Abstract
The complex infection process of parvoviruses is not well understood so far. An important role has been attributed to a phospholipase A2 domain which is located within the unique N terminus of the capsid protein VP1. Based on the structural difference between adeno-associated virus type 2 wild-type capsids and capsids lacking VP1 or VP2, we show via electron cryomicroscopy that the N termini of VP1 and VP2 are involved in forming globules inside the capsids of empty and full particles. Upon limited heat shock, VP1 and possibly VP2 become exposed on the outsides of full but not empty capsids, which is correlated with the disappearance of the globules in the inner surfaces of the capsids. Using molecular modeling, we discuss the constraints on the release of the globularly organized VP1-unique N termini through the channels at the fivefold symmetry axes outside of the capsid.
For infection, nonenveloped viruses are involved in a number of interactions with macromolecular structures of the host cell that enable virus uptake, disassembly, and delivery of the genome. Such interactions not only implicate recognition of key elements required for cell entry and intracellular passage but also enable activities of viral proteins to overcome the barriers of the host cell. This is exemplified by the binding of viral capsids to cellular receptors, the active release from endocytic vesicles, the use of cellular filament systems for trafficking, and different kinds of delivery of the viral genomes to the cell nucleus (30, 60). These processes are often supported by signaling events which reflect the reaction of the cell to the attachment and uptake of the virus (16). The information for these interactions is maintained within the structure of the virus shell.
The infection process of adeno-associated virus type 2 (AAV-2) is initiated by attachment to the cell surface via binding to heparan-sulfate proteoglycan (53). Upon interaction with a secondary receptor, either fibroblast growth factor receptor 1 or αVβ5 integrin (41, 52), virions enter the cell by means of clathrin-coated pits (5) into the endosomal pathway. While a large number of viral particles seem to accumulate in perinuclear vesicular compartments (4), they have to escape into the cytoplasm for successful infection (21, 63). Rac signaling and interaction with cellular filament systems have an impact on trafficking toward the nucleus (44). There is increasing evidence that virions can enter the cell nucleus (5, 45, 63), possibly by an as yet unknown pathway which is independent of passage through the nuclear pores (22, 63). According to this scenario, the final uncoating reaction has to take place in the nucleus.
The AAV-2 capsid harbors a 4.7-kb linear single-stranded genome which contains two open reading frames coding for four nonstructural proteins and three capsid proteins (48). They are flanked by two identical inverted terminal repeats. The nonstructural proteins have functions in the control of gene expression, in DNA replication, and in genome encapsidation (38). The three structural proteins VP1, VP2, and VP3 are expressed from the p40 promoter via alternative splicing and an alternative start codon for VP2 in an approximate stoichiometry of 1:1:10 (8, 28, 43). They share most of their amino acid sequences. VP1 and VP2 differ from VP3 only by N-terminal extensions of 65 amino acids common to VP1 and VP2 and further by 137 amino acids which are unique for VP1. While VP2 is dispensable for capsid assembly and infectivity of the virus in vitro (57), deletion or mutation of VP1 leads to strongly reduced infectivity although capsid assembly and packaging of DNA is not impaired (26, 54, 57). At least part of the decreased infectivity can be attributed to the loss of a phospholipase A2 (PLA2) catalytic domain and its activity. This domain has recently been identified in most parvoviral VP1 unique sequences (13, 14, 34, 66). Mutation of amino acids in the catalytic center of the PLA2 does not inhibit perinuclear accumulation in late endosomes or lysosomes, which suggests that this activity might be required in a following step of infection, possibly for endosomal release (14, 51, 66). In addition, other signals affecting infectivity might be located on the VP1-unique region of AAV-2 in analogy to several autonomous parvovirus capsid proteins (36, 56).
The icosahedral AAV-2 capsid is built up by 60 subunits of the three capsid proteins VP1, VP2, and VP3. Although the structure of the capsids has been established by X-ray crystallography (64), one of the unresolved questions of AAV-2 as well of other parvoviral structures (1, 2, 46, 47, 65) is the localization of the VP1 N termini which harbor the PLA2 domain. Based on the fact that globules in the interior of empty AAV-2 capsids showed a weaker protein density than the remaining capsid and had fuzzy edges, we speculated that these globules might represent at least part of the N termini of VP1 and VP2 (32). Here, we provide genetically based structural evidence that the N termini of VP1 and VP2 are essential for forming these globules inside the capsid. Furthermore, we describe at the structural and biochemical level conformational changes that occur when the N termini of VP1 and VP2 become exposed to the outside.
MATERIALS AND METHODS
Immunological assays.
Western blot analysis was performed according to standard methods (23) using the monoclonal antibody B1 (61), a peroxidase-coupled secondary antibody, and an enhanced chemiluminescence detection kit (Amersham, Buckinghamshire, United Kingdom).
For the dot blot assay, a nitrocellulose membrane was soaked in phosphate-buffered saline (PBS; 18 mM Na2HPO4, 11 mM KH2PO4, 125 mM NaCl) and inserted into a dot blot gadget (BRL, Pomona, Calif.). A total of 5 × 109 capsids were incubated in 50 μl of buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM MgCl2) at 37, 65, and 75°C, respectively, for 30 min and afterward applied to the nitrocellulose membrane. After sucking of the particles onto the membrane, they were incubated overnight in blocking solution (PBS, 6% milk powder, 0.05% Tween 20) at 4°C. Then the membrane was cut into strips and incubated for 1 h with hybridoma supernatant of the three antibodies A1 (recognizing the epitope KRVLEPLGL in VP1), A69 (binding to the epitope LNFGQTGDADSV in VP1 and VP2), and A20 (recognizing intact capsids) (62) diluted in blocking solution (A1 and A20 at 1:10, A69 at 1:20). After washing of the stripes with PBS-0.05% Tween 20 three times for 15 min, they were incubated with a secondary peroxidase-coupled goat anti-mouse antibody (dilution of 1:5,000 in blocking solution) for 1 h at room temperature (RT). Proteins were detected by means of the enhanced chemiluminescence kit (Amersham).
Preparation and purification of wild-type AAV-2 capsids.
AAV-2 virus was produced as described previously (61) and purified from freeze-thawed lysates of infected cells according to a published protocol (24). The purified viral capsids were concentrated up to 1 ml by using a concentrator (Vivaspin; 10 kDa cutoff), refilled twice up to 5 ml with PBS to change the buffer, and concentrated again to a volume of 150 to 200 μl.
Alternatively AAV-2 particles were produced by transfection of 293T cells with the plasmid pDM in which the rep-cap open reading frame of plasmid pDG (18) had been replaced by the full-length AAV-2 genome. Viruses were purified as described above, and the number of full capsids was further enriched by using a continuous iodixanol gradient (40). Gradient fractions containing predominantly full capsids were collected based on their refraction index, diluted 1:5 with buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.5], 1 mM MgCl2), and concentrated in a Vivaspin concentrator up to 1 ml. The concentrator was refilled twice up to 5 ml and concentrated again to 1 ml. The number of particles in the sample was determined by a capture enzyme-linked immunosorbent assay using microtiter plates precoated with the monoclonal antibody A20 as described previously (16).
Construction and production of recombinant adenoviruses expressing the AAV-2 cap gene.
To create the ΔVP1 and ΔVP2 mutants, the plasmid pJ407 containing AAV-2 rep and cap sequences was used (31). The mutations were introduced by means of the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, Calif.). Thereby, the VP1 start codon was point-mutated from ATG to TTG and the VP2 start codon from ACG to GCG, respectively, by using the following primers: ΔVP1A, 5′-CAATAAATGATTTAAATCAGGTTTGGCTGCCGATGGTTATCTTCC-3′; ΔVP1B, 3′-GTTATTTACTAAATTTAGTCCAAACCGACGGCTACCAATAGAAGG-5′; ΔVP2A, 5′-GGTTGAGGAACCTGTTAAGGCGGCTCCGGGAAAAAAGAGG-3′; and ΔVP2B, 3′-CCAACTCCTTGGACAATTCCGCCGAGGCCCTTTTTTCTCC-5′. To construct pTAV2.0 ΔVP1 and pTAV2.0 ΔVP2, a HindIII-EcoNI and an EcoNI-BsiWI DNA fragment of the pJ407 plasmid containing the ΔVP1 and ΔVP2 mutations, respectively, were cloned into pTAV2.0 (25) (kindly provided by J. King, German Cancer Research Centre, Heidelberg, Germany).
To produce adenoviral transfer plasmids, a 1,064-bp SwaI-BsiWI fragment of pTAV2.0 ΔVP1 respective of pTAV2.0 ΔVP2 was cloned into pADRSVVP. The plasmid pAdRSVVP was generated before from pAdRSVβgal (50) by replacing the lacZ cassette by the AAV-2 cap gene. Therefore, a HindIII-SmaI fragment including the AAV-2 cap gene was cloned from the plasmid pBS ΔTR18 (58) into the bluescript plasmid pBSIISK (Stratagene). A ClaI-SmaI fragment from this new plasmid containing the cap gene was then cloned into the plasmid pAdRSVβgal which had been digested with ClaI and EcoRV resulting in the plasmid pADRSVVP (kindly provided by D. Grimm, German Cancer Research Centre).
Recombinant adenoviral genome plasmids were generated by homologous recombination between the adenoviral transfer plasmids pAdRSVVP ΔVP1 and pAdRSVVP ΔVP2 and pTG3602ΔE3, a plasmid harboring the wild-type adenovirus type 5 genome except for the E3 region (10). The recombination was done in Escherichia coli BJ5183 cells (20). Therefore, pTG3602ΔE3 was digested with ClaI to linearize the plasmid and the transfer plasmids were digested with MscI. The 5.5-kb MscI fragment contained the mutated AAV-2 cap region and the adenoviral sequence necessary for the recombination. Recombinant viruses were produced by standard techniques (15).
To determine the amount of infectious adenoviral particles, 293T cells (104 cells per well in a 96-well plate) were infected with 10-fold serial dilutions of recombinant adenoviruses. After 24-h incubation at 37°C and 5% CO2, the medium was removed and the cells were washed (150 μl of PBS/well). Afterward, the cells were treated with 100 μl of precooled methanol and dried at RT. The cells were incubated overnight with 100 μl of hybridoma supernatant of antibody A30 per well (61) and washed three times with PBS before incubation with goat anti-mouse cy3 antibody (1:200 diluted in PBS). After two more PBS washings, 50 μl of H2O was added to each well before detection of the infected cells by immunofluorescence microscopy. The virus was propagated until the lysate contained at least 108 virus particles per ml.
Preparation and purification of mutant empty AAV-2 capsids.
AAV-2 mutant empty capsids were produced and purified as described previously (17, 49) with the following modifications. Four pellets of approximately 107 293T cells, infected with a recombinant adenovirus expressing either VP1 and VP3 or VP2 and VP3 of AAV-2, were thawed and each was resuspended in 1 ml of PBS with 0.035 mg of PMSF/ml, 0.009 mg of pepstatin/ml, and 0.006 mg of leupeptin/ml. After the first sucrose cushion, the pellet was resuspended in 300 μl of buffer (50 mM Tris [pH 7.5], 5 mM MgCl2, 5 mM MnCl2), shaken for 30 min at RT, and then digested again for 30 min with DNase (final concentration, 50 μg/ml) and RNase (25 μg/ml). The reaction was stopped with EDTA (final concentration, 10 mM). Afterward, the extract was loaded again onto a double sucrose cushion of 200 μl of 50% sucrose and 200 μl of 30% sucrose in 10 mM Tris-HCl-1 mM EDTA, pH 8.0, and centrifuged for 2.5 h at 130,000 × g at 4°C (Combi Plus ultracentrifuge, Beckmann rotor TLS-55). The final pellet was resuspended in 0.1 M NaCl-10 mM Tris (pH 7.5)-mM MgCl2 and stored at 4°C or frozen at −20°C for long-term storage.
Sucrose gradient centrifugation.
A total of 1011 capsids in 50 μl of PBS-MK (18 mM Na2HPO4, 11 mM KH2PO4, 125 mM NaCl, 1 mM MgCl2, 2.5 mM KCl) were treated at different temperatures (37, 65, and 75°C) for 30 min before storing them for 5 min on ice. After the addition of 100 μl of PBS-MK containing 10 mM EDTA, the samples were loaded onto 10 to 30% sucrose gradients (sucrose dissolved in PBS-MK), which were produced by means of a gradient mixer. The gradients were centrifuged at 160,000 × g (rotor SW41; Beckman). Fractions of 500 μl were collected and transferred onto a nitrocellulose membrane via a dot blot gadget as described above. However, Tween 20 was used in neither the blocking nor the washing solution.
Electron microscopy and image processing.
Freezing of the samples, electron cryomicroscopy, and image processing were carried out as described earlier (32) with some modifications as follows. Initial orientations of particles were determined using the map of empty capsids (32) as a reference. For the reconstruction of wild-type AAV-2 (incubation at RT and 65°C), no further refinements of orientations were performed. For the reconstructions of the empty mutant capsids ΔVP1 and ΔVP2, two refinements of orientations by using the present best map as reference were done. Resolutions were estimated by Fourier shell correlation. The different parameters for the various datasets are summarized in Table 1.
TABLE 1.
Summary of parameters for the different datasets
| Sample | No. of micrographs | No. of particles | Defocus (nm) | No. of orientational refinements | Resolution estimate (Å)a |
|---|---|---|---|---|---|
| Virus at RT | 12 | 13,970 | 710-1,790 | 0 | 10.1 |
| Virus at 65°C | 7 | 7,400 | 1,130-1,600 | 0 | 10.3 |
| ΔVP1 | 7 | 3,140 | 1,210-1,640 | 2 | 10.4 |
| ΔVP2 | 13 | 2,730 | 640-1,710 | 2 | 10.1 |
Fourier shell correlation = 0.5.
Sorting particles.
To distinguish empty and full particles in virus preparations, particle images were sorted into four classes according to their density profile by using IMAGIC 5 (55). Particle images were centered by translational alignment to a common reference. The reference was generated by rotational averaging of the sum of all particle images. Centering of particles was repeated once. After centering, particle images were rotationally averaged, giving a representation of their density profile. These rotationally averaged particle images were classified into four classes by using the first two eigen images of the multivariate statistical analysis. The number of four different profiles was chosen arbitrarily and took into account that, besides empty and completely filled particles, intermediates were observed, which appeared to be only partly filled. These partly filled particles were predominantly sorted into the two groups of particles which had the intermediate density profile. Finally, icosahedral reconstructions of particle images grouped into the same class were calculated using the Medical Research Council package (12). Spatial orientations were assigned as determined using cross common lines to the map of the empty capsid (see image processing).
Standard deviation maps.
For comparison of different reconstructions, standard deviations were used as described earlier (7). In order to calculate standard deviation maps, the data were divided into four parts. From each part, an independent image reconstruction was calculated. This was done for wild-type, ΔVP1, and ΔVP2 empty capsids. Standard deviations were calculated for the partial datasets of the wild type, ΔVP1, and ΔVP2, as well as for the combinations of the wild type plus ΔVP1 and the wild type plus ΔVP2. The calculated standard deviations were used to color code the surface representations of the individual reconstructions by using AVS version 5.3 (Advanced Visual Systems Inc.).
Comparison of image reconstruction and atomic model.
The size of the image reconstruction of empty capsids (32) was adjusted according to the catalase-calibrated magnification of the electron microscope. Then, the atomic model of the capsid of AAV-2 (64) (1LP3) was translated to the same origin and rotated into the same orientation as the image reconstruction. For comparison, a surface representation of the image reconstruction and the Cα chains of the capsid proteins were explored using the graphics program O (29).
Molecular modeling.
The available X-ray structure of AAV-2 (64) contains only coordinates of the VP3 domain (residues 217 through 735). Since the spatial structure of the VP1 and VP2 domains is unknown, here, comparative methods modeling homologous proteins were applied to generate putative three-dimensional (3D) models of the missing VP1 part of AAV-2 (residues 1 through 216). For a 59-amino-acid-long part of VP1 (residues 44 through 103), a similarity to the PLA2 fold has been described previously (66). Based on this alignment and using the SWISS Model environment for comparative protein modeling (19), a spatial model was generated and subsequently relaxed using the GROMACS (35) package for molecular simulation. ProCheck (33), Whatcheck (27), and an atomic nonlocal environment assessment procedure (37) were applied to check the reliability of the generated structures. The Protein Data Bank was searched for suitable 3D templates taking the residues 1 to 44 of AAV-2 as unknown sequence. The human immunodeficiency virus type 1 Nef protein (3) was the best matching template showing a sequence identity of 36% in 44 amino acids. For the AAV-2-VP1 sequence from residues 118 through 200, a sequence identity of 31% with the human immunodeficiency virus type 1 Tat protein was found. Both models were generated using the approach described above for the PLA2 fold and subsequently manually connected and relaxed.
The VIPER service (42) has been used to construct a part of the virus capsid, where domains forming a threefold-twofold-threefold symmetry were included. The remaining 16 amino acids between the Tat-like domain and VP3 (sequence from residues 201 through 216, NTMATGSGAPMADNNE) were constructed using the peptide builder option of INSIGHTII. Arbitrarily, one of the four VP3 domains forming the twofold symmetry was chosen and the hexadecapeptide was manually connected.
To demonstrate that it is geometrically possible, that the PLA domain can reach the outside of the capsid, the Tat-like domain was virtually unfolded by applying an additional force between its first (amino acid 104) and last (amino acid 200) residue. Thus, a slow unfolding of the domain extending a distance of about 100 Å—the roughly estimated distance, which has to be covered so that the PLA domain can be displayed outside the capsid—during a 1-ns-long simulation was achieved. This structure was manually attached to the same VP3 domain at the twofold symmetry as used for the inside model. The unfolded part of the Tat-like domain was manually oriented in such a way that it fit through the channel at the fivefold symmetry.
RESULTS
Deletion of VP1 or VP2 results in loss of the globules in the inner surface.
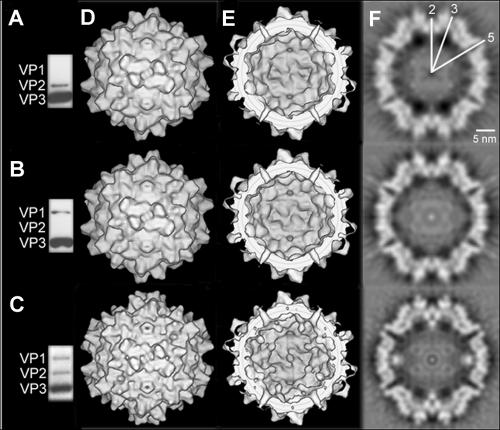
To support the assumption that the globules visible at the twofold symmetry axes inside the capsids represent structures containing the N termini of VP1 and/or VP2 in AAV-2 capsids, mutants were generated by site-directed mutagenesis in which the expression of either VP1 or VP2 was suppressed. By means of recombinant adenoviruses, we isolated empty ΔVP1 and ΔVP2 capsids and analyzed their structures by electron cryomicroscopy. Western blot analysis of purified capsid preparations confirmed the complete deletion of one of the VP proteins in each mutant (Fig. 1A and B).
FIG. 1.
Western blot analysis and 3D image reconstruction of empty ΔVP1, ΔVP2, and wild-type capsids. The presence of the VP proteins in wild-type and mutant empty capsids was determined by Western blot analysis using the antibody B1, which detects the C termini of all three VP proteins. The ΔVP1 capsids (A), the capsids devoid of VP2 (B), and wild-type empty capsids (C) are shown. The outer surfaces of the 3D image reconstructions of ΔVP1- (top), ΔVP2- (middle), and wild-type particles (bottom) are viewed down a twofold symmetry axis (D), as well as the inner surfaces (E). Equatorial slices of the reconstructions are also shown (F). Some of the symmetry axes are marked in white.
For more detailed structural information, we calculated 3D maps of both ΔVP1 and ΔVP2 capsids from electron micrographs. There was no significant difference visible regarding the outer surfaces of empty ΔVP1 and ΔVP2 capsids compared to wild-type capsids (Fig. 1D). However, the globules at the twofold symmetry axes found inside empty wild-type capsids which were tentatively attributed to the N termini of VP1 and VP2 (32) were missing in both of the mutant particles (Fig. 1E and F).
The relevance of these differences was checked by calculating standard deviations, which were used to color code the surfaces of different image reconstructions (Fig. 2). The reconstructions of wild-type and ΔVP2 capsids had significantly less variability than the reconstruction of ΔVP1 (Fig. 2A through C). In order to highlight differences between the different maps, standard deviations of wild-type empty capsids together with reconstructions of mutant empty capsids (either ΔVP1 or ΔVP2) were calculated. Here, differences were considered significant if the standard deviation calculated from wild-type and mutant reconstructions were larger than the standard deviations of the individual reconstructions. For both mutants, structural changes occurred mainly on the inside of the capsid while the outer surface remained more rigid. For both mutants, the largest differences to the wild-type empty capsids were the missing globules at the twofold symmetry axes (Fig. 2D and E, indicated in red and yellow). Furthermore, the entrance to the channels at the fivefold axes at the inner surface changed (indicated in yellow) and, according to difference maps (data not shown), was narrower in the mutants.
FIG. 2.
Surface representations of 3D image reconstructions of empty wild-type, ΔVP1, and ΔVP2 capsids including standard deviations. Standard deviations were calculated for wild-type, ΔVP1, and ΔVP2 empty capsids each, as well as for the combinations wild type with ΔVP1 and wild type with ΔVP2 as described in Materials and Methods. These standard deviations were used to color code the surface representations of the individual reconstructions (A through C) or, in the case of combinations, the reconstruction of wild-type capsids (D and E). Features with larger differences appear red, whereas smaller differences are shown in yellow. Mapped are part of the outer surface (left) and part of the inner surface (right). The view is always direct down a twofold symmetry axis.
Heat induces exposure of the N-terminal stretches of VP1 and possibly VP2 in DNA-containing wild-type AAV-2 capsids.
The N terminus of VP1 of autonomous parvoviruses can be exposed by treatment at a high temperature or with urea without disintegration of the capsid (9, 11, 56, 59). The exposure of VP1 and possibly also VP2 N termini upon heat treatment was analyzed for full and empty AAV-2 capsids. To investigate the accessibility of the N termini, the capsids were applied to nitrocellulose membranes in a dot blot assay using the antibodies A1 (anti-VP1), A69 (anti-VP1 and VP2), and A20 (detecting intact capsids) (Fig. 3). Empty capsids did not react with the N-terminal antibodies A1 and A69 after incubation at 37 or 65°C, whereas the A20 antibody was able to bind to its epitope (Fig. 3A). Incubation at 75°C led to denaturation of the capsids, which resulted in loss of the A20 antibody binding and a strong reaction with the A1 and A69 antibodies. Full capsids showed a slight reaction with the N-terminus-specific antibodies already at 37°C. This reaction strongly increased after incubation at 65°C, whereas the reaction of the A20 antibody was maintained (Fig. 3B). At 75°C, the A20 antibody could not detect its epitope anymore while binding of the antibodies A1 and A69 did not change. Thus, treatment of DNA-containing capsids at 65°C is followed by accessibility of the N termini of VP1 and possibly also VP2. However, it is not clear from this experiment if intact capsids expose the N termini or if populations of native capsids, subfragments, and single subunits coexist at 65°C and generate the observed experimental results.
FIG. 3.
Full capsids undergo a conformational change after treatment at 65°C. AAV-2 capsids were incubated at 37, 65, and 75°C for 30 min. Thereafter, empty (A) and full (B) particles were applied to a nitrocellulose membrane. N-terminal epitopes were detected using antibodies A1 (anti-VP1) and A69 (anti-VP1 and VP2), and capsids were detected using antibody A20. Enriched full AAV-2 capsids were fractionated after continuous sucrose gradients (10 to 30%) and analyzed as described above (C). Full capsids sedimented at about 110S, empty particles sedimented at 60S, and single subunits sedimented below 20S.
To distinguish between these two possibilities, viral DNA containing capsids were analyzed by using continuous sucrose gradients. Such gradients allow the simultaneous detection of full and empty capsids, as well as individual subunits sedimenting at 110S and 60S and below 20S respectively. The accessibility of the N-terminal stretches of VP1 and VP2 at the different levels can be tested by using a dot blot assay with the antibodies A1 and A69. Capsids were detected after incubation at 37°C in fractions 4 through 13 (A20 reaction), corresponding to full capsids which sediment at about 110S (39). Antibody A69 also showed a faint reaction detectable in fractions 5 through 11, while the reaction with A1 was below the detection limit. Incubation at 65°C led to an increase in the reaction with A69 and A1 at the 110S position clearly indicating the accessibility of the N-terminal epitopes in full capsids. This supports the interpretation that the majority of DNA-containing capsids undergo a conformational change where the N termini of VP1 and possibly of VP2 become accessible. An additional weak reaction of antibody A69 in fractions 19 through 21 (20S) indicated that some of the capsids were already destroyed. After treatment at 75°C, antibody A20 could not bind to its epitope anymore, while antibody A69 showed a reaction all over the gradient indicating not only dissociation but also aggregation of the capsid proteins.
Heat shock induces disappearance of the globules in DNA-containing AAV-2 capsids.
The observation of a difference in the accessibility of VP1 and VP2 N termini in empty and DNA-containing AAV capsids after heat treatment evoke the question of whether there is a difference at the structural level. Therefore, a mixture of wild-type full and empty AAV-2 capsids was investigated by electron cryomicroscopy. To distinguish between full and empty particles, the selected capsids from the micrographs were divided into four classes according to their density profiles (see Materials and Methods) (Fig. 4A). Afterward, 3D reconstructions were calculated for each class (Fig. 4B). The equatorial slices of the four reconstructions showed almost the same overall features. In particular, the globules at the twofold symmetry axes inside the capsid were present in empty (Fig. 4B, left) as well as in DNA-containing capsids (Fig. 4B, right).
FIG. 4.
Equatorial slices of wild-type AAV-2 3D image reconstructions after incubation at RT and 65°C. Capsids were incubated for 30 min at RT and analyzed by electron cryomicroscopy. Selected particles were sorted into four classes according to their density profile (A) before 3D image reconstructions were calculated for each class (B). Wild-type AAV-2 particles were treated at 65°C, and image reconstructions were calculated as described above (C). Several of the symmetry axes are marked in white.
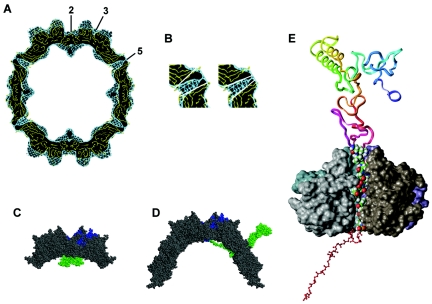
To understand the differences between empty and DNA-containing capsids in more detail, we calculated difference maps. Here, the imaging of the two types of capsids on the same micrograph was advantageous because modulations of the image information introduced by the contrast transfer of the microscope and the background of the carbon support film were the same for both types of capsids. Therefore, these effects cancelled out in a difference map. A surface representation of the resulting difference map from DNA-containing capsid and empty capsid (blue) was superimposed to a surface representation of the empty capsid (yellow) (Fig. 5). This superposition revealed that the globules at the twofold axes, which in empty capsids are formed only by protein, were deeply immersed in the difference density inside the capsid (Fig. 5, arrowhead). We think that this difference density corresponds to DNA, which is not present in the empty capsids. In addition, at this level of resolution, the protrusions at the fivefold symmetry axes on the outer surface extended further in DNA-containing capsids than in empty capsids, indicating a slight rearrangement in the protein shell.
FIG. 5.
Surface representation of the difference map between DNA-containing and empty AAV-2 capsids. A surface representation of the difference map from DNA-containing capsids and empty capsids (blue) was superimposed to a surface representation of half of the empty capsid (yellow). A direct view down a twofold symmetry axis (A) and a side view (B) are shown. One of the globules at the twofold symmetry axes inside the capsid is labeled by a white arrowhead.
In order to see whether the different reactions of full and empty capsids after treatment at 65°C (Fig. 3) correlated with a change in the structure, particles were analyzed by electron cryomicroscopy. After incubation at 65°C, viral capsids were still intact and no subfragments were visible (data not shown). For 3D reconstructions, the selected particles were again grouped according to their density profiles. In empty capsids, the globules at the twofold symmetry axes were maintained (Fig. 4C). In DNA-containing capsids, the globules were reduced, indicating a structural reorganization at 65°C. This result is in accordance with the accessibility of the VP1 and probably also VP2 N termini to specific antibodies (Fig. 3).
Comparison of the image reconstruction of AAV-2 with the atomic model of AAV-2.
Electron microscopy and image reconstruction is a valuable tool for obtaining structural information of variable less-rigid domains, which are not accessible to structural investigations by X-ray crystallography. Such variable domains are the N termini of the three structural proteins VP1, VP2 and VP3, which are not resolved in the crystallographic analysis of AAV-2 (64). Superposition of the reconstruction of the empty capsid with the atomic model of the capsid (64) showed that indeed the globules at the twofold symmetry axes and their suspensions stretching toward the fivefold symmetry axes were not matched by the atomic model (Fig. 6A). This lent further support to the hypothesis that the globules were formed by the N termini of the three structural proteins. Another obvious difference was the organization of the channels at the fivefold axes, which in the electron microscopy reconstruction, opened to the inside of the capsid (Fig. 6B, right), whereas in the atomic model, they opened toward the outside (Fig. 6B, left).
FIG. 6.
Image reconstruction compared to atomic model of AAV-2. Molecular modeling of the VP1 N-terminal region. The reconstruction of the empty capsid (blue) was superimposed with the atomic model of the capsid (1LP3) (yellow). (A) An equatorial slice of the capsids is shown. (B) Sections of a fivefold symmetry axis are marked with white lines to demonstrate the difference of the opening of the channels at the fivefold symmetry axes in the atomic model (left) and the 3D image reconstruction (right). (C) Part of an equatorial slice through an AAV-2 capsid where the N-terminal stretch of the VP1 protein (green) was modeled and linked to one of the two VP3 subunits (blue) at the twofold symmetry axis is illustrated. (D) Partial defolding of the VP1 terminus which allows its exposure through a channel at a fivefold symmetry axis where the PLA2 domain which is located within the N terminus becomes accessible on the capsid surface. (E) The partly defolded VP1 domain was fitted into the channel at the fivefold symmetry. The backbone of residues located outside the capsid (amino acids 1 to 168) is represented as a ribbon and color coded with sequence succession from blue (residues 1 through 10) to red (residues 160 through 170). The atoms of residues modeled within the channel (amino acids 169 to 185) are given as space filled model representation using an atom color code (green, carbon; white, hydrogen; blue, nitrogen; red, oxygen). The backbones of residues inside the capsid (amino acids 186 to 209) are colored red and represented by a ball and stick model. The solvent-accessible surface of the four displayed VP3 units forming the channel is depicted in various grey tones. The fifth VP3 domain is not displayed to enable a view of how the defolded VP1 strand fits into the channel.
Molecular modeling of the N-terminal region.
To get an idea of the possible fold of the N-terminal domains of VP1, molecular modeling was used. We could show that the VP1 model can occupy a space at the twofold symmetry axes similar to the globules (Fig. 6C) observed in the image reconstructions of wild-type capsids. To answer the question, if it could be geometrically possible that the PLA2 domain is exposed outside the capsid, the Tat-like part of the VP1 model (see Materials and Methods) was virtually defolded and oriented in such a way that a stretched part of this domain fit through a channel at the fivefold symmetry axes (Fig. 6D). It could be shown that the channel provided sufficient space to allow the passage of one stretched protein chain of the N-terminal region (Fig. 6E). This model would require a complete unfolding of the PLA2 domain and would not allow the transit of the helical fold which is present in the PLA2 domain. However, the different topology found for the channels at the fivefold symmetry axes in the image reconstruction and the atomic model (Fig. 6A) indicates that conformational exchanges can take place which may allow the thoroughfare of the helical folds.
DISCUSSION
We have shown that under physiological conditions, the globules on the inner surface of the AAV-2 capsid are genuine structural elements of empty and full capsids. The integrity of these structures requires the presence of VP1 and VP2 as demonstrated by the investigation of deletion mutants. According to the proposed stoichiometry of VP1:VP2:VP3 of 1:1:10, maximally one-third of the positions at the twofold symmetry axes could be occupied by VP1 or VP2 N termini. However, since the core density of the globules was comparable to the density of the protein shell (Fig. 1F and 4B), occupancy of the twofold axes close to 100% can be expected. This requires also that parts of the N termini of VP3, which are unresolved in the atomic model, are a component of the globules. Taking low occupancy and possible flexibility of the unique N termini of VP1 and VP2 into account, the size of the globules might still underestimate the true size required for accommodating the N termini.
The presence of the globules on the inside of the capsid is linked to no or very low accessibility of the N-terminal domains on the outside of the capsids. Yet the N-terminal domain of VP1 carries a PLA2 domain (66), for which accessibility is required for effective infection with AAV-2 (14). Therefore, as in other parvoviruses like minute virus of mice, canine parvovirus, or porcine parvovirus, this N-terminal domain has to become exposed. For some parvoviruses, the release of the VP1 N-terminal domain can be triggered by heat shock or urea treatment (11, 56) in vitro. Here, we used heat shock to initiate the release of the N-terminal domain of AAV-2. We could show that the structural integrity of the capsids was maintained while the N termini became accessible and the globules on the inside disappeared (Fig. 1D and 4). As in minute virus of mice, this conformational change occurred only in DNA-containing capsids (11). Comparison of empty and full capsids revealed that the DNA might be in intimate contact with the globules in the virus (Fig. 5).
It has been suggested for other parvoviruses that the N-terminal domains of VP1 and VP2 escape via the channels at the fivefold symmetry axes of the capsids (11, 56). Mutation of several conserved channel-forming amino acids prevents exposure of VP1 N termini and leads to the strongly reduced infectivity of AAV-2 (6). Based on the atomic model and assuming no conformational change of the VP3 backbone, our molecular dynamic simulations have shown that the passage of the VP1 N termini is in principle possible but requires the unfolding of the PLA2 domain (Fig. 6E). Spatial considerations lead to the assumption that only one N-terminal domain can penetrate one channel. This accounts for a maximum of 12 N-terminal domains (either VP1 or VP2) which can be released via the 12 channels. At the reported molar ratios of VP1:VP2:VP3 of 1:1:10 (28), the expected copy number of VP1 and VP2 is five copies each. Accordingly, the 12 channels would be sufficient to allow a complete release of all N-terminal domains of VP1 and VP2. It has been shown already that N-terminal extensions of VP2 of up to 30 kDa do not prevent capsid assembly and these extensions were detectable on the capsid surface by means of immuno reactions (57). The limited size of the channel at the fivefold symmetry axes makes it unlikely that the VP2 N terminus containing large folded domains transits the channel unless it becomes transiently unfolded. Alternatively, it might be that the N terminus and the insertion already become exposed during assembly.
In the mutants, the disappearance of the globules is coupled to a narrowing of the entrance to the channels on the inner surfaces of the capsids (Fig. 1F). A similar narrowing also occurred in full virus particles after heat shock (Fig. 4C). Interestingly, this region is also the region which, apart from the globules, differed most between the atomic model (64) and the image reconstruction of the empty capsid (Fig. 6A and B). Additionally, the highest temperature factors are reported for the residues forming the outer part of the channel. The narrower channel observed in the atomic model seemed more similar to the capsid structures where the globules were not observed. Preparation of capsids by repeated cesium chloride gradient fractionation and possible stress conditions during crystallization could have an influence on the localization of the flexible N termini within the capsid. This leads to the assumption that some rearrangement in the channel structure might facilitate the passage of the N-terminal domain. Unfortunately, it is unknown for the capsids used to calculate the atomic model whether the N termini of VP1 and/or VP2 are accessible on the outside.
In conclusion, we have provided the first structural insights into the underlying conformational changes by which the N-terminal domains of VP1 and possibly also VP2 can become accessible on the outsides of AAV-2 capsids. Such an alteration of accessibility is shared by most of the other parvoviruses and plays a major role in infectivity. It remains to be shown how, when, and where in the infection process the PLA2 domain becomes exposed by a conformational change of the capsid. It has recently been shown that, already 2 h after infection with CPV VP1, N termini are detectable by indirect immunofluorescence in a perinuclear compartment (51). Ongoing work suggests that the conformational change is indeed a prerequisite for efficient infection with AAV-2. This will pave the way to design more accurate experiments to probe the release of the N-terminal domains in vivo and in vitro.
Acknowledgments
We thank Dirk Grimm and Jason King for kindly providing plasmids, Petra Poberschin for technical support, and Barbara Leuchs for production of enriched full wild-type AAV-2 capsids.
REFERENCES
- 1.Agbandje, M., S. Kajigaya, R. McKenna, N. S. Young, and M. G. Rossmann. 1994. The structure of human parvovirus B19 at 8 A resolution. Virology 203:106-115. [DOI] [PubMed] [Google Scholar]
- 2.Agbandje-McKenna, M., A. L. Llamas-Saiz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 6:1369-1381. [DOI] [PubMed] [Google Scholar]
- 3.Arold, S., P. Franken, M. P. Strub, F. Hoh, S. Benichou, R. Benarous, and C. Dumas. 1997. The crystal structure of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complex in altered T cell receptor signaling. Structure 5:1361-1372. [DOI] [PubMed] [Google Scholar]
- 4.Bantel-Schaal, U., B. Hub, and J. Kartenbeck. 2002. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J. Virol. 76:2340-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 79:2528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Böttcher, B., N. Tsuji, H. Takahashi, M. R. Dyson, S. Zhao, R. A. Crowther, and K. Murray. 1998. Peptides that block hepatitis B virus assembly: analysis by cryomicroscopy, mutagenesis and transfection. EMBO J. 17:6839-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buller, R. M., and J. A. Rose. 1978. Characterization of adenovirus-associated virus-induced polypeptides in KB cells. J. Virol. 25:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canaan, S., Z. Zadori, F. Ghomashchi, J. Bollinger, M. Sadilek, M. E. Moreau, P. Tijssen, and M. H. Gelb. 2004. Interfacial enzymology of parvovirus phospholipases A2. J. Biol. Chem. 279:14502-14508. [DOI] [PubMed] [Google Scholar]
- 10.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., M. D'abramo, A., Jr., C. M. Ticknor, and P. Tattersall. 1999. Controlled conformational transitions in the MVM virion expose the VP1 N-terminus and viral genome without particle disassembly. Virology 254:169-181. [DOI] [PubMed] [Google Scholar]
- 12.Crowther, R. A., R. Henderson, and J. M. Smith. 1996. MRC image processing programs. J. Struct. Biol. 116:9-16. [DOI] [PubMed] [Google Scholar]
- 13.Dorsch, S., G. Liebisch, B. Kaufmann, P. von Landenberg, J. H. Hoffmann, W. Drobnik, and S. Modrow. 2002. The VP1 unique region of parvovirus B19 and its constituent phospholipase A2-like activity. J. Virol. 76:2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girod, A., C. E. Wobus, Z. Zadori, M. Ried, K. Leike, P. Tijssen, J. A. Kleinschmidt, and M. Hallek. 2002. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 83:973-978. [DOI] [PubMed] [Google Scholar]
- 15.Graham, F. L., and L. Prevec. 1991. Manipulation of adenovirus vectors. Methods Mol. Biol. 7:109-128. [DOI] [PubMed] [Google Scholar]
- 16.Greber, U. F. 2002. Signalling in viral entry. Cell. Mol. Life Sci. 59:608-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm, D., A. Kern, M. Pawlita, F. Ferrari, R. Samulski, and J. Kleinschmidt. 1999. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 6:1322-1330. [DOI] [PubMed] [Google Scholar]
- 18.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 19.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 20.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 21.Hansen, J., K. Qing, and A. Srivastava. 2001. Adeno-associated virus type 2-mediated gene transfer: altered endocytic processing enhances transduction efficiency in murine fibroblasts. J. Virol. 75:4080-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, J., K. Qing, and A. Srivastava. 2001. Infection of purified nuclei by adeno-associated virus 2. Mol. Ther. 4:289-296. [DOI] [PubMed] [Google Scholar]
- 23.Harlow, E., and F. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Hauswirth, W. W., A. S. Lewin, S. Zolotukhin, and N. Muzyczka. 2000. Production and purification of recombinant adeno-associated virus. Methods Enzymol. 316:743-761. [DOI] [PubMed] [Google Scholar]
- 25.Heilbronn, R., A. Burkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermonat, P. L., and N. Muzyczka. 1984. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc. Natl. Acad. Sci. USA 81:6466-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hooft, R. W., G. Vriend, C. Sander, and E. E. Abola. 1996. Errors in protein structures. Nature 381:272. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, F. B., H. L. Ozer, and M. D. Hoggan. 1971. Structural proteins of adenovirus-associated virus type 3. J. Virol. 8:860-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 30.Kasamatsu, H., and A. Nakanishi. 1998. How do animal DNA viruses get to the nucleus? Annu. Rev. Microbiol. 52:627-686. [DOI] [PubMed] [Google Scholar]
- 31.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kronenberg, S., J. A. Kleinschmidt, and B. Böttcher. 2001. Electron cryo-microscopy and image reconstruction of adeno-associated virus type 2 empty capsids. EMBO Rep. 2:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 34.Li, Y., Z. Zadori, H. Bando, R. Dubuc, G. Fediere, J. Szelei, and P. Tijssen. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82:2821-2825. [DOI] [PubMed] [Google Scholar]
- 35.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS3.0:A package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306-317. [Google Scholar]
- 36.Lombardo, E., J. C. Ramírez, J. Garcia, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 76:7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melo, F., and E. Feytmans. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141-1152. [DOI] [PubMed] [Google Scholar]
- 38.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication. Lippincott Williams and Wilkins, New York, N.Y.
- 39.Myers, M. W., and B. J. Carter. 1980. Assembly of adeno-associated virus. Virology 102:71-82. [DOI] [PubMed] [Google Scholar]
- 40.Potter, M., K. Chesnut, N. Muzyczka, T. Flotte, and S. Zolotukhin. 2002. Streamlined large-scale production of recombinant adeno-associated virus (rAAV) vectors. Methods Enzymol. 346:413-430. [DOI] [PubMed] [Google Scholar]
- 41.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 42.Reddy, V. S., P. Natarajan, B. Okerberg, K. Li, K. V. Damodaran, R. T. Morton, C. L. Brooks III, and J. E. Johnson. 2001. Virus Particle Explorer (VIPER), a website for virus capsid structures and their computational analyses. J. Virol. 75:11943-11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose, J. A., J. V. Maizel, Jr., J. K. Inman, and A. J. Shatkin. 1971. Structural proteins of adenovirus-associated viruses. J. Virol. 8:766-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by Rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seisenberger, G., M. U. Ried, T. Endress, H. Buning, M. Hallek, and C. Brauchle. 2001. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science 294:1929-1932. [DOI] [PubMed] [Google Scholar]
- 46.Simpson, A. A., V. Chandrasekar, B. Hebert, G. M. Sullivan, M. G. Rossmann, and C. R. Parrish. 2000. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses. J. Mol. Biol. 300:597-610. [DOI] [PubMed] [Google Scholar]
- 47.Simpson, A. A., P. R. Chipman, T. S. Baker, P. Tijssen, and M. G. Rossmann. 1998. The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 A resolution. Structure 6:1355-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinbach, S., A. Wistuba, T. Bock, and J. A. Kleinschmidt. 1997. Assembly of adeno-associated virus type 2 capsids in vitro. J. Gen. Virol. 78:1453-1462. [DOI] [PubMed] [Google Scholar]
- 50.Stratford-Perricaudet, L. D., I. Makeh, M. Perricaudet, and P. Briand. 1992. Widespread long-term gene transfer to mouse skeletal muscles and heart. J. Clin. Investig. 90:626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suikkanen, S., M. Antila, A. Jaatinen, M. Vihinen-Ranta, and M. Vuento. 2003. Release of canine parvovirus from endocytic vesicles. Virology 316:267-280. [DOI] [PubMed] [Google Scholar]
- 52.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. AlphaVbeta5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 53.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tratschin, J. D., I. L. Miller, and B. J. Carter. 1984. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J. Virol. 51:611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Heel, M., G. Harauz, E. V. Orlova, R. Schmidt, and M. Schatz. 1996. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116:17-24. [DOI] [PubMed] [Google Scholar]
- 56.Vihinen-Ranta, M., D. Wang, W. S. Weichert, and C. R. Parrish. 2002. The VP1 N-terminal sequence of canine parvovirus affects nuclear transport of capsids and efficient cell infection. J. Virol. 76:1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warrington, K. H., Jr., O. S. Gorbatyuk, J. K. Harrison, S. R. Opie, S. Zolotukhin, and N. Muzyczka. 2004. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 78:6595-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weger, S., A. Wistuba, D. Grimm, and J. A. Kleinschmidt. 1997. Control of adeno-associated virus type 2 cap gene expression: relative influence of helper virus, terminal repeats, and Rep proteins. J. Virol. 71:8437-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weichert, W. S., J. S. Parker, A. T. Wahid, S. F. Chang, E. Meier, and C. R. Parrish. 1998. Assaying for structural variation in the parvovirus capsid and its role in infection. Virology 250:106-117. [DOI] [PubMed] [Google Scholar]
- 60.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 61.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wobus, C. E., B. Hügle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao, W., K. H. Warrington, Jr., P. Hearing, J. Hughes, and N. Muzyczka. 2002. Adenovirus-facilitated nuclear translocation of adeno-associated virus type 2. J. Virol. 76:11505-11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 A resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]
- 66.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]