Abstract
Foamy viruses (FV) are unusual among retroviruses since they require both Gag and Env structural proteins for particle egress. Recently significant progress has been made towards the mechanistic understanding of the viral release process, in particular that of retroviruses, and the viral domains and cellular pathways involved. However little is currently known about domains of FV structural proteins and cellular proteins engaged in this process. By mutational analysis of sequence motifs in prototype FV (PFV) Gag, bearing homology to known late assembly (L) domains, a PSAP motif with L domain function that was functionally interchangeable by heterologous L domains was identified. In contrast the inactivation of a PPPI motif had no significant influence on PFV particle release, although mutant viral particles displayed reduced infectivity. Similarly mutation of an evolutionary conserved YXXL motif revealed no classical L-domain function but resulted in release of noninfectious viruslike particles. Biochemical and electron microscopy analysis demonstrated that these mutant particles incorporated all viral structural proteins but contained aberrantly capsid structures, suggesting a role in capsid assembly for this PFV Gag sequence motif. In line with the mutational analysis, overexpression of dominant negative (DN) mutants and wild-type TSG101 but not the DN mutant of AIP-1/ALIX reduced PFV particle release and infectivity. Furthermore, DN mutants of Vps4A, Vps4B, and CHMP3 inhibited PFV egress and infectivity. Taken together these results demonstrate that PFV, like other viruses, requires components of the vacuolar protein sorting (VPS) machinery for egress and enters the VPS pathway through interaction with TSG101.
Particle release from infected cells is one of the last steps in the retroviral replication cycle, which is accomplished by budding of viral particles across cellular membranes. On the viral side the orthoretroviral Gag polyprotein contains all the essential structural information for particle egress, and Gag expression by itself is sufficient for the release of viruslike particles (VLP). Although for some orthoretroviruses, such as Mason-Pfizer monkey virus (MPMV), coexpression of Env enhances viral particle release (37). In recent years significant advances have been made in the understanding of the mechanistic processes involved in the retroviral budding process. In particular the interaction domains of the viral Gag protein with essential cellular factors required during late stages of the budding process and involved in pinching off the viral particle from the cellular membrane have been identified. To date three consensus sequences of so-called viral late-budding or late-assembly (L) domains have been characterized. A P(T/S)AP L-domain motif originally identified in the human immunodeficiency virus type 1 (HIV-1) Gag p6 domain (13), a PPXY L-domain motif first found in the Rous sarcoma virus (RSV) Gag p2b cleavage product (47) and a YPXL motif present in the equine infectious anemia virus (EIAV) Gag p9 domain (33). More recently a LXXL sequence motif present in HIV-1 p6 and in EIAV p9, here overlapping the YPXL motif, has been characterized (40, 44). The cellular interaction partners for different L-domain sequence motifs have also been identified, linking the budding process to the ubiquitylation machinery and the vacuolar protein sorting (VPS) pathway responsible for sorting of cargo to the multivesicular body. The PPXY-type L domains have been shown to bind to WW domain-containing proteins, in particular the Nedd4 family of ubiquitin ligases (10). Binding of TSG101, a member of ESCRT-I (endosomal sorting complex required for transport I) of the VPS machinery (reviewed in reference 17), has been demonstrated for PSAP L domains of different viruses (3, 11, 27, 42), whereas the YPXL and LXXL L-domain motifs are interacting with AIP-1/ALIX (25, 40, 44), a cellular factor that apparently links ESCRT-I and ESCRT-III complexes. Dominant negative (DN) mutants of and small interfering RNAs specific for TSG101 or AIP-1/ALIX inhibit P(S/T)AP and YPXL L-domain-mediated particle release, respectively (3, 11, 25, 40). DN mutants of the AAA-type ATPase Vps4, a class E VPS protein that normally disassembles and thereby recycles the ESCRT machinery, potently inhibit the release of viruses that use all three types of L domains, indicating that elements of the class E VPS sorting machinery are required for the function of all types of L domains (11, 28).
Spumaretroviruses or foamy viruses (FV) use a replication pathway with features distinctive from orthoretroviruses (reviewed in reference 35). The particle release process in particular is unique among retroviruses since coexpression of FV Gag and Env proteins is essential for this process. The FV Env protein cannot be functionally replaced by heterologous viral glycoproteins, suggesting a specific interaction of FV Gag and Env proteins during budding (reviewed in reference 21). The Env domain required for this interaction has recently been identified and resides at the N terminus of the Env leader peptide (LP) containing an essential, conserved WXXW sequence motif (22, 46). The crucial interaction domains in FV Gag are less well characterized, although sequence-specific binding of bacterially expressed N-terminal feline FV (FFV) Gag and Env LP has been demonstrated biophysically (46). The requirement of Env coexpression for particle release indicates that Env itself may contain structural information for this process that in orthoretroviruses is normally associated with the Gag protein. This is further supported by the recent observation that expression of prototype FV (PFV) Env by itself in the absence of other FV proteins leads to the release of subviral particles into the supernatant (38). A similar subviral particle release is observed upon expression of the vesicular stomatitis virus (VSV) glycoprotein G or the hepatitis B virus S protein (4, 45). However, in the case of hepatitis B virus, subviral particles are secreted in vast excess over infectious viral particles, whereas subviral particle release of PFV and VSV seems to be quite inefficient (29, 38, 45).
Morphologically, FV preassemble their capsids similarly to B/D-type retroviruses in the cytoplasm of the infected cells. However, in contrast to orthoretroviruses little information about specific FV Gag domains involved in particle assembly and budding across membranes is available (reviewed in reference 24). The FV Gag protein contains a sequence homologous to the cytoplasmic targeting retention signal of B/D-type retroviruses that is thought to be responsible for intracytoplasmic capsid assembly. However, unlike the report for MPMV (36), mutation of the FV cytoplasmic targeting retention signal completely abolishes particle assembly and does not lead to a redirection to the plasma membrane (7). The current model suggests that membrane association of FV capsids is mediated by the specific interaction with the Env protein (reviewed in reference 21). Consistent with this, expression of the FV Gag protein in the absence of the homologous Env protein leads to an accumulation of naked capsids in the cytoplasm, and no association of viral capsids with and budding across cellular membranes is observed (1, 8). However, recently Eastman and Linial (7) demonstrated an FV-Env-independent particle release for a PFV Gag mutant, having the N-terminal 10 amino acids replaced by the myristoylation signal sequence of the cellular Src protein. Though, for currently unknown reasons the secreted VLP were noninfectious. This suggests that if targeted to cellular membranes, PFV Gag contains all necessary structural information required for particle egress.
In this study we intended to characterize domains in the PFV Gag protein that bear homology to known L-domain sequence motifs and analyze their role in the viral replication cycle, in particular with respect to the particle release process.
MATERIALS AND METHODS
Cells.
The human kidney cell line 293T (5) and the human fibrosarcoma cell line HT1080 (34) were cultivated in Dulbecco′s modified Eagle′s medium supplemented with 10% fetal calf serum and antibiotics.
Expression constructs.
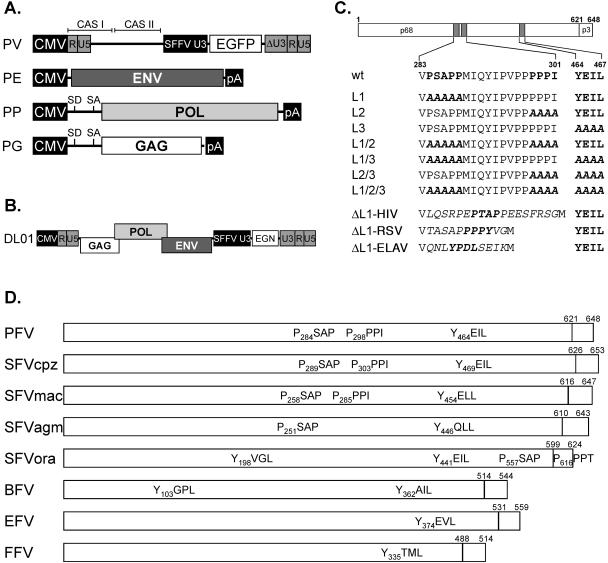
A schematic outline of the constructs used in this study is shown in Fig. 1. The four-plasmid PFV vector system consisting of the PFV Gag expression vector pcziPG (pcziGag2), the PFV polymerase (Pol) expression vector pcziPol, the PFV Env expression construct pczHFVenvEM002, and the enhanced green fluorescent protein (EGFP) expressing PFV transfer vector pMD9 has been described previously (12). The putative PFV Gag L-domain consensus sequences in the pcziPG expression vector were mutated using standard PCR cloning techniques and mutagenesis primers. Details are available on request. The mutants were termed pcziPG L1 to L3 and combination mutants thereof accordingly (e.g., pcziPG L1/2). In mutant L1 the PSAPP motif starting at amino acid 284 was changed to AAAAA, in L2 the PPPI motif starting at 298 was changed to AAAA, and in the L3 mutant the YEIL motif starting at amino acid 464 was changed to AAAA. Furthermore, the mutants L1/2, L1/3, L2/3, and L1/2/3 were generated, containing combinations of the individual mutants. The ΔL1 L-domain reconstitution constructs have the PSAPP motif in PFV Gag deleted and replaced by heterologous L-domain sequence motifs. The ΔL1-HIV construct has the amino acid sequence LQSRPEPTAPPEESFRSG of HIV-1 Gag p6 inserted, ΔL1-RSV contains the complete RSV Gag p2b peptide (TASAPPPPYVG), and ΔL1-EIAV harbors the peptide QNLYPDLSEIK with the L domain of EIAV Gag p9. The replication-deficient PFV vector pDL01 expressing PFV Gag, Pol, and Env from a chimeric 5′ long terminal repeat (LTR) having the U3 region replaced by the cytomegalovirus promoter and an EGFP-neomycin (EGN) marker gene from an internal spleen focus-forming virus U3 promoter has been described previously (31). Variants thereof containing mutations in the putative L-domain sequences were termed pDL01 L1 to L3, and combination mutants were named accordingly, e.g., pDL01 L1/2, and were generated by exchanging an XbaI/SwaI restriction fragment in pDL01 with the corresponding fragment of the respective pcziPG construct. For analysis of cellular proteins involved in PFV particle release, the following previously described expression constructs were used: full-length TSG101 (TSG-F), N-terminal truncation mutant (TSG-3′), C-terminal truncation mutant (TSG-5′) (3, 41); GFP-VPS4A, GFP-VPS4A(K173Q), GFP-VPS4A(E228Q) (11); DsRed-VPS4B, DsRed-VPS4B(K180Q), DsRed-VPS4B(E235Q) (44); CHMP3-RFP (40); YFP-d1-176AIP-1/ALIX (26).
FIG. 1.
Schematic illustration of the expression constructs and PFV Gag mutants. (A) Schematic outline of the four-plasmid PFV vector system consisting of a gene transfer vector (PV) as well as Env (PE), Pol (PP), and Gag (PG) expression vectors. (B) Schematic outline of the one-plasmid PFV vector (DL01) system. (C) Schematic organization of the PFV Gag precursor protein and processing products p68 and p3. The locations of the putative L-domain motifs are indicated as hatched boxes. Below, the sequences of the indicated specific regions of wild-type and mutant PFV Gag proteins are shown. Putative L-domain sequence motifs are highlighted in bold, and amino acids altered in the mutant constructs are marked by italic letters. (D) Putative L-domain consensus motifs in Gag proteins of different FV species. GenBank accession numbers for the cited viral genomes are NC001736 for PFV, U04327 for chimpanzee simian FV (SFVcpz), X54482 for macaque simian FV (SFVmac), M74895 for African green monkey simian FV (SFVagm), AJ544579 for orangutan simian FV (SFVora), U94514 for bovine FV (BFV), AF201902 for equine FV (EFV), and U85043 for FFV. Abbreviations: CAS I and II: cis-acting sequences I and II; CMV: cytomegalovirus promoter; R: LTR repeat region; U5: LTR unique 5′ region; U3: LTR unique 3′ region; ΔU3: enhancer-deleted U3 region; SFFV U3: spleen focus-forming virus U3 promoter; EGN: EGFP-Neo fusion gene; SD: splice donor; SA: splice acceptor; pA: polyadenylation signal sequence.
Generation of viral supernatants and analysis of transduction efficiency.
FV supernatants containing recombinant viral particles were generated essentially as described earlier (20, 23) by cotransfection of 293T cells either with pMD9, pczHFVenvEM002, pcziPol, and pcziPG or with mutants thereof as indicated. Alternatively, cells were transfected with the proviral vector construct pDL01 and mutants thereof. Extra- and intracellular viral particles were harvested as described previously (22) at 48 h posttransfection for analysis of L-domain mutations or 24 h posttransfection upon cotransfection of expression constructs for DN mutants of cellular proteins. Transductions of recombinant EGFP- or EGN-expressing PFV vector particles were performed by infection of 2 × 104 HT1080 cells plated 24 h in advance in 12-well plates for 4 to 6 h using 1 ml of viral supernatant or dilutions thereof. The percentage of GFP-positive cells was determined by fluorescence-activated cell sorter analysis 48 to 72 h after infection. All transduction experiments were performed at least three times, and in each independent experiment the values obtained with wild-type pcziPG or pDL01 were arbitrarily set to 100%.
Antisera, Western blot expression analysis, and quantification of particle release.
Western blot expression analysis of cell- and particle-associated viral proteins was performed essentially as described previously (22). Polyclonal antisera used were specific for PFV Gag (2) or the LP of PFV Env, amino acids 1 to 86 (22). Furthermore, hybridoma supernatants specific for PFV reverse transcriptase (clone 15E10) or PFV integrase (clone 3E11) were employed in some experiments (14). The chemiluminescence signal or the fluorescent signal was digitally recorded using an LAS-3000 imager (Fujifilm) or an FLA-2000 phosphoimager (Fujifilm), respectively, and quantified using the Image Gauge software package (Fujifilm). Particle release was determined in independent experiments at relative levels compared to the wild-type PFV Gag control, and values were corrected for differential intracellular Gag expression levels in the individual samples.
Electron microscopy analysis.
At 48 h posttransfection, the 293T cells were harvested and processed for electron microscopy analysis as described previously (18).
RESULTS
Identification of late domain consensus sequences in the PFV Gag protein.
Currently three major L-domain consensus sequences are known: P(S/T)AP, PPXY, and YPXL. Inspection of the PFV Gag sequence revealed the presence of sequences closely or completely matching all three L-domain consensus sequences, a PSAP motif, a PPPI motif, and a YEIL motif starting at amino acids 284, 298, and 464, respectively (Fig. 1C and D). As illustrated schematically in Fig. 1D the YXXL motif is conserved between FV of different species and is duplicated in bovine FV and the recently characterized SFVora isolate (43). All primate FV isolates contain a PSAP motif, and a PPPX motif is found in all primate FV Gag proteins except for SFVagm, whereas both motifs are absent in FFV, bovine FV, and equine FV.
Generation of putative L-domain mutants and analysis of viral infectivities.
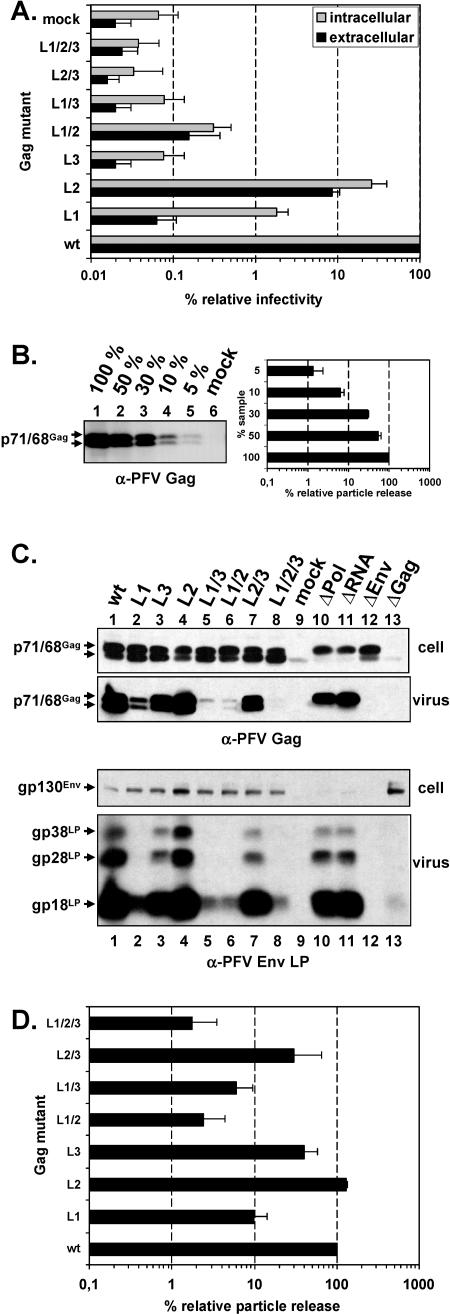
In order to elucidate the potential function of the individual PFV sequence motifs L1 (PSAPP), L2 (PPPI), and L3 (YEIL) as L-domains, we generated individual and combination mutants having the amino acids of the putative L-domain motifs changed to alanine. We used a four-plasmid PFV vector system (Fig. 1A) in which the Gag proteins are synthesized from a separate expression vector to avoid interference by effects of the mutations introduced into PFV Gag on viral genome packaging or Pol expression with the L-domain activity. In particular, the L3 mutation in which the YXXL motif was altered is partially overlapping the natural splice acceptor used for Pol expression in the proviral context. Wild-type and mutant Gag expression constructs were cotransfected in 293T cells and subsequently secreted, and cell-bound infectivities were determined in a GFP marker transfer assay on HT1080 target cells. The result is shown in Fig. 2A. This analysis revealed a 1,500-fold decrease of extracellular infectivity and a 50-fold decrease in cell-bound infectivity relative to the wild type for the PFV Gag L1 (PSAP) mutant. In contrast, the secreted and cell-bound infectivities of the PFV Gag L2 (PPPI) mutant were decreased only 10-fold and 4-fold, respectively. For the PFV Gag L3 (YEIL) mutant no infectivity, neither cell-associated nor in the supernatant, could be detected. The infectivities of the combination mutants PFV Gag L1/2, L1/3, L2/3, and L1/2/3 were comparable to that of the respective individual mutants with the strongest phenotype. In particular, all mutants affecting L3 were noninfectious.
FIG. 2.
Analysis of PFV Gag mutants using the four-plasmid vector system. Indicated mutant PFV particles were generated by transienttransfection of 293T cells using the four-plasmid PFV vector system. (A) Relative infectivity of cell supernatants (extracellular) and freeze-thaw cell lysates (intracellular) using the GFP marker gene transfer assay. The values obtained using the wild-type PFV Gag expression plasmid (wt) were arbitrarily set to 100%. The mean values and standard deviations of results from three independent experiments are shown. (B) Western blot analysis and quantification (n = 3) of serial dilution of wild-type PFV Gag particle lysates using polyclonal anti-PFV Gag (α-PFV Gag) antiserum. (C) Representative Western blot analysis of 293T cell lysates (cell) and viral particles (virus) purified by ultracentrifugation through 20% sucrose using polyclonal anti-PFV Gag (α-PFV Gag)- and anti-PFV Env LP (α-PFV Env LP)-specific antisera. Viral proteins are indicated on the left. (D) Quantification of PFV particle release. Mean values and standard deviation of particle-associated PFV Gag protein corrected for intracellular expression levels (n = 3) are shown. Lanes 1 to 9, cotransfection of pMD9, pcziPol, and pczHFVenvEM002 and pcziPG (wt), pcziPG L1 (L1), pcziPG L3, pcziPG L2 (L2), pcziPG L1/3 (L1/3); pcziPG L1/2 (L1/2), pcziPG L2/3 (L2/3); pcziPG L1/2/3 (L1/2/3), and pcDNA3.1+zeo (mock). Lanes 10 to 13, cotransfection of pMD9, pcziGag, pcziPol, and pczHFVenvEM002, replacing the indicated plasmid with pcDNA3.1+zeo as follows: pcziPol (ΔPol), pMD9 (ΔRNA), pczHFVenvEM002 (ΔEnv), and pcziPG (ΔGag).
Next we examined particle release of the different PFV Gag mutants by Western blot analysis of cell lysates and pelleted viral particles from 293T cells transfected with the four-plasmid PFV vector system. Western blot analysis and quantification of serial dilutions of purified wild-type PFV particles revealed a good linearity of the signal over a 20-fold concentration range (Fig. 2B). A typical analysis result for the PFV Gag L-domain mutants is shown in Fig. 2C. Cellular expression levels of most mutants were similar to those of the wild type except for the L2 and L1/2 mutants that displayed a somewhat diminished expression (Fig. 2C, lanes 4 and 6). PFV particle release was most affected by the L1 mutation having the PSAP motif altered (Fig. 2C, lane 2), showing a 10-fold decrease (Fig. 2D). For the L2 mutant a particle release similar to that of the wild type was observed (Fig. 2C, lane 4). Surprisingly, the L3 mutation led to only a threefold reduction in particle secretion (Fig. 2C, lane 3), although no infectivity could be detected (Fig. 2A). The combination mutants resulted in a further fourfold, twofold, and sixfold decrease for the L1/2, L1/3 and L1/2/3 mutants in comparison to the L1 mutant, respectively (Fig. 2C, lanes 2, 5, 6, and 8). The L2/3 mutant displayed a particle release similar to that of the L3 mutant (Fig. 2C, lanes 3 and 7).
PFV morphogenesis and particle release are different from these processes in orthoretroviruses insofar as coexpression of Gag and PFV Env (Fig. 2C, lanes 12 and 13) is essential for release of VLP. Omitting either the Pol protein (Fig. 2C, lane 10) or a packable vector genome (Fig. 2C, lane 11) leads to secretion of VLP, in which the p71 Gag precursor protein is not processed into p68. The analysis of the PFV Gag mutants revealed that the Gag processing was not affected, although the mutants released viral particles at different levels (Fig. 2C). This indicated that the mutations were only affecting particle egress but not viral genome packaging or Pol incorporation. Western blot analysis of Pol incorporation and RNase protection analysis of viral particle-associated RNA confirmed this assumption (data not shown). In addition, FV Env-specific Western blot analysis of particle lysates suggested a correlation of Gag and Env secretion for the individual mutants, indicating that they incorporate similar relative amounts of Env per particle (Fig. 2C). The greater molecular weight forms gp28LP and gp38LP are posttranslational modifications of the major particle-associated Env LP cleavage product gp18LP (6; N. Stanke and D. Lindemann, unpublished data).
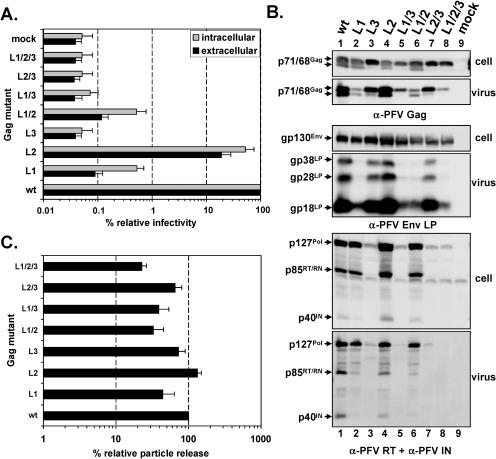
In addition we analyzed the putative L-domain mutants in the context of a one-plasmid FV vector system, in which Gag, Pol, Env mRNAs, and the EGN marker gene containing genomic RNA are expressed from the same promoter (Fig. 1B). Although the effects on particle release were not as pronounced as observed using the four-plasmid vector system, the analysis revealed similar phenotypes for the individual mutants in respect to particle release and infectivity (Fig. 3). However, as mentioned earlier the YEIL sequence motif, which is altered in all vector constructs harboring the L3 mutation, partially overlaps the Pol splice acceptor site in the Gag open reading frame, and indeed these mutants showed an almost complete block in FV Gag processing as a result of strongly reduced cellular Pol expression in the one-plasmid vector system (Fig. 3B).
FIG. 3.
Analysis of PFV Gag mutants using the one-plasmid vector system. Indicated mutant PFV particles were generated by transient transfection of 293T cells using the one-plasmid PFV vector system. (A) Relative infectivity of cell supernatants (extracellular) and freeze-thaw cell lysates (intracellular) using the GFP marker gene transfer assay. The values obtained using the wild-type PFV Gag expression plasmid (wt) were arbitrarily set to 100%. The mean values and standard deviations of results from at least three independent experiments are shown. (B) Representative Western blot analysis of 293T cell lysates (cell) and viral particles (virus) purified by ultracentrifugation through 20% sucrose using polyclonal anti-PFV Gag (α-PFV Gag)- or anti-PFV Env LP (α-PFV Env LP)-specific antisera or a mixture of anti-PFV reverse transcriptase (α-PFV RT) and anti-PFV integrase (α-PFV IN) monoclonal antibodies. Viral proteins are indicated on the left. (C) Quantification of PFV particle release. Mean values and standard deviation of particle-associated PFV Gag protein corrected for intracellular expression levels (n = 3) are shown. Cells were transfected with the following: pDL01 (wt), pDL01 L1 (L1), pDL01 L2 (L2), pDL01 L3 (L3), pDL01 L1/2 (L1/2), pDL01 L1/3 (L1/3), pDL01 L2/3 (L2/3), pDL01 L1/2/3 (L1/2/3), and pcDNA3.1+zeo (mock).
Taken together these data indicate that the PSAP motif in PFV Gag contains the major L-domain activity for PFV particle release. The YEIL and PPPI motifs seem to play only a minor role for PFV particle release, with the effect of the PPPI motif only becoming apparent in the absence of a functional PSAP motif if at all.
Electron microscopy analysis of particle morphogenesis and release of PFV Gag L-domain mutants.
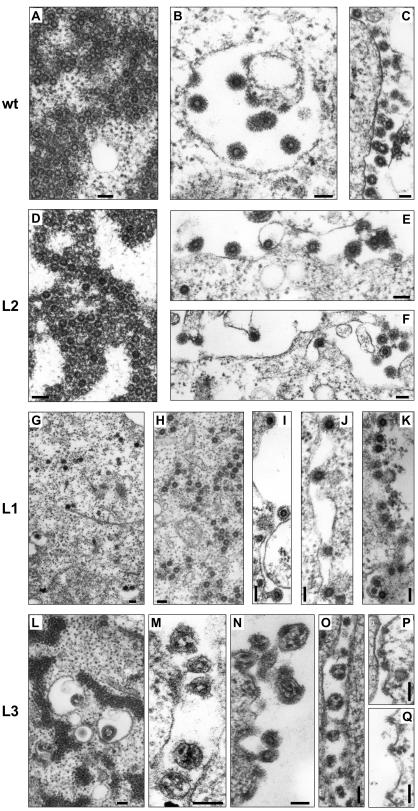
In order to examine the particle morphogenesis and particle release process of the individual mutants in more detail we performed electron microscopy analysis of 293T cells transfected with the expression constructs for the PFV Gag mutants using either the four-plasmid or the one-plasmid vector system. In wild-type PFV Gag samples budding and release of PFV particles were readily observed at intracellular membranes and the plasma membrane (Fig. 4B and C). In addition accumulated naked capsids embedded in electron-dense material were frequently observed in the cytoplasm (Fig. 4A). The L2 mutant samples were indistinguishable from that of the wild type (Fig. 4D to F). In contrast, very little particle release could be observed in L1-mutant-expressing cells. In these samples many capsids were found to be associated with cellular membranes, containing the prominent Env glycoprotein spike structures and apparently blocked in early steps of the budding process (Fig. 4I to K). Occasionally the typical stalk structures that have been reported for L-domain mutants of other viruses were observed (Fig. 4I). Furthermore, in these cells the aggregation of naked capsids in the cytoplasm was greatly diminished, and quite often isolated capsids that were never observed in wild-type Gag samples were detected (Fig. 4G and H). More importantly, the electron-dense staining observed around the aggregated wild-type (Fig. 4A) or L2 capsids (Fig. 4D) was almost completely absent in the L1 mutant samples (Fig. 4G and H).
FIG. 4.
Electron microscopy analysis of transfected 293T cells. Electron micrographs showing representative thin sections of transiently transfected 293T cells using the four- (A, C-F, H-I, L-N) or one-plasmid (B, G, J-K, O-Q) vector system. (A-C) Wild-type PFV Gag expression constructs. (D-F) L2 mutant PFV Gag expression constructs. (G-K) L1 mutant PFV Gag expression constructs. (L-Q) L3 mutant PFV Gag expression constructs. Magnifications: (A) ×52,000, (B) ×64,000, (C) ×40,000, (D-E) ×55,000, (F) ×42,000, (G) ×23,500, (H) ×33,000, (I) ×52,000, (J) ×50,000, (K) ×45,500, (L) ×34,000, (M) ×62,000, (N) ×100,000, (O) ×54,000, (P-Q) ×62,000. Bar size, 100 nm.
The L3 mutant showed a very unique phenotype. Regular capsid structures were not observed in L3-expressing samples, neither at cellular membranes nor in the cytoplasm (Fig. 4L to Q). On the other hand, in the cytoplasm quite frequently aggregates of electron-dense material, perhaps containing mutant Gag proteins, were seen (Fig. 4L). Particulate structures were observed at cellular membranes that included the typical FV Env glycoprotein spike structures; however, they were lacking the normal ring-shaped electron-dense FV capsid structures (Fig. 4M to Q). Instead, these particles enclosed electron-dense punctuated structures, which might represent aberrantly assembled Gag proteins. Thus, the electron microscopy analysis supports the view that the PSAP domain is the functional L domain in PFV Gag, whereas the YEIL sequence motif seems to be involved in capsid assembly.
Rescue of PSAP-deficient PFV Gag mutants by insertion of heterologous L domains.
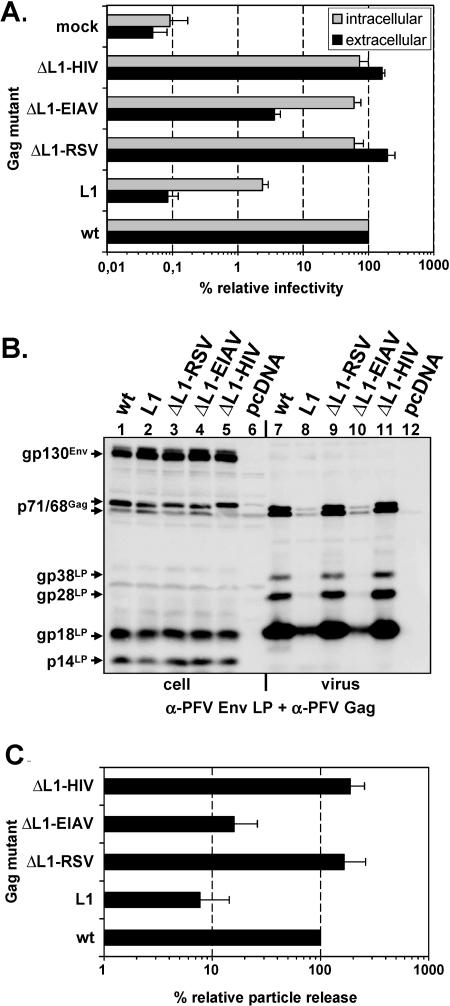
It has been reported previously that L domains of different viruses are functionally interchangeable (19, 27, 30, 49) although there seems to exist a certain context or position dependence, at least in some cases (25). The experiments presented above indicated that the PSAP motif in Gag represents the main functional L domain of PFV. To address the functional interchangeability of the PFV PSAP L domain, we generated PFV Gag mutants with the PSAP motif deleted and peptides inserted spanning the PTAP L-domain motif of HIV-1 p6, the PPPY L-domain motif of RSV p2b, or the YPDL L-domain motif of EIAV p9 (Fig. 1C). Analysis of the infectivity of these mutants revealed that the HIV-1 p6 PTAP or RSV p2b PPPY domain completely restored supernatant infectivity to wild-type levels (Fig. 5A). Cell-bound infectivity for both mutants was restored to 70 to 80% of that of the wild type (Fig. 5A). In contrast, insertion of the EIAV p9 YPDL L domain resulted in a similar increase of cell-bound infectivity to nearly wild-type levels, but extracellular infectivity was only partially restored, to about 15% of that of wild-type PFV Gag (Fig. 5A). Quantification of mutant particle release showed that the HIV-1 p6 PTAP or the RSV p2b PPPY motif in addition to infectivity also restored particle release to wild-type levels (Fig. 5C). In contrast, insertion of the EIAV YPDL motif led to only a marginal twofold increase in particle release in comparison to the L1 mutant, to about 15% of that of wild-type PFV Gag (Fig. 5C). Interestingly, for this mutant a direct correlation in the level of decrease of infectivity and particle release compared to wild-type PFV Gag could be observed, whereas for the L1 mutant infectivity was always much more reduced than particle release (Fig. 5A and C).
FIG. 5.
Analysis of PFV Gag ΔL1 L-domain replacement mutants. Indicated mutant PFV particles were generated by transient transfection of 293T cells using the four-plasmid PFV vector system. (A) Relative infectivity of cell supernatants (extracellular) and freeze-thaw cell lysates (intracellular) using the GFP marker gene transfer assay. The values obtained using the wild-type PFV Gag expression plasmid (wt) were arbitrarily set to 100%. The mean values and standard deviations of results from at least three independent experiments are shown. (B) Representative Western blot analysis of 293T cell lysates (cell, lanes 1 to 6) and viral particles (virus, lanes 7 to 12) purified by ultracentrifugation through 20% sucrose using a mixture of polyclonal anti-PFV Gag (α-PFV Gag)- and anti-PFV Env LP (α-PFV Env LP)-specific antisera. (C) Quantification of PFV particle release. Mean values and standard deviation of particle-associated PFV Gag protein corrected for intracellular expression levels (n = 3 to 5) are shown. Cotransfection of pMD9, pcziPol, and pczHFVenvEM002 and pcziPG (wt); pcziPG L1 (L1); pcziPG ΔL1-RSV (ΔL1-RSV); pcziPG ΔL1-EIAV (ΔL1-EIAV); pcziPG ΔL1-HIV (ΔL1-HIV); pcDNA3.1+zeo (mock).
Taken together these data suggest that the function of the PFV PSAP L domain can be completely replaced by an analogous HIV-1 p6 PTAP domain or the heterologous RSV p2b PPPY domain. In contrast to that the EIAV p9 YPDL domain motif is only capable to correct the infectivity defects of PSAP deleted intra- and extracellular PFV particles but not the particle release defect. Because there was a direct correlation of the reduction infectivity and particle release measured for this mutant, this suggests that the released mutant particles have a relative infectivity per particle similar to that of wild-type particles.
Inhibition of PFV infectivity by dominant-negative cellular proteins.
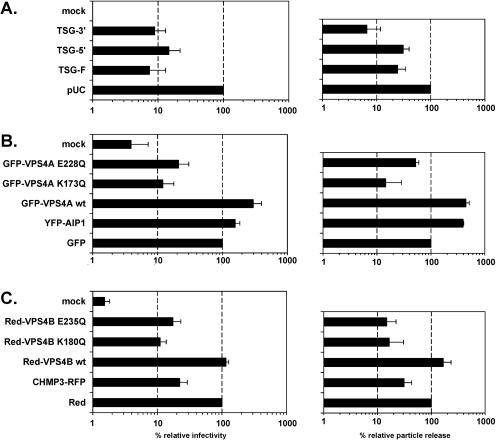
Recently, several cellular proteins that either directly interact with different types of viral L domains or which are essential at late stages of the cellular VPS pathway exploited by different viruses for particle release have been characterized (reviewed in references 9 and 32). DN mutants for a number of these proteins that inhibit viral particle release have been described (3, 11, 27, 40, 44). In order to analyze which cellular proteins are required for PFV particle release and might interact with PFV L domains, we cotransfected the EGN marker gene expressing PFV vector pDL01 together with various DN mutants and wild-type controls of cellular proteins into 293T cells and subsequently determined the supernatant infectivity and particle release. The results of these experiments are summarized in Fig. 6. Similar to the result reported for HIV-1 (3), a 7- to 14-fold reduction in infectivity and 3- to 15-fold lower particle release were observed upon cotransfection with TSG-F or TSG-5′ or TSG-3′ (Fig. 6A). Furthermore, the ATPase-deficient DN mutants of Vps4a, GFP-VPS4A E228Q, and GFP-VPS4A K137Q (Fig. 6B), or Vps4B, Red-VPS4B E235Q, and Red-VPS4B K180Q inhibited PFV particle release and infectivity (Fig. 6C). Cotransfection of the corresponding wild-type constructs, GFP-VPS4A wt or Red-VPS4B wt, had no detrimental effect on PFV infectivity and particle release. On the contrary, the wild-type Vps4a protein even increased PFV particle release fourfold and infectivity threefold (Fig. 6B). In addition, coexpression of a DN AIP-1/ALIX protein (YFP-AIP1), which had previously been shown to inhibit EIAV p9-mediated particle release (28), led to a slight but consistent threefold increase in PFV particle release associated with a marginally elevated infectivity detected in the culture supernatant (Fig. 6B). Similar to the DN forms of Vps4A and Vps4B, acting late in the VPS pathway, PFV particle release and infectivity were inhibited by a DN CHMP3 protein (CHMP3-RFP) that also inhibits HIV-1 and EIAV particle release (40). This indicates that an intact ESCRT-III complex is required for the PFV release process and that PFV links to the VPS pathway through an interaction with TSG101.
FIG. 6.
Inhibition of PFV infectivity by dominant negative mutants of cellular proteins. Wild-type PFV particles were generated in conjunction with overexpression of various cellular proteins by cotransfection of cells with the PFV vector pDL01 and different expression constructs as indicated at a 1:1 DNA weight ratio. Viral supernatants were harvested 24 h posttransfection. Relative infectivity (left panel) was determined by using the GFP marker gene transfer assay and calculated by arbitrarily setting the values obtained upon cotransfection of control expression constructs (pUC, GFP, Red) to 100%. The mean values and standard deviation of results from at least three independent experiments are shown. Similarly, the relative particle release (right panel) with respect to the same control samples was determined with adjustment of differential intracellular Gag expression levels in individual samples. The mean values and standard deviation of results from at least three independent experiments are shown. (A) Cotransfection of different TSG101 expression constructs. (B) Cotransfection of various GFP or YFP fusions of Vps4A or AIP-1/ALIX. (C) Cotransfection of different Red fusions of Vps4B or CHMP3. mock: uninfected target cells.
DISCUSSION
FV display highly unusual particle egress among retroviruses since both Gag and Env structural proteins contribute to this process (reviewed in references 21 and 24). The PFV Gag protein contains sequences homologous to all three currently known L-domain sequence motifs, whereas such sequence motifs are absent from the intracellular or membrane-spanning domains of the PFV glycoprotein. PFV Gag harbors a PSAP motif that exactly matches the P(S/T)AP consensus sequence. However, while it is found in all primate FV, this sequence motif is absent in all nonprimate FV isolates. Furthermore, a PPPI motif present in most primate FV and a YEIL sequence conserved in all FV isolates have high homology to the prototypic PPXY and YPXL L domains, although the tyrosine at position 4 of the PPXY motif and the proline at position 2 of the YPXL motif were shown to be critical for L-domain function (16, 33, 47, 48). Mutation analysis of the individual PFV Gag sequence motifs strongly suggests that the PSAP motif represents the functional PFV L domain, since both infectivity and particle release were strongly reduced upon inactivation. This is further supported by the particle release analysis of C-terminal PFV Gag truncation mutants showing that deletion of the YEIL and PPPI motifs had no influence on particle export, whereas mutants also lacking the PSAP motif displayed a severe reduction in VLP release (M. Cartellieri, unpublished observations). The PPPI motif contributes only marginally if at all to L-domain function since a slight influence on particle release was detectable only in the context of a nonfunctional PSAP L domain using the four-plasmid but not the one-plasmid vector system. Nevertheless, infectivity was reduced about fivefold in both systems, indicating that this sequence motif affects the viral viability, although this function seems to be clearly separated from L-domain function. Interestingly, in addition, inactivation of the PSAP motif always resulted in a much higher reduction in viral infectivity than in particle release, suggesting that it too might have an additional function for viral fitness beside its function as an L domain. In this respect it is remarkable that replacement of the PFV Gag PSAP motif by the minimal L domains of HIV-1 p6 or RSV p2b completely restored both particle release and infectivity, whereas introduction of an EIAV p9 YPDL L-domain sequence motif only partially rescued viral infectivity but had no significant positive effect on particle release. Intriguingly, for this mutant a direct correlation in the level of reduction in particle release and infectivity compared to the wild-type PFV Gag protein was observed, indicating that the relative infectivity per particle of these mutant particles and wild-type particles was similar. This indicates that the PFV PSAP L domain and perhaps other L-domain motifs may have dual functions, one on viral particle release and another on the infectivity of the individual particles, and that only the infectivity and not the release function of the EIAV p9 protein is compatible with PFV particle morphogenesis. However, an alternative explanation for the lack of rescue of PFV particle release by the inserted EIAV p9 peptide that cannot formally excluded is that it does not comprise the full EIAV L domain. Interestingly, a similar phenomenon has been reported for HIV-1 Gag chimeras in which PTAP was replaced by sequences from EIAV spanning the YPDL motif, although the authors did not show the actual data on particle release and infectivity or the sequence of EIAV p9 used (39).
It was recently suggested that in contrast to most viruses analyzed so far, VSV does not require late components of the VPS machinery since it was not inhibited by DN Vps4 expression constructs (15). Our results clearly demonstrate that PFV requires late components of the VPS machinery for particle egress. DN mutants of both AAA ATPases Vps4A and Vps4B but not the respective wild-type proteins inhibited PFV particle release and viral infectivity. In addition, a DN mutant of CHMP3, a component of the ESCRT-III complex, also displayed an inhibitory effect on both processes. The role of the PSAP motif as the predominant functional L domain in PFV Gag was further supported by the specific inhibition of various TSG101 proteins that have previously been shown to inhibit PTAP-dependent release of HIV but not PPPY-mediated egress of murine leukemia virus or MPMV (3). In addition, the lack of functional activity of an AIP-1/ALIX DN mutant inhibiting EIAV particle release supports our view that the YEIL motif in PFV Gag has no L-domain function. Interestingly, only the YXXL motif is present in all FV isolates from different species identified to date. The PSAP motif which seems to be the functional L domain of PFV is present only in the Gag protein of primate isolates, whereas bovine, equine, and feline FV lack both PSAP- and PPXY-like sequence motifs. This of course raises the question of which are the functional L domains for these FV isolates and whether they utilize the VPS machinery or exploit different cellular pathways for particle release.
From the data presented in this work it appears that the YEIL sequence motif has no classical L-domain function. Rather, it seems to be involved in PFV capsid assembly, since the electron microscopy analysis revealed the presence of aberrantly accumulated electron-dense material within viruslike structures containing the typical FV Env spikes. Furthermore, in the cytoplasm no spherical capsid structures were observed, but instead aggregates, which may represent mutant Gag proteins, were seen. However, this assumption awaits formal proof for example by immunoelectron microscopy. Surprisingly, VLP containing this mutant Gag protein were released into the supernatant and could be pelleted through 20% sucrose. Furthermore, this mutant Gag protein was processed by the FV protease. Because PFV Gag processing is dependent on both Pol and viral genome incorporation into the particle, this indicates that the mutant particles contain all essential viral structural constituents, and the loss of infectivity is most probably a result of the aberrant capsid assembly.
Acknowledgments
We thank Uta von Schwedler, Eric Freed, and Paul Bieniasz for helpful discussions and for providing expression constructs and B. Hub for excellent technical assistance.
This work was supported by grants from the DFG (Li621/3-1) and the BMBF (01ZZ0102) to D.L. and from the DFG (Re627/6-3) to A.R.
REFERENCES
- 1.Baldwin, D. N., and M. L. Linial. 1998. The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72:3658-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baunach, G., B. Maurer, H. Hahn, M. Kranz, and A. Rethwilm. 1993. Functional analysis of human foamy virus accessory reading frames. J. Virol. 67:5411-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubois, M. F., C. Pourcel, S. Rousset, C. Chany, and P. Tiollais. 1980. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc. Natl. Acad. Sci. USA 77:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du Bridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duda, A., A. Stange, D. Luftenegger, N. Stanke, D. Westphal, T. Pietschmann, S. W. Eastman, M. L. Linial, A. Rethwilm, and D. Lindemann. 2004. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J. Virol. 78:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eastman, S. W., and M. L. Linial. 2001. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, N., M. Heinkelein, D. Lindemann, J. Enssle, C. Baum, E. Werder, H. Zentgraf, J. G. Müller, and A. Rethwilm. 1998. Foamy virus particle formation. J. Virol. 72:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freed, E. O. 2003. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 11:56-59. [DOI] [PubMed] [Google Scholar]
- 10.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 12.Heinkelein, M., M. Dressler, G. Jarmy, M. Rammling, H. Imrich, J. Thurow, D. Lindemann, and A. Rethwilm. 2002. Improved primate foamy virus vectors and packaging constructs. J. Virol. 76:3774-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imrich, H., M. Heinkelein, O. Herchenröder, and A. Rethwilm. 2000. Primate foamy virus Pol proteins are imported into the nucleus. J. Gen. Virol. 81:2941-2947. [DOI] [PubMed] [Google Scholar]
- 15.Irie, T., J. M. Licata, J. P. McGettigan, M. J. Schnell, and R. N. Harty. 2004. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J. Virol. 78:2657-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 18.Kräusslich, H. G., M. Fäcke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindemann, D., and P. A. Goepfert. 2003. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 277:111-129. [DOI] [PubMed] [Google Scholar]
- 22.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linial, M. L., and S. W. Eastman. 2003. Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 277:89-110. [DOI] [PubMed] [Google Scholar]
- 25.Martin-Serrano, J., D. Perez-Caballero, and P. D. Bieniasz. 2004. Context-dependent effects of L domains and ubiquitination on viral budding. J. Virol. 78:5554-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nassal, M. 1996. Hepatitis B virus morphogenesis. Curr. Top. Microbiol. Immunol. 214:297-337. [DOI] [PubMed] [Google Scholar]
- 30.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 33.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027-1033. [DOI] [PubMed] [Google Scholar]
- 35.Rethwilm, A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 36.Rhee, S. S., and E. Hunter. 1990. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell 63:77-86. [DOI] [PubMed] [Google Scholar]
- 37.Sfakianos, J. N., and E. Hunter. 2003. M-PMV capsid transport is mediated by Env/Gag interactions at the pericentriolar recycling endosome. Traffic 4:671-680. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, K. L., D. Lindemann, M. J. Mulligan, and P. A. Goepfert. 2003. Foamy virus envelope glycoprotein is sufficient for particle budding and release. J. Virol. 77:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shehu-Xhilaga, M., S. Ablan, D. G. Demirov, C. Chen, R. C. Montelaro, and E. O. Freed. 2004. Late domain-dependent inhibition of equine infectious anemia virus budding. J. Virol. 78:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Göttlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 41.Sun, Z., J. Pan, W. X. Hope, S. N. Cohen, and S. P. Balk. 1999. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer 86:689-696. [DOI] [PubMed] [Google Scholar]
- 42.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verschoor, E. J., S. Langenhuijzen, S. van den Engel, H. Niphuis, K. S. Warren, and J. L. Heeney. 2003. Structural and evolutionary analysis of an orangutan foamy virus. J. Virol. 77:8584-8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Kräusslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 45.Whitt, M. A., L. Chong, and J. K. Rose. 1989. Glycoprotein cytoplasmic domain sequences required for rescue of a vesicular stomatitis virus glycoprotein mutant. J. Virol. 63:3569-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]