Abstract
Human cytomegalovirus (HCMV) is a paradigm for mechanisms subverting antigen presentation by major histocompatibility complex (MHC) molecules. Due to its limited host range, HCMV cannot be studied in animals. Thus, the in vivo importance of inhibiting antigen presentation for the establishment and maintenance of infection with HCMV is unknown. Rhesus cytomegalovirus (RhCMV) is an emerging animal model that shares many of the features of HCMV infection. The recent completion of the genomic sequence of RhCMV revealed a significant degree of homology to HCMV. Strikingly, RhCMV contains several genes with low homology to the HCMV US6 gene family of inhibitors of the MHC I antigen presentation pathway. Here, we examine whether the RhCMV US6 homologues (open reading frames Rh182, -184, -185, -186, -187, and -189) interfere with the MHC I antigen-processing pathway. We demonstrate that Rh182 and Rh189 function similarly to HCMV US2 and US11, respectively, mediating the proteasomal degradation of newly synthesized MHC I. The US3 homologue, Rh184, delayed MHC I maturation. Unlike US3, MHC I molecules eventually escaped retention by Rh184, so that steady-state surface levels of MHC I remained unchanged. Rh185 acted similarly to US6 and inhibited peptide transport by TAP and, consequently, peptide loading of MHC I molecules. Thus, despite relatively low sequence conservation, US6 family-related genes in RhCMV are functionally closely related to the conserved structural features of HCMV immunomodulators. The conservation of these mechanisms implies their importance for immune evasion in vivo, a question that can now be addressed experimentally.
Human cytomegalovirus (HCMV) is highly prevalent in the human population and establishes persistent infection of immunocompetent hosts (47). This life-long infection occurs despite a significant CMV-specific cellular immune response, with up to 10% of the total T-cell population being CMV specific (22). Moreover, seropositive individuals can be reinfected with a different strain of CMV even in the presence of preexisting immunity (8). Thus, the immune system is unable to eradicate CMV upon primary infection or to prevent reinfection. Continuous immune surveillance is required to keep the viral infection in an asymptomatic state, since CMV disease is mostly observed during immunodeficiency, particularly in cell-mediated immunity, related to either immunologic immaturity, pharmacologic immunosuppression (transplantation) (53), or the progressive immunodeficiency of human immunodeficiency virus infection (52). Thus, a balance is established between immunological control of the viral infection and immune evasion by the virus.
Immunomodulatory mechanisms encoded by CMV are thought to be central to maintaining this balance. It is conceivable that a large portion of the CMV genome, containing >250 open reading frames (ORFs), is dedicated to manipulating various aspects of the host defense. One of the major obstacles to elucidating these mechanisms is the fact that HCMV can infect only humans. This extreme host restriction of β-herpesviruses resulted in their coevolution with the host organisms. The most closely related nonhuman CMV is the chimpanzee cytomegalovirus (16). However, chimpanzees are not readily available for experimentation. Rodents are ideal for experimentation, but the genomic sequences of murine cytomegaloviruses show that the vast majority of the HCMV immunomodulatory genes are not conserved (49). Therefore, there is a need to establish CMV infection models in animals closely related to humans. Such an alternative animal model is rhesus macaque infection by rhesus cytomegalovirus (RhCMV) (3). RhCMV and HCMV have similar epidemiologies and patterns of infection in immunocompetent and immunodeficient hosts (43, 54, 58). In addition, the immune response to RhCMV is similar to that to HCMV, with a high percentage of T cells being CMV specific (5, 35-37, 48). The recently completed sequence of the RhCMV genome revealed that, in addition to genes with no obvious homology in HCMV, RhCMV encodes homologues of most of the known immunomodulators of HCMV (26). This list includes homologues of the viral inhibitor of caspase activation, UL36; the viral mitochondrial inhibitor of apoptosis, UL37; the interleukin-10 homologue, UL111; the Fc receptors, UL117/UL118; the viral CXC chemokine homologue, UL147; and the tumor necrosis factor receptor homologue, UL144 (26).
The RhCMV genomic region Rh182 to -189 contains six genes with homology to the genes US2 to US11 in the unique short (US) region of HCMV. This region in HCMV encodes a group of eight glycoproteins that were originally grouped into two families, US2 and US6 (11), and later into three families, US2, US3, and US11 (9). Primary structure alignments, as well as their functional relationship, suggests that all of the US2, US3, and US6 family genes arose by gene duplication and thus their products represent a single family of proteins (1, 20), which we will refer to as the US6 family. Six members of this family have been shown to interfere with the major histocompatibility complex class I (MHC I) antigen-processing pathway (42), and deletion of this region restores MHC I expression in HCMV-infected cells (33). Since MHC I molecules present virus-derived peptides to cytotoxic T cells, it is assumed that these proteins play an important role in viral immune evasion in vivo. Each of the US6 family proteins interferes at a distinct step during the assembly and intracellular transport of MHC I heterotrimers consisting of heavy chain (HC), β2 microglobulin (β2m), and antigen-derived peptides. Immediately upon heavy-chain synthesis, US2 and US11 extract newly synthesized MHC I molecules from the lumen of the endoplasmic reticulum (ER) and send them for degradation by the proteasome (59, 60). US11 achieves this by simultaneously interacting with MHC I molecules and the cellular quality control protein derlin 1, which extracts misfolded proteins from the ER (41, 61). US2 seems to act by a different mechanism, since it differs from US11 in several respects, e.g., it does not require the presence of a cytosolic tail in its target molecules and does not interact with derlin 1 (4, 41, 61). US6 inhibits peptide translocation across the lumen of the ER membrane by the peptide transporter TAP, preventing the association of antigen-derived peptides with nascent MHC I molecules (2, 28, 30, 40). US3 retains MHC I molecules in the ER (1, 25, 34, 39) by binding to and interfering with tapasin, an MHC-specific chaperone that controls peptide loading (46). US10 has been shown to delay MHC I exit (18). At the cell surface, US8 binds to MHC I molecules that are endocytosed (55). However, both US8 and US10 seem to be relatively inefficient in their activities, since they do not affect the steady-state levels of MHC I at the cell surface (2) or antigen presentation to T cells (32). The remaining two US6 family members, US7 and US9, have no known function (32).
In contrast to the extensive studies performed on the molecular functions of the HCMV proteins in vitro, their roles in the establishment and maintenance of virus infection in vivo are unknown. Provided that the RhCMV homologues are also functionally related to the HCMV US6 family, RhCMV could represent a model to evaluate the in vivo function of this protein family. In this report, we examine whether the RhCMV homologues interfere with the assembly and maturation of MHC I. We demonstrate that RhCMV contains functional homologues for US2, US3, US6, and US11. In the presence of RhCMV Rh182 and Rh189, nascent MHC I molecules were found to be degraded in a proteasome-dependent manner in tissue culture, suggesting that Rh182 and Rh189 are homologues of HCMV US2 and US11. The RhCMV ORF Rh185 represents a homologue of HCMV US6, since Rh185 inhibited peptide transport by TAP and, consequently, peptide loading of MHC I molecules. The US3 homologue, Rh185, was less efficient than HCMV US3 in retaining MHC I molecules in the ER but rather delayed MHC I exit, similar to the HCMV gene US10. No significant effect on MHC I assembly, transport, or surface expression was observed for the remaining US6 family-related molecules, Rh186 and Rh187. Given the low sequence similarity, the functional conservation of these molecular mechanisms is remarkable, indicating an important role for them in natural infection by primate CMVs. Our data further suggest that the RhCMV model can be applied as a homologous model to study the functions of the modulators of antigen presentation of HCMV.
MATERIALS AND METHODS
Virus, cells, and antibodies.
RhCMV strain 68-1 was obtained from Scott Wong (26). HeLa-Tet-Off cells were obtained from Clontech (Palo Alto, Calif.), and telomerized rhesus fibroblasts (TRFs) and U373-MG (human glioblastoma) and 293 cells were obtained from Jay Nelson. All cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 1× Pen-Strep-Glutamine (Invitrogen) unless otherwise noted. MHC I molecules were detected with the following antibodies: the monoclonal antibody W6/32, which is specific for MHC I heterodimers (10) (American Type Culture Collection); the monoclonal antibody HC-10, which recognizes free heavy chain (obtained from Hidde Ploegh); and antiserum K455, which recognizes both free and assembled human HLA and β2m (obtained from Per Peterson). Anti-hemagglutinin (HA) antibody was purchased from Sigma, and anti-protein-disulfide isomerase (PDI) antibody was from Stressgen. Conjugated secondary antibodies were purchased from Molecular Probes.
Plasmids.
RhCMV ORFs Rh182, -184, -185, -186, -187, and -189 were amplified by PCR from viral genomic DNA of RhCMV strain 68-1 (obtained from Scott Wong) using synthetic primers that spanned the start and stop codons of the respective ORFs and appropriate restriction sites (BglII or EcoRI at the 5′ end and Asp718 at the 3′ end). The amplified PCR products were inserted into vector pUHD10.1 (24) or pCDNA3.1 (Invitrogen) for expression in mammalian cells. HA-tagged versions of Rh182 to -189 were constructed using 3′-terminal PCR primers encoding the HA epitope in frame with the ORF and inserted into pCDNA3.1. Recombinant adenovirus (rAd) was generated using a plasmid-based recombinant system (27) with the following modifications. The promoter in the shuttle vector was replaced by a tetracycline-regulatable promoter (tet-off system) derived from plasmid pUHG10-3 (24) inserted as an AatII-XbaI fragment. In addition, a new synthetic polylinker (XhoI, SalI, ClaI, HindIII, EcoRV, BglII, SpeI, NotI, EagI, and XbaI) was introduced upstream of the XbaI restriction site. The resulting vector was named pShuttle-tetDXN.
ORFs Rh182, Rh185, Rh186, Rh187, and Rh189 were inserted into the BglII and XbaI sites of pShuttle-tetDXN. The integrity of each of the clones was confirmed by sequence analysis. Linearized pShuttle-tetDXN constructs, along with the plasmid carrying the adenovirus backbone, pAdEasy-1, were coelectroporated into electrocompetent Escherichia coli BJ8153 (a gift from Mike O'Conner, University of Minnesota) as described previously (27). Recombinant pAdEasy clones were selected on kanamycin (50 μg/ml) at 37°C overnight. The clones thus selected were analyzed for genomic integrity and recombination by restriction digestion.
Recombinant adenovirus.
For heterologous expression of RhCMV US6 homologues, we used the recombinant adenovirus system developed by He et al. (27). The replication-deficient adenovirus constructs lack the E1 and E3 regions and thus do not interfere with MHC I expression. Recombinant adenoviruses expressing Rh182, Rh185, Rh186, Rh187, and Rh189 were reconstituted in 293 cells as described previously (27). Briefly, 5 nmol of adenovirus plasmid DNA was digested with PacI (New England Biochemicals) and purified by phenol-chloroform extraction and ethanol precipitation. This vector was transfected into 293 cells using Effectene reagent (QIAGEN), and the infection was allowed to proceed until ∼70% of the cells showed a cytopathic effect. Virus was harvested from the cells by repeated freeze-thaw cycles. The virus stocks were amplified for a minimum of three rounds. The resulting virus was purified either over a 30% sucrose cushion (32) or by cesium chloride gradient centrifugation (27). Recombinant viral genomes were verified by restriction analysis and by PCR. Virus titers were determined on 293 cells by limiting dilution.
Flow cytometry and immunofluorescence.
MHC I surface expression was determined by flow cytometry using antibody W6/32. Transfected HeLa cells or TRFs were analyzed 42 h posttransfection, RhCMV-infected rhesus fibroblasts were analyzed up to 96 h postinfection, and recombinant adenovirus-transduced cells were analyzed 36 h posttransduction. Approximately 106 trypsinized cells were collected and washed by centrifugation prior to incubation with W6/32 (1:100 dilution in phosphate-buffered saline [PBS]-1% fetal calf serum) for 30 min on ice. After being washed three times, bound antibodies were detected with goat-anti-mouse phycoerythrin-conjugated secondary antibody (1:500; Dako). The stained cells were analyzed on a FACScalibur flow cytometer (BD Biosciences).
For immunofluorescence analysis, HeLa cells were transfected with HA-tagged ORFs. The cells were washed with PBS 40 h posttransfection, fixed with 2% paraformaldehyde for 20 min at room temperature, quenched with NH4Cl for 10 min, and permeabilized with 0.2% Triton X-100 for 5 min at room temperature. Nonspecific binding sites were blocked with 3% bovine serum albumin and 0.5% fish gelatin in PBS for 1 h at room temperature, followed by incubation with primary and secondary antibodies. Coverslips were mounted on slides and covered with Vectashield H-1200 plus DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories, Burlingame, Calif.). Fluorescence was visualized using an Axiovert-2 light microscope (Zeiss, Thornwood, N.Y.). All pictures were taken in monochrome, contrast enhanced, and false colored using Openlab software (Improvision, Lexington, Mass.).
Metabolic labeling and immunoprecipitation.
Cells were grown to 80 to 90% confluency in 60- or 100-mm-diameter tissue culture dishes (∼6 × 105 to 6 × 106 cells) and transfected as described above or infected with recombinant adenovirus at multiplicities of infection (MOI) as indicated for each experiment. The cells were incubated in serum-free and methionine-free medium for 1 h and metabolically labeled with [35S]cysteine-[35S]methionine (Amersham), either 60 or 100 μCi/dish, for the indicated times for each experiment. Proteasome inhibitors, either ZL3VS (15 to 50 μM) (kindly provided by Hidde Ploegh) or MG132 (10 μM) (Peptide International), were included in the starvation medium where indicated. After being labeled, the cells were washed three times with PBS and lysed immediately in PBS containing 1% Triton X-100 or 1% digitonin and protease inhibitors (Roche, Indianapolis, Ind.). The cell lysate was precleared with 20 μl of protein A-G agarose beads (Santa Cruz Biotechnology) for 1 h or overnight. The molecules of interest were immunoprecipitated by incubation with appropriate antibodies either for 1 h or overnight, and the immune complexes were captured by incubation for 1 h with 30 μl of protein A-G beads. The precipitated proteins were washed five times with either 0.1% NP-40 or 0.25% digitonin. All samples were boiled in Laemmli buffer, resolved on a 12% acrylamide gel, dried, and visualized by exposing them to X-ray film (Kodak BioMax MR).
Peptide transport assay.
HeLa cells or TRFs either mock transfected or transfected with Rh186 or HCMV US6 were collected 24 h posttransfection. The cells were permeabilized by adding ∼1 IU of activated Streptolysine O (Murex Diagnostics, Dartford, United Kingdom) in transport buffer (5 mM HEPES, pH 7.3, 30 mM KCl, 10 mM NaCl, 1 mM CaCl2, 2 mM EGTA, 2 mM MgCl2) and incubating them at 37°C for 10 to 20 min. The efficiency of permeabilization was assessed to be >90% by stain exclusion with Trypan blue dye (Sigma). About 5 × 106 permeabilized cells from each set were incubated at 37°C for 10 min with 5 μl of fluorescein-labeled peptide with the sequence CVNKTERAY(∼200 pM/μl) (a generous gift from Emmanuel Wiertz) in the presence of ATP (final concentration, 1 mM) or EDTA (final concentration, 1 mM). The transport reaction was terminated by adding ice-cold CI buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 500 mM NaCl, 2 mM MgCl2, 1% Triton X-100). The cells were lysed for 30 min at 4°C, and the nuclear debris was spun out by centrifugation at 11,000 × g at 4°C. The supernatant was incubated with 100 μl of concanavalin A Sepharose beads (Sigma) preequilibrated with the CI lysis buffer either for 2 h or overnight in the dark. The beads were washed twice with CI buffer, followed by two washes with wash buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl). The glycosylated peptide was eluted in 500 μl of elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, and 500 mM mannopyranosidase) by incubation with shaking for 1 h at room temperature. The recovered peptide was quantified by measuring fluorescence with 485-nm excitation and 530-nm emission filters (Bio-Rad).
RESULTS
RhCMV homologues of the HCMV US6 family.
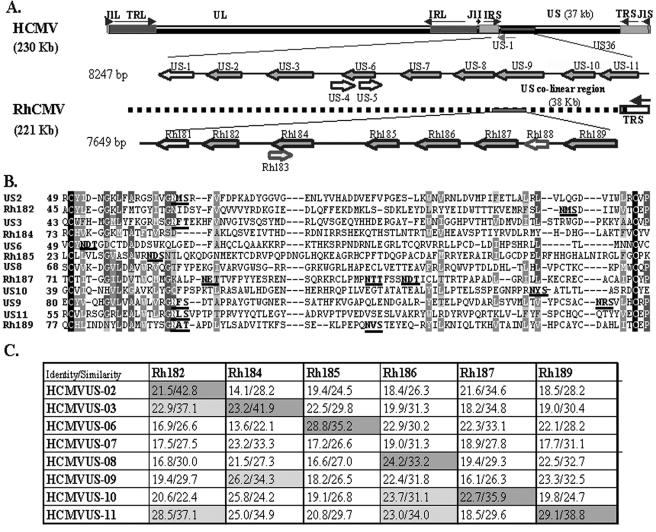
Hansen et al. reported that the genes Rh182, Rh184, Rh185, Rh186, Rh187, and Rh189 display homology to individual members of the US6 family of HCMV (26). All six ORFs encode predicted type I transmembrane-spanning proteins with a signal sequence, transmembrane domain, and C-terminal cytosolic domain. The relative locations of these genes in the genomes of RhCMV and HCMV are shown in Fig. 1A. RhCMV lacks internal repeats and thus does not have a unique short region. However, the location and orientation of ORFs Rh181 to -189 is similar to that of the region US1 to US11 in the HCMV genome. ClustalW alignment reveals that the central regions of all six RhCMV polypeptides share several conserved residues with HCMV US6 family members (Fig. 1B). Most importantly, all of the proteins share conserved cysteine residues that bracket an immunoglobulin (Ig)-like domain, similar to the structure of HCMV US2 (20). These signature sequences support the notion that all six RhCMV ORFs belong to the US6 family. To determine the relationship between the six US6-like glycoproteins encoded in region Rh181 to -189 and the eight US6 family members of HCMV (Fig. 1A), we aligned each of the HCMV ORFs with each of the RhCMV ORFs using the GAP analysis algorithm (14, 29). Identity scores were <30% in all cases, and similarity scores were <43% (Fig. 1C). These overall low scores rendered differences between individual alignments difficult to interpret and, in several instances, RhCMV ORFs showed the highest identity with one HCMV protein but the highest similarity with another. For instance, Rh182 displayed the highest similarity with HCMV US2 but the highest identity with HCMV US11. Rh184 demonstrated the highest similarity with US3 but higher identity with HCMV US9, US10, and US11. However, taking genome location into account, these alignments indicated that each RhCMV ORF showed either the highest percent identity or percent similarity with the US6 family member located in a similar position in the HCMV genome (Fig. 1C). The combination of location and sequence similarity suggests that Rh182, Rh184, Rh186, Rh187, and Rh189 are homologues of US2, US3, US6, US8, US10, and US11, respectively, and that the genome of RhCMV does not contain homologues for US7 and US9. This interpretation implies that all six ORFs of HCMV that display interference with MHC I assembly or transport (see the introduction) might be conserved in the RhCMV genome. To examine whether these functions are indeed conserved in RhCMV, or if the proteins have acquired different functions, we characterized the functions of all six RhCMV ORFs with respect to the assembly and transport of MHC I.
FIG. 1.
The US6 family of HCMV and RhCMV. (A) Schematic representation of HCMV and RhCMV genomes depicting the relative locations and orientations of US6 family members in the genome of HCMV and their homologues in RhCMV. US6 family genes are shown in dark gray. (B) Multiple sequence alignment (ClustalW) of the predicted Ig folds of all US6 family members of HCMV and RhCMV. Identical residues are shown as white on black background. Similar residues are shown as white on gray background. Conserved residues are represented as light gray on gray background, and weakly similar residues are dark gray on a light gray background. The N-linked glycosylation sites are in boldface letters and are underlined. (C) Pairwise sequence alignment between US6 family ORFs of HCMV and RhCMV. The percent similarity and identity were calculated by GAP alignment (PAM120 amino acid substitution matrix) with a gap penalty of 14 and an extension penalty of 2.
RhCMV Rh182 to Rh189 are glycoproteins that are located in the endoplasmic reticulum.
The RhCMV US6 family of genes encodes predicted type I transmembrane glycoproteins with a predicted amino-terminal signal sequence and a carboxy-terminal transmembrane domain that is followed by a short cytoplasmic domain. Each of the RhCMV ORFs also encodes at least one N-linked glycosylation site in its putative extracellular domain, with the exception of Rh184, which lacks a glycosylation site. The only ORF predicted to have more than one glycosylation site is Rh187, which encodes four such sites. The predicted molecular masses of the nonglycosylated Rh182 to -189 proteins (including the signal sequences) are as follows: Rh182, 23.1 kDa; Rh184, 20.6 kDa; Rh185, 18.9 kDa; Rh186, 28.2 kDa; Rh187, 25.5 kDa; and Rh189, 26.9 kDa.
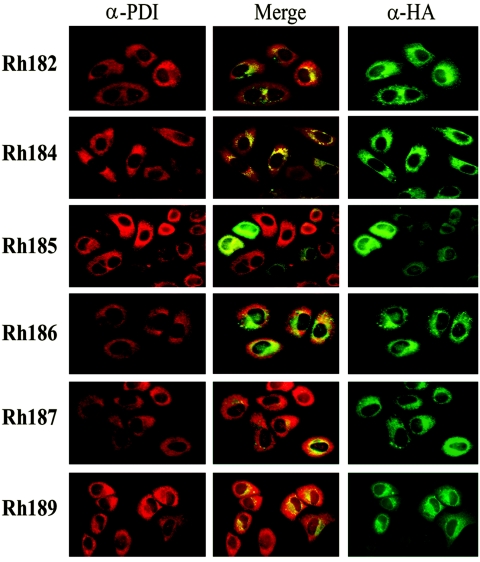
To determine the subcellular localizations of the US6 family-related ORFs of RhCMV, we generated epitope-tagged versions of all six of the ORFs and transfected the resulting constructs into HeLa cells. Immunofluorescence staining revealed a perinuclear staining pattern in all cases, consistent with localization in subcellular membranes of the exocytic pathway (Fig. 2). To examine whether this staining pattern corresponds to localization in the ER, we costained the cells with antibodies specific for PDI, an ER-resident protein. In all cases, we observed partial to complete costaining, indicating that these proteins are located, at least partially, in the ER.
FIG. 2.
Subcellular localization of RhCMV US6 family proteins. HA-tagged versions of RhCMV US6 family proteins were expressed in HeLa cells after transient transfection. The transfectants were costained with anti-HA antibody (green; right column) and with anti-PDI antibody (red; left column), which is located in the ER. The middle column shows the merger of the red and green channels.
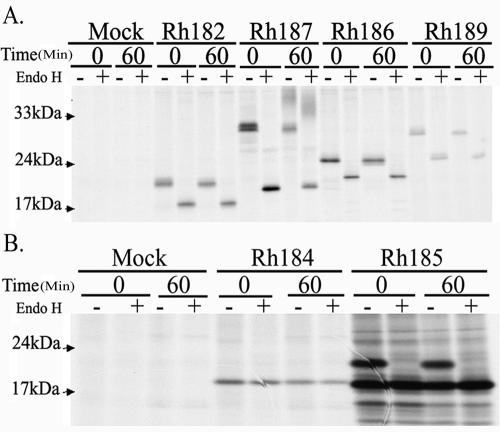
Since the high-mannose-type N-linked glycosylation of ER-resident glycoproteins is sensitive to digestion with endoglycosidase H (EndoH), we further examined whether the RhCMV proteins were sensitive to EndoH. HeLa cells were transfected with the HA-tagged constructs and metabolically labeled for 20 min, followed by a 60-min chase. Upon immunoprecipitation with anti-HA, the precipitates were treated with EndoH prior to electrophoretic separation. Rh182, Rh186, and Rh189 remained completely EndoH sensitive during the chase, suggesting that these proteins remain in the ER. The apparent molecular masses of Rh182 and Rh189 were also consistent with the predicted mass, considering that removal of a signal sequence reduces the mass by ∼2 kDa whereas the HA tag adds 1.1 kDa (Fig. 3A). However, the apparent mass of 24 kDa of the Rh186 protein was considerably lower than the predicted mass of 28.2 kDa for unknown reasons. The only protein that displayed any EndoH resistance was Rh187. At the beginning of the chase, this protein demonstrated a large shift in molecular mass upon EndoH treatment, consistent with predicted multiple glycosylation sites (Fig. 3A). During the chase, a high-molecular-mass species appeared that was only partially reduced in size by EndoH treatment (Fig. 3A), consistent with a highly glycosylated protein that reaches the Golgi apparatus but with only a fraction of oligosaccharides processed. This finding is reminiscent of the murine CMV glycoprotein M4, which has three glycosylation sites, two of which remain EndoH sensitive after reaching the surface (38). These data suggest that Rh187 is partially located in or recycles through a post-ER compartment. However, a large proportion of the Rh187 molecules seem to reside in the ER, as suggested both by immunofluorescence staining and by the fact that a significant portion of the molecules remained EndoH sensitive after 1 h of chase. The Rh185 protein occurred in two species during both the pulse and chase periods (Fig. 3B). The higher-mass form represents an EndoH-sensitive protein that comigrates with the lower-mass form upon EndoH treatment, suggesting that Rh185 is only partially glycosylated. As expected, the mobility of Rh184 did not change upon EndoH treatment, since the protein does not contain a glycosylation site. Thus, the subcellular localization of Rh184 could be deduced only from its colocalization with PDI in immunofluorescence assays, which suggests that it is located in the ER (Fig. 2). Rh184 also seemed to have a relatively short half-life, as indicated by the fact that less protein was immunoprecipitated at the end of the chase than at the beginning (Fig. 3B). After 2 h of chase, the protein could no longer be detected (data not shown). This short half-life could be related to the lack of glycosylation. Taken together, these data suggest that all US6-related proteins of RhCMV are located in the ER, with a subpopulation of Rh187 additionally reaching a post-ER compartment.
FIG. 3.
Glycosylation of RhCMV US6 family proteins. HeLa cells were transiently transfected with HA-tagged versions of the US6-related genes of RhCMV and labeled with [35S]cysteine-[35S]methionine for 30 min, followed by a 60-min chase where indicated. US6 family proteins were immunoprecipitated from 1% NP-40 lysates with monoclonal anti-HA antibody (Sigma). The immunoprecipitates were either left untreated (−) or treated (+) with EndoH prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography.
Reduction of MHC I surface levels by Rh182, Rh185, and Rh189.
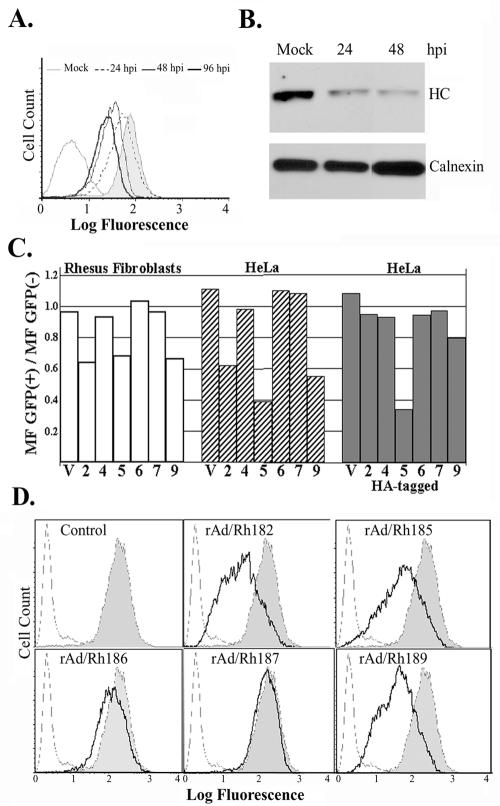
To determine whether MHC I steady-state levels are reduced in RhCMV-infected cells, surface and total levels of MHC I were examined in TRFs infected with RhCMV. Fibroblasts were mock infected or infected with RhCMV (MOI, 2) for 24, 48, or 96 h prior to flow cytometry with antibody W6/32. As shown in Fig. 4A, MHC I surface levels were reduced at 24 h and even further reduced at the later time points. A significant reduction of MHC I expression was also observed when total MHC I levels were measured in immunoblotting using a polyclonal MHC I-specific antiserum. These data are consistent with a mechanism by which RhCMV interferes with newly expressed MHC I molecules but does not remove surface-expressed MHC I molecules synthesized prior to infection.
FIG. 4.
MHC I expression in RhCMV-infected cells and in the presence of individual RhCMV US6 family proteins. (A) Flow cytometry of RhCMV-infected fibroblasts. TRFs were infected with RhCMV (MOI, 2) for the indicated times (hpi, hours postinfection), and surface levels were monitored with W6/32 antibody. (B) Immunoblot of MHC I and calnexin using cell lysates from TRFs infected with RhCMV (MOI, 2) for the indicated times. MHC I was detected with antiserum K455. Anti-calnexin antiserum was obtained from Stressgen. (C) RhCMV US6 family genes, either wild type or HA tagged, were cotransfected with a GFP expression vector into either HeLa cells or TRFs. MHC I surface expression was monitored by flow cytometry using the monoclonal antibody W6/32 at 42 h posttransfection. Transfection efficiencies, determined by GFP fluorescence, were ∼80 and 30% for HeLa cells and TRFs, respectively. The bars in the graph represent the ratio of the mean W6/32 fluorescence (MF) of GFP-positive cells to that of GFP-negative cells. The numbers (2, 4, 5, 6, 7, and 9) on the x axis represent Rh182, Rh184, Rh185, Rh186, Rh187, and Rh189, respectively, and V is the vector control. (D) MHC I surface expression upon transduction with adenovirus constructs. HeLa cells were infected with recombinant adenovirus (MOI, 25) for 36 h. As a control, cells were infected with tet-transactivator-expressing adenovirus (rtetA) (shaded). The cells were either stained with W6/32 (solid line) or unstained (broken line). Note the reduction of surface levels in cells transduced with rAd/Rh182, rAd/185, or rAd/Rh189.
To examine whether the RhCMV homologues interfere with the MHC I pathway, we initially analyzed cell surface expression of MHC I molecules by flow cytometry of cells transiently transfected with the RhCMV ORFs or transduced with recombinant adenovirus. Interference with assembly or transport is expected to reduce MHC I steady-state levels. Two different cell types were analyzed: rhesus fibroblasts, life extended by transfection with telomerase (TRF cells), and human HeLa cells. In addition to the native proteins, we also tested the HA-tagged versions. For transient-transfection experiments, cells were cotransfected with a plasmid encoding green fluorescent protein (GFP) at a ratio of 8:1 to distinguish between transiently transfected cells and nontransfected cells. The results are graphically depicted in Fig. 4C as the ratio between the mean W6/32 fluorescences of GFP-negative and GFP-positive cells. The efficiencies of transfection were 77 to 81% for HeLa cells and 30 to 32% for TRFs, as indicated by GFP fluorescence. As shown in Fig. 4C, in both TRFs and HeLa cells, transfection of Rh182, Rh185, and Rh189 resulted in a significant reduction of MHC I cell surface expression, suggesting that these three molecules inhibit expression of human and nonhuman primate MHC I. Interestingly, the HA-tagged version of Rh182 was nonfunctional despite excellent expression in transient transfection (Fig. 2 and 3). Also, the HA-tagged version of Rh189 seemed to be less active. Proteins encoded by ORFs Rh184, Rh186, and Rh187 had no effect on the surface expression of MHC I molecules. These proteins also did not interfere with MHC I steady-state levels when transfected in combination (data not shown). Reduction of MHC I surface levels was also observed when HeLa cells were transduced with recombinant adenovirus expressing Rh182, Rh185, and Rh189, whereas Rh186 and Rh187 had no significant effect (Fig. 4D). Rh184 has not yet been examined in an adenoviral expression system. Similar results were obtained with rhesus fibroblasts (data not shown). Thus, both transient transfection and adenovirus expression suggest that Rh182, Rh185, and Rh189 interfere with assembly or transport of human and rhesus MHC I molecules and that they are the RhCMV homologues of US2, US6, and US11.
Rh184 transiently retains MHC I molecules.
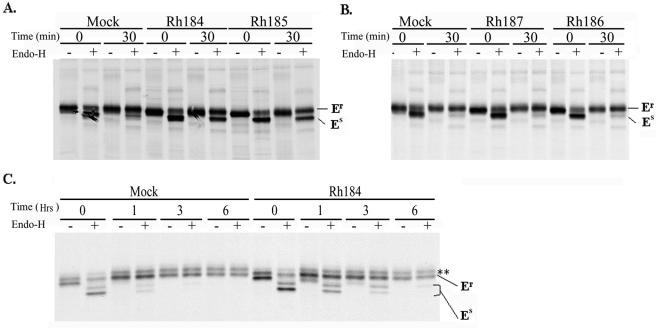
At least two of the HCMV US6 family molecules, US8 and US10, bind to MHC I but do not affect MHC I surface levels (2, 18, 32, 55). Examining the steady-state abundance of MHC I molecules at the cell surface might therefore fail to detect transient effects of US6-related proteins on newly synthesized MHC I molecules. To examine whether Rh184, Rh186, and Rh187 affect the maturation of newly synthesized MHC I molecules, we examined the acquisition of EndoH resistance by MHC I molecules in transiently transfected HeLa cells. No difference was observed between cells transfected with Rh186 or Rh187 and cells transfected with vector only (Fig. 5B). Thus, Rh187 does not seem to delay maturation of MHC I molecules, as observed for US10, its closest HCMV homologue (18). In contrast, a substantial portion of MHC I molecules were still in the EndoH-sensitive state in Rh184-transfected cells after a 30-min chase. At that time, all of the MHC I molecules in control cells had acquired EndoH resistance (Fig. 5A). Since these results were obtained in transiently transfected cells, it is likely that a substantially higher proportion of MHC I molecules, if not the entire population of MHC molecules, was retained in the ER in Rh184-transfected cells. This observation was reminiscent of HCMV US3, which retains MHC I in the ER of transfected cells (1, 34). Rh184 has the highest percentage of amino acid residues that are either identical or similar to US3 (Fig. 1B). However, unlike Rh184, MHC I surface expression is significantly reduced in cells transiently or stably transfected with US3 (1, 2). Therefore, we examined the fate of MHC I over longer chase periods. As shown in Fig. 5C, the EndoH-sensitive population of MHC I disappeared during 3- and 6-h chase periods in Rh184-transfected cells. In contrast, HCMV US3 retains MHC I molecules in an EndoH-sensitive state for prolonged periods (1). While the EndoH-resistant pool was diminished at the end of the chase, this was also observed in the mock-infected cells, presumably as a result of natural turnover. We conclude that Rh184 is unable to prevent the exit of MHC I molecules from the ER, as observed for HCMV US3, but delays MHC I maturation, as described for HCMV US10 (18).
FIG. 5.
Rh184 delays intracellular transport of MHC I molecules. HeLa cells were transiently transfected with the constructs shown and pulse-chase labeled as indicated. MHC I heterodimers were immunoprecipitated with W6/32, and the maturation state of N-linked glycosylation was determined by digestion with EndoH. (A) Retention of MHC I molecules in an EndoH-sensitive state by Rh184 and Rh185. (B) Rh186 and Rh187 do not interfere with acquisition of EndoH resistance by MHC I. (C) Pulse-labeled Rh184-transfected HeLa cells chased for a longer time. MHC I molecules become EndoH resistant after 6 h of chase. The asterisks indicate a nonspecific protein band. Er, EndoH resistant; Es, EndoH sensitive. The two Es bands in panel C indicate incomplete digestion by EndoH.
Inhibition of peptide translocation and peptide loading by Rh185.
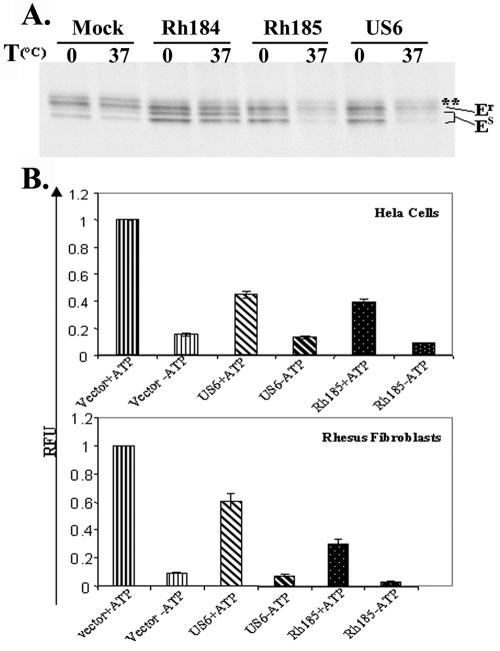
We observed a population of EndoH-sensitive MHC I molecules when HeLa cells were transiently transfected with Rh185, similar to Rh184 (Fig. 5A). Rh185 is most similar to HCMV US6 (Fig. 1B), which inhibits peptide loading of MHC I molecules by inhibiting TAP (2, 28, 40). Since empty MHC I molecules are retained by the MHC I-specific chaperone tapasin in the ER (15, 51), it was possible that intracellular transport of MHC I molecules was delayed as a consequence of a defect in peptide loading. To examine whether MHC I molecules are loaded with peptides, we subjected metabolically labeled lysates of Rh185-transfected cells to a temperature challenge for 1 h at 37°C. Empty MHC I heterodimers decay under such conditions and are no longer recognized by the dimer-specific antibody W6/32. As shown in Fig. 6A, a drastic reduction of W6/32-reactive MHC I molecules was observed in both HCMV US6- and Rh185-transfected cells. In contrast, MHC I molecules were resistant to temperature challenge in Rh184 transfectants. To examine whether MHC I instability was caused by inhibition of peptide translocation, we measured MHC-independent import of fluoresceinated peptides into the lumen of the ER (6). Since the peptides employed carried a glycosylation signal, transported peptides could be recovered with the lectin concanavalin A (45). Human HeLa and rhesus TRF cells were transfected with Rh185 or US6, and peptide import into the ER was measured in the absence or presence of ATP. As expected, depletion of ATP resulted in ∼90% reduction of recovered fluorescence in both cell types compared to cells transfected with vector plasmid (Fig. 6B). Transient transfection with HCMV US6 resulted in a 55% reduction of recovered fluorescence in HeLa cells and a 45% reduction in TRFs (the mean of three independent experiments). Similarly, a 67 or 72% reduction in peptide transport was observed in HeLa cells or TRFs, respectively, transfected with Rh185 (Fig. 6B). Since transfection of GFP plasmids indicated 70% transfection efficiency (not shown), these results indicate a complete inhibition of TAP transport by Rh185.
FIG. 6.
Rh185 is a TAP inhibitor. (A) MHC I molecules are unstable in Rh185 transfectants. Lysates from cells transfected with Rh184, Rh185, or HCMV US6 were collected after being pulse labeled for 20 min, followed by a 30-min chase. The lysates were subjected to a temperature (T) challenge of 1 h at 37°C. MHC I heterodimers were immunoprecipitated with W6/32. MHC I molecules were unstable in the presence of Rh185 and HCMV US6 but stable in Rh184 transfectants. Er, EndoH resistant; Es, EndoH sensitive. The asterisks indicate a nonspecific protein band. (B) Inhibition of peptide translocation by Rh185. Translocation of the fluoresceinated peptide CVNKTERAY into the ER of HeLa cells or TRFs transfected with Rh185 or HCMV US6 was measured at 24 h posttransfection in the presence (+) or absence (−) of ATP. The results are presented as the mean of three independent experiments with standard deviation. RFU, relative fluorescence units.
Rapid degradation of MHC I molecules in cells expressing Rh182 and Rh189.
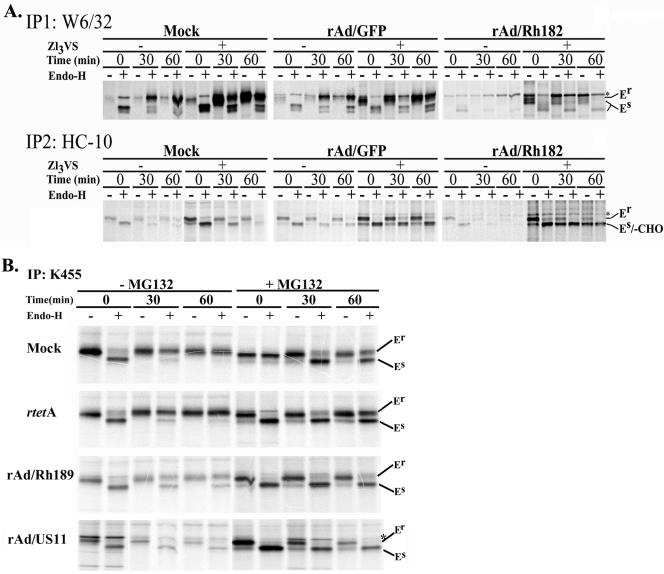
To examine how Rh182 and Rh189, the putative homologues of US2 and US11, achieve the reduction of MHC I surface expression (Fig. 4), we used an adenoviral expression system that allows efficient expression under the control of a tetracycline-regulatable promoter (see Materials and Methods). The effect of Rh182 and Rh189 expression on newly synthesized MHC I molecules was studied in U373-MG human glioblastoma cells, since these cells were used previously to examine the functions of HCMV US2 and US11 (59, 60). For controls, cells were either mock infected or infected with a GFP or tetracycline transactivator expressing adenovirus (rtetA). rAd/Rh182-transduced cells were metabolically labeled for 15 min and chased for 30 or 60 min prior to immunoprecipitation with the heterodimer-specific antibody W6/32. Compared to mock-infected and rAd/GFP-infected cells, significantly fewer MHC I heterodimers were recovered in cells transduced with rAd/Rh182 (Fig. 7A). Since this is reminiscent of the degradation of MHC I molecules observed in US2 transfectants (60), we examined whether MHC I molecules could be stabilized with the proteasomal inhibitor. Pretreatment with the proteasome inhibitor ZL3VS (60) resulted in increased recovery of assembled MHC I heterodimers from rAd/Rh182-transduced U373-MG cells. An increase in W6/32-reactive MHC I complexes was also observed in mock- or rAd/GFP-infected cells. However, this increase, which was likely caused by inhibiting the natural turnover of incompletely assembled molecules, was less pronounced than in rAd/Rh182-infected cells. To examine whether proteasome inhibitors stabilized a degradation intermediate that is not recoverable with the heterodimer-specific W6/32 antibody, we used the antibody HC-10, which specifically recognizes free MHC I heavy chains (50). HC-10-reactive heavy chains were immunoprecipitated from lysates that had been precleared with W6/32. In the absence of proteasome inhibitors, HC-10-reactive MHC I molecules of control transfectants decreased during the chase due to heterodimer formation. In Rh182-transduced cells, however, very little HC-10-reactive MHC I was observed during the pulse and almost no free heavy chain was present during the chase, consistent with efficient degradation mediated by Rh182. In the presence of proteasome inhibitors, we observed a stabilization of EndoH-sensitive free heavy chains in the control cells. As discussed above, this is most likely due to the inhibition of natural protein turnover. However, a much more pronounced stabilization was observed in rAd/Rh182-infected cells. Importantly, a low-molecular-weight species of MHC I that comigrated with EndoH-sensitive heavy chains was stabilized during the chase period. This species might correspond to a deglycosylated intermediate of MHC I degradation described previously for HCMV US2- and US11-transfected cells (59, 60). Similar results were obtained with rhesus TRFs (data not shown). The increased stabilization of free heavy chains compared to that of assembled heterodimers could indicate that Rh182 predominantly attacks MHC I molecules that have not yet assembled with β2m. Alternatively, MHC I heterodimers are disassembled during the degradation process mediated by Rh182. The occurrence of a deglycosylated intermediate also indicates that Rh182 reverse translocates the heavy chains into the cytosol, since the N-glycanase activity required for degradation resides in the cytosol (31). Taken together, these data indicate that, similar to HCMV US2, Rh182 destabilizes MHC I heavy chains for proteasomal destruction in the cytosol.
FIG. 7.
Degradation of nascent MHC I by Rh182 and Rh189. (A) Degradation of MHC I by Rh182. U373 cells were transduced with rAd/Rh182 (MOI, 100) plus rtetA (MOI, 20) or rAd/GFP (MOI, 120)for 18 h prior to metabolic labeling with [35S]Met for 15 min and chasing for the times indicated either in the absence (−) or the presence (+) of 25 μM proteasomal inhibitor ZL3VS (7). Newly assembled MHC I heterodimers were immunoprecipitated (IP) with the antibody W6/32. Remaining free heavy chains were precipitated with the monoclonal antibody HC-10 as shown. The maturation of MHC-I was assessed by EndoH digestion where indicated prior to electrophoretic separation. The gel containing rAd/Rh182 samples was exposed for twice as long as the mock and rAd/GFP samples. Note the appearance of a deglycosylated HC-reactive species in the presence of proteasome inhibitor (-CHO). The asterisks indicate a nonspecific band that comigrates with EndoH-resistant (Er) MHC I in some instances. EndoH sensitivity is labeled as in Fig. 5. (B) Degradation of MHC I molecules by Rh189. U373MG cells were transduced with rAd/US11 and rAd/Rh189 (each at an MOI of 100), together with rtetA (MOI, 20). Control cells were either mock treated or infected with rtetA (MOI, 120). Labeling conditions were as for panel A, except that the cells were pretreated for 4 h with (+) 10 μM MG132 prior to starvation and pulse-chase labeling. Both free and assembled MHC I molecules were immunoprecipitated with the polyclonal antiserum K455. The cells were treated with EndoH as indicated, except for lane 8 of the mock control, which was mistakenly not EndoH treated.
In a similar series of experiments, we compared the fates of MHC I molecules in U373-MG cells transduced with rAd/Rh189 or rAd/US11 (kindly provided by D. Johnson). The cells were metabolically labeled as described above, but MHC I molecules were immunoprecipitated with the antiserum K455, which recognizes both free and assembled MHC I heavy and light chains. As expected, the recovery of MHC I was reduced in U373-MG cells transduced with rAd/US11 compared to the mock-transduced or control adenovirus-transduced cells, particularly during the chase period (Fig. 7B). Heavy chains remained EndoH sensitive in US11-expressing cells prior to degradation, consistent with ER degradation of newly synthesized MHC I molecules (59), whereas they acquired EndoH resistance in mock-infected and control adenovirus-infected cells. Similar to US11, there was an overall reduction of MHC I recovery from rAd/Rh189-infected cells, particularly during the chase period, consistent with degradation of MHC I. In addition, MHC I also remained increasingly EndoH sensitive in Rh89-expressing cells, suggesting that Rh189 retained MHC I molecules prior to mediating their degradation. Importantly, increased recovery of MHC I molecules was observed in the presence of the proteasomal inhibitor MG132 in both rAd/US11- and rAd/Rh189-infected cells. Unlike Rh182, however, we did not observe deglycosylated HC degradation intermediates in the presence of proteasome inhibitors in cells transduced with either HCMV US11 or Rh189. Instead, proteasomal inhibition stabilized the EndoH-sensitive form of HCs. As discussed above, proteasome inhibition also prevented the natural turnover of MHC I in control cells, resulting in an increased EndoH-sensitive population during the chase. Thus, a portion of the EndoH-sensitive HC population recovered from US11- and Rh189-expressing cells in the presence of proteasome inhibitors could be a consequence of inhibiting normal MHC turnover. However, the difference between the total amounts of HC recovered in the presence of proteasome inhibition and in its absence was much more pronounced in Rh189- and US11-infected cells than in control cells. The increased HC recovery thus suggests that the US11- and Rh189-mediated HC degradation was prevented or decreased by proteasome inhibitors. Therefore, we conclude that Rh189 acts similarly to US11 by mediating the proteasomal degradation of MHC I molecules.
DISCUSSION
The β-herpesviruses are thought to have emerged prior to mammalian radiation, and therefore, viral evolution has accompanied mammalian speciation, so that CMV phylogeny generally parallels species phylogeny (44). Thus, the differences between the primary structure sequences in US6 family members of human and rhesus CMVs have accumulated over a period of 14 million years that separates humans from Old World nonhuman primates (23). During this time, considerable divergence has occurred in the corresponding genes of the two virus species, resulting in low primary structure homology (Fig. 1). In part, this divergence is probably a coadaptation to the significant divergence that occurred in the rhesus MHC compared to the human MHC (17). In addition, mutations might have occurred as a result of random drift. Nevertheless, our data suggest that the functions of the major viral modulators of MHC I antigen presentation identified in HCMV are conserved in RhCMV. For US2, US3, US6, and US11, we observed that each of the respective positional and sequence homologues interferes with MHC I assembly or transport in a manner similar to that of its HCMV homologue. Differences between HCMV and RhCMV were observed for Rh184, which did not reduce the steady-state levels of MHC I as efficiently as its HCMV counterpart. Interestingly, all RhCMV ORFs retained the ability to interfere with the maturation of human MHC I molecules, suggesting that they recognize conserved features within MHC molecules or within essential molecules of the peptide-loading machinery, such as TAP and tapasin. We conclude that Rh182, Rh184, Rh185, and Rh189 are orthologous to US2, US3, US6, and US11, respectively. Therefore, we suggest that these genes be referred to as RhUS2, RhUS3, RhUS6, and RhUS11. The relationship of Rh186 and Rh187 to the remaining US6 family members of HCMV has not been firmly established. US8 was shown to bind to MHC I and to be partially located in endosomes (55). However, Rh186, which showed the closest homology to US8, seemed to be an ER-resident glycoprotein. Rh187 became partially EndoH resistant but did not seem to be located in the endosomal compartment. Neither Rh186 nor Rh187 interfered with MHC I maturation.
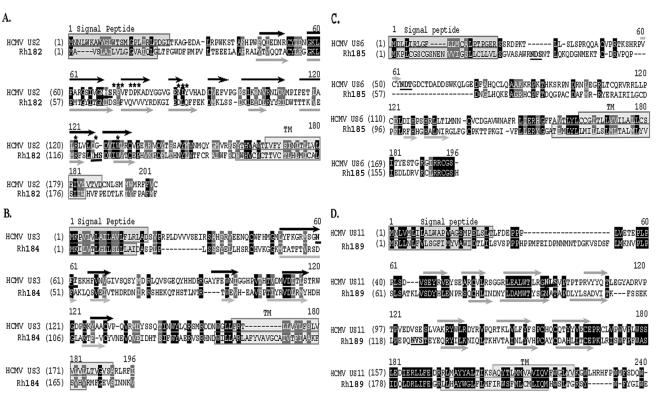
The observed functional conservation despite considerable sequence divergence might help to delineate functionally important residues in the orthologous sequences. Alignments between each pair of orthologues are shown in Fig. 8. For HCMV US2, it was previously shown that the lumenal domain is sufficient to bind to MHC I (12, 21). However, the cytosolic domain and the transmembrane domain were shown to be absolutely necessary for US2 to mediate MHC I degradation (12, 19). A function of the cytosolic tail is also supported for Rh182, since carboxy-terminal tagging rendered the molecule nonfunctional (Fig. 4). Interestingly, replacement of the US3 carboxy-terminal domain with that of US2 transfers the ability to mediate MHC I degradation to US3, suggesting that the lumenal domain mediates binding whereas the carboxy-terminal tail mediates degradation (13). Therefore, it is expected that very few residues in the carboxy terminus (including the membrane-proximal domain on the lumenal side and the transmembrane domain, as well as the cytosolic tail) of Rh182 are conserved compared to HCMV US2 (Fig. 8A). This difference renders it less likely that this region of US2 interacts with a highly conserved cellular protein, as observed for US11 (see below), and is consistent with different mechanisms of action in US2 and US11. In contrast, the lumenal domain of US2 contains several conserved sequence motifs, including residues that were found to be in direct contact with MHC I, as well as residues that were defined as “core” residues, i.e., >90% buried in US2 (20). Secondary-structure predictions further suggest that the Ig-like fold of six beta-sheets is also conserved between HCMV US2 and RhCMV Rh182. Notably, in HCMV US2, the glycosylation site is located between beta-sheets B and C, which places it well outside the MHC I interaction domain (20), whereas the glycosylation site is located between beta-sheets F and G in Rh182, thus overlapping with residues that were previously mapped to interact with MHC I (Fig. 8A). The consequences of this different placement of the glycosylation site for the interaction of Rh182 with MHC I remain to be investigated.
FIG. 8.
Sequence comparisons of orthologous immune modulator pairs of RhCMV and HCMV. Sequences of Rh182 (RhUS2), Rh184 (RhUS3), Rh185 (RhUS6), and Rh189 (RhUS11) were aligned with their corresponding orthologue of HCMV using GAP alignment (PAM120) with a gap penalty of 14 and an extension penalty of 2. Signal peptides and transmembrane regions are boxed, identical sequences are white on black, and conservative changes are white on gray. Glycosylation sites are boldface and underlined. The asterisks indicate HCMV US2 residues that interact with MHC I (20). The black arrows indicate experimentally confirmed beta-sheets in US2 (A, B, C, C′, D, E, F, and G) (20). The gray arrows indicate predicted beta-sheets.
The alignments of US3 and US6 to their RhCMV orthologues revealed a surprisingly low homology in the Ig-like domains of these molecules, suggesting that the overall structure rather than sequence motifs is responsible for their function. Interestingly, sequence conservation occurred in regions of the molecules that are not expected to be important for their function as MHC I modulators. For instance, sequences adjacent to and within the signal peptide of Rh184 are conserved (Fig. 8B). Since this sequence is cleaved upon translocation, this conserved sequence is not expected to play a role in the interaction with MHC I and tapasin. Potentially, the signal peptide itself could be involved in other immunomodulatory mechanisms, as described for the signal peptide of UL40 (56, 57). Also unexpected is the conservation of the carboxy-terminal amino acids in Rh185. Seven of the eight amino acids in HCMV US6 are also found in Rh185 (Fig. 8C). However, previous data suggested that the carboxy-terminal tail of US6 is dispensable for MHC I downregulation (2). It is possible that this sequence motif optimizes US6 function, which would have been missed in the previous study, which relied on overexpression and was not performed in the context of viral infection. Given the high conservation of this amino acid stretch, it can be assumed that the region plays an important, but yet to be defined, role in the TAP-inhibitory function of US6, or US6 could perform an as-yet-unknown function that involves its cytosolic tail.
The highest sequence conservation among the orthologues was observed for US11. Pairwise alignment clearly indicates multiple sequence motifs that are conserved within the lumenal domain, as well as the transmembrane domain. Among the conserved intramembrane residues is a central glutamine that is essential for the interaction of US11 with the cellular protein derlin 1 (41, 61) (Fig. 8C). This cellular protein is conserved in yeast and belongs to an ancient mechanism that disposes of misfolded proteins. The high conservation of the cellular mechanism used by US11 might explain its high sequence conservation. Moreover, the comparably close sequence relationship renders it highly likely that RhUS11 functions similarly to HCMV US11, despite our inability to observe a degradation intermediate.
Our study suggests that interference with MHC I assembly and transport by multiple viral proteins is highly conserved between RhCMV and HCMV. The functional conservation despite structural divergence further suggests important structural characteristics of individual members of the US6 family that can be tested experimentally in the future. Furthermore, the capability of studying RhCMV infections in nonhuman primates will further enable us to evaluate the importance of individual MHC I modulatory mechanisms for viral immune evasion in vivo.
Acknowledgments
We are grateful to Hidde Ploegh, Per Peterson, Emmanuel Wiertz, Bert Vogelstein, Mike O'Conner, David Johnson, Nag Hedge, Jay Nelson, and Scott Wong for providing reagents used in this study. We also thank David Johnson for critical reading of the manuscript.
This work was supported by a grant-in-aid from the American Heart Association (0255977Z) and Oregon National Primate Center Grant RR00163 (to K.F.) and a grant from the Korean Ministry of Health and Welfare, 03-PJ1-PG3-21200, 0003 (to K.A.).
REFERENCES
- 1.Ahn, K., A. Angulo, P. Ghazal, P. A. Peterson, Y. Yang, and K. Früh. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, K., A. Gruhler, B. Galocha, T. R. Jones, E. J. Wiertz, H. L. Ploegh, P. A. Peterson, Y. Yang, and K. Früh. 1997. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity 6:613-621. [DOI] [PubMed] [Google Scholar]
- 3.Asher, D. M., C. J. Gibbs, Jr., D. J. Lang, D. C. Gajdusek, and R. M. Chanock. 1974. Persistent shedding of cytomegalovirus in the urine of healthy Rhesus monkeys. Proc. Soc. Exp. Biol. Med. 145:794-801. [DOI] [PubMed] [Google Scholar]
- 4.Barel, M. T., M. Ressing, N. Pizzato, D. van Leeuwen, P. Le Bouteiller, F. Lenfant, and E. J. Wiertz. 2003. Human cytomegalovirus-encoded US2 differentially affects surface expression of MHC class I locus products and targets membrane-bound, but not soluble HLA-G1 for degradation. J. Immunol. 171:6757-6765. [DOI] [PubMed] [Google Scholar]
- 5.Bitmansour, A. D., S. L. Waldrop, C. J. Pitcher, E. Khatamzas, F. Kern, V. C. Maino, and L. J. Picker. 2001. Clonotypic structure of the human CD4+ memory T cell response to cytomegalovirus. J. Immunol. 167:1151-1163. [DOI] [PubMed] [Google Scholar]
- 6.Blevitt, J. M., K. Früh, C. Glass, M. R. Jackson, P. A. Peterson, and S. Huang. 1999. A fluorescence-based high throughput screen for the transporter associated with antigen processing. J. Biomol. Screen. 4:87-91. [DOI] [PubMed] [Google Scholar]
- 7.Bogyo, M., M. Gaczynska, and H. L. Ploegh. 1997. Proteasome inhibitors and antigen presentation. Biopolymers 43:269-280. [DOI] [PubMed] [Google Scholar]
- 8.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 9.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa.
- 10.Brodsky, F. M., P. Parham, C. J. Barnstable, M. J. Crumpton, and W. F. Bodmer. 1979. Monoclonal antibodies for analysis of the HLA system. Immunol. Rev. 47:3. [DOI] [PubMed] [Google Scholar]
- 11.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Browne, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169, p. 125-171. In J. K. McDougall (ed.), Cytomegaloviruses. Springer-Verlag, Berlin, Germany. [DOI] [PubMed]
- 12.Chevalier, M. S., G. M. Daniels, and D. C. Johnson. 2002. Binding of human cytomegalovirus US2 to major histocompatibility complex class I and II proteins is not sufficient for their degradation. J. Virol. 76:8265-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevalier, M. S., and D. C. Johnson. 2003. Human cytomegalovirus US3 chimeras containing US2 cytosolic residues acquire major histocompatibility class I and II protein degradation properties. J. Virol. 77:4731-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen, A. C., and S. Henikoff. 1992. Fact and fiction in alignment. Nature 358:271. [DOI] [PubMed] [Google Scholar]
- 15.Cresswell, P., N. Bangia, T. Dick, and G. Diedrich. 1999. The nature of the MHC class I peptide loading complex. Immunol. Rev. 172:21-28. [DOI] [PubMed] [Google Scholar]
- 16.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 17.Daza-Vamenta, R., G. Glusman, L. Rowen, B. Guthrie, and D. E. Geraghty. 2004. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 14:1501-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furman, M. H., N. Dey, D. Tortorella, and H. L. Ploegh. 2002. The human cytomegalovirus US10 gene product delays trafficking of major histocompatibility complex class I molecules. J. Virol. 76:11753-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furman, M. H., H. L. Ploegh, and D. Tortorella. 2002. Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules. J. Biol. Chem. 277:3258-3267. [DOI] [PubMed] [Google Scholar]
- 20.Gewurz, B. E., R. Gaudet, D. Tortorella, E. W. Wang, H. L. Ploegh, and D. C. Wiley. 2001. Antigen presentation subverted: structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. USA 98:6794-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gewurz, B. E., E. W. Wang, D. Tortorella, D. J. Schust, and H. L. Ploegh. 2001. Human cytomegalovirus US2 endoplasmic reticulum-lumenal domain dictates association with major histocompatibility complex class I in a locus-specific manner. J. Virol. 75:5197-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gillespie, G. M., M. R. Wills, V. Appay, C. O'Callaghan, M. Murphy, N. Smith, P. Sissons, S. Rowland-Jones, J. I. Bell, and P. A. Moss. 2000. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8+ T lymphocytes in healthy seropositive donors. J. Virol. 74:8140-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman, M., C. A. Porter, J. Czelusniak, S. L. Page, H. Schneider, J. Shoshani, G. Gunnell, and C. P. Groves. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9:585-598. [DOI] [PubMed] [Google Scholar]
- 24.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruhler, A., P. A. Peterson, and K. Früh. 2000. Human cytomegalovirus immediate early glycoprotein US3 retains MHC class I molecules by transient association. Traffic 1:318-325. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, S. G., L. I. Strelow, D. C. Franchi, D. G. Anders, and S. W. Wong. 2003. Analysis of the complete DNA sequence of rhesus cytomegalovirus. J. Virol. 77:6620-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengel, H., J. O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 29.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewitt, E. W., S. S. Gupta, and P. J. Lehner. 2001. The human cytomegalovirus gene product US6 inhibits ATP binding by TAP. EMBO J. 20:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirsch, C., D. Blom, and H. L. Ploegh. 2003. A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22:1036-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huber, M. T., R. Tomazin, T. Wisner, J. Boname, and D. C. Johnson. 2002. Human cytomegalovirus US7, US8, US9, and US10 are cytoplasmic glycoproteins, not found at cell surfaces, and US9 does not mediate cell-to-cell spread. J. Virol. 76:5748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, T. R., L. K. Hanson, L. Sun, J. S. Slater, R. M. Stenberg, and A. E. Campbell. 1995. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 69:4830-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones, T. R., E. J. Wiertz, L. Sun, K. N. Fish, J. A. Nelson, and H. L. Ploegh. 1996. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc. Natl. Acad. Sci. USA 93:11327-11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaur, A., M. D. Daniel, D. Hempel, D. Lee-Parritz, M. S. Hirsch, and R. P. Johnson. 1996. Cytotoxic-T-lymphocyte responses to cytomegalovirus in normal and simian immunodeficiency virus-infected rhesus macaques. J. Virol. 70:7725-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaur, A., C. L. Hale, B. Noren, N. Kassis, M. A. Simon, and R. P. Johnson. 2002. Decreased frequency of cytomegalovirus (CMV)-specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J. Virol. 76:3646-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kern, F., T. Bunde, N. Faulhaber, F. Kiecker, E. Khatamzas, I. M. Rudawski, A. Pruss, J. W. Gratama, R. Volkmer-Engert, R. Ewert, P. Reinke, H. D. Volk, and L. J. Picker. 2002. Cytomegalovirus (CMV) phosphoprotein 65 makes a large contribution to shaping the T cell repertoire in CMV-exposed individuals. J. Infect. Dis. 185:1709-1716. [DOI] [PubMed] [Google Scholar]
- 38.Kleijnen, M. F., J. B. Huppa, P. Lucin, S. Mukherjee, H. Farrell, A. E. Campbell, U. H. Koszinowski, A. B. Hill, and H. L. Ploegh. 1997. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 16:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, S., J. Yoon, B. Park, Y. Jun, M. Jin, H. C. Sung, I. H. Kim, S. Kang, E. J. Choi, B. Y. Ahn, and K. Ahn. 2000. Structural and functional dissection of human cytomegalovirus US3 in binding major histocompatibility complex class I molecules. J. Virol. 74:11262-11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lehner, P. J., J. T. Karttunen, G. W. Wilkinson, and P. Cresswell. 1997. The human cytomegalovirus US6 glycoprotein inhibits transporter associated with antigen processing-dependent peptide translocation. Proc. Natl. Acad. Sci. USA 94:6904-6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lilley, B. N., and H. L. Ploegh. 2004. A membrane protein required for dislocation of misfolded proteins from the ER. Nature 429:834-840. [DOI] [PubMed] [Google Scholar]
- 42.Loenen, W. A., C. A. Bruggeman, and E. J. Wiertz. 2001. Immune evasion by human cytomegalovirus: lessons in immunology and cell biology. Semin. Immunol. 13:41-49. [DOI] [PubMed] [Google Scholar]
- 43.London, W. T., A. J. Martinez, S. A. Houff, W. C. Wallen, B. L. Curfman, R. G. Traub, and J. L. Sever. 1986. Experimental congenital disease with simian cytomegalovirus in rhesus monkeys. Teratology 33:323-331. [DOI] [PubMed] [Google Scholar]
- 44.McGeoch, D. J., S. Cook, A. Dolan, F. E. Jamieson, and E. A. Telford. 1995. Molecular phylogeny and evolutionary timescale for the family of mammalian herpesviruses. J. Mol. Biol. 247:443-458. [DOI] [PubMed] [Google Scholar]
- 45.Neefjes, J. J., F. Momburg, and G. J. Hämmerling. 1993. Selective and ATP-dependent translocation of peptides by the MHC-encoded transporter. Science 261:769-771. [DOI] [PubMed] [Google Scholar]
- 46.Park, B., Y. Kim, J. Shin, S. Lee, K. Cho, K. Früh, and K. Ahn. 2004. Human cytomegalovirus inhibits tapasin-dependent peptide loading and optimization of the MHC class I peptide cargo for immune evasion. Immunity 20:71-85. [DOI] [PubMed] [Google Scholar]
- 47.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2705. In P. M. Howley, D. M. Knipe, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 48.Pitcher, C. J., S. I. Hagen, J. M. Walker, R. Lum, B. L. Mitchell, V. C. Maino, M. K. Axthelm, and L. J. Picker. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168:29-43. [DOI] [PubMed] [Google Scholar]
- 49.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rein, R. S., G. H. Seemann, J. J. Neefjes, F. M. Hochstenbach, N. J. Stam, and H. L. Ploegh. 1987. Association with beta 2-microglobulin controls the expression of transfected human class I genes. J. Immunol. 138:1178-1183. [PubMed] [Google Scholar]
- 51.Schoenhals, G. J., R. M. Krishna, A. G. Grandea III, T. Spies, P. A. Peterson, Y. Yang, and K. Früh. 1999. Retention of empty MHC class I molecules by Tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. 18:743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sepkowitz, K. A. 2002. Opportunistic infections in patients with and patients without Acquired Immunodeficiency Syndrome. Clin. Infect. Dis. 34:1098-1107. [DOI] [PubMed] [Google Scholar]
- 53.Söderberg-Naucler, C., and V. C. Emery. 2001. Viral infections and their impact on chronic renal allograft dysfunction. Transplantation 71:SS24-SS30. [PubMed] [Google Scholar]
- 54.Tarantal, A. F., M. S. Salamat, W. J. Britt, P. A. Luciw, A. G. Hendrickx, and P. A. Barry. 1998. Neuropathogenesis induced by rhesus cytomegalovirus in fetal rhesus monkeys (Macaca mulatta). J. Infect. Dis. 177:446-450. [DOI] [PubMed] [Google Scholar]
- 55.Tirabassi, R. S., and H. L. Ploegh. 2002. The human cytomegalovirus US8 glycoprotein binds to major histocompatibility complex class I products. J. Virol. 76:6832-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tomasec, P., V. M. Braud, C. Rickards, M. B. Powell, B. P. McSharry, S. Gadola, V. Cerundolo, L. K. Borysiewicz, A. J. McMichael, and G. W. Wilkinson. 2000. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287:1031. [DOI] [PubMed] [Google Scholar]
- 57.Ulbrecht, M., S. Martinozzi, M. Grzeschik, H. Hengel, J. W. Ellwart, M. Pla, and E. H. Weiss. 2000. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164:5019-5022. [DOI] [PubMed] [Google Scholar]
- 58.Vogel, P., B. J. Weigler, H. Kerr, A. G. Hendrickx, and P. A. Barry. 1994. Seroepidemiologic studies of cytomegalovirus infection in a breeding population of rhesus macaques. Lab. Anim. Sci. 44:25-30. [PubMed] [Google Scholar]
- 59.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 60.Wiertz, E. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 61.Ye, Y., Y. Shibata, C. Yun, D. Ron, and T. A. Rapoport. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429:841-847. [DOI] [PubMed] [Google Scholar]