Abstract
Alpha/beta interferon (IFN-α/β) is a key mediator of innate antiviral responses but has little effect on the established replication of dengue viruses, which are mosquito-borne flaviviruses of immense global health importance. Understanding how the IFN system is inhibited in dengue virus-infected cells would provide critical insights into disease pathogenesis. In a recent study analyzing the ability of individual dengue virus-encoded proteins to antagonize the IFN response, nonstructural (NS) protein 4B and possibly NS2A and NS4A were identified as candidate IFN antagonists. In monkey cells, NS4B appeared to inhibit both the IFN-α/β and IFN-γ signal transduction pathways, which are distinct but overlapping (J. L. Munoz-Jordan, G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre, Proc. Natl. Acad. Sci. USA 100:14333-14338, 2003). For this study, we examined the effects of dengue virus on the human IFN system, using cell lines that were stably transfected with self-replicating subgenomic dengue virus RNA (replicons) and that expressed all of the dengue virus nonstructural proteins together. We show here that in replicon-containing cells dengue virus RNA replication and the replication of encephalomyocarditis virus, an IFN-sensitive virus, are resistant to the antiviral effects of IFN-α. The presence of dengue virus replicons reduces global IFN-α-stimulated gene expression and specifically inhibits IFN-α but not IFN-γ signal transduction. In cells containing replicons or infected with dengue virus, we found reduced levels of signal transducer and activator of transcription 2 (STAT2), which is a key component of IFN-α but not IFN-γ signaling. Collectively, these data show that dengue virus is capable of subverting the human IFN response by down-regulating STAT2 expression.
Dengue viruses are mosquito-borne flaviviruses of immense global public health importance, causing tens of millions of human infections worldwide each year (11). The intensity of viral replication in the first days of infection determines the clinical outcome, which ranges from benign febrile illness to life-threatening disease (dengue hemorrhagic fever) (39). During this critical early phase, prior to the full recruitment of antigen-specific defenses, innate cellular antiviral mechanisms mediated by alpha/beta interferon (IFN-α/β) are potentially the most important pathways of the host defense limiting viral replication. Virus infection classically induces the secretion of IFN-α/β, which binds to cell surface IFN-α receptors (IFNAR, comprising IFNAR1 and IFNAR2 subunits) on infected and nearby cells. The binding of IFN-α/β to IFNAR leads to the activation of Jak1 and Tyk2 kinases via tyrosine phosphorylation (4). In turn, signal transducer and activator of transcription 2 (STAT2) and then STAT1 are phosphorylated and form heterodimers, which then associate with p48/IRF-9 to form ISGF3 complexes (12). ISGF3 complexes translocate to the nucleus and initiate the transcription of interferon-stimulated genes (ISGs) by binding interferon-stimulated response elements, leading to the transcriptional up-regulation of hundreds of cellular genes and the induction of an antiviral state (35).
Experimental evidence suggests that the IFN system plays an important role in limiting dengue virus replication, since knockout mice that lack IFN-α/β receptors develop severe infections after a challenge with dengue virus (15, 34). Also, the pretreatment of cultured cells with IFN-α/β dramatically reduces dengue virus replication (5, 6). This occurs primarily through the inhibition of translation of input-strand dengue virus RNA by an unknown mechanism (5). In contrast, IFN-α/β has little effect on dengue virus replication after viral replication has been established (5, 6), suggesting that the IFN system cannot fully engage in dengue virus-infected cells. In keeping with this observation, dengue virus can achieve high titers (<109 infectious doses per ml) in humans despite the induction of high levels of circulating IFN-α (21, 36, 39). It therefore seems likely that dengue virus has evolved mechanisms to counter the IFN response, although not absolutely, which is a characteristic that may be shared by many pathogenic viruses (9, 42).
Muñoz-Jordan and colleagues recently published an in vitro study that analyzed the ability of individual dengue virus proteins to block the IFN system, in which they concluded that NS4B and possibly NS2A and NS4A act as IFN signaling inhibitors (25). They showed that NS4B and dengue virus infection blocked signal transduction in response to both IFN-β and IFN-γ in a monkey kidney cell line, suggesting that the target for NS4B-mediated inhibition of IFN signaling may be a component (possibly phosphorylated STAT1 [STAT1-P]) that is common to these distinct but overlapping signal transduction pathways (25). We adopted a complementary experimental approach specifically aimed at studying the effect of dengue virus replication downstream of the translation of input-strand RNA on the human IFN system. We first established human cell lines that continuously expressed self-replicating subgenomic dengue virus RNA (replicons). Flavivirus replicons express all of the viral nonstructural proteins together in a way that mimics expression during authentic viral infection, and they have proved to be powerful tools for studying the functional roles of nonstructural proteins in RNA and virus replication (16-18, 24). We show here that the presence of dengue virus replicons in human cell lines inhibits the antiviral effect of IFN-α by blocking early events in IFN-α signal transduction, resulting in reduced levels of STAT1-P. In contrast, STAT1-P levels in replicon-containing cells are increased rather than reduced in response to IFN-γ. We show that steady-state levels of STAT2 are reduced in cells containing dengue virus replicons, which is consistent with the observed responses to IFN-α and IFN-γ. Reduced STAT2 levels are also found in cells infected with dengue virus, suggesting that dengue virus is capable of subverting the human IFN response by down-regulating STAT2 expression.
MATERIALS AND METHODS
Cell lines stably expressing dengue virus replicons.
A series of cell lines that continuously express dengue virus replicons have been established in our laboratory (unpublished data). For this study, K562 (human chronic myeloid leukemia) and THP-1 (human monocytic) cell lines stably expressing the dengue virus replicon ΔCprME-PAC2A were used (designated K562.ΔCprME-PAC2A and THP-1.ΔCprME-PAC2A, respectively). The plasmid pDENΔCprME-PAC2A was used for in vitro transcription of ΔCprME-PAC2A replicon RNAs. pDENΔCprME-PAC2A was derived from pDVWS601 (29), which contains a genome-length dengue virus type 2 (New Guinea C strain) cDNA clone, by the introduction of a large in-frame deletion in the structural region, retaining only the first 27 codons of the C gene and the last 24 codons of the E gene. In addition, pDENΔCprME-PAC2A contains an antibiotic selection cassette encoding puromycin N-acetyltransferase (PAC) followed by an artificial protein cleavage site (foot-and-mouth disease virus protein 2A) in place of the deleted structural genes (Fig. 1). Cells stably expressing dengue virus replicon RNA were generated by transfection with ΔCprME-PAC2A RNA and then propagation in RPMI containing 10% fetal bovine serum (FBS) and 3 μg of puromycin (Sigma)/ml. Cells were removed from puromycin selection and checked for replicon expression before use by indirect immunofluorescence of the dengue virus NS1 protein with a specific monoclonal antibody (5H5.4) (7). K562 and THP-1 cells without replicons were continuously maintained in the same medium without puromycin.
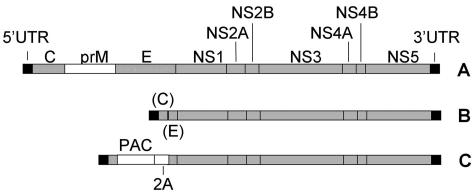
FIG. 1.
Schematic showing construction of plasmid pDENΔCprME-PAC2A. (A) Dengue virus type 2 infectious clone cDNA (in plasmid pDVWS601 [29]), showing a single open reading frame carrying three structural genes (C, core; prM, premembrane; E, envelope), seven nonstructural (NS) genes, and flanking 5′ and 3′ untranslated regions (UTR). (B) pDENΔCprME. A large in-frame deletion was introduced within the region carrying the structural genes. (C) pDENΔCprME-PAC2A. An antibiotic selection cassette encoding PAC and the foot-and-mouth disease virus protein 2A was cloned in place of the deleted structural genes.
Cured K562 cell line.
K562 cells that stably expressed ΔCprME-PAC2A were removed from puromycin selection and passaged continuously in RPMI containing 10% FBS and 500 μg of glycyrrhizic acid (Fluka Chemicals)/ml, which has activity against RNA viruses through an unknown mechanism (3). At intervals, cells were checked for replicon expression by indirect immunofluorescence of the dengue virus NS1 protein and by reverse transcription-PCR (RT-PCR) for dengue virus RNA (see below). Once the cell line had been cured of the replicon, it was subsequently grown in RPMI containing 10% FBS, without glycyrrhizic acid, and checked for the continued absence of replicons as described above.
Analysis of dengue virus RNA levels and NS1 expression.
K562.ΔCprME-PAC2A cells were grown in the presence of 0, 10, 100, 1,000, or 10,000 IU of IFN-α2a (Roferon-A; Roche)/ml for 24 h, and then the total cellular RNAs were extracted by the use of Trizol (Invitrogen). Extracted RNAs were treated with RQ1 RNase-free DNase (Promega) and reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Promega) using random decamer primers. PCRs were performed and analyzed on a Rotorgene instrument (Corbett Research) by the use of custom primers and a fluorescent probe specific for dengue virus NS1 (forward primer, 5′CTGAAGTGTGGCAGTGGGATT; reverse primer, 5′CTTCAAAGCTAGCTTCAGCTATCCA; probe, 5′CACAGACAACGTGCACACATGGACAGA). The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was analyzed in the same samples by the use of specific primers (forward primer, 5′ACAGTCCATGCCATCACTGCC; reverse primer, 5′GCCTGCTTCACCACCTTCTTG) and QuantiTect SYBR green (QIAGEN). In parallel experiments, K562.ΔCprME-PAC2A cells were grown in the presence of 100 IU of IFN-α2a/ml, and cell-associated and secreted dengue virus NS1 proteins were analyzed by immunoblotting and an enzyme-linked immunosorbent assay (ELISA), respectively, as previously described (14, 44).
EMCV trans rescue assay.
K562 cells that did and did not contain dengue virus replicons were grown in RPMI containing 10% FBS with 0, 10, and 100 IU of IFN-α2a/ml for 24 h. The cells were washed in RPMI, and 106 cells were then infected with 5 × 105 PFU of encephalomyocarditis virus (EMCV) in RPMI containing 2% FBS for 1 h. The cells were washed and then cultured for a further 24 h in RPMI plus 10% FBS. The culture supernatants were then harvested, and serial dilutions were plated onto confluent A549 cells in a 96-well plate for 1 h before replacing the inoculum with RPMI containing 10% FBS. After a further 24 h, the A549 cells were fixed and stained with methyl violet, and the optical density in each well was read at 570 nm in an automated plate reader (27).
MxA and PKR gene expression.
K562 and K562.ΔCprME-PAC2A cells were grown in the presence or absence of 100 IU of IFN-α2a/ml for 6 and 24 h. RNA extraction and reverse transcription were performed as described above. PCRs were performed and analyzed on a Rotorgene instrument by the use of SYBR green as described above, using primers specific for the MxA gene (forward primer, 5′AACAACCTGTGCAGCCAGTA; reverse primer, 5′AAGGGCAACTCCTGAGAGTG) or the protein kinase R (PKR) gene (forward primer, 5′TCTCTGGCGGTCTTCAGAAT; reverse primer, 5′ACTCCCTGCTTCTGACGGTA). The housekeeping gene GAPDH was analyzed in the same samples as described above.
ISG expression profiling by macroarray analysis.
K562 and K562.ΔCprME-PAC2A cells (2 × 107 per reaction) were treated with 100 IU of IFN-α2a/ml for 24 h before extraction of the total cellular RNAs by the use of Trizol. Radiolabeled cDNAs were generated from 20 μg of total RNA by reverse transcription with Superscript II (Gibco) in the presence of [32P]dCTP. Residual RNAs were hydrolyzed by an alkaline treatment at 70°C for 20 min, and the cDNAs were purified through G-50 columns (Amersham Pharmacia). Before hybridization to the macroarrays, the labeled cDNAs were mixed with 50 μg of COT-DNA (Gibco) and 10 μg of poly(A) DNA (Sigma), denatured at 95°C for 5 min, and hybridized for 1 h to minimize nonspecific binding. Preparation of the macroarrays (representing 150 genes, including many that are known to be stimulated by interferon), hybridization of the radioactive cDNAs, and scanning and analysis of the macroarrays were performed as described previously (33).
Immunoblotting.
K562 cells (2.5 × 105 per reaction) that did and did not contain replicons were stimulated with 100 IU of IFN-α2a or IFN-γ (R&D Systems)/ml for 30 min. Unstimulated cells were included for comparison. Cells were harvested and lysed in 250 μl of sodium dodecyl sulfate (SDS) loading buffer (0.0625 M phosphate [pH 7.0], 10% glycerol, 2% SDS, 0.001% bromophenol blue) that had been prewarmed to 60°C. Ten microliters of each sample was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were then transferred to a polyvinylidene difluoride membrane (Hybond-P; Amersham). Mouse monoclonal antibodies to STAT1, phosphorylated STAT1 (Tyr-701) (both Zymed), or STAT2 (BD Transduction Laboratories), a rabbit polyclonal antibody to phosphorylated STAT2 (Upstate Biotechnology), a goat polyclonal antibody to IFNAR1 (Abcam), and a rabbit polyclonal antibody to IFNAR2 (PBL Biomedical Laboratories) were used as primary antibodies. Detection was performed by the use of relevant horseradish peroxidase-conjugated secondary antibodies (Jackson Immunochemicals) and enhanced chemiluminescence reagents (ECL+; Amersham).
Fluorescence-activated cell sorting (FACS) analysis.
K562 and K562.ΔCprME-PAC2A (106 per reaction) cells were stained with an anti-IFNAR2 anti-body in RPMI containing 2% FBS at 4°C. Detection was performed by use of a phycoerythrin-conjugated donkey anti-rabbit secondary antibody (Jackson Immunochemicals), and samples were analyzed on a Becton Dickinson FACScan instrument. Data analysis was performed with WinMDI software.
Dengue virus infection of K562 cells.
K562 cells were incubated with dengue virus type 2 (New Guinea C strain) at a multiplicity of infection of 4 and then grown in RPMI containing 10% FBS. After 48 h, the cells were air dried on glass slides and fixed in cold methanol-acetone (50:50 [vol/vol]). The cells were dually labeled with a mouse anti-dengue virus NS1 antibody (5H5.4) and a rabbit anti-STAT2 antibody (C20; Santa Cruz Biotechnology). Fluorescein isothiocyanate-conjugated goat anti-rabbit and Texas Red-conjugated horse anti-mouse antibodies (both from Vector Laboratories) were used for detection. Images were analyzed under a Bio-Rad Radiance 2100 confocal microscope. In parallel experiments, cells (2 × 106 per reaction) were lysed and analyzed by immunoblotting as described above.
RESULTS
Dengue virus replicon RNA replication is resistant to IFN-α.
Previous studies have shown that the antiviral effect of IFN-α on dengue virus infection in cell cultures is markedly inhibited if the treatment is delayed a few hours after infection (5, 6), suggesting that dengue virus can counter the IFN response once replication has been established. We tested directly whether IFN could inhibit established dengue virus RNA replication in the form of the dengue virus replicon ΔCprME-PAC2A, which is stably maintained in K562.ΔCprME-PAC2A cells (unpublished data). K562.ΔCprME-PAC2A cells were grown in the presence of 0, 10, 100, 1,000, or 10,000 IU of IFN-α2a/ml for 24 h, and dengue virus replicon RNA levels were measured by quantitative RT-PCR. In addition, the effect of 100 IU of IFN-α/ml on the levels of cell-associated and secreted NS1 protein was analyzed by Western blotting and ELISA, respectively. Figure 2 shows that IFN-α had no significant effect on dengue virus replicon RNA levels or NS1 expression. These data confirm previous evidence suggesting that established dengue virus RNA replication is resistant to IFN-α (5).
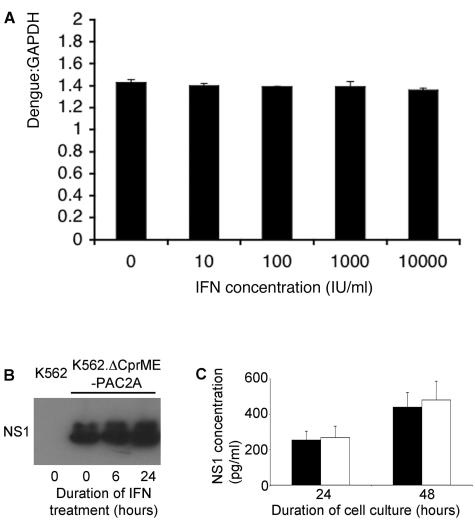
FIG. 2.
Dengue virus replicon RNA replication is resistant to IFN-α. (A) K562.ΔCprME-PAC2A cells were grown in the presence of various concentrations of IFN-α2a, as indicated, for 24 h. Dengue virus replicon RNA levels were measured by quantitative PCR and normalized to GAPDH mRNA levels. (B) K562.ΔCprME-PAC2A cells were grown in the presence of 100 IU of IFN-α2a/ml for 0, 6, and 24 h. Cell lysates (2 × 105 cells per reaction) were separated by SDS-PAGE, and then the dengue virus NS1 protein was analyzed by immunoblotting. K562 cells were included as a negative control. (C) K562.ΔCprME-PAC2A cells were grown in the absence (black bars) or presence (white bars) of 100 IU of IFN-α2a/ml for 24 and 48 h. The cumulative concentrations of NS1 in the culture supernatants at each time point were measured by ELISA.
Antiviral action of IFN-α is blocked by dengue virus RNA replication.
We next tested whether the presence of dengue virus replicons inhibits the general antiviral action of IFN in cells. K562 cells that did and did not contain replicons were first treated with IFN-α2a and then infected with an IFN-sensitive virus, EMCV. The basis of this technique is that the inhibition of the antiviral action of IFN by dengue virus replicons results in a rescue of EMCV replication, which is detected in a modified plaque assay on A549 cells. Figure 3 shows that in the absence of IFN-α2a, EMCV replication was equal in cells that did and did not contain replicons. As expected, a pretreatment of K562 cells with either 10 or 100 IU of IFN-α2a/ml dramatically inhibited the replication of EMCV. In contrast, pretreatments of K562.ΔCprME-PAC2A cells with the same concentrations of IFN-α2a had no effect on EMCV replication (Fig. 3). In order to prove that dengue virus RNA replication inhibited the IFN response, we repeated the EMCV rescue assay, using K562.ΔCprME-PAC2A cells that had previously been cured of the replicon by continuous growth in the presence of glycyrrhizic acid. Cured K562 cells were negative for the presence of dengue virus NS1 protein and dengue virus RNA by indirect immunofluorescence and RT-PCR, respectively (unpublished data). Cured K562 cells reverted to the IFN-responsive phenotype of the original K562 cells (data not shown). Taken together, these data show that the antiviral activity of IFN is blocked in the presence of dengue virus replicons.
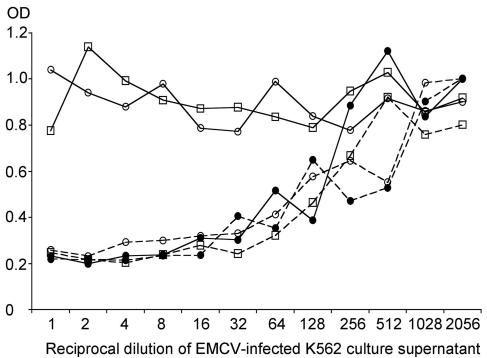
FIG. 3.
Antiviral effect of IFN-α is blocked in dengue virus replicon-containing cells. K562 cells (solid lines) and K562.ΔCprME-PAC2A cells (broken lines) were treated with 0 (•), 10 (□), or 100 (○) IU of IFN-α2a/ml for 24 h. The cells were then infected with EMCV, and after a further 24 h, the supernatants were harvested and serially diluted on confluent A549 cells. After 24 h, the A549 cells were fixed and stained with methyl violet. The amount of staining was quantified by measuring the optical density (OD) of each well at 570 nm. More EMCV replication resulted in increased cell death and lower optical density readings.
IFN-α-induced gene expression is inhibited in replicon-containing cells.
Different viruses have evolved a diverse range of molecular mechanisms that act on different cellular targets to inhibit IFN-mediated antiviral pathways (9, 13). In order to test whether the observed inhibition of the antiviral effect of IFN in replicon-containing cells was due at least in part to an inhibition of IFN signal transduction, we first measured the induction of MxA and PKR gene transcription. These genes are classical ISGs that contain an interferon-stimulated response element within the promoter region (20) and encode proteins that are key mediators of the antiviral effects of IFN (13). K562 cells that did and did not contain replicons were stimulated with IFN-α2a for 6 and 24 h, and MxA and PKR gene transcription was analyzed by quantitative RT-PCR. As expected, IFN induced the transcription of both of these genes in K562 cells. In contrast, the IFN induction of MxA and PKR gene transcription was dramatically inhibited in K562.ΔCprME-PAC2A cells (Fig. 4). Having shown that the induction of two genes, MxA and PKR, was inhibited in replicon-containing cells, we examined the global pattern of IFN-inducible gene transcription in cells that did and did not contain replicons by using a custom macroarray. We found that the IFN response was profoundly suppressed in K562.ΔCprME-PAC2A cells compared with K562 cells. Table 1 lists the ISGs that were most up-regulated (more than fourfold) in K562 cells in response to IFN and shows comparative data for K562.ΔCprME-PAC2A cells. These data indicate that IFN-α-induced gene expression is inhibited in cells containing dengue virus replicons, though not absolutely, and suggest that IFN-α signal transduction is inhibited.
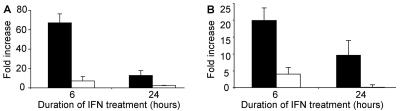
FIG. 4.
Induction of classical ISGs by IFN-α is inhibited in dengue virus replicon-containing cells. K562 cells (black bars) and K562.ΔCprME-PAC2A cells (white bars) were stimulated with 100 IU of IFN-α2a for 6 and 24 h. MxA (A) and PKR (B) gene transcription was measured by real-time PCR and normalized to the housekeeping gene GAPDH.
TABLE 1.
ISG transcription in response to IFN-α in cells that do and do not contain dengue virus repliconsa
| ISG product | Fold induction
|
|
|---|---|---|
| K562 cells | K562 ΔCprME-PAC2A cells | |
| IFN-α-induced protein 27 | 55.8 | 11.5 |
| VCAM-1 | 30.6 | 2.6 |
| MxA | 17.9 | 1.8 |
| IFN-α-induced protein (clone IFI-616) | 12.9 | 1.1 |
| Met proto-oncogene product (hepatocyte growth factor) | 7.9 | 1.8 |
| PSMB9 | 7.9 | 0.4 |
| IFN-induced protein 17 | 7.5 | 1.3 |
| Vipirin (Cig5) | 7.2 | 1.3 |
| Interleukin-15 | 6.5 | 1.1 |
| 9-27mrna | 6.4 | 1.2 |
| STAT1 | 6.2 | 1.1 |
| STAT4 | 6.2 | 1.5 |
| IFIT1 | 6.1 | 1.3 |
| KIAA0284 | 5.6 | 1.5 |
| STAT1 (91 kDa) | 4.9 | 1.8 |
| IFN-induced transmembrane protein 3 | 4.9 | 1.4 |
| INDO | 4.8 | 0.6 |
| Interleukin-6 | 4.8 | 0.6 |
| IFN-induced transmembrane protein 2 | 4.5 | 1.1 |
| MAP2K4 | 4.5 | 1.0 |
| IFI35 | 4.4 | 1.1 |
| Homo sapiens STAT | 4.1 | 1.6 |
ISGs up-regulated more than fourfold in K562 cells compared to K562.ΔCprME-PAC2A cells are shown.
Dengue virus RNA replication inhibits early events in IFN-α signaling by down-regulating steady-state STAT2 levels.
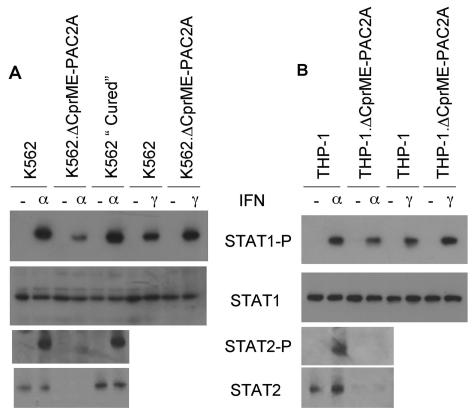
In order to determine whether early events in IFN signal transduction were inhibited in replicon-containing cells, we performed a Western blot analysis of STAT1 phosphorylation in K562 cells that did and did not contain replicons. The steady-state levels of STAT1 were similar in each cell line (Fig. 5A). As expected, a treatment with IFN-α induced STAT1 phosphorylation in K562 cells. The levels of phosphorylated STAT1 (STAT1-P) were significantly lower in K562.ΔCprME-PAC2A cells in response to IFN-α, but the response was restored to normal in cured K562 cells. In contrast, STAT1-P levels were not reduced in K562.ΔCprME-PAC2A cells in response to IFN-γ, which signals through a distinct but overlapping pathway (9); in fact, we observed a consistent increase in STAT1 phosphorylation in response to IFN-γ in K562.ΔCprME-PAC2A cells compared with K562 cells (Fig. 5A). In order to ensure that these observations were not limited to the specific cell type used, we repeated these experiments with THP-1 cells that stably expressed ΔCprME-PAC2A, with similar results (Fig. 5B). These data imply that early components of the IFN-α but not the IFN-γ signal transduction pathway are targets for dengue virus inhibition.
FIG. 5.
Dengue virus RNA replication inhibits STAT1 and STAT2 phosphorylation in response to IFN-α and reduces steady-state levels of STAT2. (A) K562, K562.ΔCprME-PAC2A, and cured K562 cells; (B) THP-1 and THP-1.ΔCprME-PAC2A cells. Cells were left untreated or treated with 100 IU of IFN-α or IFN-γ/ml for 30 min and then lysed in SDS loading buffer. Proteins were separated by SDS-PAGE and then analyzed by immunoblotting with specific antibodies for STAT1, phosphorylated STAT1, STAT2, and phosphorylated STAT2, as indicated.
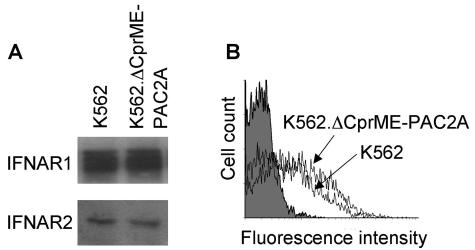
We next performed a Western blot analysis of STAT2 phosphorylation, which is a key step in IFN-α but not IFN-γ signaling (9). Figure 5 shows that the steady-state levels of STAT2 were very markedly reduced in both K562 and THP-1 cells containing dengue virus replicons compared with the parental cells. The levels of STAT2-P in response to IFN-α were also greatly reduced in both replicon-containing cell lines. Cured K562 cells reverted to the phenotype of parental K562 cells, with similar steady-state levels of STAT2 and STAT2-P in response to IFN-α (Fig. 5A). In order to confirm that cells containing dengue virus replicons did not have a generally reduced expression of proteins involved in the first part of the IFN-α signal transduction pathway, we examined the levels of IFNAR1 and IFNAR2. Total IFNAR1 and IFNAR2 protein levels were assessed by Western blotting and were similar in K562 and K562.ΔCprME-PAC2A cells (Fig. 6A). The cell surface expression of IFNAR2 was measured by flow cytometry and was similar in K562 cells that did and did not contain replicons (Fig. 6B); reagents to examine the cell surface expression of IFNAR1 by FACS analysis were not available. Similar levels of IFNAR1 and IFNAR2 were also found in THP-1 cells that did and did not contain replicons (data not shown). Taken together, these data show that the presence of dengue virus replicons specifically inhibits early events in IFN-α but not IFN-γ signal transduction by reducing STAT2 levels.
FIG. 6.
Dengue virus RNA replication does not affect IFNAR protein levels. (A) Cell lysates from K562 and K562.ΔCprME-PAC2A cells were separated by SDS-PAGE and then analyzed by immunoblotting with specific antibodies for IFNAR1 and IFNAR2, as indicated. (B) K562 and K562.ΔCprME-PAC2A cells were stained with specific anti-IFNAR2 antibodies and then analyzed by flow cytometry. Cells stained with the secondary antibody alone (gray-filled plot) were included as a negative staining control.
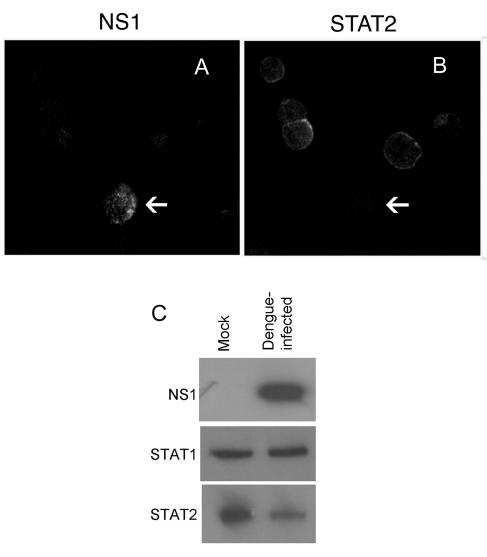
Dengue virus infection reduces STAT2 levels.
In order to determine whether STAT2 expression is also reduced in dengue virus-infected cells, we infected K562 cells with dengue virus type 2 and analyzed STAT2 expression in individual cells by dual-label immunofluorescence. We observed infected cells with markedly reduced staining for STAT2 compared with neighboring, uninfected cells (Fig. 7A and B). In order to assess STAT2 expression in the whole population of cells infected with dengue virus, we analyzed STAT2 levels at 48 h postinfection by immunoblotting. At this time point, approximately 30 to 50% of cells that had been infected stained strongly positive for NS1 protein by immunofluorescence. Figure 7C shows that STAT2 levels were markedly reduced in cells infected with dengue virus compared with mock-infected cells, whereas STAT1 levels were unchanged. Collectively, the data suggest that the down-regulation of STAT2 expression is a key component of dengue virus countermeasures against the human IFN response.
FIG. 7.
Dengue virus infection reduces STAT2 levels. K562 cells were infected with dengue virus for 48 h and then dually stained with an anti-dengue virus NS1 mouse monoclonal antibody followed by a Texas Red-labeled secondary antibody to detect dengue virus-infected cells (A) and an anti-STAT2 rabbit polyclonal antibody followed by a fluorescein isothiocyanate-labeled secondary antibody to detect STAT2 (B). Cells were visualized by confocal microscopy. Arrows show infected cells. (C) Cell lysates were separated by SDS-PAGE and then analyzed by immunoblotting with specific antibodies for dengue virus NS1, STAT1, and STAT2, as indicated. Mock-infected cells were included for comparison.
DISCUSSION
For this study, we used human cell lines that stably express dengue virus replicons to show that dengue virus RNA replication inhibits early events in IFN signaling. This work extends a related study published by Muñoz-Jordan and colleagues during the progress of our research showing that dengue virus proteins NS2A, NS4A, and NS4B have the capacity to inhibit the nuclear translocation of STAT1-P in response to IFN-β (25). We show here for the first time that dengue virus RNA replication not only reduces STAT1-P levels in response to IFN-α but also results in a marked reduction in STAT2-P levels as a consequence of reduced steady-state levels of STAT2. We first made this observation with dengue virus replicon-containing cells and subsequently with cells infected with dengue virus. Since STAT2-P recruits STAT1 for phosphorylation in response to IFN-α (22), generating STAT1/STAT2 heterodimers, all of our data can be explained if dengue virus specifically down-regulates STAT2 levels in order to counter the IFN response. In keeping with this hypothesis, dengue virus RNA replication did not reduce STAT1 phosphorylation in response to IFN-γ, which is independent of STAT2 (22). In fact, dengue virus replicon RNA replication increased rather than decreased STAT1 phosphorylation in response to IFN-γ, consistent with previous data showing increased STAT1 phosphorylation (30) and IFN-γ-mediated gene transcription (1) in the context of reduced STAT2 levels. The mechanism underlying this effect and its biological significance are as yet unknown.
We concluded that dengue virus specifically inhibits IFN-α/β signaling by down-regulating the expression of STAT2. Muñoz-Jordan and colleagues reported conflicting data showing an inhibition of both IFN-α/β- and IFN-γ-mediated STAT1 phosphorylation by dengue virus NS4B and dengue virus infection in LLCMK2 (monkey kidney) cells, and they suggested that common players involved in both signaling pathways (which do not include STAT2) were likely targets for IFN antagonism by dengue virus (25). The differences in our findings may reflect interspecies differences in IFN antagonism. Although the cell tropism of dengue virus in humans is not definitively known, the predominant targets are probably cells of hematopoietic origin, particularly dendritic cells, monocytes, and macrophages, as well as hepatocytes (2, 40, 41). For this study, we used human cell lines (K562 and THP-1) related to cells targeted by dengue virus in vivo, and our data provide the basis for further work to confirm the relevance of our observations to human dengue virus infection.
Despite the fact that IFN-α signaling was not completely blocked in dengue virus replicon-containing cells, supraphysiological concentrations of IFN-α2a had no significant effect on dengue virus replicon RNA replication or protein production. It is possible that dengue virus utilizes more than one mechanism to counter the IFN response, as suggested for other viruses, including another flavivirus, hepatitis C virus (8, 28, 37, 38). Alternatively, the inhibition of IFN signaling by reducing STAT2 levels may be sufficient for dengue virus replication to proceed faster than can be inhibited by the reduced antiviral IFN response. Future work will elucidate whether STAT2 levels are reduced as a consequence of a down-regulation of STAT2 gene transcription or protein synthesis or an enhanced degradation of the STAT2 protein. Further data are also needed to determine if the capacity to reduce STAT2 levels is conserved among all dengue virus strains (including field isolates) and in any other members of the Flaviviridae family. Recent data suggest that Japanese encephalitis virus also inhibits IFN-α signal transduction but utilizes a different mechanism that results in reduced levels of phosphorylated Tyk2 (and hence STAT2-P) without altering the expression of STAT2 (23). Two important human pathogens in the Paramyxoviridae family of enveloped, negative-strand RNA viruses, Respiratory syncytial virus and Human parainfluenza virus type 2, have been shown to subvert antiviral IFN responses by reducing STAT2 levels (26, 30). This effect is likely mediated through proteasome-mediated degradation of STAT2. However, other paramyxoviruses do not reduce steady-state STAT2 levels and instead have evolved a variety of different strategies to block IFN-α/β signaling (10, 19, 32, 43), which is a broadly effective strategy for countering the IFN response.
Future work is needed to define the precise interaction between components of the IFN signal transduction pathway and specific dengue virus proteins. Replicon-containing human cell lines are powerful tools for these studies because preliminary evidence suggests that several dengue virus nonstructural proteins may act together to produce strong, species-specific inhibition of IFN (25). We were careful to ensure that our observations did not reflect the selection of an IFN-defective subpopulation of cells during the generation of our stable replicon-containing cell lines. Several lines of evidence mitigate against this conclusion and suggest that our results accurately reflect an important host-pathogen interaction that subverts the IFN response: replicon-expressing cells were propagated from a total population of transfected cells rather than from individual cell clones; our observations were the same for two cell types, K562 and THP-1; and K562 cells that had previously expressed replicons and had been cured with glycyrrhizic acid reverted to the phenotype of parental K562 cells. Most importantly, the key observation made with our replicon model, namely, the reduction in STAT2 levels, led us to the same finding with cells infected with dengue virus. Collectively, the data suggest that the down-regulation of STAT2 expression is an important mechanism by which dengue virus subverts innate antiviral defenses mediated by IFN.
Understanding the molecular basis of the race between dengue virus replication and the IFN response early in infection would represent a critical advance, as the efficiency with which dengue virus evades the IFN response in humans is probably an important factor in early viral replication, and hence, disease pathogenesis. Further data will elucidate whether differences in IFN antagonism contribute to the differences in pathogenicity observed among dengue virus strains (31). Understanding the molecular mechanisms that dengue virus utilizes to subvert innate immune responses mediated by IFN may also inform strategies for rational attenuation in order to generate safer and more cost-effective dengue vaccine candidates.
Acknowledgments
M.J. is a Wellcome Advanced Fellow. Work in G.R.F.'s laboratory is funded by the EC. Work in J.S.'s laboratory is funded by the Deutsche Forschungsgemeinschaft.
We thank Paul Young for anti-NS1 antibodies, Pablo de Felipe for the plasmid pPDF14b (which contains the PAC2A cassette), Marion Macey for assistance with FACS analysis, and Tim Cowen and Tim Robson for assistance with confocal microscopy.
REFERENCES
- 1.Andrejeva, J., D. F. Young, S. Goodbourn, and R. E. Randall. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Couvelard, A., P. Marianneau, C. Bedel, M. T. Drouet, F. Vachon, D. Henin, and V. Deubel. 1999. Report of a fatal case of dengue infection with hepatitis: demonstration of dengue antigens in hepatocytes and liver apoptosis. Hum. Pathol. 30:1106-1110. [DOI] [PubMed] [Google Scholar]
- 3.Crance, J. M., N. Scaramozzino, A. Jouan, and D. Garin. 2003. Interferon, ribavirin, 6-azauridine and glycyrrhizin: antiviral compounds active against pathogenic flaviviruses. Antiviral Res. 58:73-79. [DOI] [PubMed] [Google Scholar]
- 4.Darnell, J. E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 5.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 6.Diamond, M. S., T. G. Roberts, D. Edgil, B. Lu, J. Ernst, and E. Harris. 2000. Modulation of dengue virus infection in human cells by alpha, beta, and gamma interferons. J. Virol. 74:4957-4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falconar, A. K., and P. R. Young. 1991. Production of dimer-specific and dengue virus group cross-reactive mouse monoclonal antibodies to the dengue 2 virus non-structural glycoprotein NS1. J. Gen. Virol. 72:961-965. [DOI] [PubMed] [Google Scholar]
- 8.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 9.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh, B., K. Takeuchi, T. Komatsu, and J. Yokoo. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 77:3360-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halstead, S. B., and J. Deen. 2002. The future of dengue vaccines. Lancet 360:1243-1245. [DOI] [PubMed] [Google Scholar]
- 12.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs, M. G., P. J. Robinson, C. Bletchly, J. M. Mackenzie, and P. R. Young. 2000. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. FASEB J. 14:1603-1610. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, A. J., and J. T. Roehrig. 1999. New mouse model for dengue virus vaccine testing. J. Virol. 73:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khromykh, A. A., M. T. Kenney, and E. G. Westaway. 1998. trans-Complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J. Virol. 72:7270-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khromykh, A. A., P. L. Sedlak, K. J. Guyatt, R. A. Hall, and E. G. Westaway. 1999. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 73:10272-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C terminal Cys-rich region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 20.Kuhen, K. L., and C. E. Samuel. 1999. Mechanism of interferon action: functional characterization of positive and negative regulatory domains that modulate transcriptional activation of the human RNA-dependent protein kinase Pkr promoter. Virology 254:182-195. [DOI] [PubMed] [Google Scholar]
- 21.Kurane, I., B. L. Innis, S. Nimmannitya, A. Nisalak, A. Meager, and F. A. Ennis. 1993. High levels of interferon alpha in the sera of children with dengue virus infection. Am. J. Trop. Med. Hyg. 48:222-229. [DOI] [PubMed] [Google Scholar]
- 22.Leung, S., S. A. Qureshi, I. M. Kerr, J. E. Darnell, Jr., and G. R. Stark. 1995. Role of STAT2 in the alpha interferon signaling pathway. Mol. Cell. Biol. 15:1312-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, R. J., C. L. Liao, E. Lin, and Y. L. Lin. 2004. Blocking of the alpha interferon-induced JAK-STAT signaling pathway by Japanese encephalitis virus infection. J. Virol. 78:9285-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, W. J., P. L. Sedlak, N. Kondratieva, and A. A. Khromykh. 2002. Complementation analysis of the flavivirus Kunjin NS3 and NS5 proteins defines the minimal regions essential for formation of a replication complex and shows a requirement of NS3 in cis for virus assembly. J. Virol. 76:10766-10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 27.Paterson, M., C. D. Laxton, H. C. Thomas, A. M. Ackrill, and G. R. Foster. 1999. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology 117:1187-1197. [DOI] [PubMed] [Google Scholar]
- 28.Pflugheber, J., B. Fredericksen, R. Sumpter, Jr., C. Wang, F. Ware, D. L. Sodora, and M. Gale, Jr. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc. Natl. Acad. Sci. USA 99:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pryor, M. J., J. M. Carr, H. Hocking, A. D. Davidson, P. Li, and P. J. Wright. 2001. Replication of dengue virus type 2 in human monocyte-derived macrophages: comparisons of isolates and recombinant viruses with substitutions at amino acid 390 in the envelope glycoprotein. Am. J. Trop. Med. Hyg. 65:427-434. [DOI] [PubMed] [Google Scholar]
- 30.Ramaswamy, M., L. Shi, M. M. Monick, G. W. Hunninghake, and D. C. Look. 2004. Specific inhibition of type I interferon signal transduction by respiratory syncytial virus. Am. J. Respir. Cell Mol. Biol. 30:893-900. [DOI] [PubMed] [Google Scholar]
- 31.Rico-Hesse, R., L. M. Harrison, R. A. Salas, D. Tovar, A. Nisalak, C. Ramos, J. Boshell, M. T. de Mesa, R. M. Nogueira, and A. T. da Rosa. 1997. Origins of dengue type 2 viruses associated with increased pathogenicity in the Americas. Virology 230:244-251. [DOI] [PubMed] [Google Scholar]
- 32.Rodriguez, J. J., C. D. Cruz, and C. M. Horvath. 2004. Identification of the nuclear export signal and STAT-binding domains of the Nipah virus V protein reveals mechanisms underlying interferon evasion. J. Virol. 78:5358-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlaak, J. F., C. M. Hilkens, A. P. Costa-Pereira, B. Strobl, F. Aberger, A. M. Frischauf, and I. M. Kerr. 2002. Cell-type and donor-specific transcriptional responses to interferon-alpha. Use of customized gene arrays. J. Biol. Chem. 277:49428-49437. [DOI] [PubMed] [Google Scholar]
- 34.Shresta, S., J. L. Kyle, H. M. Snider, M. Basavapatna, P. R. Beatty, and E. Harris. 2004. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. J. Virol. 78:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 36.Sudiro, T. M., J. Zivny, H. Ishiko, S. Green, D. W. Vaughn, S. Kalayanarooj, A. Nisalak, J. E. Norman, F. A. Ennis, and A. L. Rothman. 2001. Analysis of plasma viral RNA levels during acute dengue virus infection using quantitative competitor reverse transcription-polymerase chain reaction. J. Med. Virol. 63:29-34. [PubMed] [Google Scholar]
- 37.Taguchi, T., M. Nagano-Fujii, M. Akutsu, H. Kadoya, S. Ohgimoto, S. Ishido, and H. Hotta. 2004. Hepatitis C virus NS5A protein interacts with 2′,5′-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J. Gen. Virol. 85:959-969. [DOI] [PubMed] [Google Scholar]
- 38.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 39.Vaughn, D. W., S. Green, S. Kalayanarooj, B. L. Innis, S. Nimmannitya, S. Suntayakorn, T. P. Endy, B. Raengsakulrach, A. L. Rothman, F. A. Ennis, and A. Nisalak. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181:2-9. [DOI] [PubMed] [Google Scholar]
- 40.Wang, W. K., T. L. Sung, Y. C. Tsai, C. L. Kao, S. M. Chang, and C. C. King. 2002. Detection of dengue virus replication in peripheral blood mononuclear cells from dengue virus type 2-infected patients by a reverse transcription-real-time PCR assay. J. Clin. Microbiol. 40:4472-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, S. J., G. Grouard-Vogel, W. Sun, J. R. Mascola, E. Brachtel, R. Putvatana, M. K. Louder, L. Filgueira, M. A. Marovich, H. K. Wong, A. Blauvelt, G. S. Murphy, M. L. Robb, B. L. Innes, D. L. Birx, C. G. Hayes, and S. S. Frankel. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816-820. [DOI] [PubMed] [Google Scholar]
- 42.Young, D. F., L. Andrejeva, A. Livingstone, S. Goodbourn, R. A. Lamb, P. L. Collins, R. M. Elliott, and R. E. Randall. 2003. Virus replication in engineered human cells that do not respond to interferons. J. Virol. 77:2174-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]
- 44.Young, P. R., P. A. Hilditch, C. Bletchly, and W. Halloran. 2000. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 38:1053-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]