antibody, the major band was identified as an RTA-ISRE complex. In addition to K14 ISRE, RTA also binds specifically to these well-established ISREs, e.g., ISRE-1 from vIL-6 promoters (Fig. 3B). Almost identical patterns could be observed when ISRE from ISG-15 was used as probe (data not shown). Both vIL6 ISRE1 and ISG-15 ISRE were shown to be responsive to IFN (9, 27). These results strongly suggest that RTA can bind to ISRE directly and specifically.
RTA has sequence similarity to IRF family proteins.
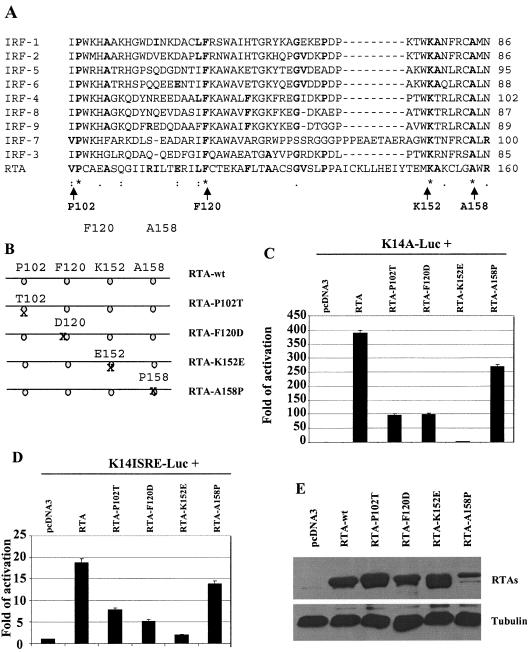
Since IRFs have the ability to bind to ISRE, whether RTA has similarity to IRF family proteins was examined using the Clustal W program (25, 63). RTA and all human IRF protein sequences were aligned, and a region with some degree of similarity was identified (Fig. 4A). The similar regions are located within the DNA-binding domain (amino acids 1 to 272) of RTA as well as in the DNA-binding domains of IRFs. The similarity between RTA and IRF family proteins suggests that RTA and IRFs may target ISRE by a similar mechanism. The conserved amino acids in the region with similarity could be involved in the DNA binding.
FIG. 4.
Mutations in the DNA-binding domain of RTA affect activation of the K14-ORF74 promoter. A. RTA has certain similarities to IRF proteins. The Clustal W program was used for the alignment of RTA with all human IRF members. Gaps were introduced to allow best alignment. The identity and coordinates of mutated amino acids in RTA are shown at the bottom with arrows. B. Schematic diagram of RTA mutant constructs. The coordinates and identities of the amino acids are shown. The positions of amino acids are not on scale. C. Mutations in the DNA-binding domain of RTA reduce activation of the K14-ORF74 promoter. 293T cells were transfected with reporter construct, K14A-luc, along with CMV-β-Gal and equal amounts (50 ng) of RTA or various mutant expression plasmids. The reporter activity is expressed relative to the vector control. Luciferase activity was normalized by β-galactosidase activity. Standard deviations are shown. D. Mutations in the DNA-binding domain of RTA reduce activation of K14-ISRE. 293T cells were transfected with reporter construct, K14ISRE-luc, along with CMV-β-Gal and equal amounts (0.3 μg) of RTA or various mutant expression plasmids. The reporter activity is expressed relative to the vector control. Standard deviations are shown. E. Expression levels of RTA mutants. Western blotting with RTA as well as tubulin antibodies was performed. The cell lysates from transfected cells shown in panels C or D are indicated. The identities of proteins are shown.
The similarity between IRF and RTA was not particularly high (Fig. 4A). In order to determine the relevance of the comparison in Fig. 4A, mutations were made in all of the completely conserved residues of RTA individually (Fig. 4B), and their effects on the activation of the K14-ORF74 promoter reporter constructs were examined upon transfection into 293T cells (Fig. 4C and D). RTA K152E had almost completely lost its ability to activate the K14-ORF74 promoter and the K14ISRE promoter reporter constructs (Fig. 4C and D). The expression levels of these mutant proteins were similar to the wild-type RTA in 293T cells, except for the RTA-A158P mutant (Fig. 4E). The RTA-A158P mutant always expressed lower levels of proteins in our systems, even when threefold more DNA was used for transfection (data not shown). Thus, the mutation in A158 may influence the protein expression of RTA, but it may not affect the transactivation potential of RTA. Also, these mutants are all localized in the nucleus in a similar pattern to wild-type RTA (data not shown). These results suggest that the comparison in Fig. 4A may be relevant.
A mutation in RTA reduced its DNA-binding affinity.
Because RTA K152E and wild-type RTA were both localized in the nuclei of transfected cells (data not shown), it is possible that the K152E mutation could have disrupted the DNA-binding activity of RTA. The RTA-K152E protein was partially purified from bacteria via a similar procedure as for the wild-type RTA. The input levels of RTA and RTA K152E proteins were adjusted to similar levels (Fig. 5C and D), and their ability to bind to DNA was then tested by EMSA (Fig. 5A and B). Binding to ISREs by RTA was reproducibly reduced (Fig. 5A). In addition, RTA K152E could only marginally bind to two well-studied RREs, PAN and ORF57 RREs (Fig. 5B). Thus, the mutation at K152 may have affected RTA DNA-binding activity in general.
RTA K152E failed to activate RTA-responsive genes.
Since RTA K152E has a reduced ability to bind to DNA, we tested its ability to activate RTA-responsive promoter constructs. RTA K152E was unable to activate several RTA-responsive promoters (Fig. 6A). The results suggested that RTA K152E might be unable to activate the lytic replication of HHV-8. HHV-8-positive BCBL-1 cells were transfected with RTA or RTA K152E to determine their abilities to induce HHV-8 lytic gene expression. Due to the low transfection efficiency of BCBL-1 cells, we used CD4 selection procedures to enrich the transfected cells (see Materials and Methods for details). RTA K152E could not induce the expression of several lytic genes, such as K8, vIRF1, K8.1, and vIL6, all of which were inducible by wild-type RTA (Fig. 6B). The wild-type RTA was apparently expressed at higher levels than the K152E mutant in BCBL-1 cells (Fig. 6B), which could be due to the autoregulation of RTA expression (15, 49). However, the expression levels of RTA K152E were comparable to the level of RTA induced by TPA. Thus, RTA K152E is ineffective in the induction of HHV-8 lytic gene expression at a level similar to that induced by TPA (Fig. 6B).
RTA activated several ISG promoter reporter constructs.
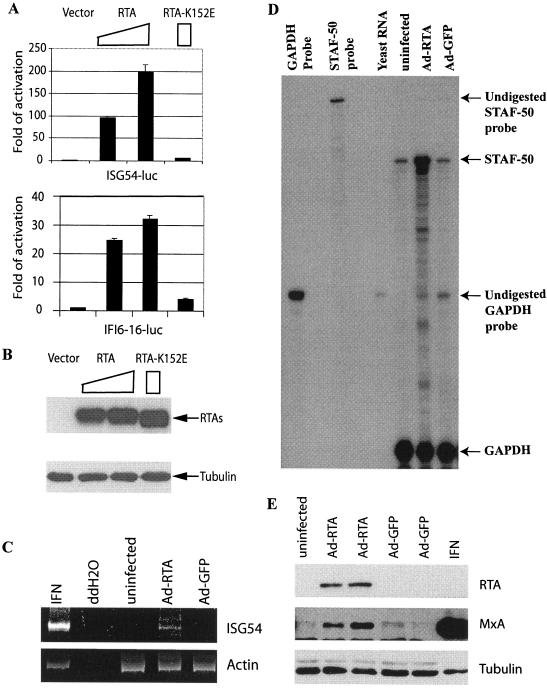
Since RTA can bind to ISRE, which is present in all ISGs (50, 55), it is possible that RTA activates ISGs. To test this possibility, two different ISG promoter reporter constructs (ISG-54-luc and IFI6-16-luc) that contain ISREs and are responsive to IFN were studied (20). RTA activated both ISG-54 and IFI6-16 promoter reporter constructs, while the RTA-K152E mutant could not (Fig. 7A). In addition, the expression levels of RTA and its mutants were similar in these experiments (Fig. 7B). Thus, RTA was able to activate different ISG promoter reporter constructs. Since these promoter reporter constructs contain ISREs, RTA may at least partially activate ISG promoters via ISRE.
FIG. 7.
RTA selectively induces the expression of IFN-stimulated genes. A. RTA activates IFN-inducible promoter reporter constructs. ISG54-luc or IFI6-16-luc promoter constructs were transfected into 293T cells along with various amounts of RTA expression plasmids (0.1 or 0.3 μg) or with RTA K152E expression plasmid (0.3 μg). Luciferase activity was normalized by β-galactosidase activity. The relative reporter activities are shown with standard deviations. B. Expression levels of RTA mutants. Western blot assays with RTA as well as tubulin antibodies were performed. The cell lysates from transfected cells are indicated. The identities of proteins are shown. C. RTA induces ISG-54 in HUVECs. RNAs from uninfected HUVECs or the cells infected with recombinant adenovirus with RTA or GFP, respectively, were used for RT-PCR analyses. IFN-α (100 IU/ml)-treated 293 cells were used as a positive control. D. RTA induces STAF-50 in HUVECs. STAF-50 and GAPDH probes were labeled with [α-32P]UTP and used for RPA. Specific protections of STAF-50 and GAPDH mRNAs and undigested probes are indicated. Relative STAF-50 RNA levels (STAF-50/GAPDH) were increased 5.3 ± 0.9-fold. The data were obtained by normalizing STAF-50 to GAPDH levels from three independent experiments with the use of a Gene Genius Bioimaging system. E. RTA induces the expression of MxA protein. Lysates from uninfected, AdRTA-infected, AdGFP-infected, or IFN-α (100 IU/ml)-treated HUVECs were used for Western blot analysis. The same membrane was stripped and reprobed with other antibodies. The identities of proteins are shown.
RTA selectively induced endogenous ISGs.
The binding to ISRE and activation of the ISG promoter reporter constructs by RTA suggested that it might also induce the expression of endogenous ISGs. Because HHV-8 infects endothelial cells efficiently and the cellular ISGs are induced upon HHV-8 lytic replication (46), HUVECs were chosen to test the hypothesis. Five ISGs (ISG-15, and -54, IRF-7, myxovirus resistance protein 1 [MxA], and STAF-50) were examined. Due to the low transfection efficiency of HUVECs, the recombinant adenoviruses containing RTA (AdRTA) or GFP (AdGFP) were used to infect HUVECs and the effects of RTA in these cells were examined. RTA expression was confirmed by Western blot analysis (Fig. 7E), and the expression of GFP was confirmed by the presence of green fluorescent cells under a fluorescence microscope (data not shown). Endogenous ISG-54 and STAF-50 RNA expression was increased in RTA-expressing cells as determined by RT-PCR and RPA (Fig. 7C and D). The expression of the MxA protein also increased, as determined by Western blotting (Fig. 7E). However, expression of neither IRF-7 nor ISG-15 was found to increase significantly in the presence of RTA (data not shown). Therefore, RTA selectively induces endogenous ISGs. It is of note that wild-type adenovirus infection induces ISG-15 (43a); the results here suggested that the induction of these ISGs was not due to the contamination of wild-type RTA.
DISCUSSION
The structure and functions of RTA have been extensively studied (16, 68). However, the exact mechanism involved in RTA functions remains incompletely understood. RTA appears to activate transcription through two distinct mechanisms, either by direct binding to DNA or via association with other DNA-binding transcription factors. RREs appear to be heterogeneous and ill defined (14, 17, 30, 47, 49, 57, 59).
In this study, we have shown that ISRE could serve as an RRE and that RTA may regulate the K14-ORF74 genes via an ISRE-like sequence, K14 ISRE. However, K14 ISRE may not be the sole element in the promoter responsible for RTA activation: a functional RRE was identified and a putative RRE has been proposed in the K14-ORF74 promoter region (Fig. 1A) (26, 30). Indeed, the ISRE-deficient K14-ORF74 promoter reporter construct (K14B-luc) (Fig. 1A), which contains the putative RRE, could also be activated by RTA and TPA stimulation but at a low efficiency (Fig. 1B and C). Thus, K14 ISRE, K14 RRE, and an additional RRE(s) may be responsible for the optimal activation of K14-ORF74 genes by RTA. Nevertheless, our results demonstrated that K14 ISRE may play a role in the activation.
Other than K14 ISRE, at least seven other ISRE-like sequences can be identified in the HHV-8 (BC-1 strain) genome. ORF-K2 (vIL-6) is an IFN-inducible viral gene which can also be activated by RTA, and the vIL-6 promoter contains two ISREs and one RRE (9, 14). The presence of ISREs suggests that RTA may activate the vIL-6 promoter via one or both vIL-6 ISREs in the absence of the identified RRE. As expected, RTA could efficiently activate vIL-6 promoter reporter constructs that lacked the identified RRE (data not shown), and RTA could bind to at least one of the two vIL6 ISREs (Fig. 3 and 5). These results thus suggest that the binding of RTA to ISRE may play a regulatory role in the control of different viral gene expressions.
A previous report suggested that RTA binds to multiple, phased A/T triplets in RRE to bring about potent transcriptional activation (Table 1). ISREs are loosely conserved, but one conserved feature is the two consecutive A triplets, i.e., AAANNNAAA, in the consensus sequence (Table 1) (12, 55, 62). Thus, our data in this report are in agreement with the published results (30). It is likely that ISRE is a variant of consensus RREs except for the spacer regions between the two A triplets.
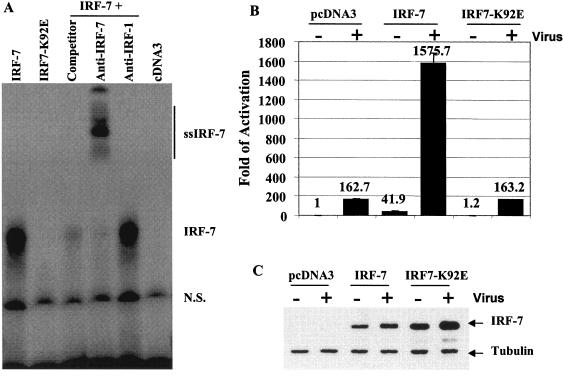
Lysine 152 of RTA, which is conserved among RTA and IRFs, plays a critical role in RTA functions. In addition, the same lysine in IRF-1 (K78) was implicated in IRF-1-DNA contact (18). Thus, the lysine residue may also be a critical one in the biological functions of the IRF family. To examine the functional role of the lysine in the IRF family, we have mutated the lysine residue (K92) in IRF-7 because IRF-7 and RTA have similar lengths in the similarity region (Fig. 4A). We then examined the properties of the particular mutant (IRF7-K92E) on DNA binding and on activation of the IFN-β promoter, because IRF-7 is a critical determinant for the induction of IFN-α/β genes in response to viral infection (3, 33, 37, 67, 75). IRF7-K92E failed to bind to DNA efficiently (Fig. 8A), and the IRF7-K92E failed to activate the IFN promoter (Fig. 8B); however, the infection of transfected cells with Sendai virus led to activation of IRF7 K92E, as judged by the nuclear translocation after viral infection (data not shown). These results strongly suggested that the comparison between RTA and IRFs in Fig. 4A is relevant.
FIG. 8.
IRF7-K92E failed to bind to DNA and activate IFN promoters. A. IRF7-K92E failed to bind to DNA. Equal amounts of in vitro-translated IRF-7 and IRF7 K92E proteins were used for an EMSA with ISRE from ISG-15 as probe. The expression levels of IRF-7 and IRF7-K92E were similar in the in vitro-translated lysates (data not shown). Cold competitors (concentrated 100 times) and antibodies used are indicated on top. IRF-7 and IRF-1 antibodies were used to detect the specificity of the protein-DNA complex. The IRF-7-DNA complex is shown. ss-IRF7, supershifted IRF7-DNA complex; n.s., nonspecific. B. IRF7-K92E failed to activate the IFN-β promoter. The IFN-β-promoter reporter construct was transfected into 293T cells along with the same amounts of IRF-7 wild-type or IRF7-K92E expression plasmid. At 12 h after transfection, cells were infected with (+) or without (-) Sendai virus at 20 hemagglutinin U/ml. After 24 h of infection, cells were lysed and luciferase activities were measured. Luciferase activity was normalized by β-galactosidase activity. The relative reporter activities are shown with standard deviations. C. Expression levels of IRF-7. Western blotting with IRF-7 as well as tubulin antibodies was performed. The cell lysates from transfected 293T cells are indicated. The identities of proteins are shown.
IRFs are capable of binding to DNA by a helix-turn-helix motif, and the corresponding lysine K78 in IRF-1 is located in the DNA recognition helix of IRF-1 (18). The corresponding region of RTA is predicted to form a helix (data not shown). Our study has shown that the conserved lysine plays a critical role in both RTA and IRF-7. These data collectively suggest that RTA might use a similar helix-turn-helix motif to bind to DNA as IRF, and lysine 152 of RTA may be an important residue involved in the RTA-DNA interaction. Due to the fact that RTA K152E fails to bind to DNA efficiently and to induce lytic gene expression, it suggests that the DNA-binding activity of RTA is a prerequisite for its transactivation function and disruption of viral latency.
ISRE is present in the promoters of all ISGs (50, 55, 62). Our data demonstrated that two ISG promoters could be activated by RTA, and RTA could induce some endogenous ISGs, such as ISG-54, MxA, and STAF-50 (Fig. 7). The selective induction of ISGs by RTA was not unexpected, as not all IRFs can induce ISGs (44, 50, 55, 62) and IFN induces different sets of ISGs in various cell lines (53). The selectivity may also be due to differences in the affinities of RTA to various ISREs, or cell type-specific factors. The induction of ISGs upon HHV-8 lytic replication has been reported (46), and STAF-50 RNA was shown to be highly expressed in KS specimens (48). Since many KS cells undergo spontaneous lytic replication (16, 39, 68), the induction of STAF-50 may be relevant to lytic replication and might be mediated in part by RTA. It is of note that the induction of ISGs appears to be a common phenomenon upon herpesvirus infection (5, 41, 42, 46, 56, 79). Although the exact functions of these induced ISGs are currently unknown, it is likely that these genes may play a role in viral replication and pathogenic processes.
Because ISG induction is a major outcome upon IFN treatment, one possibility is that the activation of ISGs by RTA is indirect via the induction of IFNs. However, it is unlikely for several reasons. The selective induction of ISGs suggests that IFN may not be involved, since IFN induces IRF-7 and ISG15 in HUVECs (data not shown). The culture medium from RTA-transfected cells could not induce ISGs when transferred to fresh cells. No significant induction of IFN by RTA was observed by RT-PCR analyses (data not shown). Moreover, the induction of ISGs during HHV-8 lytic replication was shown to be independent of IFNs (10). Finally, IFN was not involved in the induction of ISGs upon infections by other herpesviruses (5, 6, 42, 56, 79). Thus, it is likely that the observed activation of ISGs is a direct effect of RTA rather than an indirect effect via IFN.
HHV-8 has been known to contain several different cellular homologues, including four vIRFs that appear to play an important role in HHV-8 pathogenesis (39, 40). These vIRFs have some homology to the IRF family proteins, but none of these vIRFs has been shown to bind to ISRE. Our data suggest that RTA has a region with sequence similarity to IRFs and binds to ISRE. In addition, the activation of ISG promoters, induction of endogenous ISGs, and similarity between RREs and ISREs (Table 1) provide additional evidence that RTA and IRFs have similar functional properties. Because RTA is loosely conserved in the gammaherpesvirus family, RTAs of other gammaherpesviruses might also have similar properties. Indeed, EBV RTA was also able to induce at least two ISGs (data not shown). Thus, our findings may not only shed new light on the mechanisms of HHV-8 RTA but also other RTAs in the regulation of both viral and cellular genes.
Acknowledgments
We thank Byrd Quinlivan, David Lukac, Gary Hayward, Lingjun Zhao, Jae Jung, Yuan Chang, John Nicholas, Ernest Bordin, Mark McNiven, John Hiscott, Rongtuan Lin, and Ahmet Civac for providing various reagents for this work. We also thank John West for helpful discussions, Evan Burkala for the use of the Gene Genius Bioimaging System, and the UNL Microscopy and Sequencing Facility for their assistance.
This work is supported by PHS grants CA75903 and CA76958, Fogarty International Training grant TW01492, and NCRR COBRA grant RR15635 to C.W. and grants from the Tobacco Settlement Biomedical Research Enhancement Fund and the Layman Foundation to L.Z.
REFERENCES
- 1.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvanitakis, L., E. A. Mesri, R. G. Nador, J. W. Said, A. S. Asch, D. M. Knowles, and E. Cesarman. 1996. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88:2648-2654. [PubMed] [Google Scholar]
- 3.Au, W. C., P. A. Moore, D. W. LaFleur, B. Tombal, and P. M. Pitha. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J. Biol. Chem. 273:29210-29217. [DOI] [PubMed] [Google Scholar]
- 4.Bais, C., A. Van Geelen, P. Eroles, A. Mutlu, C. Chiozzini, S. Dias, R. L. Silverstein, S. Rafii, and E. A. Mesri. 2003. Kaposi's sarcoma associated herpesvirus G protein-coupled receptor immortalizes human endothelial cells by activation of the VEGF receptor-2/ KDR. Cancer Cell 3:131-143. [DOI] [PubMed] [Google Scholar]
- 5.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, K. L., E. Cahir-McFarland, and E. Kieff. 2002. Epstein-Barr virus-induced changes in B-lymphocyte gene expression. J. Virol. 76:10427-10436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., and P. S. Moore. 1996. Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect. Agents Dis. 5:215-222. [PubMed] [Google Scholar]
- 9.Chatterjee, M., J. Osborne, G. Bestetti, Y. Chang, and P. S. Moore. 2002. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298:1432-1435. [DOI] [PubMed] [Google Scholar]
- 10.Chiou, C. J., L. J. Poole, P. S. Kim, D. M. Ciufo, J. S. Cannon, C. M. ap Rhys, D. J. Alcendor, J. C. Zong, R. F. Ambinder, and G. S. Hayward. 2002. Patterns of gene expression and a transactivation function exhibited by the vGCR (ORF74) chemokine receptor protein of Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3421-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, Y. H., R. E. Means, J. K. Choi, B. S. Lee, and J. U. Jung. 2002. Kaposi's sarcoma-associated herpesvirus OX2 glycoprotein activates myeloid-lineage cells to induce inflammatory cytokine production. J. Virol. 76:4688-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darnell, J. E., Jr. 1997. STATs and gene regulation. Science 277:1630-1635. [DOI] [PubMed] [Google Scholar]
- 13.Deng, H., J. T. Chu, M. B. Rettig, O. Martinez-Maza, and R. Sun. 2002. Rta of the human herpesvirus 8/Kaposi sarcoma-associated herpesvirus up-regulates human interleukin-6 gene expression. Blood 100:1919-1921. [DOI] [PubMed] [Google Scholar]
- 14.Deng, H., M. J. Song, J. T. Chu, and R. Sun. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252-8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, H., A. Young, and R. Sun. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043-3048. [DOI] [PubMed] [Google Scholar]
- 16.Dourmishev, L. A., A. L. Dourmishev, D. Palmeri, R. A. Schwartz, and D. M. Lukac. 2003. Molecular genetics of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) epidemiology and pathogenesis. Microbiol. Mol. Biol. Rev. 67:175-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan, W., S. Wang, S. Liu, and C. Wood. 2001. Characterization of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF57 promoter. Arch. Virol. 146:403-413. [DOI] [PubMed] [Google Scholar]
- 18.Escalante, C. R., J. Yie, D. Thanos, and A. K. Aggarwal. 1998. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature 391:103-106. [DOI] [PubMed] [Google Scholar]
- 19.Gao, S. J., C. Boshoff, S. Jayachandra, R. A. Weiss, Y. Chang, and P. S. Moore. 1997. KSHV ORF K9 (vIRF) is an oncogene which inhibits the interferon signaling pathway. Oncogene 15:1979-1985. [DOI] [PubMed] [Google Scholar]
- 20.Genin, P., P. Morin, and A. Civas. 2003. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J. Virol. 77:7113-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gwack, Y., H. Byun, S. Hwang, C. Lim, and J. Choe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gwack, Y., S. Hwang, H. Byun, C. Lim, J. W. Kim, E. J. Choi, and J. Choe. 2001. Kaposi's sarcoma-associated herpesvirus open reading frame 50 represses p53-induced transcriptional activity and apoptosis. J. Virol. 75:6245-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 25.Higgins, D. G. 1994. CLUSTAL V: multiple alignment of DNA and protein sequences. Methods Mol. Biol. 25:307-318. [DOI] [PubMed] [Google Scholar]
- 26.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler, D. S., S. A. Veals, X. Y. Fu, and D. E. Levy. 1990. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 4:1753-1765. [DOI] [PubMed] [Google Scholar]
- 28.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. USA 100:8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao, W., Y. Tang, Y. L. Kuo, B. Y. Liu, C. J. Xu, and C. Z. Giam. 2003. Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 transcriptional activator Rta is an oligomeric DNA-binding protein that interacts with tandem arrays of phased A/T-trinucleotide motifs. J. Virol. 77:9399-9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 77:3809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, Jr., G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 33.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320-34327. [DOI] [PubMed] [Google Scholar]
- 34.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 75:6786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montaner, S., A. Sodhi, A. Molinolo, T. H. Bugge, E. T. Sawai, Y. He, Y. Li, P. E. Ray, and J. S. Gutkind. 2003. Endothelial infection with KSHV genes in vivo reveals that vGPCR initiates Kaposi's sarcomagenesis and can promote the tumorigenic potential of viral latent genes. Cancer Cell 3:23-36. [DOI] [PubMed] [Google Scholar]
- 39.Moore, P., and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus, p. 2803-2833. In D. M. Knipe et al. (ed.), Virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 40.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 41.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen, H., J. Hiscott, and P. M. Pitha. 1997. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 8:293-312. [DOI] [PubMed] [Google Scholar]
- 43a.Nielsch, U., R. Pine, S. G. Zimmer, and L. E. Babiss. 1992. Induced expression of the endogenous beta interferon gene in adenovirus type 5-transformed rat fibroblasts. J. Virol. 66:1884-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pine, R., T. Decker, D. S. Kessler, D. E. Levy, and J. E. J. Darnell. 1990. Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 10:2448-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pitha, P. M., W. C. Au, W. Lowther, Y. T. Juang, S. L. Schafer, L. Burysek, J. Hiscott, and P. A. Moore. 1998. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochimie 80:651-658. [DOI] [PubMed] [Google Scholar]
- 46.Poole, L. J., Y. Yu, P. S. Kim, Q. Z. Zheng, J. Pevsner, and G. S. Hayward. 2002. Altered patterns of cellular gene expression in dermal microvascular endothelial cells infected with Kaposi's sarcoma-associated herpesvirus. J. Virol. 76:3395-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlivan, E. B., E. A. Holley-Guthrie, M. Norris, D. Gutsch, S. L. Bachenheimer, and S. C. Kenney. 1993. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein-Barr virus promoter, BMRF1. Nucleic Acids Res. 21:1999-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saveliev, A., F. Zhu, and Y. Yuan. 2002. Transcription mapping and expression patterns of genes in the major immediate-early region of Kaposi's sarcoma-associated herpesvirus. Virology 299:301-314. [DOI] [PubMed] [Google Scholar]
- 53.Schlaak, J. F., C. M. Hilkens, A. P. Costa-Pereira, B. Strobl, F. Aberger, A. M. Frischauf, and I. M. Kerr. 2002. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J. Biol. Chem. 277:49428-49437. [DOI] [PubMed] [Google Scholar]
- 54.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 55.Sen, G. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 56.Simmen, K. A., J. Singh, B. G. Luukkonen, M. Lopper, A. Bittner, N. E. Miller, M. R. Jackson, T. Compton, and K. Fruh. 2001. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc. Natl. Acad. Sci. USA 98:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song, M. J., H. J. Brown, T. T. Wu, and R. Sun. 2001. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:3129-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, M. J., H. Deng, and R. Sun. 2003. Comparative study of regulation of RTA-responsive genes in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 77:9451-9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Song, M. J., X. Li, H. J. Brown, and R. Sun. 2002. Characterization of interactions between RTA and the promoter of polyadenylated nuclear RNA in Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 76:5000-5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, S., S. Liu, M. Wu, Y. Geng, and C. Wood. 2001. Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 ORF50 gene product contains a potent C-terminal activation domain which activates gene expression via a specific target sequence. Arch. Virol. 146:1415-1426. [DOI] [PubMed] [Google Scholar]
- 65.Wang, S., S. Liu, M. H. Wu, Y. Geng, and C. Wood. 2001. Identification of a cellular protein that interacts and synergizes with the RTA (ORF50) protein of Kaposi's sarcoma-associated herpesvirus in transcriptional activation. J. Virol. 75:11961-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, S. E., F. Y. Wu, M. Fujimuro, J. Zong, S. D. Hayward, and G. S. Hayward. 2003. Role of CCAAT/enhancer-binding protein alpha (C/EBPα) in activation of the Kaposi's sarcoma-associated herpesvirus (KSHV) lytic-cycle replication-associated protein (RAP) promoter in cooperation with the KSHV replication and transcription activator (RTA) and RAP. J. Virol. 77:600-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 68.West, J. T., and C. Wood. 2003. The role of Kaposi's sarcoma-associated herpesvirus/human herpesvirus-8 regulator of transcription activation (RTA) in control of gene expression. Oncogene 22:5150-5163. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, L. 2000. Gradient temperature hybridization using a thermocycler for RNase protection assays. Mol. Biotechnol. 14:73-75. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, L., and J. S. Pagano. 1999. Interferon regulatory factor 2 represses the Epstein-Barr virus BamHI Q latency promoter in type III latency. Mol. Cell. Biol. 19:3216-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, L., and J. S. Pagano. 2000. Interferon regulatory factor 7 is induced by Epstein-Barr virus latent membrane protein 1. J. Virol. 74:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7 mediates the activation of Tap-2 by Epstein-Barr virus latent membrane protein 1. J. Virol. 75:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, L., and J. S. Pagano. 2001. Interferon regulatory factor 7: a key cellular mediator of LMP-1 in EBV latency and transformation. Semin. Cancer Biol. 11:445-453. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, L., and J. S. Pagano. 2002. Review: structure and function of IRF-7. J. Interferon Cytokine Res. 22:95-101. [DOI] [PubMed] [Google Scholar]
- 76.Zhang, L., L. H. Wu, K. Hong, and J. S. Pagano. 2001. Intracellular signaling molecules activated by Epstein-Barr virus for induction of interferon regulatory factor 7. J. Virol. 75:12393-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhu, F. X., S. M. King, E. J. Smith, D. E. Levy, and Y. Yuan. 2002. A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation. Proc. Natl. Acad. Sci. USA 99:5573-5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]