Abstract
Due to the cardiotoxicity of doxorubicin (DOX), its clinical application is limited. Lipid peroxidation caused by excessive ferrous iron is believed to be a key molecular mechanism of DOX-induced cardiomyopathy (DIC). Dexrazoxane (DXZ), an iron chelator, is the only drug approved by the FDA for reducing DIC, but it has many side effects and cannot be used as a preventive drug in clinical practice. Single-nucleus RNA sequencing (snRNA-seq) analysis identified myocardial and epithelial cells that are susceptible to DOX-induced ferroptosis. The glutathione peroxidase 4 (GPX4) activator selenomethione (SeMet) significantly reduced polyunsaturated fatty acids (PUFAs) and oxidized lipid levels in vitro. Consistently, SeMet significantly decreased DOX-induced lipid peroxidation in H9C2 cells and mortality in C57BL/6 mice compared to DXZ, ferrostatin-1, and normal saline. SeMet can effectively reduce serum markers of cardiac injury in C57BL/6 mice and breast cancer patients. Depletion of the GPX4 gene in C57BL/6 mice resulted in an increase in polyunsaturated fatty acid (PUFA) levels and eliminated the protective effect of SeMet against DIC. Notably, SeMet exerted antitumor effects on breast cancer models with DOX while providing cardiac protection for the same animal without detectable toxicities. These findings suggest that pharmacological activation of GPX4 is a valuable and promising strategy for preventing the cardiotoxicity of doxorubicin.
Keywords: DOX, Cardiotoxicity, GPX4, scRNA-seq, Selenium
Abbreviations
- DOX
doxorubicin
- DIC
DOX-induced cardiomyopathy
- DXZ
dexrazoxane
- scRNA-seq
single-cell RNA sequencing
- snRNA-seq
single-nucleus RNA sequencing
- DC
dendritic cells
- SMC
smooth muscle cells
- UFAs
unsaturated fatty acids
- PUFAs
polyunsaturated fatty acids
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PC
phosphatidylcholine
- AA
arachidonic acid
- GSH
glutathione
- GPX4
glutathione peroxidase 4
- SeMet
selenomethione
- Se
selenium
- NS
normal saline
- DCM
dilated cardiomyopathy
- FDA
Food and Drug Administration
- UMAP
uniform manifold approximation and projection
- DEGs
differentially expressed genes
- CM
cardiomyocytes
- ECG
electrocardiogram
- GPX4hetko
heterozygous GPX4-knockout
- LVEF
left ventricular ejection fraction
- LVFS
left ventricular fraction shortening
- HR
heart rate
- VERP
ventricular effective refractory period
- VF
ventricular fibrillation
- AF
atrial fibrillation
- ICP‒MS
inductively coupled plasma‒mass spectrometry
- Nrf2
nuclear factor erythroid 2-related factor 2
1. Introduction
Doxorubicin (DOX) is a foundation of chemotherapy regimens for some malignant tumors, including breast cancer, lymphoma, and soft tissue sarcoma [1]. However, cardiotoxicity caused by DOX is one of the most serious adverse effects and has dose-dependent and cumulative characteristics, which greatly limits its clinical application. The incidence of DIC is as high as 17% and is mainly characterized as dilated cardiomyopathy, which is progressive and irreversible and can lead to heart failure in severe cases [2]. Cardiovascular disease is one of the most common complications among cancer survivors and a major cause of death, especially in the long-term follow-up of child cancer survivors [3,4]. Dexrazoxane (DXZ) is the only drug approved by the US Food and Drug Administration (FDA) to reduce the cardiotoxicity of DOX. However, it still has some shortcomings: it can aggravate the bone marrow suppression caused by chemotherapy drugs, affect the antitumor effect of DOX, and may lead to secondary malignant tumors after long-term use [5,6]. Therefore, there is an urgent need to explore new strategies to prevent DIC in clinical practice. DOX-induced excessive accumulation of lipid peroxides in mitochondria is believed to play a critical role in the progression of DIC [7,8]. Thus, the pharmacological activation of GPX4 to suppress lipid peroxidation by selenium compounds is potentially valuable for preventing DIC.
2. Results
2.1. Single-cell characterization of cardiac cells in DIC
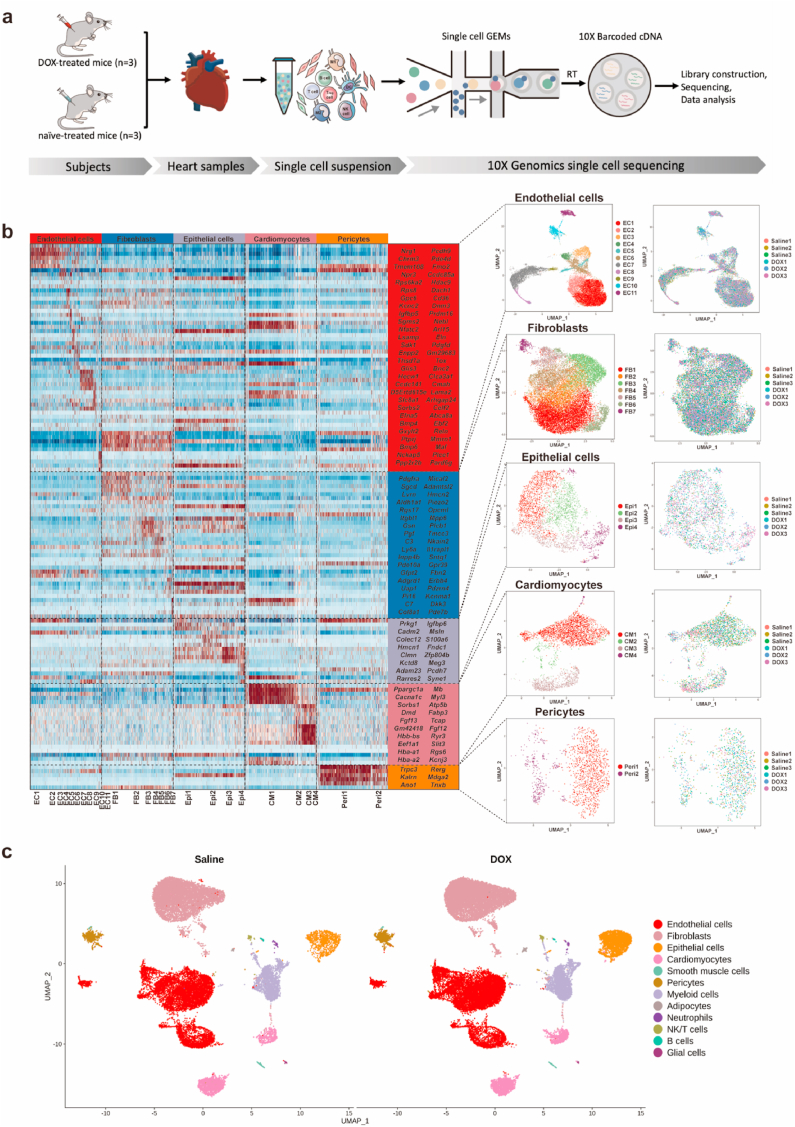
To elucidate the mechanism of DOX-induced myocardial damage and screen new drugs with few side effects to prevent DIC, we conducted single-nucleus RNA sequencing (snRNA-seq) on heart samples from DOX-treated mice (DOX 20 mg/kg, n = 3) and untreated mice (n = 3) (Fig. 1a). Using unbiased clustering analysis, we identified and visualized 32 clusters, which included twelve different cell types (Fig. 1b, Extended Data Fig. 1a–d). Five nonimmune cardiac cell populations were identified by examining the expression of known lineage markers in cardiomyocytes (Ryr2, Trdn, Myh7, Myh6, Ttn), fibroblasts (Pdgfra, Ckap4, Col1a1), endothelial cells (Pecam1, Vwf, Cdh5, Cd93, Ldb2, Tie1), adipocytes (Plin1, Pparg, Adam12), smooth muscle cells (Acta2, Tagln, Myh11), epithelial cells (Wt1, Tbx18, Krt18, Msln), and pericytes (Pdprb) (Fig. 1c). Most studies on DOX-related cardiotoxicity are based on cardiomyocytes. Using unsupervised dimensionality reduction and clustering, we identified four cardiomyocyte subclusters that exhibited different gene expression profiles (Fig. 1c). To further study the characteristics of cardiomyocytes after chemotherapy, we compared their transcriptional profiles and pathway enrichment between the two groups. Here, we found that after DOX treatment, hypertrophic cardiomyopathy, dilated cardiomyopathy, adrenergic signaling in cardiomyocytes, cardiac muscle contraction, arrhythmogenic right ventricular cardiomyopathy, and ferroptosis pathways were significantly enriched (Fig. 2a, Extended Data Fig. 1e). To determine which populations were most susceptible to ferroptosis, pathway enrichment of DEGs was conducted in four cardiomyocyte (CM) subclusters. We found that ferroptosis pathways were significantly enriched in the CM1 and CM3 clusters (Extended Data Fig. 3a). We further examined the expression levels of ferroptosis-related genes that were previously reported [9]. We confirmed that after DOX treatment, ferroptosis-related genes such as Gclm (glutamate-cysteine ligase modifier subunit), Fth1 (ferritin heavy chain 1), Hmox1 (heme oxygenase-1), Slc39a14 (solute carrier family 39 member 14), Ftl1 (ferritin light polypeptide 1), Nqo1(NAD(P)H quinone oxidoreductase 1), Cp (ceruloplasmin), Slc39a8 (solute carrier family 39 member 8), Map1lc3b (microtubule-associated protein 1 light chain 3 beta), and Ncoa4 (nuclear receptor coactivator 4) were significantly upregulated in cardiomyocytes and epithelial cells (Fig. 2b–g, Extended Data Fig. 3a and b, Extended Data Tables 1–4). To further address the alterations in immune cell types, seven subclusters were identified based on their genetic markers, including four myeloid cell populations (Wt1, Fcgr1), one neutrophil population (S100a8, S1009a), one NK/T-cell population (Cd3d, Cd3e), and one B-cell population (Ms4a1, Cd79a). Among them, myeloid cells were further divided into subgroups, including one monocyte population (Itgal), two dendritic cell populations (H2-Eb1, H2-Ab1), four macrophage populations (Adgre1), and one proliferating cell population (Top2a, Mki67) (Extended Data Fig. 2). The infiltration and activation of neutrophils contribute to DIC [10], and pharmacological inhibition of neutrophils significantly prevents the cardiotoxic effects of DOX [11]. The acute inflammatory response to DOX is associated with the apoptosis of monocytes and macrophages [12,13]. DOX treatment disrupts the cardiac M1/M2 macrophage balance and suppresses M2 macrophage differentiation [14], which plays a critical role in cardiac injury and repair. M2 macrophage transplantation alleviated DOX-induced cardiac apoptosis and remodeling [15,16]. More studies are needed to illuminate the relationship between cardiac immune cells and DIC in the future. Taken together, our snRNA-seq analysis identified cardiomyocytes and epithelial cells as the main specific cell types that contribute to DOX-induced ferroptosis.
Fig. 1.
Single-nucleus profiling of cardiac cells during DOX treatment. (a) Schematic diagram showing the single nucleus RNA-sequencing (snRNA-seq) experimental design and analytical workflow. (b) Heatmap of the expression of cell type-specific markers in the no-immune cell atlas (left) and uniform manifold approximation and projection (UMAP) plots of cardiac no-immune cells, colored by clusters (center) and treatment (right). (c) UMAP plots of cardiac cells from mice treated with DOX (20 mg/kg) or saline.
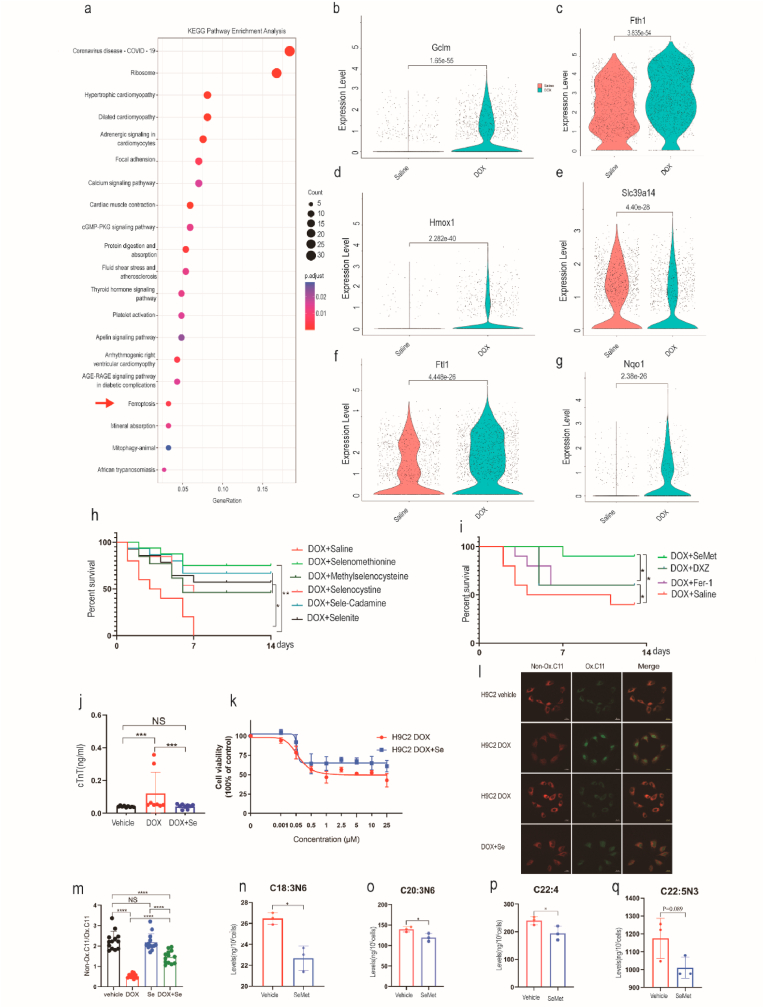
Fig. 2.
Ferroptosis is associated with DOX-induced cardiotoxicity. (a) Dot plots showing the top 20 biological terms for the DEG processes of cardiomyocytes by KEGG pathway enrichment analysis. Red arrows indicate ferroptosis-related pathways. (b–g) Violin plots showing the expression of ferroptosis-related genes in cardiomyocytes after DOX (20 mg/kg) treatment. (h) Kaplan‒Meier survival curves of mice pretreated with saline (control), SeMet, l-selenocystine, Se-methylselenocysteine, sodium selenite, and Se-rich cardamine enshiensis (0.75 mg/kg, by gavage, weekly) before DOX (20 mg/kg) administration and after the start of DOX (0.375 mg/kg, by gavage, weekly). (i) Kaplan‒Meier survival curves of mice pretreated with saline (control), Fer-1 (a ferroptosis inhibitor, 1 mg/kg), DXZ (dexrazoxane, an iron chelator, 50 mg/kg), or SeMet (a GPX4 activator), followed by DOX (15 mg/kg, i.p.) on day 0 (n = 10 mice per group). (j) Serum cTnT activity in mice on day 2 after DOX injection. DOX (20 mg/kg DOX, i.p), n = 10 mice for each group. (k) Cell viability of H9C2 cells treated with various concentrations of DOX (0 μM, 0.001 μM, 0.05 μM, 0.5 μM, 1 μM, 2.5 μM, 5 μM, 10 μM, 25 μM) or DOX + SeMet for 24 h. (l–m) C11-BODIPY 581/591 staining showing lipid peroxidation levels in H9C2 cells treated with vehicle, DOX (2 μM, 6 h), or DOX (2 μM, 6 h) + SeMet (0.1 μM). Scale bar, 20 μm. A quantitative image analysis of fluorescence median intensity was performed with ImageJ software. (n–q) Levels of selected PUFAs with the indicated treatment (n = 4). Groups were compared using one-way ANOVA or Student’s t-test. Data show the mean ± s.d. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
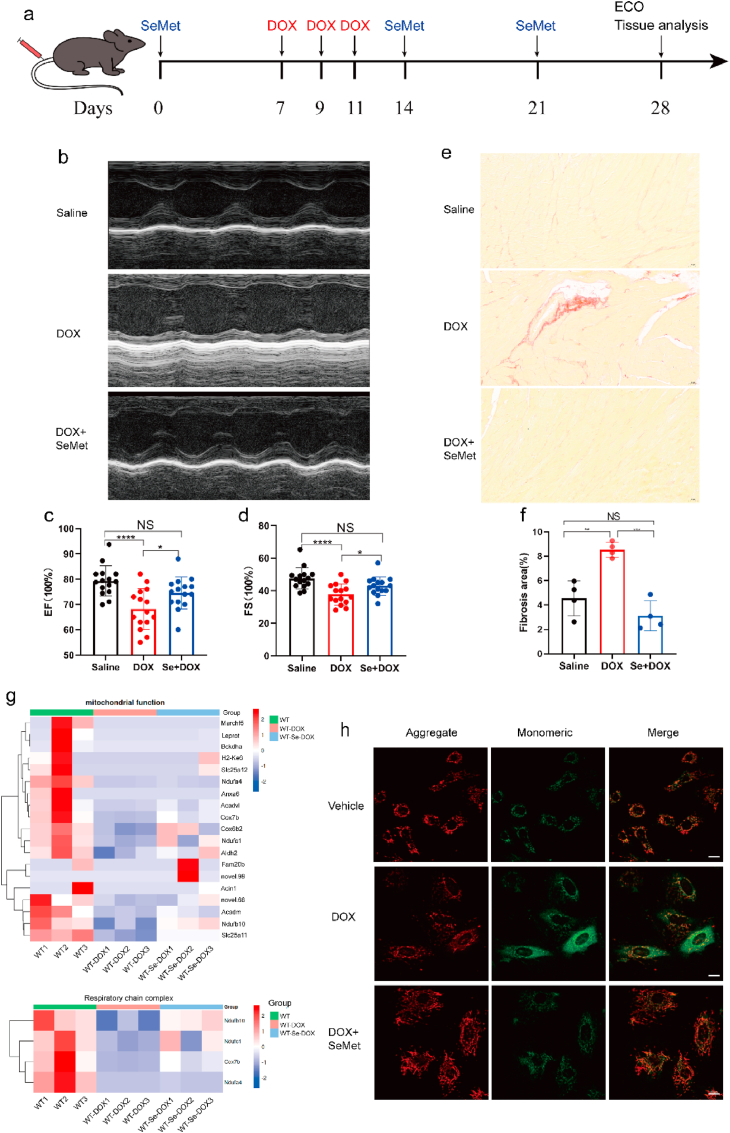
Fig. 3.
Pharmacological activation of GPX4 protects mice from DOX-induced cardiotoxicity. (a) The experimental protocol for testing the protective effect of SeMet in the DOX-induced acute cardiotoxicity model. DOX (6 mg/kg, body weight) was administered to mice via the tail vein on days 7, 9, and 11. SeMet (0.75 mg/kg, by gavage, weekly) was administered before DOX administration and after the start of DOX (0.375 mg/kg, by gavage, weekly). (b) Representative M-mode echocardiographic images of mice on day 14 after DOX treatment. (c, d) Ejection fraction (EF) and fractional shortening (FS) of the heart by electrocardiography on day 28 after DOX administration (n = 20 mice per group). (e, f) Representative images of picrosirius red (PSR) staining at 28 days (n = 4 mice) after DOX treatment. Scale bar, 1000 μm. (g) Heatmaps showing the expression profile of genes related to mitochondrial function and respiratory chain events in the heart tissues from mice in saline, DOX, and SeMet + DOX. (h) Representative images of cardiac JC-1 fluorescence in H9C2 cells treated with vehicle, 0.1 μM SeMet and/or 1 μM DOX for 24 h. Scale bar, 20 μm. Groups were compared using one-way ANOVA. Data are presented as the mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. SeMet ameliorates DIC by suppressing lipid peroxidation
Glutathione peroxidase 4 (GPX4) is a selenoprotein that plays a crucial role in the progression of ferroptosis by inhibiting the peroxidation of unsaturated fats [17], and selenium supplementation boosts the activity of GPX4. Regarding ferroptosis as a potential mechanism of DIC, we hypothesized that pharmacological activation of GPX4 might prevent DOX-induced cardiac injury. We tested 5 selenium compounds and found that selenomethionine (SeMet) had the highest protective effect against DIC in an acute DOX model (Fig. 2h). SeMet supplementation resulted in a 60% survival rate compared with no survival of DOX alone at 14 day (Fig. 2h). To further confirm whether SeMet was superior to ferrostatin-1 and DXZ, we conducted survival experiments with lower doses of DOX (15 mg/kg). Notably, there was a significant difference among the groups, indicating that DIC involves ferroptosis and that SeMet had a better cardioprotective effect than ferrostatin-1 and DXZ (Fig. 2i). Furthermore, SeMet prominently inhibited DOX-induced increases in cardiac biomarkers in the blood (Fig. 2j). We further evaluated whether SeMet supplementation could alleviate DOX-induced ferroptosis in H9C2 cells. In the H9C2 cell model, ferrostatin-1, a ferroptosis inhibitor, rescued DOX-induced cell death [7]. Consistently, we found that SeMet could reduce cell death and lipid peroxidation induced by DOX in H9C2 cells (Fig. 2k–m).
To further explore the mechanism by which SeMet suppresses lipid peroxidation, we assessed the levels of PUFAs, which are substrates for peroxidation. SeMet supplementation significantly decreased the levels of seven PUFAs (C18:3N6, C20:3N6, C20:4, C22:4, C22:5N3) in H9C2 cardiomyocytes compared to the vehicle controls (Fig. 2n–q). DIC resulted in dilated cardiomyopathy and heart failure. DIC could result in dilated cardiomyopathy and heart failure. Ventricular function tended to normalize in the period following cessation of DOX, and left ventricular ejection fraction (EF) and fraction shortening (FS) was the best parameters to evaluate the cardiotoxicity of DOX [18]. Upon injury, cardiac fibroblasts (CF) in the heart begin to remodel the myocardium via extracellular matrix deposition, resulting in increased tissue stiffness and reduced compliance. Excessive cardiac fibrosis is an important factor in the progression of various forms of cardiac disease and heart failure [19]. Hence, EF and FS were used to evaluate the heart function after DOX treatment, and cardiac fibrosis was used to assess the severity of myocardial injury after DOX treatment. Cardioprotective effects were consistently observed in another model based on cardiac systolic function measured four weeks after DOX treatment (Fig. 3a–d). Notably, SeMet treatment also significantly blocked fibrosis in mouse heart tissue after DOX administration (Fig. 3e and f). To assess the overall impact of SeMet on cardiac transcriptome reprogramming during DIC, RNA sequencing (RNA-seq) of cardiac tissue after DOX administration was performed. DEG analysis showed that genes that were induced in the DOX group were significantly enriched in the muscle cell apoptotic process, striated muscle cell apoptotic process, and cardiac muscle cell apoptotic process, while genes that were upregulated in the SeMet + DOX group were significantly enriched in apoptosis and the MAPK signaling pathway (Extended Data Fig. 3c). DOX induces cardiac apoptosis by many mechanisms, including upregulation of BAX and activation of calcineurin and MAPK signaling pathways [20]. Inflammatory cell infiltration is believed to be directly associated with DIC [21]. However, SeMet showed no effects on inflammatory cell infiltration after DOX treatment (Extended Data Fig. 3d). Mitochondria-dependent ferroptosis plays a pivotal role in DIC [7,8]. In this report, we found that SeMet treatment has extensive effects on mitochondrial function and respiratory chain complex cellular pathways (Fig. 3g). Statistical analysis of mitochondrial gene expression showed only 9 gene significant differences between the DOX and Se + DOX treatment groups (Extended Data Table 5). Furthermore, DOX treatment resulted in more JC-1 monomers than vehicle treatment, and SeMet supplementation significantly reduced this DOX-induced effect (Fig. 3h). In summary, these results show that SeMet has the potential to inhibit DOX-induced ferroptosis and ameliorate DOX-induced cardiomyopathy.
2.3. The protective effect of SeMet depends on GPX4
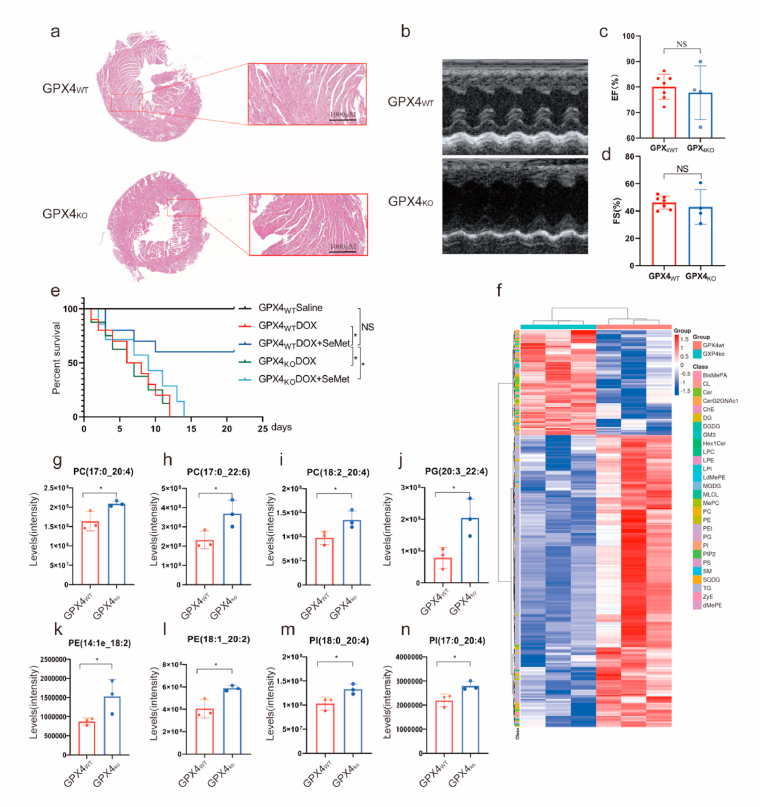
To investigate whether the heart protection of SeMet was dependent on GPX4, we generated heterozygous GPX4-knockout (GPX4hetko) mice to evaluate the impacts of GPX4 on mouse cardiac morphology, cardiac function and electrophysiology (Extended Data Fig. 4a). Compared to GPX4wt mice, the effects of heterozygous ablation of GPX4 on cardiac morphology and function, including EF and FS were negligible (Fig. 4a–d). On the other hand, the most consistent electrocardiogram (ECG) changes observed after DOX treatment were reversible prolongation of the QRS complex and Q alpha progressive extension of the T interval [22] in clinic. We also assessed whether the ablation of GPX4 has effect on the electrical signals in the heart in mice. There were no significant differences in heart rate (HR), RR intervals, P-wave duration, PR intervals, QT intervals, heart rate corrected QT intervals (QTc), or ventricular effective refractory period (VERP) between the two groups. Furthermore, in GPX4hetko mice, the atrial fibrillation (AF) induction rate was higher than that in WT mice (Extended Data Fig. 4b–j).
Fig. 4.
The cardioprotective effect of SeMet depends on GPX4. (a) Representative HE staining of GPX4WT and GPX4KO mouse hearts (n = 4). Scale bar: 1000 μm. (b–d) Representative cardiac electrocardiographs of GPX4WT and GPX4KO mice. Ejection fraction (EF) and fractional shortening (FS) of the heart by electrocardiography (n = 4–6). (e) Kaplan‒Meier survival curves of the indicated mice treated with SeMet followed by DOX (20 mg/kg, i.p.). SeMet (0.75 mg/kg, by gavage, weekly) was administered before DOX administration and after the start of DOX (0.375 mg/kg, by gavage, weekly). (n = 7–10 mice per group). (f) Heatmap of significantly changed lipid species (PUFAs) (one-way ANOVA, FDR corrected p value < 0.05, n = 4 biologically independent samples) between two groups. Each row represents z score-normalized intensities of the detected lipid species. Each column represents a sample. The relative abundance of each lipid is color-coded, with red indicating high signal intensity and blue indicating low signal intensity. CE, cholesteryl ester; PE, phosphatidylethanolamine; PA, phosphatidic acid; PC, phosphatidylcholine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; FA, free fatty acid; TAG, triacylglycerol; DAG, diacylglycerol. (g–n) Levels of selected PUFAs with the indicated treatment. Data show the mean ± s.d. (*P < 0.05; **P < 0.01), n = 4. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To further verify whether SeMet reduced DOX-induced cardiotoxicity in a GPX4-dependent manner, we administered a single high dose of DOX to male wild-type and GPX4hetko mice. Survival analysis revealed that pretreatment with the GPX4 activator SeMet significantly reduced DOX-induced mortality compared to the control group, and this effect was dependent on GPX4 expression (Fig. 4e). To confirm the role of GPX4 in maintaining the homeostasis of lipid metabolism in mouse heart tissue, we performed a nontargeted lipidomic assay to identify the differential metabolites between GPX4wt and GPX4hetko mouse heart tissues. Metabolomic analysis clearly separated GPX4hetko samples from WT samples, indicating global changes in cardiac lipid metabolism due to GPX4 deficiency (Fig. 4f). Compared to WT mice, GPX4 knockout resulted in a significant decrease in the levels of 220 lipid species (Extended Data Table 6). GPX4 deficiency resulted in markedly higher levels of 79 lipid species (Extended Data Table 7). The accumulation of PUFAs such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylinositol (PI) plays an essential role in driving ferroptosis [[23], [24], [25]]. Accordingly, our lipidomic analysis showed that GPX4 deficiency significantly increased the levels of 52 PUFAs, including PC (17:0_20:4), PC (17:0_22:6), PC (18:1_20:3), PC (18:2_20:4), PC (19:1_18:2), PC (19:1_22:6), PC (40:7e), PE (14:1e_18:2), PE (18:1_20:2), PI (17:0_20:4), PI (18:0_18:2), PI (18:0_20:4), PG (20:3–20:4), ceramides and triglycerides (Fig. 4g–n, Extended Data Fig. 5a–e). In contrast, SeMet treatment resulted in a significant decrease in the levels of five PUFAs in H9C2 cardiomyocytes compared to the vehicle controls (Fig. 2n–r, Extended Data Fig. 5f). In summary, SeMet, which is a GPX4 activator, mediates PUFA levels in the heart and has the ability to inhibit DIC in the presence of GPX4.
Fig. 5.
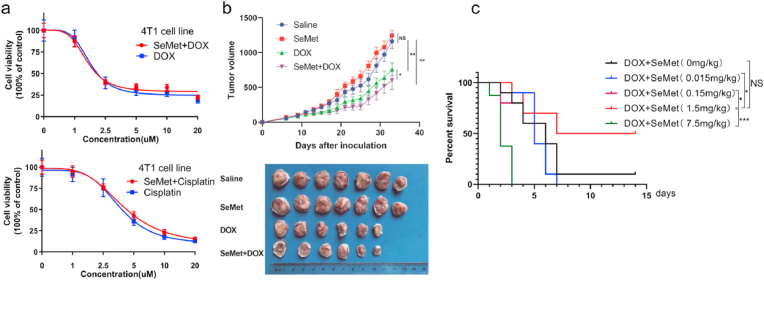
SeMet enhances the antitumor activity of DOX. (a) SeMet alone does not affect the viability of H9C2 cells treated with various concentrations of DOX or cisplatin. Cell viability was measured by Cell-Quanti-Blue. H9C2 cells grown with or without 100 nM SeMet supplementation in the growth medium were treated for 24 h, and then cells were treated with DOX for 24 h. (b) Tumor volumes at the indicated times (n = 6–7 females per group). Groups were compared using one-way ANOVA (*P < 0.05; **P < 0.01). DOX (4 mg/kg) was administered to mice via the tail vein on days 0, 7, and 14. SeMet (0.75 mg/kg, by gavage, weekly) was administered before DOX administration and after the start of DOX (0.375 mg/kg, by gavage, weekly). (c) We subjected different concentrations of Se in Se-rich oyster mushrooms to conform effective dosage ranges that can protect against doxorubicin-induced cardiomyopathy. Kaplan‒Meier survival curves of the indicated mice treated with Se-rich oyster mushrooms (counted as SeMet) followed by DOX (20 mg/kg, i.p.) (n = 10 mice per group). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. SeMet does not interfere with the chemotherapeutic efficacy of DOX
Theoretically, an ideal drug for preventing DIC should not compromise the antitumor efficacy of DOX. We further explored the use of SeMet as a cardioprotective agent during DOX treatment and evaluated the combined effects of SeMet and DOX on tumor growth. In the 4T1 breast cancer cell model, the combination of SeMet and DOX exhibited favorable antitumor effects ex vivo (Fig. 5a). SeMet did not decrease the antitumor efficiency of DOX and cisplatin, and the combination therapy using SeMet and DOX achieved a better antitumor effect than therapy with SeMet or DOX alone (Fig. 5b). DXZ was approved by the FDA as an iron chelator to prevent DIC in cancer patients. However, the disadvantages of DXZ, such as bone marrow suppression, impair the therapeutic efficacy of chemotherapy and limit its clinical application [26]. Consistently, DXZ aggravated the inhibitory effect of bone marrow function induced by DOX in mice. (Extended Data Fig. 6). We further investigated whether supplementation with SeMet could affect bone marrow function in mice. Importantly, blood tests and H&E staining demonstrated that SeMet treatment had no impact on bone marrow function or tissues (Extended Data Figs. 7 and 8). Taken together, these data suggest that SeMet can prevent cardiomyopathy without compromising the antitumor efficacy and additional adverse effects of DOX.
Fig. 6.
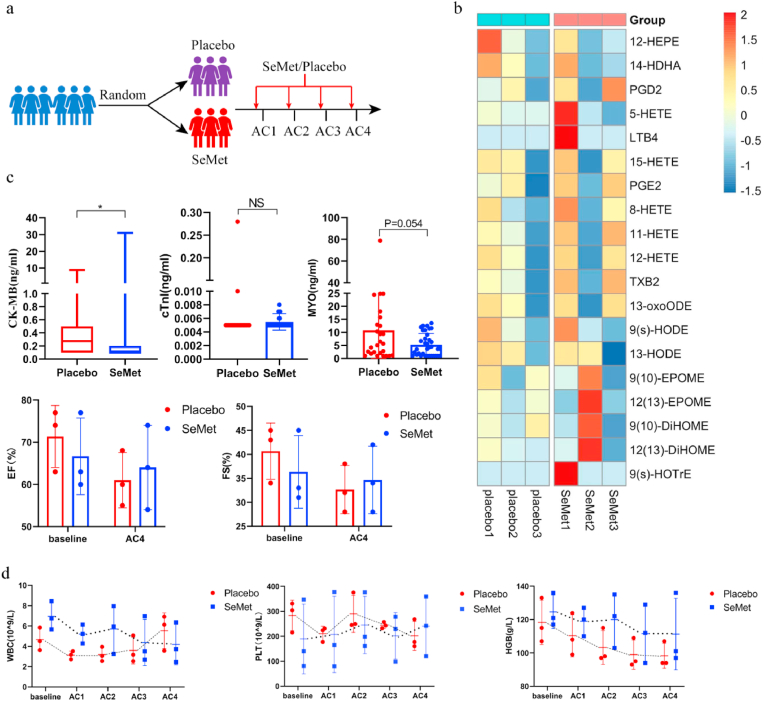
SeMet reduces myocardial enzymes in breast cancer patients. (a) Schematic of the participant workflow in an oral Se-enriched mushroom pilot study (ChiCTR2200056796). Six patients with breast cancer were enrolled and randomized into Se-rich oyster mushroom (SeMet 300 μg/d) and placebo groups. Peripheral blood was collected at baseline, day 2, and day 3 after the AC regimen (DOX: 60 mg/m2, cyclophosphamide 600 mg/m2) administration. (b) Serum contents of peroxidized lipids on day 2 after the two cycles of the AC regimen. (c) Cardiac CK-MB, cTnT and MYO activities were measured with a highly sensitive assay (upper panels), with samples taken from the patients' four treatment cycle nodes. Representative electrocardiograph results of patients at baseline and after the fourth treatment (bottom panels). Groups were compared using the Mann‒Whitney U test (CK-MB) and Student’s t-test (cTnT, MYO activities). Data are presented as the mean ± SEM, *P < 0.05. (d) Se-rich oyster mushroom (SeMet) treatment does not affect hematological parameters. AC regimen: doxorubicin at a dose of 60 mg/m2 and cyclophosphamide at a dose of 600 mg/m2.
3.1. SeMet reduces myocardial enzymes in breast cancer patients
SeMet has not been approved by the FDA or cFDA for clinical use. To explore the clinical application of SeMet, we used a selenium (Se)-rich oyster mushroom as a resource for Se supplementation. The main form of Se present in this mushroom is SeMet, as confirmed by inductively coupled plasma‒mass spectrometry (ICP‒MS) (Extended Data Fig. 4k). Next, we used Se-rich oyster mushrooms with different concentrations calculated based on SeMet to meet the effective dose range, thereby preventing DIC. Consistent with previous results (Fig. 2h), 1.5 mg/kg SeMet in mushrooms exhibited the highest protective effect (Fig. 5c). To investigate the safety of SeMet in patients with breast cancer, we conducted an exploratory trial using Se-enriched oyster mushroom tablets and oyster mushroom tablets as placebo (ChiCTR2200056796). In the clinical trial, the dose of Se was 300 μg/day based on the human-mouse conversion rate. Six patients with stage II breast cancer after surgery were enrolled in the study and randomly received placebo (oyster mushroom tablets, n = 3) or Se-enriched oyster mushroom tablets (300 μg SeMet per day) (n = 3) (Fig. 6a). All enrolled patients did not undergo chemotherapy or radiotherapy before surgery and received four cycles of the AC regimen (doxorubicin at a dose of 60 mg/m2 and cyclophosphamide at a dose of 600 mg/m2) as an adjuvant therapy. A 7-day regimen of Se-enriched oyster mushroom tablets/placebo started 3 days before chemotherapy initiation. Cardiac enzyme levels were detected on the second and third days after chemotherapy. All six patients in the trial completed the study. There was no significant difference between the two groups in baseline parameters (Extended Data Table 8). We further explored the changes in lipid oxides after DOX treatment. Targeted metabolic analysis showed that Se-rich oyster mushrooms decreased the levels of 12-HEPE, 14-HDHA, and 13-HODE after the second AC regimen administration, but there was no significant difference between the placebo and Se supplementation groups (Fig. 6b). Remarkably, we found that the administration of Se-rich oyster mushrooms could effectively reduce serum markers of cardiac injury, including CK-MB and MYO (Fig. 6c). Intriguingly, consistent with our previous study [27], Se supplementation tended to reduce the decrease in platelets and hemoglobin after chemotherapy (Fig. 6d). The pleiotropic effects of SeMet have resulted in therapeutic benefits and protective effects on drug toxicities in several xenograft models [28]. However, in our phase II clinical trial [27], Se supplement decreased the incidence of acute side effects during concurrent chemoradiotherapy, but did not improve the efficacy of chemoradiotherapy in patients with cervical cancer. More clinical trials are needed in the future to determine whether Se can effectively improve the therapeutic efficacy of anticancer drug.
4. Discussion
The mechanism of DIC is still unclear and may be mainly related to the following two factors: (1) DOX causes cardiac injury through the production of oxygen free radicals mediated by enzymatic and nonenzymatic pathways [20]; and (2) topoisomerase Ⅱβ (TopⅡβ) is a key mediator of DOX-induced cardiotoxicity [29,30]. There are two subtypes of topoisomerase: type Ⅱα and type Ⅱβ. TopⅡβ is mainly located in cardiomyocytes, while TopⅡα mainly exists in proliferating cells and is essential for DNA replication by altering the topological structure of DNA during replication, chromosome condensation, and double-strand separation of sister chromosomes, which is considered the molecular basis of the antitumor activity of DOX [31]. DOX nonselectively inhibits TopⅡβ in cardiomyocytes, leading to DNA double bond breakage and cardiomyocyte death [32]. Specific knockout of TopⅡβ can protect mice from DOX-induced myocardial damage and progressive heart failure [30]. However, the partial reduction in reactive oxygen species (ROS) in cardiomyocytes by TopⅡβ knockout suggests that ROS may be produced by other pathways independent of TopⅡβ, indicating that there are other pathogenic links that may interfere with the development of DOX-induced cardiotoxicity but have not yet been discovered. Recent studies have shown that DOX induces lipid peroxidation and ferroptosis in cardiomyocytes by inhibiting glutathione peroxidase 4 (GPX4) in the cytoplasm and mitochondria [33,34]. In this study, the snRNA-seq results revealed that DOX could induce ferroptosis not only in cardiomyocytes but also in epithelial cells. Targeting ferroptosis is a valuable and promising strategy for preventing DIC.
Ferroptosis is a type of programmed death that is catalyzed by iron and mediated by polyunsaturated fatty acid (PUFA) peroxidation [[35], [36], [37]]. In our study, SeMet regulated the level of PUFAs in cardiomyocytes, reducing lipid peroxidation and ferroptosis. Iron chelators, including DFO and DPD, can inhibit ferroptosis induction in cells [38]. DOX and its metabolites can affect iron homeostasis [39], and DXZ has been approved by the FDA to prevent DIC in cancer patients. However, the side effects of DXZ limit its clinical use, and it can only be started after a cumulative dose of DOX of 300 mg/m2 in patients. Thus, there is an urgent need to develop pharmaceuticals to prevent DIC based on the molecular mechanism of ferroptosis. Protective mechanisms against cell membrane peroxidation damage, including glutathione peroxidase 4 (GPX4) and FSP1, can prevent the occurrence of ferroptosis [40]. GPX4 catalyzes the reduction of lipid peroxides in a glutathione-dependent pathway. N-acetylcysteine (NAC) increases the levels of intracellular cysteine with a subsequent increase in GSH and was proven to be effective in the prevention of DIC [41]. The low toxicity and unlimited availability of NAC make it a potential drug to prevent DIC in the clinic. Further clinical trials to evaluate NAC’s effect on DOX toxicity and whether it affects the antitumor properties of DOX in cancer patients are warranted. In addition to the evaluation of NAC alone, NAC and Se-Met could also be tested together in the future. DOX downregulates GPX4 expression through the DOX-Fe2+ complex and induces lipid peroxidation in mitochondria, leading to mitochondria-dependent ferroptosis [7]. In this study, the depletion of GPX4 showed no effect on the cardiac function of mice but exacerbated DOX-induced myocardial damage, as indicated by significant impairment of left ventricular ejection fraction (LVEF) [7], indicating that GPX4 plays a critical mediating role in suppressing DIC and is a key molecular target for the development of drugs to prevent and treat DIC. Nuclear factor erythroid 2-related factor 2 (Nrf2) is a transcription factor that regulates enzymes required for GSH synthesis and plays an important regulatory role in ferroptosis [42]. Yang et al. reported that Se treatment effectively alleviated DOX-induced myocardial dysfunction by regulating the Nrf2-NLRP3 pathway, and the suppression of Nrf2 abolished the cardioprotective effects of Se [43]. However, Yang reported that the levels of Nrf2 mRNA were significantly decreased in the DOX group, which was inconsistent with previous studies showing that DOX treatment led to an increase in nuclear Nrf2 protein levels and myocardial Nrf2 mRNA levels [44,45]. Hence, further work is needed to illuminate and confirm whether Se prevents DIC via the Nrf2 pathway. Nrf2 plays a critical role in suppressing oxidative stress. However, DOX-induced cardiac oxidative stress is modestly enhanced by knockout of Nrf2, indicating that Nrf2 mediates DIC independent of ROS formation [44]. DOX promotes heme degradation and ferrous ion release through Nrf2-mediated upregulation of HMOX1, and the excessive accumulation of free iron in mitochondria leads to lipid peroxidation [45,46]. Considering the protective functional role of Nrf2 activation in the pathogenesis of DIC [44], inhibiting the expression of HMOX1 by zinc protoporphyrin IX (ZnPP) is a good strategy to alleviate DIC [45]. In view of this, the NRF2/GPX4 axis is a key target for the development of drugs to alleviate DOX-induced ferroptosis.
The use of selenium (Se) by GPX4 is indispensable for inhibiting peroxide-induced ferroptosis [47]. Se-containing biological agents are widely used in the treatment of diseases, but too much dietary selenium can cause side effects and even lead to selenosis [48]. Because the threshold between essential and toxic concentrations of inorganic Se is rather narrow, we screened organic seleno-compounds to discover effective cardioprotective effects. We found that SeMet significantly improved the survival rate of an acute DOX mouse model and improved cardiac function in another model. In the clinical trial, the dose of SeMet was 300 μg/day based on the human-mouse conversion rate, which is safe for patients and lower than the tolerable upper intake level (400 μg, daily). Among the functionally characterized selenoproteins are five glutathione peroxidases (GPXs) and three thioredoxin reductases (TrxR/TXNRD), which protect cells from oxidative damage and can be functionally replaced by other oxidoreductases. There is also crosstalk between GPX4 and the selenoprotein thioredoxin reductase1 [49]. Although SeMet can activate TXNRD1/2 to attenuate oxidative stress, to date, no studies have reported the association of TXNRD1/2 with DIC. Thus, we generated GPX4hetKO mice to confirm whether the protective effect of GPX4 against SeMet was dispensable. However, the protective effect of SeMet depended entirely on the GPX4 gene. In addition, GPX4 deficiency resulted in higher levels of PUFAs in heart tissue compared with WT mice, and SeMet significantly reduced the levels of PUFAs in vivo. Our findings extend this basic physiological perspective of selenium biology and demonstrate that increasing GPX4 activity and decreasing PUFA levels through selenium supplementation protects against DIC.
An ideal cardioprotective factor should not act at the expense of antitumor efficacy. DXZ is only used in the clinic when the cumulative dose of DOX reaches 300 mg/m2 and in patients with symptoms of myocardial injury due to reduced antitumor efficacy. SeMet supplementation does not offset the antitumor efficacy of DOX in vivo and in vitro. In our clinical trial, the limited side effects and good tolerance of SeMet provided protection against DIC without much concern for toxicity. However, our clinical trial had several limitations, including the following: (1) The cardioprotection of SeMet was dependent on fine tuning of the dosage and bioavailability in mice. However, the tuning of dosage in the animal model observed in this study could be different in human. Regarding dose-escalation, clinical trial should be carried out very carefully and thoughtfully to warrant the safety of SeMet in the human. (2) There was a limited number of subjects and a short follow-up period, but these limitations can be improved through longer and larger trials to verify this protective effect in the future. SeMet has the characteristics of low toxicity and high efficiency and has potential clinical application prospects for cardioprotection.
5. Conclusion
SeMet inhibits DOX-induced ferroptosis in a GPX4-dependent manner and does not compromise the therapeutic effect of chemotherapy. This agent may be a promising therapeutic treatment for preventing DIC.
6. Materials and methods
Clinical study design and specimen collection. This study was approved by the Ethics Committee of the Central Hospital of Enshi Tujia and Miao Autonomous Prefecture, and all patients with breast cancer signed informed consent forms to participate in the study. The trial was registered at http://www.chictr.org.cn/(ChiCTR2200056796). Se-enriched oyster mushroom tablets and placebo were provided by Shenfeng Co., Ltd., China. In October 2019, the Central Hospital of Enshi Tujia and Miao Autonomous Prefecture recruited 6 breast cancer patients and randomly divided them into a selenium supplementation group (n = 3) and a placebo group (n = 3) (Extended Data Table 8). The inclusion criteria were as follows: (1) The age was between 18 and 70 years old; (2) The patients with primary breast cancer diagnosed by histopathology after surgery were stage II and stage III a [according to the TNM staging system of AJCC, 8th Edition 2017]; (3) ECOG physical fitness score 0–1; (4) The functional level of major organs must meet the following requirements: white blood count ≥3.5*109/L, neutrophils (ANC) ≥ 1.5*109/L (no growth factors used within 14 days), platelet count (PLT) ≥ 100*109/L (no correct treatment within 7 days), hemoglobin (Hb) ≥ 90 g/L (no correct treatment within 7 days), total bilirubin (TBIL) ≤ 1.5*upper limit of normal (ULN), alanine aminotransferase (ALT) and aspartate aminotransferase (AST) ≤ 3*ULN, urea nitrogen and creatinine ≤1.5*ULN and creatinine clearance ≥50 mL/min (Cockcroft-Gault formula), cardiac color Doppler ultrasound: left ventricular ejection fraction (LVEF) ≥ 50%; (5) The patients have good compliance with the planned treatment and are able to understand the research process of this The exclusion criteria were as follows: (1) complications of other serious diseases; (2) cardiac dysfunction, including but not limited to congestive heart failure (CHF), transmural myocardial infarction, angina requiring medication, clinically significant valvular disease, and high-risk cardiac arrhythmia, or QTc abnormalities in the ECG during screening (at rest, QTc >450 ms for men or 470 ms for women after correction in the ECG); and (3) patients with serious systemic infections or other serious diseases during pregnancy or lactation. All blood samples were processed and analyzed by investigators blinded to the clinical data of the patients. All samples were used in accordance with approved guidelines and regulations.
Animal studies. All animal experiments were approved by the Animal Care and Use Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology. Wild-type C57BL/6 mice (10 weeks old) were supplied by Hunan Silaike Jingda Laboratory Animal Co., Ltd., kept in a temperature- and humidity-controlled room, fed a commercial diet, and provided free access to water. Heterozygous GPX4-knockout (GPX4hetko) mice were generated by Shanghai Model Organisms Center, Inc.
In vivo doxorubicin treatment. For the acute model, 10-week-old male mice received a single i.p. injection of DOX (Cat#S1208, Selleck Chemicals, 15 mg/kg or 20 mg/kg, body weight) or saline. Where indicated, mice were given a daily i.p. injection of Fer-1 (1 mg/kg, Cat#S7243, Selleck Chemicals), dexrazoxane (50 mg/kg, Cat#S5651, Selleck Chemicals), or vehicle 24 h before DOX treatment. SeMet(Cat#HY-B1000, MCE), L-Selenocystine(Cat#HY-129960, MCE), Se-Methylselenocysteine(Cat#HY-114245, MCE), Sodium Selenite (Cat#71950, Sigma‒Aldrich) was administered by gavage, 0.75 mg/kg, once a week before DOX administration, 0.375 mg/kg once a week after the start of DOX. Se-rich C. enshiensis is a local Se-rich natural plant provided by the Enshi Autonomous Prefecture Academy of Agricultural Sciences (Enshi, China). All the Se-containing compounds were dissolved in water at room temperature.
For cardiac function examination, a DIC model was established as previously described [7]. Briefly, DOX (6 mg/kg, body weight) was administered to mice via the tail vein on days 0, 2, and 4. SeMet was administered by gavage at 0.75 mg/kg once a week before DOX administration and 0.375 mg/kg once a week after the start of DOX. Four weeks after DOX administration, cardiac function in mice was measured by echocardiogram. Then, the mice were sacrificed, and heart tissues were collected for immunohistochemistry (IHC) and transcriptome analysis.
Mouse echocardiography for cardiac function. Transthoracic echocardiography was performed to evaluate the cardiac function of mice through echocardiography imaging using the Vevo2100 High-Resolution Imaging System (Visual Sonics, Toronto, ON, Canada) equipped with a 10-MH2 phased-array transducer with M-mode recording. The mice were anesthetized with Avertin (200 mg/kg i.p., Sigma‒Aldrich), and the anterior chest area was depilated using NairTM depilatory cream (Church & Dwight Co., Inc., Princeton, NJ, USA). They were then placed on an electric heating pad (part of the Visual Sonics Vevo Integrated Rail System II) to maintain body temperature at 37 °C. Medical ultrasound gel (Tianjin Yajie Medical Material Co., Ltd., Tianjin, China) was used as a coupling agent between the ultrasound scanning head and skin. The two-dimensional targeted M-mode traces were recorded from the parasternal short-axis view at the level of the mid-papillary muscles and from the parasternal long-axis view at the level immediately under the papillary muscle. Left ventricular parameters were measured based on M-mode recordings, including left ventricular end diastolic volume (LVEDV), left ventricular end systolic volume (LVESV), left ventricular internal dimension at end diastole (LVIDd), and left ventricular internal dimension at systole (LVIDs). The data are presented as the average of the measured values of three consecutive beats. The ejection fraction (EF) was calculated as EF=(LVEDV-LVESV)/LVEDV × 100%, and fractional shortening (FS) was calculated as (LVIDd-LVIDs)/LVIDd × 100%. After measurement, the mice were sacrificed with an overdose of pentobarbital sodium, and their hearts were quickly removed and placed in liquid nitrogen or 4% paraformaldehyde for subsequent experiments.
Mouse electrocardiography. Electrocardiograms (ECGs) obtained from mice at rest. Briefly, after mice were lightly anesthetized with isoflurane vapor (0.5–1%), the mice were placed on a heating pad (28 °C), and subcutaneous needle electrodes were applied to the left upper limb, right upper limb and right lower limb for ECG recording (BIOPAC MP System, Goleta, CA). The ECG waveforms were continuously monitored under anesthesia until the heart rate stabilized. Baseline ECG was recorded for 5 min. ECG traces were analyzed using the AcqKnowledge 3.9.1 program (BIOPAC, Goleta, CA).
Mouse Electrophysiology. Unipolar and bipolar electrogram recordings were obtained from the right atrium and right and left ventricles via the epicardial route. The mice were anesthetized with Avertin (200 mg/kg i.p., Sigma‒Aldrich, St Louis, USA), and the operative field (approximately from neck to midsternal region) was shaved using hair clippers, or hair removal cream was applied. Then, they were placed on an electric heating pad to maintain body temperature at 37 °C. Disinfect the operative field. Place the catheter with all its electrodes into a syringe (10 mL volume) filled with 0.9% sodium chloride. Scissors were used to make a T-shaped incision to the incisura jugularis and the long leg extending below the chin. Fat and lymphatic tissue were expanded and dissected to visualize the right external jugular vein. A suture was placed around the cranial and caudal end of the vein. The catheter was inserted into the opening without loosening the tension in the vein. The catheter was advanced through the vein until the catheter electrodes were correctly positioned in the right atrium and ventricle, as confirmed by endocardial electrograms (EGMs). Signals were amplified and filtered (EVR recorder, E for M Corp) for oscilloscopic display at a speed of 50–200 mm/s. Pacing thresholds (in milliamperes) were determined for each lead, and stimulation was performed for 1.0-ms pulse widths at twice the diastolic capture threshold. Cardiac rhythm was continuously monitored and recorded (at 100 mm/s), and all ECG frontal axes (P and QRS) and time intervals (PR, QRS, QT, JT, QTc, JTc, RR) were calculated for each animal in standard fashion. Standard clinical electrophysiological pacing protocols were used to determine all basic electrophysiological parameters.
4T1 breast cancer xenograft mouse model. A total of 5 × 105 mouse 4T1 breast cancer cells were injected into the right inferior mammary fat pad of BALB/c mice. When the tumor volume reached 50 mm3, the mice were randomly divided into four groups: normal saline control group, SeMet (2 mg/kg) group, DOX group (4 mg/kg on days 0, 7, and 14, i.p.), and SeMet + DOX group. The tumor size was measured every 3 days, and the tumor volume was calculated (mm3 = 0.52 × length × width2 [2]). Once the tumor volume reached 1500 mm3, the mice were sacrificed, and the liver, heart, kidney, spleen, and tumor tissues were collected for H&E staining. Blood samples from mice were also collected for routine blood, liver, and kidney function tests.
Cell culture. The H9C2 cardiac myoblast cell line was obtained from ATCC. H9C2 cells were grown in DMEM (HyClone) containing 10% fetal bovine serum and 100 U/ml penicillin and streptomycin.
Cell viability. According to the manufacturer’s instructions, cell viability was quantified using the QUANTI-Blue (Nordic Diagnostica, CQBL-10K) method. Briefly, 3000 cells were inoculated overnight in a 96-well plate and exposed to various concentrations of compounds for a specified time. QUANTI-Blue Reagent (10 μl) was added to each well and incubated in 5% CO2 at 37 °C for 3 h. The plates were then measured at 540 nm/600 nm (Ex/Em) using a Tecan Microplate Reader (Morrisville NC, USA).
BODIPY 581/591C11 analysis. For confocal imaging, 10,000 cells were seeded per well in a 6-well plate containing glass slides. Replaced with medium supplemented with or without 2 μM Se for 72h. Cells treated with 1 μM DOX for 24h or DMSO were used as negative controls. Removed medium and wash cells with HBSS for 5 min. The samples were incubated with 5 μM BODIPY 581/591C11 dye (Cat. #D3861; Thermo Fisher Scientific) at 37 °C for 30 min. Removed mixture, washed cells with HBSS for 5 min. Washed cells 3 times with PBS (cells are live) and added 4% Paraformaldehyde at RT for 20 min (cells are fixed). After 3 times washing with PBS, the slides were mounted using Prolong (R) Gold Antifade with DAPI Molecular Probes (Cat.8961S; Cell Signaling Technology). The cells were imaged at 63 × oil magnification using a Zeiss LSM 900 confocal laser scanning microscope. Image analysis was performed using ImageJ software. At least six randomly selected fields were analyzed per sample.
Analysis of mitochondrial membrane potential (ΔΨm). The fluorescent probe 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) (T3168, Invitrogen) was used to measure ΔΨm. Briefly, H9C2 cells pretreated with vehicle or SeMet were seeded at a density of 100,000 cells/well in a 6-well plate and treated as indicated. The following day, the cells were treated with DMSO (vehicle) or DOX (1 μM) for 12 h and stained with 2.5 mg/mL JC-1 (100 μl) at room temperature for 20 min in the dark. The stained slides were washed three times with PBS and then analyzed immediately with an LSM-900 confocal microscope (Zeiss).
Transcriptome profiling. RNA was extracted from the heart tissue of mice using TRIzol reagent according to the manufacturer’s instructions. Transcriptome libraries were generated and sequenced on an Illumina HiSeq 2500/x. The “HTQC” R package was used to filter low-quality bases and reads. Adapter sequences and reads with >10% N bases or >50% low-quality bases (≤5) were eliminated. The transcriptome assembly for cardiac cells was generated using Trinity with the default parameters [50]. The gene expression levels were computed as the number of reads per kilobase of gene length per million mapped reads (FPKM) using RSEM software [51].
Single cell enucleation and single nuclei (snRNA)-seq library preparation. There were six heart tissue samples from DOX-treated mice (n = 3) and naïve-treated mice (n = 3) in our study. Tissue dissociation was performed on the sample tissue to form a single-cell suspension, followed by single-cell nuclear separation. The nuclear extraction method was based on 10x Genomics official protocols for nuclei isolation from fresh tissue. Nuclei suspensions were loaded on a 10x Chromium Controller (10x Genomics) according to the manufacturer’s protocol based on the 10x Genomics proprietary technology. Single-cell transcriptomic amplification and library preparation were performed using a Chromium Single Cell 3′ v3 Reagent Kit (10x Genomics) according to the manufacturer’s instructions. The libraries were then pooled and sequenced on an Illumina NovaSeq 6000 system.
Single-nucleus RNA sequencing data processing. The sequencing data from 10x Genomics were aligned and quantified using the CellRanger software package (version 6.1.1) against the mouse reference genome (mm10). Subsequently, high-quality cells were kept using the following criteria: single-cell gene expression greater than 500 and less than 5000, UMI greater than 800, and mitochondrial ratio less than 20%. Then, the R package “DoubletFinder” was used for dual cell identification and elimination. We used the R package “Seurat” to create a Seurat object for the remaining cells belonging to the snRNA-seq gene expression matrix for further analysis, including the SCTransform function for normalization of expression, identification of highly variable genes (HVGs) and scaling expression data, and data integration of six samples for batch effect removal using the algorithm RunHarmony. Then, RunPCA functions were performed based on the top 3000 HVGs to obtain the number of principal components (PCs) and the first 40 principal components (40 PCs) for dimensionality reduction. The FindNeighbors and FindClusters commands were used to achieve cell clustering, and RunUMAP was used to visualize UMAP clustering. The function FindAllMarkers or FindMarkers (wilcox.test) was used to compare differences among cell types and identify differential genes. The enrichment analysis of GO and KEGG was conducted using the “clusterprofiler” R package, and gene sets with adjusted p values < 0.05 were considered significantly enriched. All cells were annotated using the “SingleR” R package and manual methods, and clusters with a cell count less than 50 were discarded. Unbiased clustering generated 32 main clusters and annotated 5 nonimmune cardiac cell populations based on canonical marker genes.
Serum cTnT, CK-MB, and MYO activities. The serum cTnT activity of mice was measured using a cTnT Activity Kit (Sangon Biotech Co., China). Briefly, blood was collected from the inferior vena cava on the second day after DOX administration. The serum was isolated by centrifugation at 1000 rpm for 10 min. Serum cTnT was measured in accordance with the manufacturer’s instructions. The serum cardiac cTnT, CK-MB, and MYO activities of humans were measured using a highly sensitive assay (Shenzhen Kingfocus Biomedical Engineering Co., Ltd., China) according to the manufacturer’s instructions. In short, blood was collected on the second and third days after DOX administration, and the serum was isolated by centrifugation at 1000 rpm for 15 min. Serum cTnT, CK-MB, and MYO were measured in accordance with the manufacturer’s instructions.
Histological and immunohistochemistry. Histological and immunofluorescence staining was performed on paraffin sections (5 μm) of mouse hearts fixed in 10% phosphate-buffered formalin. Standard H&E and PSR staining were performed on these sections to assess histopathology and collagen deposition, respectively.
CD11b immunofluorescence in cardiac tissue. For immunofluorescence analysis, paraffin-embedded mouse heart sections were deparaffinized, and antigen retrieval was performed by boiling in citrate buffer. The sections were incubated overnight with the CD11b primary antibody (Cat#17800S, CST), followed by the corresponding secondary antibody. Slides were examined using a Leica DM2500 microscope (Leica Biosystems, Germany).
Lipidomic analysis. (1) Lipid Extraction: Total lipids of each cell pellet were extracted using a modified Folch method, as described previously [52]. Briefly, 107 cells were mixed with 5 mL MeOH/CHCl3 (1:2, v/v) and 10 μL lipid internal standards. Subsequently, the mixture was vortexed at 1800 rpm for 1 h, and 1.33 mL of H2O was added to induce phase separation. The sample was centrifuged at 5000 rpm for 10 min at 4 °C, and then the lower CHCl3 phase was collected and evaporated under a nitrogen stream. The extracted lipids were resuspended in DCM/MeOH (50:50, v/v) and diluted to 1 mL. The diluent was then stored at −20 °C for MS analysis.
(2) LC‒MS Conditions: An ultrahigh-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry method was applied for untargeted lipid profiling. Lipid profiling was accomplished with a Shimadzu 30A system (Shimadzu Corporation, Kyoto, Japan) and a quadrupole time-of-flight mass spectrometer (LC-Q-TOFMS, Triple TOF 6600 system, Applied Biosystems/MDS Sciex, Concord, ON, Canada) as previously described [52]. Briefly, lipids were separated using a Phenomenex Kinetex C18 column (100 × 2.1 mm, 2.6 μm) and a Phenomenex Security Guard precolumn of the same material (Phenomenex, Torrance, CA, U.S.A.). The injection volumes for positive and negative ESI modes were 2 and 6 μL, respectively. The column temperature was maintained at 60 °C, and the flow rate was 400 μL/min. Gradient elution was started with 20% solvent B (IPA/ACN = 5:1, v/v; 5 mM ammonium acetate) and 80% solvent A (water/MeOH/CAN = 1:1:1, v/v/v; 5 mM ammonium acetate) for 0.5 min, increased to 40% B from 0.5 to 1.5 min, increased to 60% B from 1.5 to 3 min, increased to 98% B from 3 to 13 min, decreased to 20% B from 13 to 13.1 min, and maintained at 20% B from 13.1 to 17 min, and the total run time was 17 min. The chromatographic and mass spectrometry data were acquired by Analyst 1.6 software (Applied Biosystems) in both ESI+ and ESI modes with information-dependent acquisition (IDA). The MS parameters were as follows: the declustering potential was set to 80 V (+) and −80 V (−); the collision energy was set to 10 V (+) and −30 V (−); and the ion spray voltage was set to 5500 V (+) and −4500 V (−), with a mass range from m/z 50 to 1200.
(3) Data preprocessing and statistical analysis: MS-DIAL (version 3.30) and Peak View, Master View and Multi Quant (SCIEX) were used for data processing. All raw data files were converted from the original SCIEX format (.wiff) into Analysis Base File format (.abf) prior to MS-DIAL data processing. All lipids were identified based on the MS/MS spectrum of each feature, such as mass accuracy, fragment ion, isotopic pattern, retention time tolerance, MS accurate mass tolerance, MS/MS accurate mass tolerance and identification score cutoff value (80%). After lipid identification, Master View (version 2.0) was used to establish a lipid quantitative method, including lipid name, retention time, and m/z, and was further applied for more in-depth quantitative analysis with Multi Quant (version 3.0.3). Group differences were calculated using a paired two-sample t-test with a fold-change threshold of 1.5. An FDR-adjusted p value of 0.05 or less was considered statistically significant.
Statistical analysis. Data were analyzed and graphed using GraphPad Prism software, and all summary data are presented as the mean ± s.e.m. The median and interquartile range (IQR) were used to identify outliers of serum CK-MB. The Mann‒Whitney U test was used to compare the differences in serum CK-MB between the placebo and SeMet groups. The chi-square test was used to compare the differences in the atrial fibrillation (AF) induction rate between the GPX4WT and GPX4KO groups. The other groups were compared using Student’s t-test or one-way ANOVA with Tukey’s post hoc test. For the Kaplan‒Meier survival plots, statistical significance was measured using the log-rank (Mantel‒Cox) test.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81660503, 82160490); Natural Science Foundation of Hubei Province (2022BCE005); Natural Science Foundation of Enshi Tujia and Miao Autonomous Prefecture Government (D20200018, D20210033, D20220052).
Ethical standards
The Ethics Committee of the Central Hospital of Enshi Tujia and Miao Autonomous Prefecture granted approval for the study, and all patients with breast cancer signed informed consent forms to participate in the study. The trial was registered at https://www.chictr.org.cn/showproj.aspx?proj=130215. All animal experiments were approved by the Animal Care and Use Committee of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology.
CRediT authorship contribution statement
Chuying Huang: Writing – review & editing, Writing – original draft, Validation, Conceptualization. Yishan Guo: Project administration, Conceptualization. Tuo Li: Supervision, Conceptualization. Guogen Sun: Project administration. Jinru Yang: Project administration. Yuqi Wang: Project administration. Ying Xiang: Software, Data curation. Li Wang: Project administration. Min Jin: Project administration. Jiao Li: Project administration. Yong Zhou: Project administration. Bing Han: Project administration. Rui Huang: Project administration. Jiao Qiu: Project administration. Yong Tan: Software, Data curation. Jiaxing Hu: Project administration. Yumiao Wei: Software, Data curation. Bo Wu: Software, Data curation. Yong Mao: Software, Data curation. Lingshan Lei: Software, Data curation. Xiusheng Song: Software, Data curation. Shuijie Li: Writing – review & editing, Validation, Conceptualization. Yongsheng Wang: Conceptualization. Tao Zhang: Conceptualization.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgments
We are grateful to the volunteers for their participation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.103024.
Contributor Information
Chuying Huang, Email: huangchuying2008@126.com.
Shuijie Li, Email: shuijie.li@hrbmu.edu.cn.
Yongsheng Wang, Email: wangys@scu.edu.cn.
Tao Zhang, Email: taozhangxh@hust.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Wenningmann N., Knapp M., Ande A., Vaidya T.R., Ait-Oudhia S. Insights into doxorubicin-induced cardiotoxicity: molecular mechanisms, preventive strategies, and early monitoring. Mol. Pharmacol. 2019;96:219–232. doi: 10.1124/mol.119.115725. [DOI] [PubMed] [Google Scholar]
- 2.Kalyanaraman B. Teaching the basics of the mechanism of doxorubicin-induced cardiotoxicity: have we been barking up the wrong tree? Redox Biol. 2020;29 doi: 10.1016/j.redox.2019.101394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancilla T.R., Iskra B., Aune G. J. Doxorubicin-induced cardiomyopathy in children. Compr. Physiol. 2019;9:905–931. doi: 10.1002/cphy.c180017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleming B., Edison P., Kenny L. Cognitive impairment after cancer treatment: mechanisms, clinical characterization, and management. BMJ (Clinical research ed.) 2023;380 doi: 10.1136/bmj-2022-071726. [DOI] [PubMed] [Google Scholar]
- 5.de Baat E.C., et al. Primary cardioprotection with dexrazoxane in patients with childhood cancer who are expected to receive anthracyclines: recommendations from the International Late Effects of Childhood Cancer Guideline Harmonization Group. Lancet. Child Adoles. Health. 2022;6:885–894. doi: 10.1016/s2352-4642(22)00239-5. [DOI] [PubMed] [Google Scholar]
- 6.Spalato Ceruso M., et al. Use of cardioprotective dexrazoxane is associated with increased myelotoxicity in anthracycline-treated soft-tissue sarcoma patients. Chemotherapy. 2019;64:105–109. doi: 10.1159/000501195. [DOI] [PubMed] [Google Scholar]
- 7.Tadokoro T., et al. Mitochondria-dependent ferroptosis plays a pivotal role in doxorubicin cardiotoxicity. JCI insight. 2020;5 doi: 10.1172/jci.insight.132747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abe K., et al. Doxorubicin causes ferroptosis and cardiotoxicity by intercalating into mitochondrial DNA and disrupting Alas1-dependent heme synthesis. Sci. Signal. 2022;15 doi: 10.1126/scisignal.abn8017. [DOI] [PubMed] [Google Scholar]
- 9.Fang X., Ardehali H., Min J., Wang F. The molecular and metabolic landscape of iron and ferroptosis in cardiovascular disease. Nat. Rev. Cardiol. 2023;20:7–23. doi: 10.1038/s41569-022-00735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng K.H., Contreras G.P., Yeh T.Y. Potential role of neutrophil extracellular traps in cardio-oncology. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23073573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhagat A., et al. Doxorubicin-induced cardiotoxicity is mediated by neutrophils through release of neutrophil elastase. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.947604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhagat A., Shrestha P., Kleinerman E.S. The innate immune system in cardiovascular diseases and its role in doxorubicin-induced cardiotoxicity. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232314649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., et al. Self-maintenance of cardiac resident reparative macrophages attenuates doxorubicin-induced cardiomyopathy through the SR-A1-c-Myc Axis. Circ. Res. 2020;127:610–627. doi: 10.1161/circresaha.119.316428. [DOI] [PubMed] [Google Scholar]
- 14.Lavine K.J., et al. The macrophage in cardiac homeostasis and disease: JACC macrophage in CVD series (Part 4) J. Am. Coll. Cardiol. 2018;72:2213–2230. doi: 10.1016/j.jacc.2018.08.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y., Wu M., Zhong C., Xu B., Kang L. M2-like macrophages transplantation protects against the doxorubicin-induced heart failure via mitochondrial transfer. Biomater. Res. 2022;26:14. doi: 10.1186/s40824-022-00260-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singla D.K., Johnson T.A., Tavakoli Dargani Z. Exosome treatment enhances anti-inflammatory M2 macrophages and reduces inflammation-induced pyroptosis in doxorubicin-induced cardiomyopathy. Cells. 2019;8 doi: 10.3390/cells8101224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao Y., et al. Contribution of ferroptosis and GPX4’s dual functions to osteoarthritis progression. EBioMedicine. 2022;76 doi: 10.1016/j.ebiom.2022.103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloom K.R., Bini R.M., Williams C.M., Sonley M.J., Gribbin M.A. Echocardiography in adriamycin cardiotoxicity. Cancer. 1978;41:1265–1269. doi: 10.1002/1097-0142(197804)41:4<1265::aid-cncr2820410408>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 19.Kong P., Christia P., Frangogiannis N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. : CMLS. 2014;71:549–574. doi: 10.1007/s00018-013-1349-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christidi E., Brunham L.R. Regulated cell death pathways in doxorubicin-induced cardiotoxicity. Cell Death Dis. 2021;12:339. doi: 10.1038/s41419-021-03614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milano G., et al. Intravenous administration of cardiac progenitor cell-derived exosomes protects against doxorubicin/trastuzumab-induced cardiac toxicity. Cardiovasc. Res. 2020;116:383–392. doi: 10.1093/cvr/cvz108. [DOI] [PubMed] [Google Scholar]
- 22.Jensen R.A., Acton E.M., Peters J.H. Doxorubicin cardiotoxicity in the rat: comparison of electrocardiogram, transmembrane potential, and structural effects. J. Cardiovasc. Pharmacol. 1984;6:186–200. [PubMed] [Google Scholar]
- 23.Wiernicki B., et al. Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell Death Dis. 2020;11:922. doi: 10.1038/s41419-020-03118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H., et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat. Cell Biol. 2020;22:225–234. doi: 10.1038/s41556-020-0461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagan V.E., et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swain S.M., et al. Cardioprotection with dexrazoxane for doxorubicin-containing therapy in advanced breast cancer. J. Clin. Oncol. : Off. J. Am. Soc. Clin. Oncol. 1997;15:1318–1332. doi: 10.1200/jco.1997.15.4.1318. [DOI] [PubMed] [Google Scholar]
- 27.Yang M., et al. Effects of selenium supplementation on concurrent chemoradiotherapy in patients with cervical cancer: a randomized, double-blind, placebo-parallel controlled phase II clinical trial. Front. Nutr. 2023;10 doi: 10.3389/fnut.2023.1094081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zakharia Y., Bhattacharya A., Rustum Y.M. Selenium targets resistance biomarkers enhancing efficacy while reducing toxicity of anti-cancer drugs: preclinical and clinical development. Oncotarget. 2018;9:10765–10783. doi: 10.18632/oncotarget.24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasinoff B.B., Patel D., Wu X. The role of topoisomerase IIβ in the mechanisms of action of the doxorubicin cardioprotective agent dexrazoxane. Cardiovasc. Toxicol. 2020;20:312–320. doi: 10.1007/s12012-019-09554-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S., et al. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 31.Uemura T., et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 32.Qiao X., et al. Uncoupling DNA damage from chromatin damage to detoxify doxorubicin. Proc. Natl. Acad. Sci. U.S.A. 2020;117:15182–15192. doi: 10.1073/pnas.1922072117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., et al. PRMT4 promotes ferroptosis to aggravate doxorubicin-induced cardiomyopathy via inhibition of the Nrf2/GPX4 pathway. Cell Death Differ. 2022;29:1982–1995. doi: 10.1038/s41418-022-00990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ta N., et al. Mitochondrial outer membrane protein FUNDC2 promotes ferroptosis and contributes to doxorubicin-induced cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 2022;119 doi: 10.1073/pnas.2117396119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X., Stockwell B.R., Conrad M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan H.F., et al. Ferroptosis: mechanisms and links with diseases. Signal Transduct. Targeted Ther. 2021;6:49. doi: 10.1038/s41392-020-00428-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dixon S.J., Pratt D. A. Ferroptosis. A flexible constellation of related biochemical mechanisms. Mol. Cell. 2023;83:1030–1042. doi: 10.1016/j.molcel.2023.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stockwell B.R., et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitakata H., et al. Therapeutic targets for DOX-induced cardiomyopathy: role of apoptosis vs. Ferroptosis. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23031414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doll S., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 41.Arica V., et al. N-acetylcysteine prevents doxorubucine-induced cardiotoxicity in rats. Hum. Exp. Toxicol. 2013;32:655–661. doi: 10.1177/0960327112467043. [DOI] [PubMed] [Google Scholar]
- 42.He R., et al. Itaconate inhibits ferroptosis of macrophage via Nrf2 pathways against sepsis-induced acute lung injury. Cell Death Disc. 2022;8:43. doi: 10.1038/s41420-021-00807-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang H.B., Lu Z.Y., Yuan W., Li W.D., Mao S. Selenium attenuates doxorubicin-induced cardiotoxicity through Nrf2-NLRP3 pathway. Biol. Trace Elem. Res. 2022;200:2848–2856. doi: 10.1007/s12011-021-02891-z. [DOI] [PubMed] [Google Scholar]
- 44.Li S., et al. Nrf2 deficiency exaggerates doxorubicin-induced cardiotoxicity and cardiac dysfunction. Oxid. Med. Cell. Longev. 2014 doi: 10.1155/2014/748524. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang X., et al. Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U.S.A. 2019;116:2672–2680. doi: 10.1073/pnas.1821022116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hou K., et al. Loss of TRIM21 alleviates cardiotoxicity by suppressing ferroptosis induced by the chemotherapeutic agent doxorubicin. EBioMedicine. 2021;69 doi: 10.1016/j.ebiom.2021.103456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingold I., et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2018;172:409–422. doi: 10.1016/j.cell.2017.11.048. e421. [DOI] [PubMed] [Google Scholar]
- 48.Hou W., Xu H. Incorporating selenium into heterocycles and natural Products─From chemical properties to pharmacological activities. J. Med. Chem. 2022;65:4436–4456. doi: 10.1021/acs.jmedchem.1c01859. [DOI] [PubMed] [Google Scholar]
- 49.Carlson B.A., et al. Glutathione peroxidase 4 and vitamin E cooperatively prevent hepatocellular degeneration. Redox Biol. 2016;9:22–31. doi: 10.1016/j.redox.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grabherr M.G., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu A., et al. Comprehensive and high-coverage lipidomic analysis of oilseeds based on ultrahigh-performance liquid chromatography coupled with electrospray ionization quadrupole time-of-flight mass spectrometry. J. Agric. Food Chem. 2021;69:8964–8980. doi: 10.1021/acs.jafc.0c07343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.