Abstract
Historically, the adenoviral E3 region was found to be nonessential for viral replication in vitro. In addition, adenoviruses whose genome was more than approximately 105% the size of the native genome were inefficiently packaged. These profound observations were used experimentally to insert transgenes into the adenoviral backbone. More recently, however, the reintroduction of the E3 region into oncolytic adenoviruses has been found to positively influence antitumor efficacy in preclinical models and clinical trials. In the studies reported here, the granulocyte-macrophage colony-stimulating factor (GM-CSF) cDNA sequence has been substituted for the E3-gp19 gene in oncolytic adenoviruses that otherwise retained the E3 region. Five viruses that differed slightly in the method of transgene insertion were generated and compared to Ar6pAE2fGmF (E2F/GM/ΔE3), a previously described E3-deleted oncolytic adenovirus encoding GM-CSF. In all of the viruses, the human E2F-1 promoter regulated E1A expression and GM-CSF expression was under the control of the adenoviral E3 promoter and the packaging signal was relocated immediately upstream from the right terminal repeat. The E3-gp19-deleted viruses had similar cytolytic properties, as measured in vitro by cytotoxicity assays, but differed markedly in their capacity to express and secrete GM-CSF. Ar15pAE2fGmF (E2F/GM/E3b), the virus that produced the highest levels of GM-CSF and retained the native GM-CSF leader sequence, was selected for further analysis. The E2F/GM/E3b and E2F/GM/ΔE3 viruses exhibited similar cytotoxic activity and GM-CSF production in several tumor cell lines in vitro. However, when compared in vivo in nude mouse xenograft tumor models, E2F/GM/E3b spread through tumors to a greater extent, resulted in higher peak GM-CSF and total exposure levels in both tumor and serum, and was more efficacious than the E3-deleted virus. Using the matched WI-38 (parental) and WI-38-VA13 (simian virus 40 large T antigen transformed) cell pair, GM-CSF was shown to be selectively produced in cells expressing high levels of E2F, indicating that the tumor-selective E2F promoter controlled E1A and GM-CSF expression.
Replication-selective adenoviruses have been studied as gene therapy viruses to treat cancers in recent years. Selective replication of adenoviruses in tumors has been achieved by replacing the native adenoviral E1a promoter with tumor-selective promoters derived from the E2F-1, PSA, kallikrein-2 and hTERT genes (24, 25, 32, 37). Therapeutic transgenes have also been inserted into the adenoviruses to enhance their antitumor efficacy (2, 3). The adenovirus genome contains more than 20 genes that are used by the virus to control the innate and acquired immune responses of the infected host. These gene products are not necessary for viral replication; however, they appear to target processes that are essential for the survival of the virus during acute or latent infections in vivo (33, 35).
The E3 region of the adenovirus genome contains seven expressed open reading frames, six of which are known to encode proteins with immunomodulatory functions that are involved in the evasion of host immune defenses (17, 28). For example, the E3-gp19 gene encodes a glycoprotein localized in the membrane of the endoplasmic reticulum, where it binds major histocompatibility complex class I antigens and prevents their export from the endoplasmic reticulum to the cell surface. Therefore, gp19 blocks the killing of infected cells by CD8+ cytotoxic T lymphocytes (1, 4).
The immunoregulatory genes of adenovirus appear to have evolved to enable the virus to persist in the face of immune responses. The prolonged presence of oncolytic adenoviruses in tumors should lead to more replication prior to lysis. Recent work demonstrating that an intact E3 region enhanced the antitumor efficacy of a replication-selective virus appears to support these hypotheses (36). Another E3-region protein, the E3-11.6 adenovirus death protein enhances lysis of adenovirus infected cells and mediates the release of progeny virus (29). Overexpression of adenovirus death protein in oncolytic viruses has been shown to increase antitumor efficacy (6, 7) and adenovirus death protein gene mutants were found to slowly release and spread virions from cell to cell (12, 29). Since the lytic function of the adenovirus maximizes antitumor efficacy, it would seem reasonable to include the adenovirus death protein gene in an oncolytic virus.
We have previously constructed and characterized an oncolytic adenovirus, Ar6pAE2fmGmF (E2F/mGM/ΔE3), an E3-deleted oncolytic virus that uses the human E2F-1 promoter to selectively activate E1a expression and thus control adenoviral replication in Rb-pathway defective cells (3). A copy of the murine granulocyte-macrophage colony-stimulating factor (GM-CSF) cDNA was included to enhance antitumor activity of the virus. In addition, the packaging signal was inserted immediately upstream of the right terminal repeat (19). The E2F/mGM/ΔE3 virus has previously been shown to be selectively oncolytic in Rb pathway-defective tumor cells and capable of significantly diminishing tumor burden when injected into xenografted tumors (3).
In the current study, we constructed adenoviruses that were based on the previous E2F/mGM/ΔE3 virus but contained E3 region genes. The human or murine GM-CSF gene was inserted into the E3 region to replace the similarly sized gp19 gene. Several viral constructs were generated that differed slightly in the area of the junction of GM-CSF with the upstream E3-6.7 gene. In each of these adenoviruses, the packaging signal was also relocated upstream of the right terminal repeat. These virus constructs were further characterized and compared with the E2F/mGM/ΔE3 virus for their ability to selectively lyse tumor cells and to support GM-CSF transgene expression. The selected viral constructs were also tested for their antitumor efficacy and the ability to spread in solid tumors. Our findings demonstrate that promoters regulating E1A expression can selectively express transgenes inserted into the E3 region. Furthermore, minor changes in position within the E3 region can have significant effects on transgene expression.
MATERIALS AND METHODS
Plasmids and viruses.
To create adenoviruses that otherwise contained the E3 region, the E3-gp19 gene of a shuttle plasmid containing 11 kb of the right end of the virus was replaced with either the mouse or human GM-CSF cDNA from the start codon to the stop codon. For each virus, three overlapping DNA fragments were generated in initial PCRs, followed by a second PCR to generate a single DNA fragment by gene SOEing (splicing by overlap extension [16]; scheme shown in Fig. 1). The DNA fragments were then digested with BsiWI and NotI and used to replace the BsiWI/NotI fragment containing the E3-gp19 sequence of the shuttle plasmid. In the native adenovirus, the E3-6.7 stop codon overlaps the initial ATG codon of the E3-gp19 gene. To optimize GM-CSF expression from the position of the E3-gp19 gene, viruses with alternative junctions between the E3-6.7 gene and the GM-CSF cDNA were generated and tested for GM-CSF expression. This method of transgene insertion differed markedly from a previous study that used convenient restriction endonuclease (14) sites in that the generation of alternative junction sequences with subtle differences was possible. These alternatives included the following.
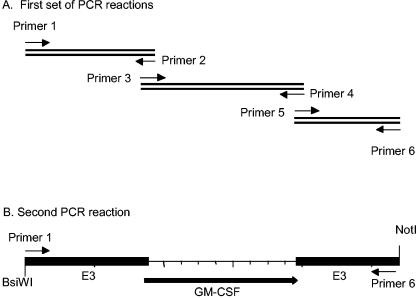
FIG. 1.
Schematic diagram of PCR and overlap PCR. The mouse GM-CSF or human GM-CSF cDNA was inserted into the E3 region, replacing the E3-gp19 open reading frame (ORF) using two steps of PCR amplification. (A) In the first step, three individual PCR amplifications were carried out using three pairs of primers and corresponding DNA templates. Primers 1 and 6 contained only viral sequence. Primers 2, 3, 4, and 5 contained sequences derived from both the virus and GM-CSF cDNA. (B) In the second step, the three DNA fragments were mixed as the template DNA for the overlap PCR amplification using primers 1 and 6. The overlap PCR product was then digested with BsiWI and NotI and used to replace the BsiWI/NotI fragment of adenovirus type 5 containing the E3-gp19 open reading frame.
(i) Δgp19a.
The GM-CSF cDNA was directly swapped for the E3-gp19 coding sequence without regard to the effect on the E3-6.7 stop codon. For the Δgp19a human GM-CSF virus, the fusion protein contains the full-length E3-6.7kDa protein with an additional 33 amino acids at the COOH-terminal end. For the Δgp19a mouse GM-CSF virus, the fusion protein includes an additional 9 amino acids at the COOH end.
(ii) Δgp19b.
A stop codon (TAA) for the E3-6.7 gene was inserted 5′ to the start codon of the GM-CSF cDNA to ensure the correct termination of the E3-6.7 protein. A Kozak consensus sequence (21) was included between the E3-6.7 gene and the GM-CSF cDNA in order to enhance the expression of GM-CSF. The resulting transgenes retained the native sequences.
(iii) Δgp19c.
The E3-6.7 stop codon (TGA) was restored by mutating the fourth nucleotide of GM-CSF to an A, resembling the overlapping ATG and TGA codons of the native E3-6.7/gp19 junction. This single nucleotide change results in a mutation to the GM-CSF signal peptide. For human GM-CSF, the second amino acid is mutated from tryptophan to arginine. For mouse GM-CSF, the second amino acid is mutated from alanine to threonine.
(iv) Δgp19d.
The E3-6.7 stop codon (TGA) was restored by overlapping with an ATGACC sequence added to the 5′ end of GM-CSF. This construct is predicted to add two amino acids (a methionine and a threonine) to the NH2 terminus of the signal sequence of both mouse and human GM-CSF.
(v) Δgp19IRES.
The GM-CSF cDNA was attached to the ATG of the 573-bp EMC IRES (9) using overlap PCR and the IRES/GM-CSF was inserted 3′ to the native stop codon (TGA) of E3-6.7. The resulting transgenes retained the native sequences.
The resulting modifications in the shuttle plasmid were introduced into the adenoviral genome by homologous recombination in Escherichia coli strain BJ5183 (5, 15). The exact sequences of the E3-6.7/GM-CSF junction regions are shown in Table 1, and a key showing the characteristics of all viruses generated is shown in Table 2.
TABLE 1.
E3-6.7/GM-CSF junction sequences
| Construct | Sequencea | Description |
|---|---|---|
| Wild type | E3-6.7...ccaagATGAttaggtac...E3-gp19 | Native sequence showing overlap between the E3-6.7 and gp19 coding sequences |
| Δgp19a | E3-6.7...ccaagATG........TGA...GM-CSF | GM-CSF added without consideration of effects on E3-6.7 stop codon. A stop codon, in-frame with E3-6.7, is reached after 33 amino acids with human GM-CSF; with the mouse GM-CSF, 9 amino acids are added to E3-6.7 |
| Δgp19b | E3-6.7...ccaagaTAAccATG...GM-CSF | E3-6.7 and GM-CSF sequences do not overlap; start and stop codons separated and Kozak sequence inserted 5′ to GM-CSF |
| Δgp19c | E3-6.7...ccaagATGA...GM-CSF | Start and stop codons of E3-6.7 and GM-CSF sequences overlap; second codon (fourth nucleotide) of GM-CSF leader sequence mutated |
| Δgp19d | E3-6.7...ccaagaTGAccATG...GM-CSF | Start and stop codons of E3-6.7 and GM-CSF sequences overlap; two amino acids added to GM-CSF leader sequence |
| Δgp19/IRES | E3-6.7...ccaagaTGA...IRES...ATG...GM-CSF | Start and stop codons of E3-6.7 and GM-CSF sequences separated by EMC IRES |
Normal font, wild-type viral sequences; bold, native GM-CSF sequence; italics, nucleotides added as spacer between E3-6.7 and GM-CSF; uppercase, start and stop codons. The Δgp19 a, b, c, and d modifications were introduced by PCR using primers that overlapped the junction between E3-6.7 and GM-CSF. The encephalomyocarditis virus IRES sequence was first attached by PCR to the initial ATG of GM-CSF prior to being joined to E3-6.7. Viruses carrying the mouse GM-CSF cDNA were generated from all of the constructs described. Human GM-CSF viruses were made using the Δgp19 a, b, and c constructs.
TABLE 2.
Description of adenoviruses used
| Virus designation | Virus name | E1A promotera | E1Ab | E3 | Transgene | Replication | Reference |
|---|---|---|---|---|---|---|---|
| E2F/ΔE3 | Ar6pAE2fF | E2F-1 | WT | Deleted | None | Competent | 19 |
| E2F/E3+ | Ar6pAE2fE3F | E2F-1 | WT | WT | None | Competent | 19 |
| E2F/E3Δgp19 | Ar6pAE2fDg19F | E2F-1 | WT | E3 positive with gp19 deletion | None | Competent | This study |
| E2F/hGM/ΔE3 | Ar6pAE2fhGMF | E2F-1 | WT | Deleted | Human GM-CSF | Competent | 3 |
| E2F/hGM/E3a | Ar6pAE2fhGMDg19aF | E2F-1 | WT | E3 positive with Δgp19a | Human GM-CSF | Competent | This study |
| E2F/hGM/E3b | Ar6pAE2fhGMDg19bF | E2F-1 | WT | E3 positive with Δgp19b | Human GM-CSF | Competent | This study |
| E2F/hGM/E3c | Ar6pAE2fhGMDg19cF | E2F-1 | WT | E3 positive with Δgp19c | Human GM-CSF | Competent | This study |
| E2F/mGM/ΔE3 | Ar6pAE2fmGMF | E2F-1 | WT | Deleted | Mouse GM-CSF | Competent | 3 |
| E2F/mGM/E3a | Ar6pAE2fmGMDg19aF | E2F-1 | WT | E3 positive with Δgp19a | Mouse GM-CSF | Competent | This study |
| E2F/mGM/E3b | Ar6pAE2fmGMDg19bF | E2F-1 | WT | E3 positive with Δgp19b | Mouse GM-CSF | Competent | This study |
| E2F/mGM/E3c | Ar6pAE2fmGMDg19cF | E2F-1 | WT | E3 positive with Δgp19c | Mouse GM-CSF | Competent | This study |
| E2F/mGM/E3d | Ar6pAE2fmGMDg19dF | E2F-1 | WT | E3 positive with Δgp19d | Mouse GM-CSF | Competent | This study |
| E2F/mGM/E3I | Ar6pAE2fmGMDg19IF | E2F-1 | WT | E3 positive with Δgp19 IRES | Mouse GM-CSF | Competent | This study |
| Addl312 | Add/312 | WT | Deleted | Deleted | None | Defective | 20 |
| Addl327 | Add/327 | WT | Deleted | None | Competent | 34 |
E2F-1 unless noted.
Wild type unless noted.
Infectious viral particles were generated following linearization of the plasmids containing the corresponding adenoviral genomes by digestion with SwaI to release the inverted terminal repeats (ITRs). Linear DNA was transfected using Lipofectamine Plus (Life Technologies, Rockville, Md.) into AE1-2A cells (10). After 7 to 10 days, crude viral lysates were generated by five freeze-thaw cycles followed by centrifugation to remove cell debris. Crude viral lysates were passaged on fresh AE1-2A cells until viral cytopathic effect was observed. Viruses were then purified on CsCl gradients (18) and particle titers were determined as previously described (22). Viruses carrying the mouse GM-CSF cDNA with all of the constructs described (Δgp19a, b, c, d, and IRES) were generated; only the Δgp19a, b, and c human GM-CSF viruses were generated. The construction of Ar6pAE2fGmF (E2F/GM/ΔE3), an E3-deleted oncolytic adenovirus that encodes GM-CSF, has been described (3). As detailed previously (19), the packaging signal has been inserted immediately upstream of the right ITR in all viruses.
Cell lines.
The human non-small cell lung carcinoma cell line H460 and human hepatocellular carcinoma cell line Hep3B were obtained from the American Type Culture Collection, Manassas, Va. H460 cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum, 2 mM l-glutamine, and 4.5 g of glucose. Hep3B cells were cultured in Earles's modified eagle medium (EMEM) containing 10% fetal bovine serum. The parental human embryonic lung fibroblast cell line WI-38 (American Type Culture Collection, Manassas, Va.), and its simian virus 40 large T antigen-transformed variant, WI-38-VA13, were cultured in Eagle's minimal essential medium containing 10% fetal bovine serum, supplemented with 1 mM sodium pyruvate, 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 1.5g of sodium bicarbonate. Under these conditions, the WI-38 and WI-38-VA13 cells proliferate normally. As detected by Northern analysis, WI-38 cells express low levels and WI-38-VA13 cells express high levels of E2F-1, as previously shown (19).
Xenograft nude mouse models.
Xenograft H460 or Hep3B tumors were established in 6- to 8-week-old female athymic outbred nu/nu mice (Harlan Sprague Dawley, Indianapolis, Ind.).
H460 tumor model.
We implanted 2 x 106 H460 tumor cells in 0.1 ml of Hanks' balanced salt solution subcutaneously in the right flanks of nude mice. When tumor volumes reached approximately 150 mm3 ([calculated by V = W2 × L π/6]; V, volume; W, width; L, length), the animals were sorted into groups with similar mean tumor volumes and intratumoral administration of adenoviruses or Hanks' balanced salt solution was initiated.
Hep3B tumor model.
We implanted 107 Hep3B tumor cells in 0.1 ml of Hanks' balanced salt solution subcutaneously in the right flanks of nude mice. When tumor volumes reached 150 to 200 mm3, about 12 to 14 days after tumor inoculation, the animals were sorted as described above and intratumoral administration of adenoviruses or Hanks' balanced salt solution was initiated.
Viral doses and treatment regimens varied depending on the model and experiment and are indicated in the figure legends. All studies were conducted in the Genetic Therapy, Inc., Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility according to institutional regulations under approved Institutional Animal Care and Use Committee protocols and in accordance with all applicable animal welfare regulations.
In vitro cytotoxicity assay.
To evaluate the ability of the E3Δgp19 viruses to kill tumor cells, MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] cytotoxicity assays were performed as previously described (3). Briefly, H460 or Hep3B tumor cells were seeded at 10,000 cells per well in 90 μl of growth medium in 96-well dishes one day prior to adenoviral infection. The next day, adenoviruses were diluted serially in the appropriate growth medium and 10 μl of each dilution was added to the wells. Cells were exposed to virus for 7 days, after which the MTS assay (Promega CellTiter 96 AQueous non-radioactive cell proliferation assay; Promega Corp., Madison, Wis.) was performed according to the manufacturer's instructions. All E3Δgp19 viral constructs, as well as E2F/GM/ΔE3 and E2F/ΔE3 (a virus lacking a transgene in which the E2F promoter activates E1a transcription) were tested in the MTS assay. Also tested in MTS assays were an E1a negative control virus Addl312 (20) and a E1a positive control virus Addl327 (35). The results were subjected to sigmoidal dose-response curve fit analysis using Prism GraphPad software to determine the LD50, the effective concentration at which 50% of the cells remained alive compared to untreated control cells.
GM-CSF ELISAs.
Mouse and human GM-CSF enzyme-linked immunosorbent assays (ELISAs) were used to screen for expression of the mouse and human GM-CSF transgenes following infection of H460 cells with the Δgp19 viruses, as described previously (3). Briefly, H460 cells (5 × 105 cells/well) were infected with adenoviruses at 10, 100, or 1,000 particles per cell in a volume of 0.5 ml of serum-free medium in a six-well dish. After 2 h, the infection medium was aspirated and the cells were cultured in 2 ml of complete medium for 24 or 48 h. GM-CSF was detected in the culture supernatant by murine or human GM-CSF ELISA according to the manufacturer's instructions (R&D Systems, Minneapolis, Minn.). The levels of GM-CSF detected in the culture supernates were used as key criteria in the selection of a Δgp19 virus backbone for further study.
Quantitative PCR.
WI-38 and WI-38-VA13 cells (4 × 105/well) were plated in six-well plates 1 day prior to virus infection. Cells were infected with viruses in 0.5 ml of serum-free medium for 1 h at 4°C, after which the infection medium was replaced with fresh complete medium. Cells were cultured for 4 h at 37°C to allow for virus internalization and gene transcription. At the end of the incubation, cells were washed with cold phosphate-buffered saline, lysed in 1 ml of RNAzol B (Tel-Test Inc, Friendwood, Tex.) and stored at −70°C until RNA isolation.
For RNA isolation, the cell lysates were extracted with 0.1 volume of chloroform, RNA was precipitated with 1 volume of isopropanol, washed with 75% ethanol, and resuspended in nuclease-free water. Each sample was treated with 5 U of DNase I (Life Technologies, Rockville, Md.) for 15 min at room temperature. The DNase I was inactivated by the addition of EDTA (to a final concentration of 2.5 mM) and incubated at 65°C for 10 min. The RNA samples were reprecipitated with 2 volumes of ethanol-0.1 volume of 3M sodium acetate, washed in 75% ethanol, and resuspended in nuclease-free water. RNA concentrations were determined spectrophotometrically (A260 and A280) using a SPECTRAmax PLUS spectrophotometer (Molecular Devices, Sunnyvale, Calif.).
First-strand cDNA was generated from 100 ng of RNA using Taqman reverse transcription reagents (Applied Biosystems, Foster City, Calif.). The reverse transcription was performed in a 70-μl reaction volume under the following conditions: 1X Taqman reverse transcription buffer, 5.5 mM MgCl2, 3.8 mM deoxynucleoside triphosphate mixture (0.96 mM each), 2.5 μM random hexamer, 1 U of RNase inhibitor, and 2.5 U of Multiscribe reverse transcriptase. The reactions were incubated for 10 min at 25°C, 30 min at 48°C, 5 min at 95°C and held at 4°C. Primers specific for the adenoviral E1a sequences were designed using the Primer Express software v. 1.0 (Applied Biosystems, Foster City, Calif.). Primer and probe sequences were: E1a Forward primer: 5′-AGCTGTGACTCCGGTCCTTCT-3′; E1a Reverse primer: 3′-GCTCGTTAAGCAAGTCCTCGA-3′; E1a Probe: 5′-FAM-TGGTCCCGCTGTGCCCCATTAAA-TAMRA-3′.
Amplification was performed in a reaction volume of 50 μl under the following conditions: 20 μl of sample cDNA, 1× Taqman Universal PCR Master Mix (Applied Biosystems, Foster City, Calif.), 300 nM forward primer, 900 nM reverse primer, and 100 nM E1a probe. Thermal cycling conditions were: 2 min of incubation at 50°C, 10-min 95°C activation step for the Amplitaq Gold, followed by 35 cycles of successive incubations at 95°C for 15 s and 60°C for 1 min. Thermal cycling was carried out with the 7700 Sequence Detection System (Applied Biosystems, Foster City, Calif.). Data were collected and analyzed using the 7700 Sequence Detection System software v1.6.3 (Applied Biosystems). Relative levels of E1a were determined based on an E1a plasmid curve; with dilutions from 1,500,000 to 15 copies placed in a background reverse transcription reaction. Input RNA was assessed by using a calibrator RNA sample on each plate. The threshold cycle (Ct) for each sample was then compared to the threshold cycle of the calibrator using the following equation: 2−ΔCt where ΔCt = sample 18S Ct value − calibrator Ct value. Reverse transcription reactions for E1a and 18S RNA were performed in duplicate and the average value for each was reported.
Adenovirus copies in the cells were measured by hexon DNA PCR. Multiple wells for each infection were plated to achieve the 3 × 106 cells required for the DNA PCR. Cells were washed and trypsinized after infection and centrifuged and the cell pellets were quickly frozen on dry ice and stored at −70°C for later extraction of DNA. DNA was isolated from approximately 3 × 106 cells using Qiamp Mini columns (Qiagen Inc., Chatsworth, Calif.), according to the manufacturer's instructions. Elution of DNA was done in 250 to 300 μl of water, and concentrations were determined spectrophotometrically (A260 and A280) on the Spectramax Plus. PCR primers and a Taqman probe specific to adenovirus hexon sequences were designed using Primer Express software v1.0. Primer and probe sequences were: Hexon Forward primer: 5′-CTTCGATGATGCCGCAGTG-3′; Hexon Reverse primer: 3′-GGGCTCAGGTACTCCGAGG-3′; Hexon Probe: 5′-FAM-TTACATGCACATCTCGGGCCAGGAC-TAMRA-3′.
Amplification was performed in a 50-μl reaction volume under the following conditions: 10 ng of sample DNA, 1X Taqman Universal PCR Master Mix, 600 nM forward primer, 900 nM reverse primer, and 100 nM hexon probe. Thermal cycling conditions were: 2 min of incubation at 50°C, 10 min at 95°C, followed by 35 cycles of successive incubations at 95°C for 15 s and 60°C for 1 min. Data were collected and analyzed using the 7700 Sequence Detection System software v.1.6.3. Quantification of adenovirus copy number was performed using a standard curve consisting of dilutions of adenovirus DNA from 1,500,000 to 15 copies in 10 ng of cellular genomic DNA. The average number of total copies was normalized to copies per cell based on the input DNA amount and a genome size of 6 × 109 bp.
Flow cytometry analysis.
For enumeration of virus containing cells from tumor xenografts, mice from each group (n = 10 per group) were sacrificed and tumors were excised 1, 4, or 7 days following a single intratumoral viral injection into H460 or Hep3B tumors growing on the flanks of nude mice. The tumors were cut into small pieces and dissociated with collagenase (Sigma, St. Louis, Mo.) treatment at 37°C for 1 h at 2 mg/ml/100 mg of tissue. Cells were then washed with phosphate-buffered saline and passed through a 100-μm nylon cell strainer (Becton Dickinson Labware, Bedford, Mass.), washed and resuspended in phosphate-buffered saline prior to staining for cell surface major histocompatibility complex class I and intracellular hexon.
Unless otherwise indicated, all monoclonal antibodies and solutions used in staining were purchased from Pharmingen, San Diego, Calif.. The tumor cells from each mouse were spun down and incubated with purified rat anti-mouse CD16/CD32 monoclonal antibody (Fcγ III/II receptor, clone 2.4G2) to reduce nonspecific background caused by Fc receptor binding. For surface major histocompatibility complex class I staining, cells were incubated with CyChrome-conjugated human HLA-A, B, and C monoclonal antibody (clone G46-2.6) for 30 min at 4°C in the dark. A CyChrom-labeled mouse immunoglobulin G1 monoclonal antibody (clone MOPC-21) was used as an isotype control.
For intracellular hexon staining, cells must be fixed and permeabilized. Cells were resuspended in 250 μl/tube of cytofix/cytoperm solution (BD PharMingen, San Diego, Calif.) and incubated at 4°C for 20 min. Cells were washed in 1 ml of 1× perm/wash (BD PharMingen) buffer and collected by centrifugation. Cells were then resuspended in 50 μl of perm/wash solution and a fluorescein isothiocyanate-labeled antihexon monoclonal antibody (clone 20/11, Chemicon International Inc., 0.5 μg/106 cells) was added to incubate with cells for 30 min at 4°C in the dark. A fluorescein isothiocyanate-labeled mouse immunoglobulin G1 monoclonal antibody (clone MOPC-21) was used as an isotype control. Cells were acquired and analyzed with the use of CellQuest software on a Becton Dickinson FACSCalibur equipped with dual laser and four-color fluorescence capability. The gate was set to the large (tumor) cell population on the side scatter versus forward scatter dot plot and the compensation was set using the single CyChrome or single fluorescein isothiocyanate-positive cells. Ten thousand gated events were collected and analyzed, and the data were displayed on CyChrome/fluorescein isothiocyanate two-color dot plots to determine hexon-positive human tumor cells.
Statistical analyses.
Statistical analyses were performed using SigmaStat software (SPSS Inc., Chicago, Ill.). Flow cytometric results were compared using one-way analysis of variance with the Student-Newman-Keuls test. In efficacy studies, average tumor volumes of different groups were compared by repeat-measures one-way analysis of variance with Tukey's test for multiple comparisons. Statistical comparisons of tumor volumes were halted when one or more groups were no longer intact due to humane termination of mice according to facility policies.
RESULTS
Construction of Δgp19 viruses.
The modified E3-6.7/GM-CSF junction sequences, viruses, and cloning scheme are summarized in Tables 1 and 2 and Fig. 1. Following cloning, the orientation of the GM-CSF inserts in donor plasmids was determined with SalI digestion and the structure was further confirmed by digestion with SpeI and FspI or SpeI and ClaI (data not shown). Similarly, the integrity of the large plasmids that were generated was confirmed by restriction enzyme digestions with EcoRV, BsrGI, NotI, and MluI (data not shown).
The Δgp19 viruses were isolated and amplified following transfection of S8 cells with SwaI-digested large plasmid DNA. Genomic DNAs from the E2F/hGM/E3a, b, and c viruses (Table 2) containing the E2F promoter and human GM-CSF cDNA in the Δgp19a, Δgp19b, or Δgp19c construct, respectively, was isolated and digested with EcoRV, SalI, BsrG, and NotI (data not shown). A similar analysis was performed on the E2F/mGM/E3a, b, c, d, and I viruses (Table 2) containing the mouse GM-CSF cDNA in the Δgp19a, Δgp19b, Δgp19c, Δgp19d, or Δgp19IRES construct, respectively. All restriction enzyme digests showed the expected patterns. The integrity of the mouse and human GM-CSF cDNA inserts and viral flanking sequences of the Δgp19b constructs (i.e., E2F/mGM/E3b and E2F/hGM/E3b, respectively) were confirmed by sequencing (data not shown).
In vitro cytotoxicity and transgene expression of Δgp19 viruses.
The Δgp19 viruses killed both H460 and Hep3B human tumor cell lines in vitro at LD50 values very close to those of the parental E2F/ΔE3 virus (Table 3). Furthermore, the LD50 values of the Δgp19 viruses, like the E2F/ΔE3 virus, were within a few fold of the values obtained for the positive control E3-deleted Addl327 virus. In other experiments, in vitro LD50 values for the Addl327 virus were indistinguishable from the LD50 values of the fully wild-type adenovirus type 5 for a number of tumor cell lines (data not shown). Thus, the cytolytic function of the viruses was maintained following the deletion of the E3-gp19 gene and its replacement, from start to stop codon, by either human or mouse GM-CSF.
TABLE 3.
Cytotoxicity assay of Δgp19 viruses on H460 and Hep3B tumor cell linesa
| Virus | LD50 (ppc)
|
|||
|---|---|---|---|---|
| H460
|
Hep3B
|
|||
| Expt 1 | Expt 2 | Expt 1 | Expt 2 | |
| Controls | ||||
| Addl312 | >10,000 | >10,000 | 361 | >625 |
| Addl327 | 324 | 323 | 0.0055 | <0.025 |
| E2F/ΔE3 | 253 | 838 | 0.0022 | <0.025 |
| Mouse GM-CSF secreting viruses | ||||
| E2F/mGM/ΔE3 | 621 | ND | 0.0093 | ND |
| E2F/mGM/E3a | 1301 | ND | 0.068 | ND |
| E2F/mGM/E3b | 513 | ND | 0.0053 | ND |
| E2F/mGM/E3c | 1127 | ND | 0.0362 | ND |
| E2F/mGM/E3d | 708 | ND | 0.0558 | ND |
| E2F/mGM/E3I | 1184 | ND | 0.0327 | ND |
| Human GM-CSF secreting viruses | ||||
| E2F/hGM/ΔE3 | ND | 624 | ND | <0.025 |
| E2F/hGM/E3a | ND | 530 | ND | <0.025 |
| E2F/hGM/E3b | ND | 589 | ND | <0.025 |
| E2F/hGM/E3c | ND | 355 | ND | 0.055 |
To evaluate the cytolytic potential of the Δgp19 GM-CSF-secreting viruses, cytotoxicity assays were performed as previously described (3). Two human tumor cell lines, H460 (non-small cell lung carcinoma) and Hep3B (hepatocellular carcinoma) were used. For each cell line, two assays were done for all viruses listed, and the derived LD50 values of a representative assay are shown. The E2F/ΔE3 virus was used for comparison of LD50s of “armed” viruses versus “unarmed” viruses. The E2F/mGM/ΔE3 and E2F/hGM/ΔE3 viruses were used to compare an E3-deleted to the E3-containing viruses with mouse or human GM-CSF in the E3-gp19 position. In addition, control viruses Addl327 (replication-competent Ad5, ΔE3-positive control virus) and Addl312 (E1a-deficient negative control) were also included in the cytotoxicity assays. The viruses with GM-CSF (mouse and human) in place of the E3-gp19 gene retained the cytolytic capacity of the E2F/ΔE3 virus in all cell lines tested. ND, not done.
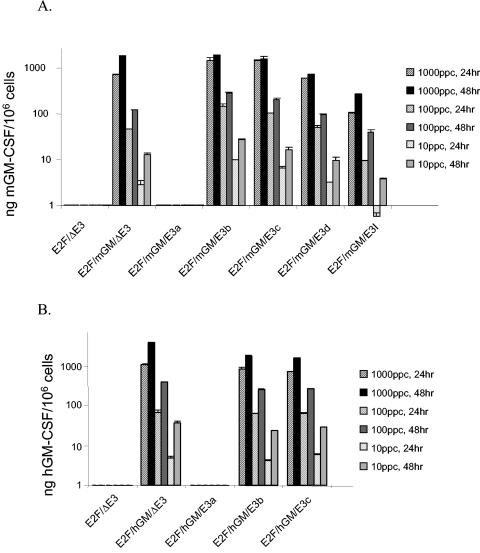
In contrast to the similarities in LD50 values, clear differences were evident in the ability of the various constructs to induce the secretion of GM-CSF in vitro by H460 cells (Fig. 2). The differences were a function of the virus modifications and were not due to the human or mouse GM-CSF transgene, since, for a given modification, similar levels of human and mouse GM-CSF were detected in the tissue culture supernates. As shown in Fig. 2A and 2B, although the Δgp19a viruses were cytolytic, as measured in the MTS assay, surprisingly, neither the human nor the mouse GM-CSF viruses produced detectable levels of GM-CSF. In contrast, the Δgp19b and Δgp19c viruses produced GM-CSF at levels that were comparable to the GM-CSF produced by the E2F/hGM/ΔE3 and E2F/mGM/ΔE3 viruses. The mouse GM-CSF-secreting Δgp19d (E2F/mGM/E3d) and the IRES-containing Δgp19IRES (E2F/mGM/E3I) viruses produced about half and 10% as much GM-CSF as the E2F/mGM/ΔE3 virus, respectively. Potentially, these data could be used to adjust transgene expression levels according to the requirements of a particular application.
FIG. 2.
GM-CSF expression by E3/Δgp19 viruses. Δgp19 viruses expressing (A) mouse GM-CSF and (B) human GM-CSF were assayed for their ability to mediate GM-CSF transgene expression in H460 cells infected with viruses. H460 cells were plated in six-well plates in 2 ml/well of culture medium at a density of 2.5 × 105 cells/well. The next day, the medium was removed and the cultured cells were infected in duplicate with viruses at 10, 100, and 1,000 particles per cell (ppc) in 500 μl of serum-free medium. After 2 h of incubation at 37°C in a 5% CO2 incubator, virus was aspirated and 2 ml of fresh complete culture medium was added to each well. At 24 and 48 h postinfection, the supernatants were collected for mouse GM-CSF ELISA and human GM-CSF ELISA.
The MTS cytotoxicity and GM-CSF production assays narrowed the viruses under consideration for further study to the Δgp19b and Δgp19c constructs. As depicted in Table 1, the Δgp19c viruses restore the sequence of the native overlap between the E3-6.7 stop codon and the initial methionine of the immediate downstream E3-gp19 gene. In order to accomplish this, it was necessary to mutate the fourth nucleotide of both human and mouse GM-CSF with subsequent mutations at the amino acid level as well. In the Δgp19b constructs, the E3-6.7 stop codon was altered to a TAA from the native TGA. With this change, there is no alteration to the E3-6.7 amino acid sequence and the gene downstream of E3-6.7 (GM-CSF) would not be expected to initiate translation at this site. To optimize GM-CSF expression, a consensus Kozak sequence was placed immediately upstream of the native GM-CSF sequence. Consequently, the GM-CSF produced by the Δgp19b viruses, in contrast to the Δgp19c viruses, is expected to have a fully native amino acid sequence. Based on these considerations, the Δgp19b viruses were chosen for further analysis.
Major histocompatibility complex class I expression following infection with Δgp19b viruses.
The product of the E3-gp19 gene, the gp19 protein, binds with major histocompatibility complex class I proteins in the endoplasmic reticulum and inhibits their transport to the cell surface (35). As a result, cells infected by wild-type adenoviruses are less visible to the immune system, are poor antigen-presenting cells, and are more likely to evade detection by cytotoxic T lymphocytes. We reasoned that substitution of the gp19 gene with GM-CSF in the vaccine setting would likely improve immunological antitumor responses.
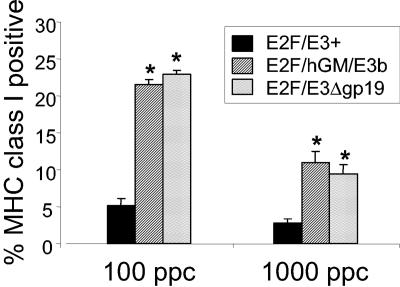
In order to analyze this, Hep3B cells were infected with E2F/hGM/E3b, E2F/E3Δgp19, or E2F/E3+ at 100 or 1,000 viral particles per cell for 2 h and cultured for 24 h, after which fluorescence-activated cell sorting analysis was performed on the infected cells to determine the levels of major histocompatibility complex class I expression (Fig. 3). The levels of major histocompatibility complex class I are significantly higher in E2F/hGM/E3b- or E2F/E3Δgp19-infected cells when compared to the E2F/E3+-infected cells (P < 0.001). These results confirm that the gp19 protein functions to inhibit cell surface expression of major histocompatibility complex class I and provide a rationale for deleting the gp19 gene. Higher levels of major histocompatibility complex class I expression in the infected cells are expected to result in increased presentation and detection of tumor-associated antigens. Increased tumor-associated antigen presentation in association with major histocompatibility complex class I should result in more robust systemic antitumor cytotoxic T lymphocyte responses.
FIG. 3.
Effect of E3-gp19 deletion on major histocompatibility complex class I expression. Hep3B tumor cells were infected with the E2F/E3+, E2F/hGM/E3b, or E2F/E3Δgp19 virus at 100 particles per cell and 1,000 particles per cell for 2 h, after which the infection medium was aspirated and cells were cultured in fresh complete medium for 24 h. The major histocompatibility complex class I expression level was measured by staining cells with a pan-class I monoclonal antibody and analyzed by fluorescence-activated cell sorting analysis. Each column represents the average of triplicates ± standard deviation. *, P < 0.001 compared with E2F/E3+-infected cells, one-way analysis of variance.
E1a and GM-CSF transgene expression in Rb pathway-functional and -deficient cell lines.
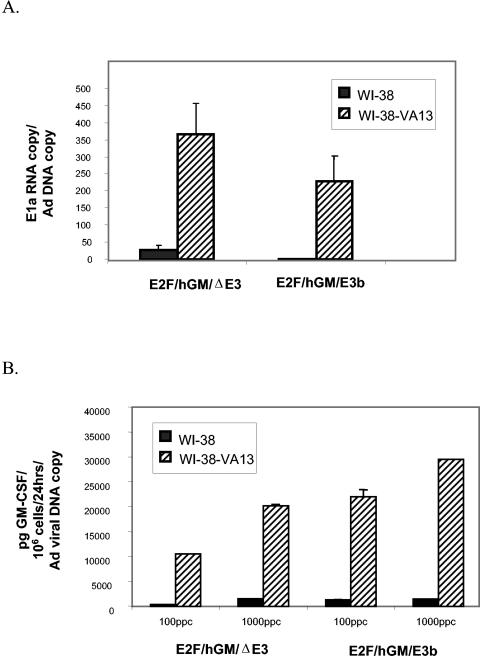
E1a gene transcription occurs following adenoviral entry into the host cells and lasts approximately 6 to 8 h, after which viral DNA replication is first detected (26). Thus, 4 h following infection, adenoviral DNA replication has yet to begin in the host cells and detection of DNA copies per cell at this time point should reflect the adenovirus particles that entered the cells during the infection period (i.e., the transduction efficiency). Preliminary studies indicated that E1a transcripts had reached a significant level 4 h following infection (data not shown). Accordingly, WI-38 and WI-38-VA13 cells were infected with 1,000 particles per cell of E2F/hGM/ΔE3 or E2F/hGM/E3b for 4 h as described in the Materials and Methods section. Quantitative E1a RNA PCR was performed to determine the viral transcription levels and is indicative of E2F promoter activity in a given infected cell.
Quantitative hexon DNA PCR was also performed 4 h postinfection to determine the number of adenoviral particles that had been internalized by WI-38 and WI-38-VA13 cells and was used to normalize E1a expression levels (Fig. 4a). The E1a RNA copies in simian virus 40-transformed WI-38-VA13 cells were significantly higher than in parental WI-38 cells infected with the same adenoviruses (P < 0.05), indicating that E1a is selectively expressed in T-antigen-transformed cells that have increased E2F levels. The E1A RNA levels were similar between viruses regardless of total E3 region deletion (in E2F/hGM/ΔE3) or partial E3 gp19 deletion (in E2F/hGM/E3b).
FIG. 4.
Tumor-selective transgene expression. (A) Selective E1a gene transcription in WI-38-VA13 cells. WI-38 and WI-38-VA13 cells were infected with adenoviruses E2F/hGM/ΔE3 and E2F/hGM/E3b at 1,000 particles per cell. Real-time PCR was performed on the infected cells at the 4-h time point to determine E1a RNA levels. To correct for differences in initial transduction efficiency, E1a RNA levels were normalized to hexon DNA copy number 4 h postinfection, ± standard deviation. (B) Selective transgene expression in WI-38-VA13 cells. Human GM-CSF levels secreted by WI-38 and WI-38-VA13 cells were determined by ELISA. Human GM-CSF was analyzed at 24 h after infection, using the quantitative human GM-CSF sandwich enzyme immunoassay kits from R&D Systems. The concentrations of human GM-CSF in the samples were calculated from the standard curve based on the optical density at 490 nm and normalized to adenoviral genome copy number determined 4 h following infection.
In order to analyze the regulation of GM-CSF expression, WI-38 and WI-38-VA13 cells were infected with the oncolytic adenoviruses E2F/hGM/ΔE3 and E2F/hGM/E3b at 100 and 1,000 particles per cell as described above. Culture supernatants collected 24 h after infection were analyzed by ELISA for human GM-CSF expression. As in the E1a analysis, the number of viruses transducing the two cell lines could vary such that differences in GM-CSF production could artifactually be a reflection of different transduction efficiencies and human GM-CSF production was therefore normalized to the 4-h postinfection hexon DNA copy number. As shown in Fig. 4b, the normalized levels of human GM-CSF produced by WI-38-VA13 cells were substantially higher than the levels produced by the parental WI-38 cells. In addition, the amounts of human GM-CSF produced in WI-38-VA13 cells by the two viruses were comparable, indicating that the structural differences between the viruses do not have obvious effects on GM-CSF expression.
These results suggest that the E2F promoter is substantially more active in WI-38-VA13 cells than in WI-38 cells and that the level of E2F in the cells regulates the promoter. It is important to note that the conditions used in these studies allowed proliferation and modest E2F-1 expression in WI-38 cells, as previously demonstrated (19). Thus, selectivity for Rb pathway-defective cells was demonstrated for both E1a and transgene expression.
In vivo intratumoral spread of oncolytic adenoviruses.
E3 region-deleted and E3-containing oncolytic adenoviruses dependent on the E2F promoter for control of E1a expression have been shown to selectively replicate in and lyse tumor cells in vitro and in vivo (9, 31). The E2F/E3+ virus contains the complete E3 region, including the adenovirus death protein that is necessary for efficient host cell lysis (29). Overexpression of adenovirus death protein has been shown to improve the efficacy of oncolytic adenoviruses (6), but increases in actual cell-to-cell spread in solid tumors due to inclusion of the adenovirus death protein gene have not previously been demonstrated in vivo.
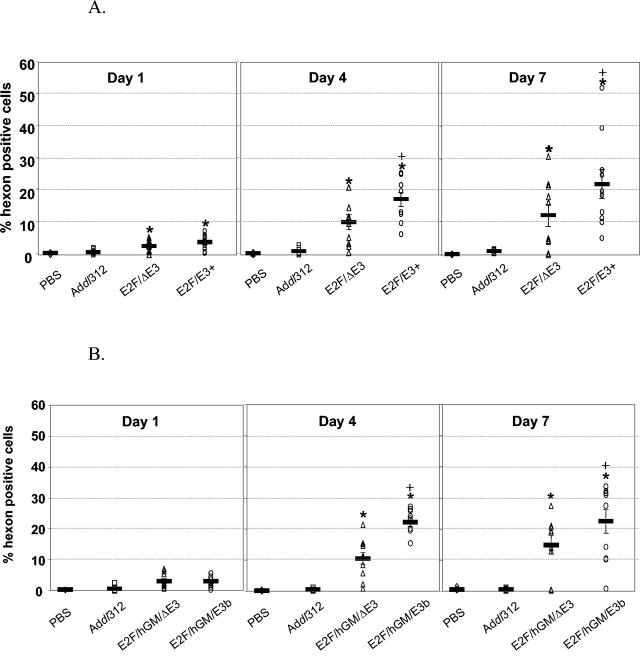
In Fig. 5, the cell-to-cell spread of E3-containing and E3-deleted viruses in xenografted H460 and Hep3B tumors is compared. The tumors, growing subcutaneously in nude mice, were directly injected with a single dose of the viruses (2 × 1010 viral particles/mouse) by intratumoral injection. Mice (n = 10 per group) were sacrificed 1, 4, or 7 days after the viral injection. Tumors from each mouse were analyzed for intracellular hexon expression by fluorescence-activated cell sorting analysis with hexon-specific monoclonal antibodies. A monoclonal antibody directed against human HLA-A, -B, and -C antigens was used to differentiate human tumor cells from mouse cells (>90% of the cells harvested from the tumors were HLA-A, -B, or -C positive).
FIG. 5.
Effect of E3 region on viral spread though tumors in vivo. A single intratumoral injection of 2 × 1010 adenoviruses, as indicated, was administered to established human (A) H460 or (B) Hep3B tumors growing on the flanks of nude mice. Tumor samples were taken on days 1, 4, and 7 after the virus injection and assayed by flow cytometry for adenoviral hexon expression. E3-containing viruses spread more efficiently in vivo, resulting in a significantly higher level of tumor cell transduction. *, P < 0.05 compared to Addl312; +, P < 0.05 compared to E2F/ΔE3, analysis of variance.
Tumor cells dissociated from either H460 or Hep3B tumors injected with replication-competent viruses showed positive staining for hexon, while tumor cells from the negative control tumors treated with phosphate-buffered saline or Addl312 did not. One day after the virus injection, significant hexon expression was detectable in 1 to 2% of the cells in H460 tumors injected with E3-deleted and E3-containing viruses (Fig. 5A, left panel). Four and 7 days after the injection, hexon-positive cells from tumors injected with replication=competent viruses were significantly higher than saline and Addl312 treatments in both H460 and Hep3B tumors (Fig. 5A and B, center and right panels), with a spread to greater than 20% of the tumor cells. Most importantly, significantly more hexon-positive cells (P < 0.05) were detected in tumors injected with E3-containing viruses than with E3-deleted viruses, indicating at a cellular level that the E3 region had further facilitated spread of the virus through the tumors. Importantly, similar transduction levels were attained with viruses that contained the full E3 region (Fig. 5A) and viruses that carried the human GM-CSF transgene and were E3-gp19 deleted but otherwise E3 positive (Fig. 5B).
Antitumor efficacy in vivo.
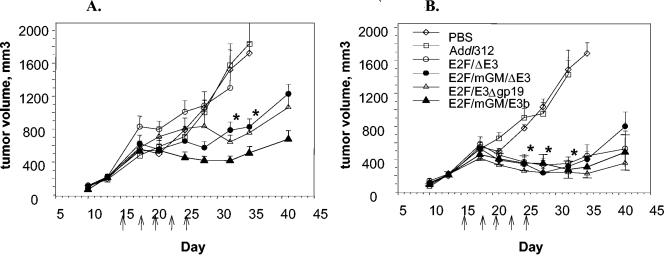
The antitumor efficacy of several viruses was tested in Hep3B xenograft tumor models. Tumor-bearing mice (n = 10 per group) were given five intratumoral viral injections at 2 × 106 or 2 × 108 particles/injection, on an every-other-day schedule, as shown in Fig. 6. Antitumor efficacy was determined by the decrease in tumor volumes following the viral injection. The results indicated a dose-response relationship, such that greater efficacy was achieved at the 2 × 108 particle per injection dose level compared to 2 × 106 particles per injection. All four treatment groups, E2F/ΔE3, E2F/mGM/ΔE3, E2F/E3Δgp19, and E2F/mGM/E3b, were significantly efficacious compared to the phosphate-buffered saline and Addl312 groups and showed remarkably similar responses at the high dose level (Fig. 6B) such that the contributions of the E3 region and GM-CSF to the observed responses were not evident.
FIG. 6.
Effect of E3 region on efficacy in Hep3B tumor model. Group average tumor volumes (+standard error of themean) of Hep3B tumors following treatment with phosphate-buffered saline (PDS, n = 6), Addl312 (n = 6), or oncolytic viruses (n = 8), as indicated in the graph legends. The treatment groups are separated by virus doses. (A) Tumors injected with 2 × 106 particles; (B) tumors injected with 2 × 108 particles. Asterisks indicate P < 0.05 compared to dose-matched Addl312-treated tumors for all groups below the asterisk.
All high-dose treatment groups showed a decline in tumor volume following the third dose that reached significance prior to administration of the fifth dose. In contrast, at 2 × 106 particles per injection, viruses that included the E3 region and/or the mouse GM-CSF transgene (i.e., E2F/mGM/ΔE3, E2F/E3Δgp19 and E2F/mGM/E3b) were significantly more efficacious than the E2F/ΔE3 virus (Fig. 6A). In fact, throughout the course of the study, tumor volumes in the E2F/ΔE3 low-dose treatment group overlapped the saline and Addl312 controls. Furthermore, the E2F/mGM/E3b virus that contained both the E3 region and the mouse GM-CSF cDNA demonstrated a clear trend towards being more efficacious than viruses that contained either the E3 region or GM-CSF alone. Thus, in terms of effects on tumor volume, the treatment groups segregated as follows: E2F/mGM/E3b > E2F/mGM/ΔE3 = E2F/E3Δgp19 > E2F/ΔE3 > Addl312 = phosphate-buffered saline.
DISCUSSION
Adenoviruses that selectively replicate in tumor cells have been developed to specifically eradicate tumor cells using the natural viral life cycle of lysing the infected host cells. Replication-selective adenoviruses have the assumed advantages of replicating in the target tumor cells with subsequent lysis, spread, and infection of adjacent tumor cells. However, one of the main difficulties encountered in such cancer gene therapy is the inability of the viruses to efficiently infect all the tumor cells and spread through the entire solid tumor to eradicate every tumor cell (11, 27). The reasons for this are not entirely clear but could include physical barriers within the tumor (e.g., normal connective tissues and endothelial cells and/or high intratumoral pressure) or the innate and acquired immune responses.
In this study, we retained the E3 region in the viral backbones to include the immunoregulatory genes that a virus uses to evade the immune system, except for the E3-gp19 gene, which was replaced by either the human or murine GM-CSF gene. The viruses that contain the E3 genes are able to persist within the tumor for longer periods of time. The adenovirus death protein in the E3 region would facilitate the lysis of infected tumor cells and release viral progeny to infect neighboring tumor cells. We demonstrated in this study that tumors injected with the E3-containing viruses had more hexon-positive tumor cells 4 and 7 days after a single viral injection compared with the E3-deleted viruses. Moreover, the deletion of the E3-gp19 gene enabled the infected tumor cells to express higher levels of major histocompatibility complex class I, potentially leading to improved recognition of tumor-associated antigens and activation of tumor-specific cytotoxic T lymphocytes. While this is not a functional mechanism in nude mouse xenograft models, it is expected to play a significant role in antitumor efficacy in immunocompetent patients.
In addition to the substitution of the E3-gp19 gene with GM-CSF, viruses in which the E3-14.7 gene has been replaced have also been constructed (not shown). The initial ATG of the E3-14.7 gene is overlapped by the terminal sequence of the upstream E3-14.5 gene in a manner similar to the junction between the E3-6.7 and E3-gp19 genes. A PCR-based strategy similar to the strategy used in creating the Δgp19b constructs was pursued; that is, a stop codon for the upstream gene was inserted and the E3-14.5 and human GM-CSF genes were separated by a consensus Kozak sequence. The resulting Δ14.7 viruses produced GM-CSF at levels very similar to the E3-deleted E2F/hGM/ΔE3 and Δgp19 E2F/hGM/E3b viruses. As with the E3-gp19 constructs, the cloning of genes into this region using restriction sites has been described (13). The PCR-based strategy for both the E3-gp19 and E3-14.7 substitutions results in greater flexibility in insertion sites, the potential of various transgene expression levels, and allows the preservation of adjacent E3 region genes.
The successful replacement of the E3-gp19 or E3-14.7 gene opens up the possibility of inserting transgenes in either or both locations, depending on the application. As an example, a virus that expresses both chains of the interleukin-12 cytokine (30) while maintaining the remainder of the E3 region, including the adenovirus death protein gene, may be produced using these strategies. Additionally, numerous combinations are possible; the cDNA of a tumor-associated antigen could be expressed from the gp19 position and an immunological costimulatory molecule (8) could be expressed from the E3-14.7 position. Alternately, GM-CSF expression could be combined with Flt3L expression (23).
The oncolytic adenoviruses constructed in this study utilize the E2F-1 promoter to control the expression of the E1a gene. E1A proteins then transactivate the E3 promoter to induce the transcription of E3 genes and of the human or murine GM-CSF transgene. Thus, the tumor-selective E2F-1 promoter ultimately controls the expression of GM-CSF. In order to determine whether the GM-CSF expression was restricted to the cells that produce high levels of E2F protein, we compared the GM-CSF levels produced by WI-38 (low E2F) and WI-38-VA13 (high E2F) cells following viral infection. The data showed that GM-CSF was selectively produced in WI-38-VA13 cells, while GM-CSF expression in WI-38 was very low, indicating that the E2F promoter is active in cells with a high free E2F level, and expression of the GM-CSF transgene is ultimately controlled by the E2F-1 promoter.
The in vivo antitumor effect of the E2F/mGM/E3b virus was evaluated and compared with that of E3-deleted viruses in existing tumors in the Hep3B xenograft nude mouse model. Viruses containing the E3 region or mouse GM-CSF were more efficacious in reducing tumor volumes than the E3-deleted E2F/ΔE3 viruses. The increased antitumor efficacy could be partially attributed to the presence of the adenovirus death protein in the E3 region, since other genes in the E3 region would not be expected to be factors in immunodeficient nude mice. In addition, as we have previously shown (3), xenograft tumors treated with mouse GM-CSF-producing viruses demonstrated increased growth inhibition compared to viruses that lacked the GM-CSF transgene, demonstrating an advantage to including the immunoenhancing cytokine GM-CSF in the virus. These findings provide a strong rationale for inclusion of both the E3 region and GM-CSF in an oncolytic adenovirus.
Acknowledgments
We thank Lynda Hawkins, Russette Lyons, D. C. Yu, and Peter Working for critical review of the manuscript, Arnold Berk for the Addl312 virus, and Bill Kaelin for the E2F-1 promoter plasmid. We also thank Michele Kaloss, Anne Pinkstaff, and Maria Theurer for the quantitative PCR studies.
Footnotes
Reprint requests should be addressed to Susan Bryan, Cell Genesys, Inc., 500 Forbes Avenue, South San Francisco, CA 94080, Susan.Bryan@cellgenesys.com.
REFERENCES
- 1.Andersson, M., S. Paabo, T. Nilsson, and P. A. Peterson. 1985. Impaired intracellular transport of class I MHC antigens as a possible means for adenoviruses to evade immune surveillance. Cell 43:215-222. [DOI] [PubMed] [Google Scholar]
- 2.Bauzon, M., D. Castro, M. Karr, L. K. Hawkins, and T.W. Hermiston. 2003. Multigene expression from a replicating adenovirus using native viral promoters. Mol. Ther. 7:526-534. [DOI] [PubMed] [Google Scholar]
- 3.Bristol, J. A., M. Zhu, J. H., M. Mina, Y. Xie, L. Clarke, S. Forry-schaudies, and D. L. Ennist. 2003. In vitro and in vivo activities of an oncolytic adenoviral vector designed to express GM-CSF. Mol. Ther. 7:755-764. [DOI] [PubMed] [Google Scholar]
- 4.Burgert, H. G., and S. Kvist. 1985. An adenovirus type 2 glycoprotein blocks cell surface expression of human histocompatibility class I antigens. Cell 41:987-997. [DOI] [PubMed] [Google Scholar]
- 5.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doronin, K., M. Kuppuswamy, K. Toth, A. E. Tollefson, P. Krajcsi, V. Krougliak, and W. S. Wold. 2001. Tissue-specific, tumor-selective, replication-competent adenovirus vector for cancer gene therapy. J. Virol. 75:3314-3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doronin, K., K. Toth, M. Kuppuswamy, P. Ward, A. E. Tollefson, and W. S. Wold. 2000. Tumor-specific, replication-competent adenovirus vectors overexpressing the adenovirus death protein. J. Virol. 74:6147-6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frauwirth, K. A., and C. B. Thompson. 2002. Activation and inhibition of lymphocytes by costimulation. J. Clin. Investig. 109:295-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghattas, I. R., J. R. Sanes, and J. E. Majors. 1991. The encephalomyocarditis virus internal ribosome entry site allows efficient coexpression of two genes from a recombinant provirus in cultured cells and in embryos. Mol. Cell. Biol. 11:5848-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorziglia, M. I., M. J. Kadan, S. Yei, J. Lim, G. M. Lee, R. Luthra, and B. C. Trapnell. 1996. Elimination of both E1 and E2 from adenovirus vectors further improves prospects for in vivo human gene therapy. J. Virol. 70:4173-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harrison, D., H. Sauthoff, S. Heitner, J. Jagirdar, W. N. Rom, and Hay. J. G. 2001. Wild-type adenovirus decreases tumor xenograft growth, but despite viral persistence complete tumor responses are rarely achieved-deletion of the viral E1b-19-kD gene increases the viral oncolytic effect. Hum. Gene Ther. 12:1323-1332. [DOI] [PubMed] [Google Scholar]
- 12.Hawkins, L. K., and T. W. Hermiston. 2001. Gene delivery from the E3 region of replicating human adenovirus: Evaluation of the ADP region. Gene Ther. 8:1132-1141. [DOI] [PubMed] [Google Scholar]
- 13.Hawkins, L. K., and T. W. Hermiston. 2001. Gene delivery from the E3 region of replicating human adenovirus: Evaluation of the E3B region. Gene Ther. 8:1142-1148. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins, L. K., L. Johnson, M. Bauzon, J. A. Nye, D. Castro, G. A. Kitzes, M. D. Young, J. K. Holt, P. Trawn, and T. W. Hermiston. 2001. Gene delivery from the E3 region of replicating human adenovirus: evaluation of the 6.7K/gp19K region. Gene Ther. 8:1123-1131. [DOI] [PubMed] [Google Scholar]
- 15.He, T. C., Z;, S., D. A. L. T. Costa, Yu, J., K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton, R. M., Z. L. Cai, Ho, S. N., and L.R. Pease. 1990. Gene splicing by overlap extension: Tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 17.Horwitz, M. S.. 2001. Adenovirus immunoregulatory genes and their cellular targets. Virology 279:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Jakubczak, J. L., M. L. Rollence, D. A. Stewart, et al.. 2001. Adenovirus type 5 viral particles pseudotyped with mutagenized fiber proteins show diminished infectivity of coxsackie B-adenovirus receptor-bearing cells. J. Virol. 75:2972-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakubczak, J. L., P. Ryan, M. Gorziglia, et al.. 2003. An oncolytic adenovirus selective for retinoblastoma tumor suppressor protein pathway-defective tumors: dependence on E1A, the E2F-1 promoter, and viral replication for selectivity and efficacy. Cancer Res. 63:1490-1499. [PubMed] [Google Scholar]
- 20.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 21.Kozak, M.. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 22.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mwangi, W., W. C. Brown, H. A. Lewin, C. J. Howard, J. C. Hope, T. V. Baszler, P. Caplazi, J. Abbott, and G. H. Palmer. 2002. DNA-encoded fetal liver tyrosine kinase 3 ligand and granulocyte macrophage-colony-stimulating factor increase dendritic cell recruitment to the inoculation site and enhance antigen-specific CD4+ T cell responses induced by DNA vaccination of outbred animals. J. Immunol. 169:3837-3846. [DOI] [PubMed] [Google Scholar]
- 24.Parr, M. J., Y. Manome, T. Tanaka, P. Wen, D. W. Kufe, W. G. Kaelin, Jr., and H. A. Fine. 1997. Tumor-selective transgene expression in vivo mediated by an E2F-responsive adenoviral vector. Nat. Med. 3:1145-1149. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez, R., E. R. Schuur, H. Y. Lim, G. A. Henderson, J. W. Simons, and D. R. Henderson. 1997. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 57:2559-2563. [PubMed] [Google Scholar]
- 26.Russell, W.C.. 2000. Update on adenovirus and its vectors. J. Gen. Virol. 81:2573-2604. [DOI] [PubMed] [Google Scholar]
- 27.Sauthoff, H., Hu, J., C. Maca, M. Goldman, S. Heitner, H. Yee, T. Pipiya, W. N. Rom, and J. G. Hay. 2003. Intratumoral spread of wild-type adenovirus is limited after local injection of human xenograft tumors: virus persists and spreads systemically at late time points. Hum. Gene Ther. 14:425-433. [DOI] [PubMed] [Google Scholar]
- 28.Sparer, T. E., R. A. Tripp, D. L. Dillehay, T. W. Hermiston, W. S. Wold, and L. R. Gooding.. 1996. The role of human adenovirus early region 3 proteins (gp19K, 14.5K, and 14.7K) in a murine pneumonia model. J. Virol. 70:2431-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tollefson, A. E., A. Scaria, T. W. Hermiston, J. S. Ryerse, L. J. Wold, and W. S. Wold. 1996. The adenovirus death protein (E3-11.6K) is required at very late stages of infection for efficient cell lysis and release of adenovirus from infected cells. J. Virol. 70:2296-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trinchieri, G.. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133-146. [DOI] [PubMed] [Google Scholar]
- 31.Tsukuda, K., WIewrodt, R., K. Molnar-kimber, V. P. Jovanovic, and K. M. Amin. 2002. An E2F-responsive replication-selective adenovirus targeted to the defective cell cycle in cancer cells: potent antitumoral efficacy but no toxicity to normal cell. Cancer Res. 62:3438-3447. [PubMed] [Google Scholar]
- 32.Wirth, T., L. Zender, B. Schulte, B. Mundt, R. Plentz, K. L. Rudolph, M. Manns, S. Kubicka, and F. Kuhnel. 2003. A telomerase-dependent conditionally replicating adenovirus for selective treatment of cancer. Cancer Res. 63:3181-3188. [PubMed] [Google Scholar]
- 33.Wold, W. S., K. Doronin, K. Toth, M. Kuppuswamy, D. L. Lichtenstein, and A. E. Tollefson. 1999. Immune responses to adenoviruses: viral evasion mechanisms and their implications for the clinic. Curr. Opin. Immunol. 11:380-386. [DOI] [PubMed] [Google Scholar]
- 34.Wold, W. S., and L. R. Gooding. 1991. Region E3 of adenovirus: a cassette of genes involved in host immunosurveillance and virus-cell interactions. Virology 184:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Wold, W. S., A. E. Tollefson, and T. W. Hermiston. 1995. E3 transcription unit of adenovirus. Curr Top Microbiol Immunol. 199:237-274. [DOI] [PubMed] [Google Scholar]
- 36.Yu, D. C., Y. Chen, M. Seng, J. Dilley, and D. R. Henderson. 1999. The addition of adenovirus type 5 region E3 enables Calydon virus 787 to eliminate distant prostate tumor xenografts. Cancer Res. 59:4200-4203. [PubMed] [Google Scholar]
- 37.Yu, D. C., G. T. Sakamoto, and D. R. Henderson. 1999. Identification of the transcriptional regulatory sequences of human kallikrein 2 and their use in the construction of Calydon virus 764, an attenuated replication competent adenovirus for prostate cancer therapy. Cancer Res. 59:1498-1504. [PubMed] [Google Scholar]