Abstract
Envelope proteins of hepadnaviruses undergo a unique folding mechanism which results in the posttranslational translocation of 50% of the large envelope protein (L) chains across the endoplasmic reticulum. This mechanism is essential for the eventual positioning of the receptor-binding domain on the surface of the virus particle and in duck hepatitis B virus (DHBV) is dependent on the small (S) envelope protein as part of the assembly process. In this study, we report the identification of a third envelope protein, St, derived from the S protein and carrying functions previously attributed to S. Antibody mapping and mutagenesis studies indicated St to be C terminally truncated, spanning the N-terminal transmembrane domain (TM1) plus the adjacent cysteine loop. We have previously shown that the mutation of two conserved polar residues in TM1 of S (SAA) eliminates L translocation and assembly. A plasmid expressing a functional equivalent of St was able to rescue assembly, demonstrating that this assembly defect is due to mutations of the corresponding residues in St and not in S per se. Immunofluorescence analysis showed that St directly affects L protein cellular localization. These results indicate that St acts as a viral chaperone for L folding, remaining associated with the DHBV envelope upon secretion. The presence of St at a molar ratio of half that of L suggests that it is St which regulates L translocation to 50%.
The processes of hepadnavirus assembly and budding are unique among enveloped viruses with respect to their mechanisms and the cellular compartments used. Particle formation is initiated early at the endoplasmic reticulum (ER) membrane and completed in an intermediate, pre-Golgi compartment (14); the lipid bilayer is not maintained during maturation, and the resulting dense complex of surface proteins displays an altered lipid content, indicating a substantial reorganization of the ER membrane (4, 26). Furthermore, particle budding can proceed in the absence of a matrix protein or nucleocapsid serving as a scaffold. The envelope curvature and particle secretion are instead determined by a single major structural component, the S protein, consisting of an extended, multitransmembrane S domain which is shared with the large surface protein (L) as well as with a third, middle-sized protein (M) in mammalian viruses (12). Subviral particles (SVPs) are produced in vast excess of virions, and in hepatitis B virus (HBV), consist of 22-nm-diameter spheres comprising the S protein and filaments, predominantly of S and M. In contrast, SVPs of duck HBV (DHBV) are similar in size and composition to the 60-nm-diameter virions, which contain L and S at an approximate ratio of 1:4.
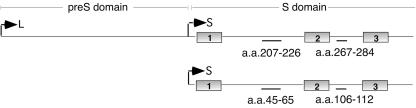
The envelope proteins are produced by differential translation initiation from a single pre-S/S open reading frame (Fig. 1). The S protein, and consequently all of the surface proteins, contains three hydrophobic transmembrane regions (TM1, TM2, and TM3), with TM2 acting as the signal anchor sequence. The L protein exhibits mixed membrane topologies due to partial posttranslational translocation of the N-terminal pre-S and TM1 domains across the ER. This translocation process, unique to the hepadnaviruses, allows translocated, external L chains on the virus particle to function in receptor binding (19, 27), while untranslocated, internal chains engage in nucleocapsid interactions (1). In DHBV, L translocation is dependent on the S protein and is therefore assumed to be associated with particle assembly (6, 9). In contrast, translocation in HBV is independent of S (18), reflecting differences in the folding pathways which are evident by the glycosylation of the mammalian and not the avian HBV envelope proteins as well as differences in their particle morphologies. One characteristic that the two viruses have in common is the regulation of posttranslational translocation to 50% of the L chains, although how this is achieved is unknown.
FIG. 1.
Linear representations of L and S proteins of DHBV, with the pre-S and S domains indicated at the top. The transmembrane regions are marked by numbered boxes. The approximate positions of the epitopes of the antipeptide antiserum to the S domain (anti-45-65 of S/anti-207-226 of L) and the monoclonal antibody 7C.12 (anti-106-112 of S/anti-267-284 of L) are indicated.
Through analyses of mature DHBV particles purified from serum and from chicken hepatoma cells transfected with L or S expression plasmids, we demonstrate here the presence of a third, truncated envelope protein, St, which is derived from S. For this study, we used a previously described S mutant (SAA) defective in L translocation and assembly, with the conserved polar residues K24 and E27 in TM1 replaced with alanine residues, to assess whether the defect is due to a nonfunctional St protein (6). Using the SAA mutant and a plasmid expressing a functional equivalent of St, we demonstrate a direct role for St in L protein folding and assembly.
MATERIALS AND METHODS
Plasmids.
Constructs encoding the wild-type DHBV envelope proteins L (pCI L) and S (pCI S) were made by use of the vector pCI-neo (Promega, Madison, Wis.) as described previously (5). The S mutant with mutations of the charged residues lysine 24 and glutamic acid 27 to alanine (SAA) was described previously (6). The R78stop construct was produced by substituting the A and G at nucleotides (nt) 1515 and 1516 of the envelope sequence with T and A by overlap extension PCR. The PCR product containing the desired mutation was digested with KpnI (nt 1294) and BstEII (nt 1847) and introduced into pCI S by subcloning into the same restriction sites via a three-way ligation strategy. The SAAR78stop mutant was constructed in two steps. First, a three-way ligation of a KpnI-SnaBI fragment (nt 1294 to 1466 of DHBV) of the SAA mutant, a SnaBI-BstEII fragment (nt 1466 to 1847 of DHBV) of R78stop, and the KpnI-BstEII vector fragment of pMDL-wt (a gift from H. Schaller, Zentrum für Molekulare Biologie, Heidelberg, Germany) was done. In the second step, the KpnI-BstEII fragment (nt 1294 to 1847 of DHBV), containing both of the alanine substitutions and the stop codon, was introduced into pCI S by subcloning into the same restriction sites via a three-way ligation strategy.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) and transferred to nitrocellulose membranes (Schleicher and Schüll) by use of a Trans-Blot SD semidry transfer cell (Bio-Rad). The membranes were blocked for 1 h with 3% skim milk in TBST (100 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.3% Tween 20). Envelope proteins were detected with a monoclonal antiserum to the pre-S or S domain (1H1 and 7C12, respectively; kind gifts from J. Pugh) (23) as indicated or with a rabbit anti-S-domain antiserum raised against a peptide of amino acids (aa) 46 to 65 of S, located in the cysteine-containing loop between the first two transmembrane domains (kindly prepared and mapped by Christa Kuhn) (9). The membranes were probed for 1 h in 1% skim milk-TBST, washed with TBST, and probed with horseradish peroxidase-conjugated species-specific antibodies (Amersham) in 1% skim milk-TBST. After a final wash in TBST (three times for 10 min each), protein bands were visualized by enhanced chemiluminescence (Amersham).
Sucrose density gradient purification and treatment of particles.
Subviral particles (SVPs) were purified from DHBV-positive duck serum as previously described (16). Briefly, 8 ml of serum was centrifuged for 3 h at 38,000 rpm in an SW40 rotor (Beckman) through 3 ml of 20% sucrose onto a 2-ml 70% sucrose cushion. The particle-containing fraction was diluted with phosphate-buffered saline (PBS) and further centrifuged for 4 h at 38,000 rpm through a 20 to 70% sucrose step gradient. Fractions were collected from the bottom of the gradient and analyzed by SDS-PAGE and silver staining. Envelope protein-containing fractions were pooled and stored in small aliquots at −70°C.
Trypsin digestion of SVPs.
A major trypsin site exists at lysines 204 and 206 in L, between the first two transmembrane domains of the S domain, but is only accessible to cleavage in the presence of a detergent. SVPs (10 μl) that had been solubilized in 0.5% NP-40 or left untreated were digested with trypsin (Sigma) (20 μg/ml) for 1 h at 37°C. Trypsin digestion was stopped by the addition of aprotinin (final concentration, 2 μg/ml) and boiling in Laemmli buffer.
Cells. (i) PDHs.
Primary duck hepatocyte (PDH) cultures were prepared by collagenase perfusion of a liver from a DHBV-positive duckling as described previously (24). The cells were seeded at a density of 2 × 106 cells per well in a six-well culture plate (Greiner, Frickenhausen, Germany).
(ii) LMH cells.
LMH cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Transfections were performed by the dextran sulfate method as described previously (10), with 5 μg of DNA per well in six-well plates (Greiner).
(iii) Preparation of microsomal membranes.
Microsomes were prepared according to the method of Prange and Streeck (22), with the following modifications. LMH cells transfected with pCI L and/or pCI S (two 30-mm-diameter wells) were washed with cold Tris-buffered saline (TBS; 50 mM Tris-HCl [pH 7.5], 150 mM NaCl). The monolayer in each well was incubated on ice with 0.4 ml of 0.1× TBS for 10 min, harvested by scraping, pooled, and dispersed by being drawn five times through a 26-gauge needle. The homogenate was adjusted to 1× TBS with 5× TBS and centrifuged for 20 min at 1,500 × g at 4°C to remove unbroken cells, plasma membranes, and nuclei. The supernatant was removed and set aside while the pellet was again dispersed in 300 μl of TBS and centrifuged as described above. Supernatants were pooled and layered onto 2.7 ml of 250 mM sucrose in TBS and centrifuged for 30 min at 38,000 rpm at 4°C in an SW-60 rotor (Beckman). The microsomal pellets were washed once with TBS and resuspended in 20 μl of TBS containing 5 μl of 5× Laemmli buffer.
Protease protection assay.
For trypsin protection analysis, the microsomal preparation described above was divided into three 20-μl aliquots. One sample was left untreated, while the remaining two were treated with 25 μg of trypsin (tosylsulfonyl phenylalanyl chloromethyl ketone treated; Worthington Biochem. Corp.)/ml, with or without 0.5% NP-40, for 1 h on ice. Proteolysis was halted by the addition of 30 μg of aprotinin (Boehringer)/ml and a further incubation on ice for 20 min. Five microliters of 5× Laemmli buffer was then added to each sample, and the samples were boiled for 5 min prior to separation by SDS-13% PAGE followed by Western blotting for the L protein.
Immunofluorescence.
LMH cells were grown on glass coverslips in 12-well Nunc tissue culture plates and transfected with 5 μg of each appropriate plasmid by the dextran sulfate method as described previously (10). After 3 days, the cells were fixed with cold 100% methanol and the L protein was detected by use of a monoclonal antibody to DHBV pre-S (1.H.1) (23) followed by a secondary antibody, Alexa 488-labeled goat anti-mouse immunoglobulin G (Molecular Probes). The cells were blocked with 2% bovine serum albumin in PBS, and antibody dilutions were made in 2% bovine serum albumin-0.25% Triton X-100 in PBS. The cells were washed for 30 min with 0.25% Triton X-100 in PBS after each antibody incubation. Propidium iodide, used to stain cell nuclei, was diluted 1:5,000 in wash solution and incubated for 5 min during the final washing step. Coverslips were removed and mounted on glass slides, and fluorescence was observed with a Bio-Rad MRC 1024 confocal microscope mounted on a Nikon e600 upright epifluorescence microscope.
RESULTS
Detection of a third, 10-kDa envelope protein derived from the S protein in DHBV.
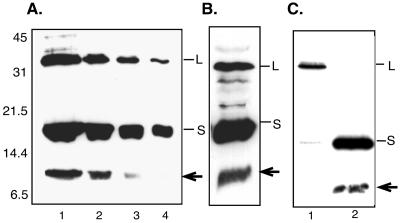
A novel envelope protein of approximately 10 kDa was detected by Western blotting of sucrose gradient-purified SVPs from a DHBV-positive duck serum by use of an antiserum to a peptide sequence (aa 46 to 65 of S) located in the cysteine-containing loop between the first two transmembrane domains (9) (Fig. 2A). The relative abundance of this species, determined by the titration of SVPs and Western blotting with anti-46-65, revealed an approximate molar ratio with L and S of 1:2:8 (Fig. 2A), which is in agreement with the predicted L-to-S ratio of 1:4 (3) and shows that this envelope protein is a significant constituent of the particles.
FIG. 2.
(A) Western blot analysis of a dilution series of SVPs showing detection of L, S, and the 10-kDa species (arrow). Lanes 1 to 4, 30, 15, 7.5, and 3.25 μl of SVPs. (B) Western blot of particles exported from PDH cultures and then methanol precipitated showing detection of the 10-kDa species (arrow). (C) Western blot of microsomes derived from LMH cells transfected with the L-expressing pCI L (lane 1) or S-expressing pCI S (lane 2) construct. The arrow indicates the 10-kDa species. Western blots were probed with anti-45-65.
We next examined whether it was present in DHBV-infected PDH cultures and transfected LMH cells. Cell culture supernatants and cell lysates were analyzed by Western blotting. The 10-kDa species was detected by methanol precipitation and Western blotting with anti-46-65 in particles exported from the PDH culture (Fig. 2B). It was not detected in the cellular lysate or the detergent-extracted membrane fraction due to the relatively low level relative to the total intracellular proteins, thus limiting the amount which could be loaded on the gel (data not shown). The presence of the 10-kDa species in the intracellular compartment was examined instead by preparing microsomal membranes (equivalent to 4 × 106 cells), thus concentrating the envelope proteins, which are synthesized in the ER, to a small sample devoid of plasma membrane and cytosolic proteins. Using this method, we examined whether the 10-kDa species was derived from the L or S protein. Microsomes from cells transfected with either an L-expressing (pCI L) or S-expressing (pCI S) plasmid were assessed by Western blotting with anti-46-65. Figure 2C shows that the 10-kDa species is derived from the S protein (lane 2) and not L (lane 1), and as such, it was designated the truncated S protein, or St.
St is formed by truncation from the C terminus of S.
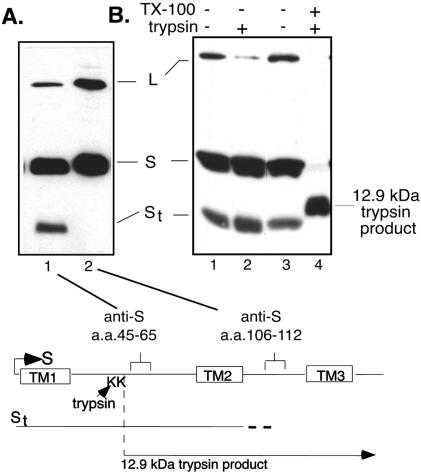
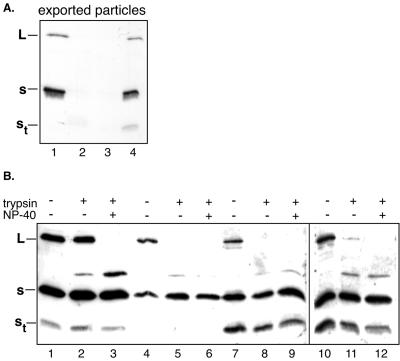
St was mapped by Western blotting with domain-specific antibodies (as indicated in the schematic diagram in Fig. 3) to a region upstream of a monoclonal antibody epitope in the second hydrophilic loop (aa 106 to 112 of S) which did not react (Fig. 3A, lane 2). To confirm that St encompasses the cysteine-containing loop region, we subjected particles to trypsin digestion in the presence of detergent, which typically generates a 12.9-kDa trypsin product from the S protein through cleavage at lysine residues (K204 and K206) within the loop (9, 11) (Fig. 3B). Cleavage within the loop by trypsin in the presence of detergent at 37°C resulted in the loss of St concomitant with the generation of the 12.9-kDa trypsin product (Fig. 3B, lane 4), while in the absence of detergent, trypsin digestion did not affect St (Fig. 3B, lane 2).
FIG. 3.
(A) Antibody mapping of St by Western blot analysis of SVPs by the use of domain-specific antibodies. Lane 1, anti-45-65; lane 2, anti-106-112. (B) Western blot analysis with anti-45-65 after incubation of SVPs for 30 min at 37°C in the absence of trypsin and detergent (lanes 1 and 3), with 20 μg of trypsin/ml (lane 2), and with trypsin in the presence of detergent (lane 4). The bottom panel is a schematic representation of S showing the positions of the epitopes of the domain-specific antisera used to map St. The estimated extent of St is indicated, as is the 12.9-kDa trypsin product generated by cleavage at lysine residues within the loop region in the presence of detergent.
The abundance of St was not affected by a prolonged incubation of particles enabling the action of endogenous or serum-derived proteases (data not shown) or by the addition of trypsin (Fig. 3B, lanes 1 and 2). Therefore, St appears to be a naturally occurring envelope component derived from S through truncation of the C terminus.
St is able to rescue an S mutant defective in assembly with L.
Since St and S have an overlapping sequence which makes identification of the role of St difficult to dissociate from that of S, and since S is the major envelope component and often not amenable to mutations, we employed a previously described S mutant (SAA) to assess the role of St. This mutant has the conserved polar residues K24 and E27 in TM1 replaced with alanine residues and was previously shown to cause a defect in the translocation of L and its assembly into particles (6). Further assessment of this mutant has revealed that although assembly with L is blocked, the assembly of SAA particles without L is not, implying that a particular property of S associated with L folding and/or translocation but not intrinsic for S assembly is involved (data not shown). Therefore, we asked whether the block in L association caused by the mutant was a result of a defect in St generated from the mutant S rather than a defect in mutant S per se.
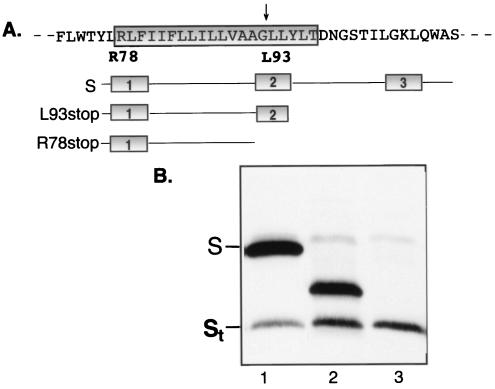
To test this hypothesis, we used a plasmid that expressed a functional equivalent of St to rescue the SAA mutant. First, a series of S mutants with stop codons at nt 1560 (L93stop) and nt 1515 (R78stop) were constructed and expressed in LMH cells, and the mobilities of the truncated S proteins were assessed in an SDS-4 to 16% gradient PAGE gel against those of full-length S and St. The choice to insert stop codons was made based on a predicted signal peptide cleavage site between A91 and G92 in TM2 (21) and the presence of a charged residue, R78, as a next possible cleavage site immediately adjacent to TM2 (Fig. 4A). Figure 4B shows the migration of the truncated S proteins generated from L93stop and R78stop in a gradient gel, which is able to further resolve these hydrophobic proteins. L93stop generated two S species, with one form migrating between the sizes of S and St and one form being St itself (Fig. 4B, lane 2), indicating that native St occurs by a truncation upstream of G92 but also demonstrating that full-length S is not required as a precursor for St. In contrast, R78stop generated a single truncated S protein, which migrated exactly with St (Fig. 4B, lane 3). The replacement of arginine 78 with alanine resulted in very poor S protein expression, confirming the difficulty of eliminating St production by mutagenesis of the S gene (data not shown).
FIG. 4.
The product of an S mutant with a stop codon at nt 1515 (R78stop) is a single truncated S species which comigrates with St. (A) Amino acid sequence of part of the S protein encompassing the second TM domain (indicated by the boxed region), indicating the amino acid positions where the S stop mutants were terminated. The lengths of the truncated S mutants are shown below. The arrow indicates the predicted signal peptide cleavage site. (B) Western blot of wild-type S protein (lane 1), truncated S protein species generated from the L93stop mutant (lane 2), and truncated species generated from the R78stop mutant (lane 3). The samples were separated in a 4 to 16% gradient gel, and the Western blot was probed with anti-45-65.
Using R78stop as a functional equivalent of St, we examined whether the expression of this protein could rescue the assembly of L and SAA. To confirm that functional St is required for assembly, we included a mutant of R78stop which carried the alanine substitutions in TM1 (R78stopAA). LMH cells were transfected with pCI L and the SAA mutant, with and without the addition of the R78stop plasmid or with the addition of R78stopAA. The export of particles was assessed by Western blotting of particles isolated from the medium by sedimentation through 20% sucrose onto a 70% sucrose cushion. Figure 5A shows the export of particles from cells expressing L and wild-type S (Fig. 5A, lane 1) but none from cells expressing either the SAA mutant or SAA with the addition of the St AA mutant (Fig. 5A, lanes 2 and 3). However, functional St expressed from R78stop was able to rescue the abortive defect in L and SAA particle production (Fig. 5A, lane 4). The translocation of L across the ER membrane was assessed by protease protection analysis of microsomes prepared from the transfected LMH cells described above. In protease protection analysis, L was protected from trypsin digestion in the presence of a functional S protein (Fig. 5B, lanes 1 to 3) but not in the presence of the assembly mutant, SAA (Fig. 5B, lanes 4 to 6), or with the addition of the St AA mutant (Fig. 5B, lanes 7 to 9). However, the inclusion of R78stop expressing functional St afforded some protection of L (Fig. 5B, lanes 10 to 12). With the L protein expressed in the presence of St alone (using the R78stop plasmid), no translocation of L was observed, indicating that translocation is still an assembly-dependent process requiring the presence of S. Similarly, R78stop was unable to allow particle formation with L in the absence of S, consistent with the role of S as the major structural entity in particles and the essential driver of particle export (data not shown) (20).
FIG. 5.
The product of R78stop is able to rescue the assembly defect of the SAA mutant. (A) LMH cells were transfected with pCI L and pCI S (lane 1), pCI L, SAA, and R78stop (lane 2), or pCI L and SAA (lane 3), and particles exported to the medium were pelleted by sedimentation through 20% sucrose and assessed by Western blotting. (B) Microsomes were prepared from the cells described above and subjected to protease protection analysis as described in Materials and Methods. Lanes 1 to 3, pCI L and pCI S; lanes 4 to 6, pCI L and SAA; lanes 7 to 9, SAA and R78stop. Microsomes were subjected to digestion with trypsin in the absence or presence of 0.5% NP-40 or left untreated as denoted above each lane. Western blots were probed with anti-45-65.
Direct effect of St on L protein cellular distribution.
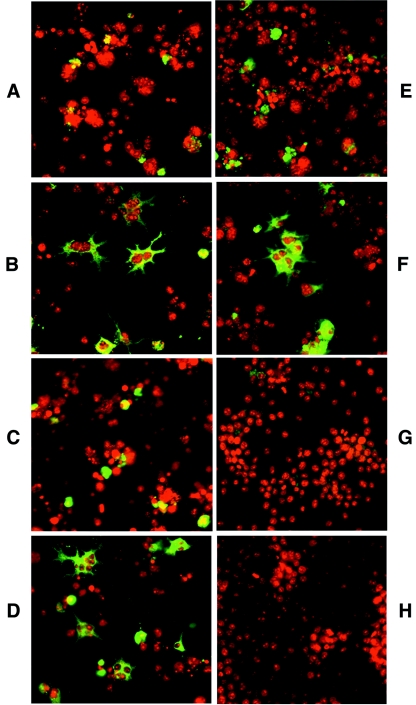
The role of St on L protein folding was further examined by immunofluorescence using a monoclonal antibody to the pre-S domain (1.H.1) (23), which was therefore specific for the L protein. Previous studies in HBV have shown that when L is expressed alone, it is predominantly localized to the perinuclear compartment, and when coexpressed with S, is distributed throughout the cytoplasm in association with intracellular membranes (2). Similarly, for DHBV L, we observed perinuclear staining in LMH cells in the absence of S (Fig. 6A) and cytoplasmic staining with S coexpression (Fig. 6B). This changed distribution of L reflects the interaction of the two proteins for particle assembly. Knowing that the SAA mutant was unable to form particles with L, we assessed the distribution pattern of L when expressed with this mutant and observed the same punctate, perinuclear staining as that seen with L alone, indicating the lack of interaction between these envelope proteins (Fig. 6C). This finding is consistent with a previous pulse-chase analysis study which showed that, even after a 24-h chase, SAA does not leave the ER membrane (6). When L and the SAA mutant were complemented with the R78stop construct, the cellular distribution of L became cytoplasmic, indicating that the block to envelope protein interactions had been overcome (Fig. 6D). This role of a functional St protein was further confirmed by the inability of St AA to affect L protein distribution (Fig. 6E). In order to assess if St has a direct affect on L protein folding, we cotransfected pCI L and R78stop into cells, and in the absence of any S expression, the distribution of L was cytoplasmic (Fig. 6F). In contrast, St AA appeared to have a more deleterious effect on L, with little or no L staining detected (Fig. 6G). These results indicate a direct association of St with L.
FIG. 6.
Intracellular distribution of DHBV L protein. LMH cells were transfected with pCI L alone (A) or with pCI L and pCI S (B), pCI L and SAA (C), pCI L, SAA, and R78stop (D), pCI L, SAA, and R78stopAA (E), pCI L and R78stop (F), or pCI L and R78stopAA (G). (H) Mock transfection. The L protein was detected by immunofluorescence staining with a monoclonal anti-pre-S antibody (green), and the nuclei were stained with propidium iodide (red).
DISCUSSION
While HBV and DHBV share many similarities in replication and morphogenesis, there are clearly some fundamental differences in particle composition and hence morphology. These differences are also apparent in the requirements for L protein translocation, a process which is common to the Hepadnaviridae and essential for the formation of infectious virions. The different folding pathways of the HBV and DHBV L proteins—HBV L translocation occurs independently of S (18), while S is a requirement for DHBV L translocation (9)—are perhaps reflected in the exclusion of HBV L from SVPs and its retention at the ER for virion formation, while DHBV L is assembled and exported equally in virions and SVPs (20).
In this study, we have identified a previously unrecognized envelope protein component of DHBV, St, which appears to play a crucial role in the unique folding strategy of the L protein. This protein has previously escaped detection due to its small size and the lack of antibodies to this region of the S domain. Through analyses of proteins after the transfection of cells with an L expression construct (pCI L) or an S expression construct (pCI S), we showed that St was derived from the S protein (Fig. 2C). Whether St arises from specific protease cleavage or through some other translational mechanism is unclear, but it is consistently found in serum-derived particles, primary duck hepatocyte cultures, transfected LMH cells (Fig. 2), and also a yeast expression system (data not shown). Somewhat surprisingly, mature St is also generated from the S protein truncated at lysine 93 (L93stop) (Fig. 4B, lane 2). The region encompassing the presumed C-terminal arginine (R78) of St does not conform to a particular protease cleavage consensus site other than one for trypsin, which requires lysine or arginine residues. It is possible that a trypsin-like protease of the proteasome complex is involved, although it is difficult to envisage how this cytoplasmic organelle could act on the S protein spanning the ER. Moreover, we have observed that although lactacystin, a proteasome inhibitor of trypsin-like enzymes, blocks the export of SVPs from PDHs, it results in the reduction of envelope proteins in the ER in general and not specifically St production (E. V. L. Grgacic, unpublished results).
The difficulty of dissociating the role of St from that of S was overcome by use of a previously described S mutant (SAA) which is defective in L translocation and L assembly (6). Using an S construct with a stop codon at nt 1515 (R78stop) which expresses a functional equivalent of St, we were able to rescue the assembly of L with SAA, demonstrating that the defect is due directly to the mutation of residues K24 and E27 in St and not in S per se (Fig. 5). Curiously, in a study assessing the effect of mutations in HBV S on hepatitis delta virus assembly, it was shown that the deletion of five amino acids from HBV TM1, including the equivalent residues R24 and T27, did not affect S particle assembly but instead eliminated hepatitis delta virus assembly (15). This implies that these residues play a role in heterologous assembly for mammalian hepadnaviruses as well as DHBV. However, it is unknown whether HBV produces a similar truncated envelope protein to DHBV St.
While the assembly of the SAA mutant could be rescued with St, the level of protection from trypsin in the protease protection assay, seen as an indication of ER translocation, was not complete. This result, coupled with the direct interaction of St with L, as seen by the changed cellular distribution and the inability of St AA to do this (Fig. 6), suggests that there is a distinct role for St in L protein folding for assembly but that a functional S protein may still be needed for efficient translocation. HBV L protein translocation does not require the S protein but instead appears to be dependent on the host cell ER transport machinery and the presence of the cellular chaperones Hsc70, Hsp40, and BiP for the regulation of this process (17). Our study shows that St has a direct effect on L protein folding, and together with S, translocates L during the assembly process. Therefore, it appears that St plays the role of a viral chaperone, preparing the L protein for translocation, while in HBV, the chaperones are provided by the cell. However, the facts that Hsc70 has also been identified as binding to the DHBV L protein and is packaged in particles (13) indicate that several factors are required for L translocation and its regulation. However, since St is present in exported serum-derived particles at approximately half the molar ratio of L (Fig. 2A) and has a direct association with the L protein, it may regulate L translocation to approximately 50%.
The study of membrane targeting, translocation across the ER, and management of folded and unfolded polypeptides has revealed that these complex processes require many factors, particularly when they occur posttranslationally and differentially, as in the case of the multifunctional L protein of the hepadnaviruses. Several factors have now been identified in the unusual biogenesis of L, with the mammalian and avian forms adopting different pathways but engaging several chaperones for the folding process. What is not understood is the order of these events and how they are regulated. On the basis of the known role of Hsc70, we propose a possible model for DHBV in which Hsc70 would bind to the unfolded L polypeptide soon after synthesis to maintain an unfolded, translocation-competent state. In DHBV, the viral protein St further interacts with L to affect its folding and/or targeting, possibly to different subdomains of the ER for interaction with S and translocation across the membrane. Once in the ER, the folded envelope proteins are stabilized by intramolecular disulfide bonding for the final assembly and secretion of the particle. The factor limiting the number of L chains attaining the all-important external topology may be the number of St chains. Other signals involved in biogenesis include the phosphorylation of L (8), which occurs in response to a buildup of S protein, probably through ER stress (7) and cellular stresses such as temperature (25), possibly for the maintenance of appropriate L protein levels.
Acknowledgments
This study was supported in part by a fellowship from the Zentrum für Molekulare Biologie, Heidelberg, Germany, the Research Fund of the Macfarlane Burnet Centre for Medical Research, and project grant number 111710 (E.V.L.G. and D.A.A.) from the National Health and Medical Research Council of Australia.
We thank Heinz Schaller for helpful comments on the manuscript.
REFERENCES
- 1.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clayton, R. F., A. Owsianka, and A. H. Patel. 2001. Evidence for structural differences in the S domain of L in comparison with S protein of hepatitis B virus. J. Gen. Virol. 82:1533-1541. [DOI] [PubMed] [Google Scholar]
- 3.Ganem, D., and H. E. Vamus. 1987. The molecular biology of the hepatitis B viruses. Annu. Rev. Biochem. 56:651-693. [DOI] [PubMed] [Google Scholar]
- 4.Gavilanes, F., A. Gonzales-Ros, and D. Peterson. 1982. Structure of hepatitis B virus surface antigen: characterization of the lipid components and their association with the viral proteins. J. Biol. Chem. 257:7770-7777. [PubMed] [Google Scholar]
- 5.Gazina, E. V., B. Lin, A. Gallina, G. Milanesi, and D. A. Anderson. 1998. Intracellular retention of duck hepatitis B virus large protein is independent of preS topology. Virology 242:266-278. [DOI] [PubMed] [Google Scholar]
- 6.Grgacic, E. V. L. 2002. Identification of structural determinants of the first transmembrane domain of the small envelope protein of duck hepatitis B virus essential for particle morphogenesis. J. Gen. Virol. 83:1635-1644. [DOI] [PubMed] [Google Scholar]
- 7.Grgacic, E. V. L. 1996. Protein modifications of the large surface protein of duck hepatitis B virus. Ph.D. thesis. University of Melbourne, Melbourne, Australia.
- 8.Grgacic, E. V. L., and D. A. Anderson. 1994. The large surface protein of the duck hepatitis B virus is phosphorylated in the pre-S domain. J. Virol. 68:7344-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grgacic, E. V. L., C. Kuhn, and H. Schaller. 2000. Hepadnavirus envelope topology: insertion of a loop region in the membrane and role of S in L protein translocation. J. Virol. 74:2455-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grgacic, E. V. L., B. Lin, E. V. Gazina, M. J. L. Snooks, and D. A. Anderson. 1998. Normal phosphorylation of duck hepatitis B virus L protein is dispensable for infectivity. J. Gen. Virol. 79:2743-2751. [DOI] [PubMed] [Google Scholar]
- 11.Guo, J.-T., and J. C. Pugh. 1997. Topology of the large surface protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J. Virol. 71:1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heermann, K. H., U. Goldmann, W. Schwartz, T. Seyffarth, H. Baumgarten, and W. H. Gerlich. 1984. Large surface protein of hepatitis B virus containing the pre-S sequence. J. Virol. 52:396-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildt, M. 1997. Zellulare funktionen waehrend der fruehen und spaeten Schritte im Infektionszyklus des enten hepatitis B virus. Ph.D. thesis. University of Heidelberg, Heidelberg, Germany.
- 14.Huovila, A.-P. J., A. M. Eder, and S. D. Fuller. 1992. Hepatitis B surface antigen assembles in a post-ER pre-Golgi compartment. J. Cell Biol. 118:1305-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenna, S., and C. Sureau. 1998. Effect of mutations in the small envelope protein of hepatitis B virus on assembly and secretion of hepatitis delta virus. Virology 251:176-186. [DOI] [PubMed] [Google Scholar]
- 16.Klingmuller, U., and H. Schaller. 1993. Hepadnavirus infection requires interaction between the viral pre-S domain and a specific hepatocellular receptor. J. Virol. 67:7414-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert, C., and R. Prange. 2003. Chaperone action in the posttranslational topological reorientation of the hepatitis B virus large envelope protein: implication for translocational regulation. Proc. Natl. Acad. Sci. USA 100:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert, C., and R. Prange. 2001. Dual topology of the hepatitis B virus large envelope protein: determinants influencing post-translational pre-S translocation. J. Biol. Chem. 276:22265-22272. [DOI] [PubMed] [Google Scholar]
- 19.Le Seyec, J., P. Chouteau, I. Cannie, C. Guguen-Guillouzo, and P. Gripon. 1999. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J. Virol. 73:2052-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nassal, M. 1996. Hepatitis B virus morphogenesis. Curr. Top. Microbiol. Immunol. 214:297-337. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, H., J. Englebrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 22.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugh, J. C., Q. Di, W. S. Mason, and H. Simmons. 1995. Susceptibility to duck hepatitis B virus infection is associated with the presence of cell surface receptor sites that efficiently bind viral particles. J. Virol. 69:4814-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigg, R. J., and H. Schaller. 1992. Duck hepatitis B virus infection of hepatocytes is not dependent on low pH. J. Virol. 66:2829-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rothman, K., M. Schnoelzer, G. Radziwill, E. Hildt, K. Moelling, and H. Schaller. 1998. Host cell-virus cross talk: phosphorylation of a hepatitis B virus envelope protein mediates intracellular signaling. J. Virol. 72:10138-10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satoh, O., H. Imai, T. Yoneyama, T. Miyamura, H. Utsumi, K. Inoue, and M. Umeda. 2000. Membrane structure of the hepatitis B virus surface antigen particle. J. Biochem. 127:543-550. [DOI] [PubMed] [Google Scholar]
- 27.Urban, S., K. M. Breiner, F. Fehler, U. Klingmueller, and H. Schaller. 1998. Avian hepatitis B virus infection is initiated by the interaction of a distinct pre-S subdomain with the cellular receptor gp180. J. Virol. 72:8089-8097. [DOI] [PMC free article] [PubMed] [Google Scholar]