Abstract
We previously reported that inhibition of endosomal/lysosomal function can dramatically enhance human immunodeficiency virus type 1 (HIV-1) infectivity, suggesting that under these conditions productive HIV-1 infection can occur via the endocytic pathway. Here we further examined this effect with bafilomycin A1 (BFLA-1) and show that this enhancement of infectivity extends to all HIV-1 isolates tested regardless of coreceptor usage. However, isolate-specific differences were observed in the magnitude of the effect. This was particularly evident in the case of the weakly infectious HIV-1SF2, for which we observed the greatest enhancement. Using reciprocal chimeric viruses, we were able to determine that both the disproportionate increase in the infectivity of HIV-1SF2 in response to BFLA-1 and its weak infectivity in the absence of BFLA-1 mapped to its envelope gene. Further, we found HIV-1SF2 to have lower fusion activity and to be 12-fold more sensitive to the fusion inhibitor T-20 than HIV-1NL4-3. Proteasomal inhibitors also enhance HIV-1 infectivity, and we report that the combination of a lysosomal and a proteasomal inhibitor greatly enhanced infectivity of all isolates tested. Again, HIV-1SF2 was unique in exhibiting a synergistic 400-fold increase in infectivity. We also determined that inhibition of proteasomal function increased the infectivity of HIV-1 pseudotyped with vesicular stomatitis virus G protein. The evidence presented here highlights the important role of the lysosomes/proteasomes in the destruction of infectious HIV-1SF2 and could have implications for the development of novel antiviral agents that might take advantage of these innate defenses.
Infection of cells by enveloped viruses requires membrane fusion. Many viruses utilize one of two pathways to gain access to the target cell's cytoplasm (24). The first is fusion directly with the plasma membrane. This event occurs at neutral pH. In the second pathway viral particles are endocytosed and subsequently induced by low pH to fuse with endosomal or lysosomal membranes. Inhibitors of vacuolar H+-ATPase have been used to directly demonstrate infection by the second pathway for vesicular stomatitis virus (VSV) and other viruses (14, 19, 29, 35).
Recently, it has been reported that treatment of cells with vacuolar H+-ATPase inhibitors at concentrations that completely block VSV infection markedly increases the infectivity of human immunodeficiency virus type 1 (HIV-1) (20, 41). Thus, it appears that not only can the endocytic pathway of HIV-1 entry be infectious under these conditions but also the normal acidification of endosomes results in extensive inactivation of HIV-1. For these reasons the entry of HIV-1 into cells cannot be described accurately by either of the two pathways described above. In fact, electron microscopy studies of HIV-1 fusion have provided clear examples of HIV-1 particles fusing at the cell surface and in endocytic vesicles (21, 22, 33). Therefore, possible routes for HIV-1 infection include fusion at the cell surface and fusion early in the endocytic pathway prior to vesicular acidification.
Whereas lysosomes serve to degrade endocytosed proteins, proteasomes primarily degrade cytoplasmic proteins (4, 11). Thus, once in the cytoplasm, viral particles could be susceptible to degradation by proteasomes. Indeed, the infectivity of one HIV-1 isolate has been reported to be enhanced by proteasome inhibition (43). In this report we confirm and extend these results to multiple HIV-1 isolates and further demonstrate that combining the inhibitors of proteasomal and lysosomal proteolysis yields larger increases in HIV-1 infectivity than any previously reported. The magnitudes of these effects demonstrate that lysosomal and proteasomal degradation represent significant cellular defenses to HIV-1 infection. Particularly noteworthy is HIV-1SF2 since the combination of lysosomal and proteasomal inhibitors yielded a synergistic increase in infectivity with this isolate. We have further characterized HIV-1SF2 and HIV-1NL4-3 entry using the BlaM-Vpr fusion assay, inhibition by T-20 (also known as enfuvirtide and Fuzeon), and genetic mapping. Our findings indicate that the extensive degradative functions of host cells place significant constraints on Env function.
MATERIALS AND METHODS
Cells, constructs, and viruses.
GHOST cells (kindly provided by V. KewalRamani and D. Littman through the NIH AIDS Research and Reference Reagent Program) and 293T cells were cultured in Dulbecco′s modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 IU penicillin/ml, 100 μg/ml streptomycin, and 2 mM glutamine. P4-R5 Magi cells (kindly provided by N. Landau through the NIH AIDS Research and Reference Reagent Program) were cultured in DMEM supplemented with 10% FBS and 1.0 μg/ml puromycin (8). HeLa-CD4+ cells (kindly provided by B. Chesebro through the NIH AIDS Research and Reference Reagent Program) were grown in RPMI supplemented with 10% FBS and 1 mg/ml G418 (10). U937 and THP-1 cells were cultured in RPMI supplemented with 10% FBS, 100 IU penicillin/ml, 100 μg/ml streptomycin, and 2 mM glutamine. Cell lines were maintained at 37°C in a humidified incubator with 5% CO2. Virus stocks of HIV-1SF162 and HIV-1JR-FL (kindly provided by J. Levy and I. Chen, respectively, through the NIH AIDS Research and Reference Reagent Program), the molecular clones HIV-1SF2 (kindly provided by P. Luciw), HIV-1NL4-3 (kindly provided by M. Martin), HIV-1LAI and HIV-1LAIΔE (kindly provided by M. Emerman), HIV-1YU2 (kindly provided by B. Hahn through the NIH AIDS Research and Reference Reagent Program), BH10ψ−E+, and RtatpEGFP all have been described previously (1, 9, 13, 26, 27, 34, 40). Modified proviral constructs of HIV-1SF2 and HIV-1NL4-3 were constructed using standard molecular biology techniques. The HIV-1SF2 BamHI-XhoI fragment was made by PCR and sequenced before cloning into HIV-1NL4-3 to generate HIV-1NL4-3BaXSF2. The VSV G protein (VSV-G) expression construct pLVSV-G was kindly provided by M. Emerman. The pCMV4-3BlaM-Vpr plasmid was kindly provided by W. Greene (6), and pAdVAntage was purchased from Promega (Madison, WI). Virus supernatants were prepared by transient transfection of 293T cells, and their p24Gag content was determined by enzyme-linked immunosorbent assay (ELISA) using a kit purchased from Coulter (Miami, FL) essentially as we have previously reported (20).
Infectivity and fusion assays.
P4-R5 Magi cells were used for the infectivity assays as previously described (20). These cells express CD4, CXCR4, and CCR5 and contain an integrated β-galactosidase gene under the control of the Tat-responsive HIV-1 long terminal repeat (LTR). Cells were preincubated with lysosomal inhibitors dissolved in dimethyl sulfoxide (DMSO) or with DMSO alone, as a control, for 1 h at 37°C. Viruses were then added to cells in the presence of DEAE-dextran (20 μg/ml final concentration) and incubated for 16 to 18 h at 37°C. Cells were washed twice with phosphate-buffered saline (PBS) and incubated for an additional 24 h in DMEM supplemented with 10% FBS. Cells were then fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Infectious units (IU; number of β-galactosidase positive cells) were counted as previously described (25). In the experiments utilizing proteasomal inhibitors, the inhibitors and viruses were added at the same time and incubated for 1 h at 37°C. Cells were then washed twice with PBS and incubated for an additional 36 to 40 h in DMEM supplemented with 10% FBS before fixation and staining. In the experiments using either the CXCR4 inhibitor AMD3100 or the fusion inhibitor T-20, the inhibitor and viruses were mixed and then added to the cells and incubated for 16 to 18 h at 37°C. Cells were washed twice with PBS, incubated for an additional 24 h in DMEM supplemented with 10% FBS, fixed, and then stained for β-galactosidase activity. GHOST cell infections were performed essentially as previously described (5). U937 and THP-1 cells were infected with BH10ψ−E+/RtatpEGFP virus, in the presence or absence of the indicated inhibitors, for 1 h and analyzed by flow cytometry 36 to 48 h after infection as we have previously described (13). The following compounds were used at the indicated final concentrations: bafilomycin A1, 30 or 100 nM (Sigma); MG132, 25 μM, and lactacystin, 20 μM (both from CalBiochem); AMD3100, 0.1, 1, 10, or 100 ng/ml (AnorMED; obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health); and T-20, 0.001, 0.003, 0.006, 0.01, 0.02, 0.1, or 1.0 μg/ml (Trimeris, Inc.). T-20 was dissolved in sterile water, and all other inhibitors were dissolved in DMSO. Individual samples were evaluated in triplicate in at least two independent experiments.
The BlaM-Vpr fusion assay was performed essentially as previously described using HeLa-CD4+ cells (6) with equal amounts of p24Gag (100 to 300 ng) per well for each virus used. Cells were analyzed by fluorescence microscopy using a Zeiss Axiovert 100 M microscope equipped with a β-lactamase filter set (Chroma Technology Corp., Rockingham, VT), and images were obtained with Openlab 3.1.5 imaging software at the UTSW Medical Center Molecular and Cellular Imaging Core Facility.
Isolation of human PBMC and HIV-1 replication assay.
Healthy donor human peripheral blood mononuclear cells (PBMC) were isolated from the buffy coat layer following Ficoll-Paque PLUS (Amersham Pharmacia, Piscataway, NJ) density gradient centrifugation. Before infection, the PBMC were stimulated with phytohemagglutinin (2 μg/ml) for 3 days at 37°C in RPMI containing 10% heat-inactivated FBS, 100 U penicillin/ml, 100 μg/ml streptomycin, 2 mM l-glutamine, and 1 mM sodium pyruvate (complete RPMI). Activated PBMC were then washed with PBS, pelleted, resuspended in complete RPMI, and incubated with HIV-1 (2 ng p24Gag per 106 cells) for 1 h in the presence of DEAE-dextran (20 μg/ml). PBMC (1 × 106) were then washed three times with PBS and resuspended in 1 ml complete RPMI supplemented with interleukin 2 (40 U/ml; Chiron Corporation, Emeryville, CA) in 12-well plates. At 3, 6, and 10 days postinfection, culture supernatants (100 μl) from each sample were collected and replaced with an equal volume of fresh complete RPMI containing interleukin 2 (40 U/ml). Culture supernatants were subsequently analyzed for the presence of p24Gag by ELISA as indicated above.
Western blot analysis of viral proteins.
Virus supernatants prepared by transient transfection of 293T cells were lysed with sodium dodecyl sulfate (SDS) gel loading buffer (62.5 mM Tris-Cl, 2% SDS, 10% glycerol, 0.1 mM dithiothreitol, pH 6.8). Supernatants containing equal amounts of p24Gag were analyzed by 10% SDS-polyacrylamide gel electrophoresis and were transferred to nitrocellulose filters. HIV-1 envelope protein was detected using goat anti-HIV-1 gp120 polyclonal antibodies (1:1,500 dilution; Fitzgerald Industries International, Inc.), and the fusion protein β-lactamase-HIV-1 Vpr was detected using rabbit anti-β-lactamase polyclonal antibodies (1:2,500 dilution; Chemicon International), followed by horseradish peroxidase conjugated anti-goat or anti-rabbit immunoglobulin G (1:10,000 dilution; Zymed). Horseradish peroxidase conjugates were visualized using enhanced chemiluminescence (Amersham).
Statistics.
Statistical analysis was performed using Student's t test (GraphPad Prism Version 4.0; GraphPad Software Inc., San Diego, CA). A value of P < 0.05 was considered significant. Data are presented as the means of experimental measurements and standard errors of the mean. Fifty percent inhibition concentrations (IC50) were calculated on a sigmoidal dose response curve with GraphPad Prism Version 4.0.
RESULTS
HIV-1SF2 has low infectivity that is dramatically enhanced in the presence of BFLA-1.
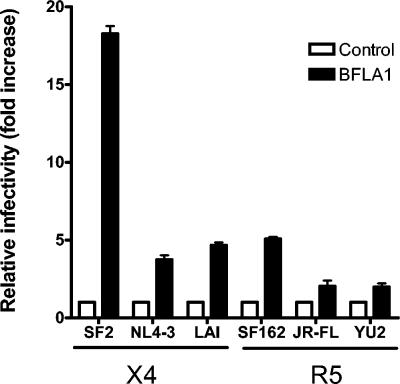
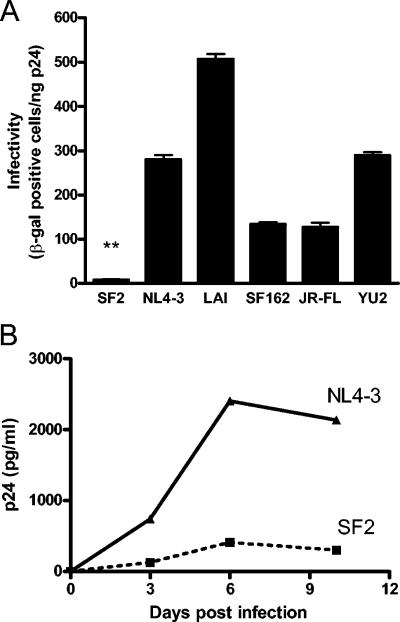
We have previously described an increase in infectivity of several HIV-1 isolates brought about by inhibitors of endosomal acidification (20). We also showed that this increase in infectivity is neither due to increased receptor or coreceptor expression nor to an increase in transcription from the viral LTR. Rather, this increase in infectivity is due to a block of virus degradation upon viral entry into the cell via endocytosis (20). Of the six different isolates we have tested in this assay, HIV-1SF2 was unique because it showed by far the highest enhancement of infectivity (20- to 50-fold) in response to the lysosomal inhibitor BFLA-1 (Fig. 1; 20). To determine if this infectivity increase in response to BFLA-1 by HIV-1SF2 was dependent on the amount of virus input, we infected GHOST indicator cells with increasing amounts of HIV-1SF2. When compared to the amount of virus input used in our standard assay conditions (5 ng p24Gag), at the highest amount of virus input used (125 ng p24Gag) there was a 30% reduction in the increase in infectivity observed. These results suggest that this effect is not saturable by the amount of input virus used (data not shown). In order to begin addressing the basis of this phenomenon, we evaluated the infectivity of these six HIV-1 isolates: HIV-1SF2, HIV-1NL4-3, HIV-1LAI, HIV-1SF162, HIV-1JR-FL, and HIV-1YU-2. Our results show that HIV-1SF2 is markedly less infectious than other HIV-1 isolates (30- to 50-fold-lower infectivity than HIV-1NL4-3 and HIV-1LAI) (Fig. 2A). Thus, HIV-1SF2, the least infectious HIV-1 isolate tested, also has the greatest response to lysosomal inhibitors such as BFLA-1.
FIG. 1.
BFLA-1 increases HIV-1 infectivity independently of the coreceptor tropism. P4-R5 Magi cells were incubated with 30 nM BFLA-1 prior to the addition of HIV-1SF2, HIV-1NL4-3, HIV-1LAI, HIV-1SF162, HIV-1JR-FL, or HIV-1YU2, as indicated in Materials and Methods. Infections were performed in triplicate in at least two independent experiments. Shown are averages of these measurements. To facilitate comparisons between viruses, the infectivity of the control sample for each virus was set to 1. X4, CXCR4-tropic; R5, CCR5-tropic.
FIG. 2.
HIV-1SF2 is significantly less infectious than other HIV-1 isolates. (A) The absolute infectivity of HIV-1SF2, HIV-1NL4-3, HIV-1LAI, HIV-1SF162, HIV-1JR-FL, or HIV-1YU2 was directly compared using P4-R5 Magi cells. Infections were performed in triplicate in at least two independent experiments. **, P < 0.001 represents the statistical difference between the infectivity of HIV-1SF2 and all the other viruses tested. (B) PBMC were activated by phytohemagglutinin stimulation prior to infection with either HIV-1SF2 or HIV-1NL4-3. Viral replication was monitored by ELISA to detect p24Gag present in the culture supernatants. Results shown are representative of three separate experiments performed with PBMC isolated from three different donors. β-gal, β-galactosidase.
To address whether HIV-1SF2 is also less infectious in cell types other than HeLa-based indicator cell lines, we infected activated human PBMC with HIV-1SF2 and HIV-1NL4-3 and measured viral replication as a function of time. Our results indicated that HIV-1SF2 replicated approximately six times less efficiently than HIV-1NL4-3 (Fig. 2B). These data show that while the difference in replication between HIV-1SF2 and HIV-1NL4-3 might vary in different systems, HIV-1SF2 remains substantially less infectious than HIV-1NL4-3 in human primary lymphocytes.
The lower infectivity of HIV-1SF2 and its response to BFLA-1 map to env.
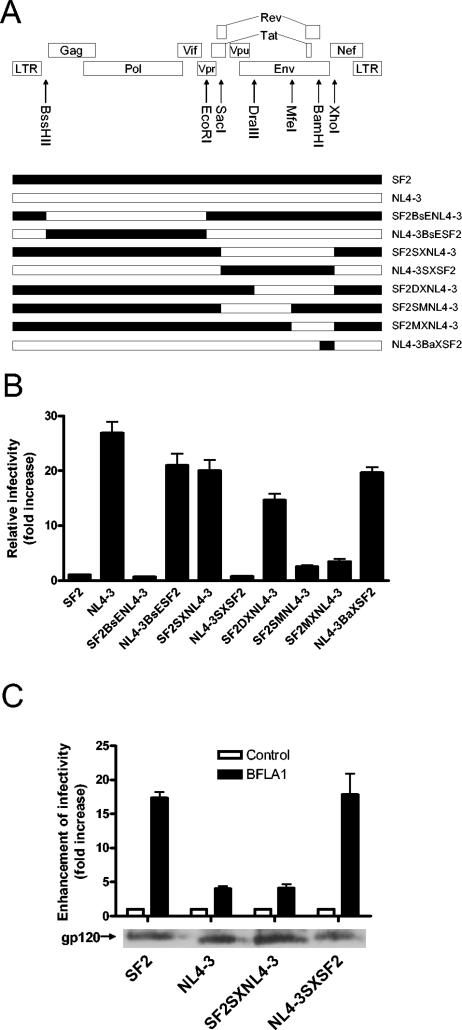
To determine the genetic basis for the lower infectivity of HIV-1SF2 shown in Fig. 2, we constructed several chimeric proviral clones and evaluated their respective infectivity. As shown in Fig. 3A and B, the first reciprocal chimeras, HIV-1SF2BsENL4-3 and HIV-1NL4-3BsESF2, mapped the low infectivity of HIV-1SF2 to the genes in the 3′ half of the genome. Results of further mapping experiments using the reciprocal chimeras HIV-1SF2SXNL4-3 and HIV-1NL4-3SXSF2 localized the low infectivity of HIV-1SF2 and the high infectivity of HIV-1NL4-3 to the env region. We then attempted to localize further this effect within env. Replacing HIV-1SF2 gp120 with HIV-1NL4-3 gp120 (HIV-1SF2SMNL4-3) or replacing HIV-1SF2 gp41 with HIV-1NL4-3 gp41 (HIV-1SF2MXNL4-3) resulted in chimeras that were only 2.5- or 3.5-fold more infectious than HIV-1SF2 (Fig. 3B). Our analysis also showed that the Env C terminus and the Nef N terminus are not responsible for the dramatic difference in the infectivities of these two viruses since replacing the HIV-1NL4-3 BamHI-XhoI fragment, which encodes the Env C-terminal 106 amino acids and the Nef N-terminal 34 amino acids, with the HIV-1SF2 BamHI-XhoI fragment (HIV-1NL4-3BaXSF2) did not decrease the infectivity of HIV-1NL4-3. Together these results map the lower infectivity of HIV-1SF2 to its env gene and suggest that the weak infectivity of HIV-1SF2 cannot be attributed to a single locus within the envelope gene; rather, they indicate that this phenotype is genetically complex.
FIG. 3.
The lower infectivity and higher responsiveness to BFLA-1 of HIV-1SF2 map to its envelope. (A) Diagram depicting the genomic structure of HIV-1 and the chimeric viruses used for this analysis. (B) Infectivities of wild-type and chimeric viruses relative to HIV-1SF2 (set as 1). Infections with each wild-type virus and the individual chimeras were performed twice in triplicate using P4-R5 Magi cells. (C) Top: effect of BFLA-1 on the infectivities of HIV-1SF2, HIV-1NL4-3, and two chimeras. The infectivity of each virus in the absence of BFLA-1 was set as 1 for comparison purposes. Bottom: similar levels of Env incorporation into virions were confirmed by Western blot analysis of HIV-1 gp120 in the four virus preparations. For both panels B and C, absolute infectivities expressed as IU/ng p24Gag for each isolate are: SF2, 13; NL4-3, 350; SF2BsENL4-3, 9; NL4-3BsESF2, 273; SF2SXNL4-3, 260; NL4-3SXSF2, 12; SF2DXNL4-3, 191; SF2SMNL4-3, 33; SF2MXNL4-3, 45; NL4-3BaXSF2, 260.
To determine if the increase in HIV-1SF2 infectivity mediated by BFLA-1 also maps to its envelope gene, the infectivities of the parental viruses and reciprocal chimeras (HIV-1SF2SXNL4-3 and HIV-1NL4-3SXSF2, in which the env genes of HIV-1SF2 and HIV-1NL4-3 are exchanged) were evaluated in the presence and in the absence of BFLA-1. As shown in Fig. 3C, the magnitudes of increase in infectivity of the HIV-1SF2SXNL4-3 and HIV-1NL4-3SXSF2 chimeras were similar to those of HIV-1NL4-3 and HIV-1SF2, respectively. These results map the increase in infectivity of HIV-1SF2 in response to BFLA-1 to its envelope gene.
The lower infectivity of HIV-1SF2 can be explained by its lower fusion activity.
One possible explanation for the low infectivity of HIV-1SF2 is that HIV-1SF2 Env is present at lower levels than HIV-1NL4-3 Env in their respective virions. We determined the relative levels of Env on the parental and two reciprocal chimeric virions by Western blot analysis. The Env expression levels for the four viruses were found to be similar, suggesting that the difference in infectivity of these viruses is not likely due to different levels of Env incorporation (Fig. 3C).
Alternatively, the difference in infectivity between HIV-1SF2 and HIV-1NL4-3 could be due to differences in their binding to the CXCR4 coreceptor molecule (17). To investigate this possibility, we compared the sensitivities of these two viruses to the CXCR4 inhibitor AMD3100 (23). Our results demonstrated that HIV-1SF2 and HIV-1NL4-3 have similar sensitivities to the inhibitory effects of AMD3100 (data not shown) as no statistical difference was found between the calculated AMD3100 IC50 values for HIV-1SF2 and HIV-1NL4-3 (0.9 and 1.4 nM, respectively; P = 0.2).
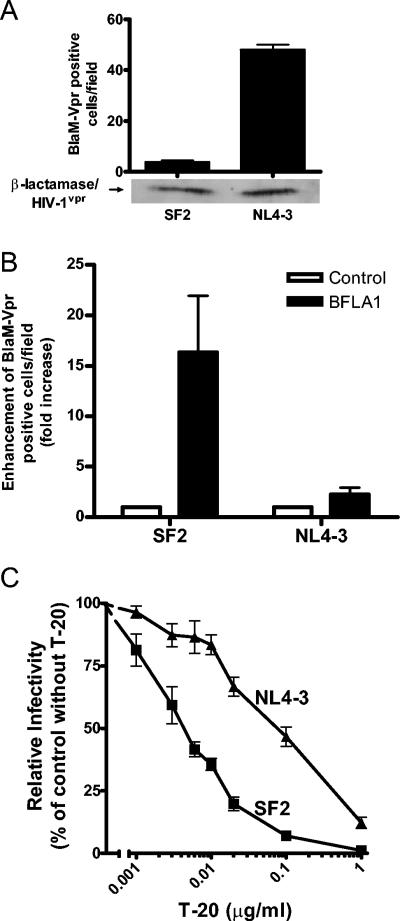
We then investigated whether the lower infectivity of HIV-1SF2 is due to a lower fusion activity. For this we used a recently described fusion assay based on the incorporation of a BlaM-Vpr fusion protein into virions (6). As shown in Fig. 4A, the fusion activity of HIV-1NL4-3 is approximately 15-fold higher than that of HIV-1SF2. It should be noted that Western blot analysis of BlaM-Vpr demonstrates that this result is not due to differences in the levels of BlaM-Vpr incorporation into the virions (Fig. 4A). In the presence of BFLA-1, the number of HIV-1SF2 viral particles capable of scoring positive in this fusion assay was increased by 16-fold whereas the number of HIV-1NL4-3 viral particles capable of fusing was increased by only 2-fold (Fig. 4B). These findings are in agreement with the single-round infectivity assays described above and suggest that differences in the fusion ability between HIV-1NL4-3 and HIV-1SF2 account for the reduced infectivity of HIV-1SF2 and the enhancement of infectivity mediated by BFLA-1.
FIG. 4.
Analysis of HIV-1SF2 and HIV-1NL4-3 fusion to target cells. (A) Top: fusion differences between HIV-1SF2 and HIV-1NL4-3 were determined using a BlaM-Vpr fusion assay essentially as described by Cavrois et al. (6). HeLa-CD4+ cells were incubated with HIV-1SF2BlaM-Vpr and HIV-1NL4-3BlaM-Vpr, and the infected blue cells were counted from acquired images. Shown are averages of triplicate determinations of one experiment representative of three independent experiments. Bottom: Western blot analysis indicates similar levels of BlaM-Vpr fusion protein incorporated into the virions. (B) Effects of BFLA-1 on fusion activities of HIV-1SF2BlaM-Vpr and HIV-1NL4-3BlaM-Vpr. The fusion activity of the control sample for each virus was set as 1 for comparison purposes. Data shown represent one of two experiments. (C) Differential sensitivities of HIV-1SF2 and HIV-1NL4-3 to inhibition by T-20 measured using a single-round infectivity assay with P4-R5 Magi cells. Results are averages from at least three determinations. The experiment was repeated three times.
If HIV-1SF2 Env fuses less efficiently than HIV-1NL4-3 Env, then HIV-1SF2 would be expected to be more sensitive than HIV-1NL4-3 to the fusion inhibitor T-20 (38, 46). Therefore, we measured the sensitivities of HIV-1SF2 and HIV-1NL4-3 to T-20. As shown in Fig. 4C, HIV-1SF2 exhibited a 12-fold-greater sensitivity to T-20 when compared to HIV-1NL4-3. The calculated T-20 IC50 values for HIV-1SF2 and HIV-1NL4-3 were 0.9 and 10.3 nM (equivalent to 0.004 and 0.047 μg/ml, P < 0.01), respectively. These data are consistent with HIV-1SF2 Env exhibiting lower fusion activity and account for its lower infectivity and responsiveness to BFLA-1.
The combination of BFLA-1 and a proteasomal inhibitor results in either synergistic or additive effects on HIV-1 infectivity.
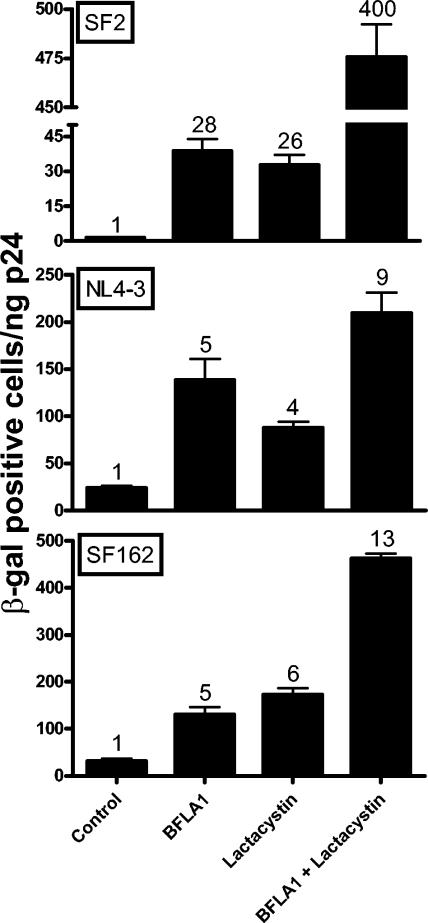
Regardless of the route of entry to the cytoplasm, HIV-1 cores could be susceptible to degradation by proteasomes. This has been demonstrated for one HIV-1 isolate (43). To investigate whether this observation extends to other HIV-1 isolates, we treated target cells with two different inhibitors of proteasomal degradation: lactacystin, a highly specific and irreversible proteasomal inhibitor (18); and MG132, which reversibly blocks all activities of the 26S proteasome (31). Since the standard length of exposure to these inhibitors (16 to 18 h) reduced the viability of P4-R5 Magi cells, we limited the time of exposure of the cells to the mixture of inhibitors with the virus to 1 h. As shown in Fig. 5, lactacystin increased the infectivities of HIV-1SF2, HIV-1NL4-3, and HIV-1SF162 by 26-, 4-, and 6- fold, respectively. Similar results were obtained with MG132 (data not shown). For a direct comparison, we also tested the effect of BFLA-1 (100 nM, 1-h exposure) in the same experiment. The infectivities of these viruses were enhanced 28-, 5-, and 4-fold, respectively, by BFLA-1. Thus, the magnitude of the effects of lactacystin and BFLA-1 are surprisingly similar for these viruses; in particular, HIV-1SF2 was again differentially impacted. Another surprise resulted when the infectivities of these three viruses were determined in the presence of both lactacystin and BFLA-1. An additive increase in infectivity was observed for HIV-1NL4-3 and HIV-1SF162 (9- and 13-fold, respectively), but a remarkable synergistic 400-fold enhancement was reproducibly observed for HIV-1SF2 (Fig. 5). The combined action of these two mechanistically different inhibitors of proteolysis yields an infectivity for HIV-1SF2 of 483 IU per ng p24Gag, which is higher than that of HIV-1NL4-3 or HIV-1SF162 under the same experimental conditions (210 IU and 419 IU per ng p24Gag, respectively, in the presence of the combined inhibitors). Similarly, we observed an 8-fold increase with MG132 alone and a synergistic 95-fold increase with the combination of MG132 and BFLA-1 in the infectivity of HIV-1SF2 (data not shown).
FIG. 5.
Effect of the combination of lactacystin and BFLA-1 on HIV-1 infectivity. P4-R5 Magi cells were incubated for 1 h with HIV-1SF2, HIV-1NL4-3, or HIV-1SF162 in the absence or presence of either 100 nM BFLA-1, 20 μM lactacystin, or both. Shown are the averages of triplicate infections with fold increases indicated above each column. Note that the increased infectivity in the presence of the combination of inhibitors is additive for HIV-1NL4-3 and HIV-1SF162 but synergistic for HIV-1SF2. The lower absolute infectivity of the different viruses when compared to results shown in Fig. 1, 2, and 3 is due to the shorter incubation time used for infection (1 h vs. 16 to 18 h). β-gal, β-galactosidase.
Proteasomal inhibitors and BFLA-1 have opposite effects on the infectivity of VSV-G-pseudotyped HIV-1.
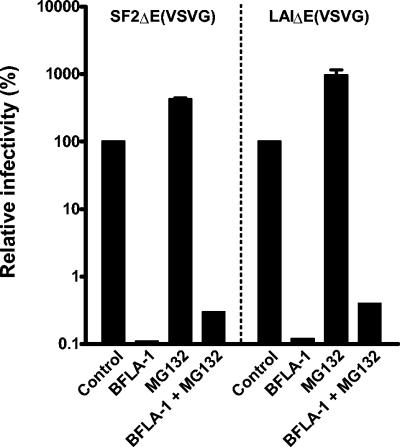
The synergistic effects of proteasomal inhibitors in the presence of BFLA-1 on HIV-1SF2 infectivity suggest the possibility that the proteasomal inhibitors and BFLA-1 might act sequentially. To look into this possibility, we determined the effect of MG132 on VSV-G-pseudotyped HIV-1SF2ΔE and HIV-1LAIΔE, which enter cells via endocytosis (2, 28, 29). Pseudotyped viruses were produced by cotransfection of 293T cells with env-defective proviral constructs and the pLVSV-G expression construct. Although wild-type HIV-1LAI is much more infectious than HIV-1SF2, this is not the case for the VSV-G pseudotypes (data not shown). As we have previously shown (28) and as predicted by the fact that VSV-G-pseudotyped particles fuse only at low pH (14, 19, 29, 35), BFLA-1 resulted in a dramatic inhibition of their infectivity (Fig. 6). However, in sharp contrast, MG132 increased the infectivity of HIV-1SF2ΔE and HIV-1LAIΔE VSV-G-pseudotyped viruses by five- and ninefold, respectively. Treatment of cells with both inhibitors resulted in a small increase in infectivity over BFLA-1 alone. Similar results were obtained when lactacystin was used in place of MG132 (data not shown). We interpret these results as suggesting that proteasomal inhibitors can act in conjunction with the lysosomal inhibitor BFLA-1, and these results provide additional evidence showing that the differential infectivities between HIV-1SF2 and other strains are due to env. Sequential actions of BFLA-1 and proteasomal inhibitors would be consistent with the observed synergistic enhancement of HIV-1SF2 infectivity.
FIG. 6.
Opposite effects of MG132 and BFLA-1 on infectivity of VSV-G-pseudotyped HIV-1. P4-R5 Magi cells were incubated for 1 h with HIV-1SF2ΔE (VSV-G) or HIV-1LAIΔE (VSV-G) in the absence or presence of either 100 nM BFLA-1, 25 μM MG132, or both. The infectivity of the control sample for each virus was set to 100% for comparison purposes. Shown are the values of two independent infections in triplicate. Note that a log scale is used to facilitate presentation of the dramatic reduction of infectivity of the VSV-G-pseudotyped viruses in the presence of BFLA-1.
Effect of BFLA-1 and lactacystin on the infectivity of human monocytic cell lines.
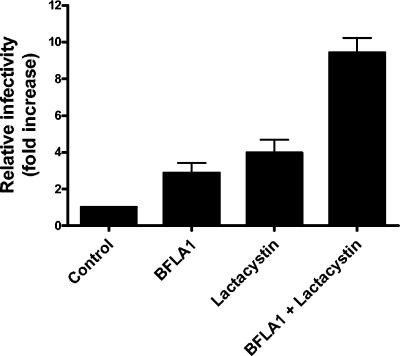
In order to determine if the infectivity enhancement mediated by lysosomal and proteasomal inhibitors extended to other cell types, we evaluated the effect of BFLA-1 and lactacystin on infectivity in human PBMC, monocytic cells, and T cells. The combination of drugs had high toxicity to most of these cells tested, which limited our investigation. However, we found that two monocytic cell lines, U937 and THP-1, were able to withstand the drugs' toxic effects. So we pursued further study using these two cell lines. Unlike the HeLa cells used above, these cells do not contain an integrated copy of the HIV-1 LTR driving β-galactosidase expression. Therefore, we used BH10ψ−E+/RtatpEGFP virus expressing enhanced green fluorescent protein under the control of the HIV-1 LTR as a marker for these experiments. Infection of U937 cells in the presence of BFLA-1 or lactacystin resulted in a reproducible threefold and fourfold increase in infectivity, respectively (Fig. 7). Infection of U937 cells in the presence of both inhibitors resulted in up to a 10-fold increase in infectivity (Fig. 7). Similar results were obtained with THP-1 cells (six-, three-, and ninefold increases in infectivity, respectively). These results demonstrate that the increase in HIV-1 infectivity mediated by lysosomal and proteasomal inhibitors extends to human monocytic cell lines.
FIG. 7.
Enhancement of infectivity by BFLA-1 and lactacystin in U937 cells. U937 cells were infected for 1 h with BH10ψ−E+/RtatpEGFP virus in the absence or presence of either 100 nM BFLA-1, 20 μM lactacystin, or both. The percentage of infected cells in each sample was determined by flow cytometry. The infectivity of the control sample was set to 1 for comparison purposes. Shown are the values of triplicate infections from one of two independent experiments.
DISCUSSION
The molecular clones HIV-1NL4-3, HIV-1LAI, and HIV-1BH10 have envelope genes derived from an early French isolate (7, 37, 45). The mechanism of infection by viruses containing Env encoded by these clones has been extensively studied. HIV-1SF2 was the first of the early isolates not related to HIV-1LAI (40). HIV-1SF2 exhibits strikingly different infectivity properties than HIV-1NL4-3 and HIV-1LAI (Fig. 2). It has a 30- to 50-fold-lower infectivity, and the enhancing effect of BFLA-1 is approximately 20-fold, in contrast to the 4-fold effect for HIV-1NL4-3 and HIV-1LAI. Similar, but smaller, effects have been reported by Schaeffer et al. (41). In contrast, Fackler and Peterlin observed that concanamycin A decreased HIV-1SF2 infectivity by 60% (16). As we have indicated previously (20), these apparent discrepancies are likely due to the multiple differences between the two different experimental approaches used. Based on the results of this study, the envelope gene is responsible for these two unique features of HIV-1SF2. This conclusion comes from the following observations: (i) swapping HIV-1NL4-3 env and HIV-1SF2 env yielded chimeric proviral clones with infectivities that matched the levels of the infectivities of the parental viruses from which the envelopes were derived (Fig. 3A and B), (ii) the enhancement of the infectivity by BFLA-1 of a chimeric virus in which the env of HIV-1NL4-3 was substituted with the HIV-1SF2 env (HIV-1NL4-3SXSF2) was the same as for HIV-1SF2 (Fig. 3C), (iii) The infectivities of HIV-1LAIΔE (VSV-G) and HIV-1SF2ΔE (VSV-G) are less than twofold different.
Since the differential infectivities of HIV-1SF2 and HIV-1NL4-3 result from altered Env function, it seemed likely that HIV-1SF2 was deficient in the delivery of viral cores to the cytoplasm of target cells. To confirm this possibility, we first showed that both HIV-1NL4-3 and HIV-1SF2 have the same sensitivity to the CXCR4 inhibitor AMD3100. Then we used the BlaM-Vpr assay described by Cavrois et al. (6) to show a near-15-fold difference in cytosolic BlaM-Vpr-positive cells between HIV-1NL4-3 and HIV-1SF2, strongly implicating a weak ability of HIV-1SF2 particles to fuse with cellular membranes as the major reason for the low infectivity of this virus. A reduced fusion activity is also supported by the respective sensitivities of the viruses to the fusion inhibitor T-20. Less efficient fusion by HIV-1SF2 could yield a longer kinetic window during which T-20 exerts its inhibitory effect (38, 39). Similarly, the greatly enhanced infectivity of HIV-1SF2 in the presence of BFLA-1 is consistent with an increased delivery of viral cores to the cytoplasm from endocytic vesicles.
Proteasomal inhibitors also increase the infectivity of HIV-1 (43 and data herein). Again, HIV-1SF2 was differentially affected, with a greater than 20-fold enhancement compared to a 4- to 6-fold enhancement for other HIV-1 isolates in the presence of lactacystin (Fig. 5). Even more striking is the 400-fold enhancement of HIV-1SF2 infectivity compared to the approximately 10-fold enhancement of other HIV-1 isolates in the presence of both BFLA-1 and lactacystin. Interpretation of these results required that we determine how proteasomal inhibitors and BFLA-1 act synergistically to enhance HIV-1SF2 infectivity. Because VSV-G-pseudotyped HIV-1 enters cells only by endocytosis, the observed enhanced infectivity of the HIV-1SF2 pseudotype in the presence of proteasomal inhibitor is consistent with a model in which the sequential activities of BFLA-1 and the proteasomal inhibitors account for the synergistic effects observed in Fig. 5.
The magnitude of the effects on HIV-1SF2 by these inhibitors is unique and suggests that the pathway of HIV-1SF2 entry is different from that of the other viruses tested. In agreement, a previous report (16) demonstrated that, under similar conditions, a greater proportion of HIV-1SF2 was endocytosed compared to HIV-1NL4-3. The overall result of these remarkable increases is that the low HIV-1SF2 infectivity is fully compensated for in the presence of lactacystin and BFLA-1. This requires a compensation for the 30-fold-lower infectivity of HIV-1SF2 relative to commonly used laboratory viruses, like HIV-1NL4-3 and HIV-1LAI, and further compensation for the 10-fold enhancement of the infectivity of these viruses in the presence of lactacystin and BFLA-1. The synergistic increase of HIV-1SF2 infectivity in the presence of BFLA-1 and lactacystin could be explained by proposing that HIV-1SF2 fusion involves two stages. In the first stage, viral particles would bind to CD4 and CXCR4 at the plasma membrane, but few if any would fuse. Because of their low fusion ability the vast majority are endocytosed. In the second stage, only a few particles would fuse early in endocytosis prior to reaching acidified endosomes/lysosomes and being degraded. Even the few particles released by fusion at the plasma membrane or in early endosomes would be mostly degraded by proteasomes. The result is a virus that is weakly infectious. HIV-1SF2, being highly deficient in fusion near the cell surface, disproportionately benefits from inhibition of viral degradation.
In addition to HeLa cells, we have been able to observe the effects of BFLA-1 and lactacystin in human monocytic cell lines U937 and THP-1. In these cases, the Env used was derived from HIV-1BH10. As with HIV-1NL4-3, severalfold increases were observed when BFLA-1 and lactacystin were used alone. However, in combination these drugs exhibited an additive effect. Thus, the results obtained with human monocytic cell lines and HeLa cells are consistent with each other.
In this paper we report that the infectivity of HIV-1SF2 and HIV-1NL4-3 can be enhanced up to 28- and 5-fold, respectively, by BFLA-1. These increases are larger than the 5- and 2.5-fold enhancements recently reported by Schaeffer et al. (41). These differences appear to be the result of the longer treatment at higher concentrations of BFLA-1 that are utilized in our experiments. The fact that the concentration of inhibitors used is limited by cell viability raises the possibility that substantial viral degradation persists in our experiments. In the presence of BFLA-1 and lactacystin the infectivity of HIV-1NL4-3 was further enhanced to 10-fold. Again, this represents a minimal estimate as a result of cell viability considerations. From these considerations, it is likely that previous estimates of the ratio of defective to infectious particles might have been overstated (25, 33). Furthermore, based on our results, we propose that the discrepancy between infectious events and the total number of HIV-1 particles is more likely the result of efficient cellular defenses to viral infection than a large number of defective particles.
The fact that proteasomal inhibitors reproducibly increase the infectivity of HIV-1 has important implications for our understanding of the role of the proteasomes in the overall viral infection process. Based on the results presented here and those previously published by other investigators (12, 32, 42, 43), the proteasome machinery seems to play two different and opposing roles during viral infection. These opposite effects occur at different stages of HIV-1 replication. The viral production stage requires the proteasome machinery for viral assembly, release, and maturation, resulting in viruses which contain ubiquitinated p6Gag (30, 32, 42, 44). When virion cores are disassembled during the infection stage, the ubiquitinated viral proteins may be what targets virions for degradation by proteasomes (3, 30). Based on the role of the proteasome during viral assembly and release, there has been significant interest in the possibility of using proteasomal inhibitors to block HIV-1 replication (15, 36). However, the fact that proteasomal inhibitors can increase the infectivity of HIV-1 particles from isolates such as HIV-1SF2 suggests that this approach could have negative and unanticipated consequences. On the other hand, identification of the proteasome as an important site for HIV-1SF2 degradation presents a yet unexplored opportunity for the development of novel inhibitors of HIV-1 infection.
Acknowledgments
We thank D. Margolis, D. Sodora, and L. Baugh and members of the Garcia laboratory for critical reading of the manuscript; F. Otieno for her technical assistance; R. Doms for his advice regarding specific aspects of the manuscript; W. Greene for the generous gift of the BlaM-Vpr plasmid; M. Emerman for the HIV-1LAI and HIV-1LAIΔE; P. Luciw for the HIV-1SF2; and M. Martin for the HIV-1NL4-3. We thank V. KewalRamani and D. Littman (for the GHOST cells), B. Chesebro (for the HeLa CD4+ cells), N. Landau (for the P4-R5 Magi cells), J. Levy (for the HIV-1SF162), B. Hahn (for the HIV-1YU2), I. Chen (for the HIV-1JR-FL), and Division of AIDS, NIAID (for the AMD3100), all obtained from the NIH AIDS Research and Reference Reagent Program, and C. J. Gilpin and the Molecular and Cellular Imaging Facility, UTSW Medical Center, for assistance with the fusion assay.
This work was supported by National Institutes of Health grant AI-33331 (J.V.G.). P.W.D. was supported in part by training grant 5T32 AI-005284 from NIAID.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiken, C. 1997. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71:5871-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, L. O., J. W. Bess, Jr., R. C. Sowder, 2nd, R. E. Benveniste, D. L. Mann, J. C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed] [Google Scholar]
- 4.Baumeister, W., J. Walz, F. Zuhl, and E. Seemuller. 1998. The proteasome: paradigm of a self-compartmentalizing protease. Cell 92:367-380. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, E. M., R. Nunez, and T. J. Hope. 2004. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J. Virol. 78:5745-5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 7.Chang, S. Y., B. H. Bowman, J. B. Weiss, R. E. Garcia, and T. J. White. 1993. The origin of HIV-1 isolate HTLV-IIIB. Nature 363:466-469. [DOI] [PubMed] [Google Scholar]
- 8.Charneau, P., G. Mirambeau, P. Roux, S. Paulous, H. Buc, and F. Clavel. 1994. HIV-1 reverse transcription. A termination step at the center of the genome. J. Mol. Biol. 241:651-662. [DOI] [PubMed] [Google Scholar]
- 9.Cheng-Mayer, C., and J. A. Levy. 1988. Distinct biological and serological properties of human immunodeficiency viruses from the brain. Ann. Neurol. 23(Suppl.):S58-S61. [DOI] [PubMed] [Google Scholar]
- 10.Chesebro, B., and K. Wehrly. 1988. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J. Virol. 62:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 12.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas, J., P. Kelly, J. T. Evans, and J. V. Garcia. 1999. Efficient transduction of human lymphocytes and CD34+ cells via human immunodeficiency virus-based gene transfer vectors. Hum. Gene Ther. 10:935-945. [DOI] [PubMed] [Google Scholar]
- 14.Drose, S., and K. Altendorf. 1997. Bafilomycins and concanamycins as inhibitors of V-ATPases and P-ATPases. J. Exp. Biol. 200:1-8. [DOI] [PubMed] [Google Scholar]
- 15.Elliott, P. J., and J. S. Ross. 2001. The proteasome: a new target for novel drug therapies. Am. J. Clin. Pathol. 116:637-646. [DOI] [PubMed] [Google Scholar]
- 16.Fackler, O. T., and B. M. Peterlin. 2000. Endocytic entry of HIV-1. Curr. Biol. 10:1005-1008. [DOI] [PubMed] [Google Scholar]
- 17.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 18.Fenteany, G., R. F. Standaert, W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 19.Fleming, D. M. 2001. Managing influenza: amantadine, rimantadine and beyond. Int. J. Clin. Pract. 55:189-195. [PubMed] [Google Scholar]
- 20.Fredericksen, B. L., B. L. Wei, J. Yao, T. Luo, and J. V. Garcia. 2002. Inhibition of endosomal/lysosomal degradation increases the infectivity of human immunodeficiency virus. J. Virol. 76:11440-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto, T., S. Harada, N. Yamamoto, and M. Nakai. 1988. Entry of human immunodeficiency virus (HIV) into MT-2, human T cell leukemia virus carrier cell line. Arch. Virol. 102:29-38. [DOI] [PubMed] [Google Scholar]
- 22.Grewe, C., A. Beck, and H. R. Gelderblom. 1990. HIV: early virus-cell interactions. J. Acquir. Immune Defic. Syndr. 3:965-974. [PubMed] [Google Scholar]
- 23.Hendrix, C. W., C. Flexner, R. T. MacFarland, C. Giandomenico, E. J. Fuchs, E. Redpath, G. Bridger, and G. W. Henson. 2000. Pharmacokinetics and safety of AMD-3100, a novel antagonist of the CXCR-4 chemokine receptor, in human volunteers. Antimicrob. Agents Chemother. 44:1667-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 25.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koyanagi, Y., S. Miles, R. T. Mitsuyasu, J. E. Merrill, H. V. Vinters, and I. S. Chen. 1987. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science 236:819-822. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 65:3973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, T., J. L. Douglas, R. L. Livingston, and J. V. Garcia. 1998. Infectivity enhancement by HIV-1 Nef is dependent on the pathway of virus entry: implications for HIV-based gene transfer systems. Virology 241:224-233. [DOI] [PubMed] [Google Scholar]
- 29.Matlin, K. S., H. Reggio, A. Helenius, and K. Simons. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 156:609-631. [DOI] [PubMed] [Google Scholar]
- 30.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, 2nd, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palombella, V. J., O. J. Rando, A. L. Goldberg, and T. Maniatis. 1994. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell 78:773-785. [DOI] [PubMed] [Google Scholar]
- 32.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pauza, C. D., and T. M. Price. 1988. Human immunodeficiency virus infection of T cells and monocytes proceeds via receptor-mediated endocytosis. J. Cell Biol. 107:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peden, K., M. Emerman, and L. Montagnier. 1991. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology 185:661-672. [DOI] [PubMed] [Google Scholar]
- 35.Perez, L., and L. Carrasco. 1994. Involvement of the vacuolar H(+)-ATPase in animal virus entry. J. Gen. Virol. 75:2595-2606. [DOI] [PubMed] [Google Scholar]
- 36.Piccinini, M., M. T. Rinaudo, N. Chiapello, E. Ricotti, S. Baldovino, M. Mostert, and P. A. Tovo. 2002. The human 26S proteasome is a target of antiretroviral agents. AIDS 16:693-700. [DOI] [PubMed] [Google Scholar]
- 37.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, L. Ivanoff, S. R. J. Petteway, M. L. Pearsons, J. A. Lautenberg, T. S. Papas, J. Ghrayeb, N. T. Chang, R. C. Gallo, and F. Wong-Staal. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 38.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves, J. D., J. L. Miamidian, M. J. Biscone, F. H. Lee, N. Ahmad, T. C. Pierson, and R. W. Doms. 2004. Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J. Virol. 78:5476-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanchez-Pescador, R., M. D. Power, P. J. Barr, K. S. Steimer, M. M. Stempien, S. L. Brown-Shimer, W. W. Gee, A. Renard, A. Randolph, J. A. Levy, D. Dina, and P. A. Luciw. 1985. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2). Science 227:484-492. [DOI] [PubMed] [Google Scholar]
- 41.Schaeffer, E., V. B. Soros, and W. C. Greene. 2004. Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J. Virol. 78:1375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wain-Hobson, S., P. Sonigo, O. Danos, S. Cole, and M. Alizon. 1985. Nucleotide sequence of the AIDS virus, LAV. Cell 40:9-17. [DOI] [PubMed] [Google Scholar]
- 46.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]