Abstract
In the presence of pathogenic bacteria, plants close their stomata to prevent pathogen entry. Intracellular nucleotide-binding leucine-rich repeat (NLR) immune receptors recognize pathogenic effectors and activate effector-triggered immune responses. However, the regulatory and molecular mechanisms of stomatal immunity involving NLR immune receptors are unknown. Here, we show that the Nicotiana benthamiana RPW8-NLR central immune receptor ACTIVATED DISEASE RESISTANCE 1 (NbADR1), together with the key immune proteins ENHANCED DISEASE SUSCEPTIBILITY 1 (NbEDS1) and PHYTOALEXIN DEFICIENT 4 (NbPAD4), plays an essential role in bacterial pathogen- and flg22-induced stomatal immunity by regulating the expression of salicylic acid (SA) and abscisic acid (ABA) biosynthesis or response-related genes. NbADR1 recruits NbEDS1 and NbPAD4 in stomata to form a stomatal immune response complex. The transcription factor NbWRKY40e, in association with NbEDS1 and NbPAD4, modulates the expression of SA and ABA biosynthesis or response-related genes to influence stomatal immunity. NbADR1, NbEDS1, and NbPAD4 are required for the pathogen infection-enhanced binding of NbWRKY40e to the ISOCHORISMATE SYNTHASE 1 promoter. Moreover, the ADR1-EDS1-PAD4 module regulates stomatal immunity in Arabidopsis (Arabidopsis thaliana). Collectively, our findings show the pivotal role of the core intracellular immune receptor module ADR1-EDS1-PAD4 in stomatal immunity, which enables plants to limit pathogen entry.
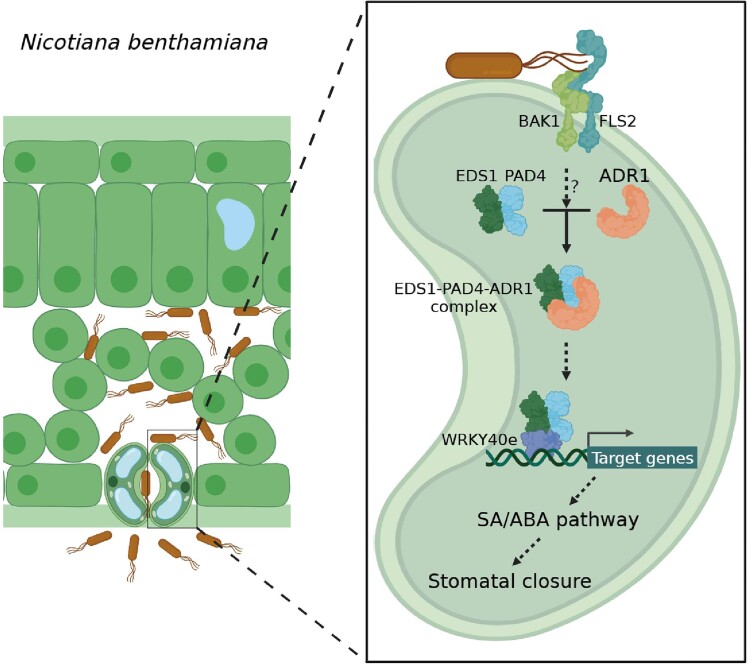
Intracellular receptor NbADR1 forms a module with immunity proteins NbEDS1 and NbPAD4, functioning together with transcription factor NbWRKY40e to mediate stomatal immunity in Nicotiana benthamiana.
Introduction
Plants are continually under threat of attack from invasive pathogens (Zhang et al. 2020; Ngou et al. 2022; Wang et al. 2022). Stomata, surrounded by a pair of guard cells, are one of the most important entry sites for pathogens into plant tissues, and they are a major battleground during plant–pathogen interactions (Ye et al. 2020; Wang et al. 2021; Liu et al. 2022). Upon sensing pathogen-associated molecular patterns (PAMPs) using cell surface pattern recognition receptors (PRRs), guard cells close the stomatal pores by modulating various signaling pathways, including those involving salicylic acid (SA) and abscisic acid (ABA), thereby mediating a PAMP-triggered immune (PTI) response (Melotto et al. 2006; Zeng et al. 2010; Murata et al. 2015; Melotto et al. 2017). Bacterial pathogens have evolved strategies to reopen stomata and facilitate bacterial entry by delivering phytotoxins (e.g. coronatine) or effectors into host cells (Melotto et al. 2006; Hu et al. 2022; Roussin-Léveillée et al. 2022).
Pathogens secrete effector proteins into host cells to dampen plant immunity (Dong and Ma 2021; Bundalovic-Torma et al. 2022; Wang et al. 2022). In turn, plants utilize intracellular nucleotide-binding leucine-rich repeat (NLR) receptors, which recognize pathogenic effectors and activate effector-triggered immune (ETI) responses, often accompanied by hypersensitive response (HR), to restrict pathogen spread (Adachi et al. 2019; Van de Weyer et al. 2019; Zhang et al. 2020; Freh et al. 2022). Plant NLRs are categorized into 3 main types based on their N-terminal domains: Toll/interleukin-1 receptor (TIR)-NLRs, coiled-coil (CC)-NLRs, and RESISTANCE TO POWDERY MILDEW 8 (RPW8)-NLRs (Peart et al. 2005; Bonardi et al. 2011; Collier et al. 2011). TIR-NLRs (TNLs) and CC-NLRs (CNLs) function mainly as sensor NLRs that directly or indirectly detect pathogen effectors during ETI signaling (Qi and Innes 2013; Wang et al. 2019a; Wang et al. 2019b; Ma et al. 2020; Martin et al. 2020; Outram et al. 2022), while RPW8-NLRs (RNLs) and a specific class of CNLs, known as NRCs, function downstream as core immune receptors to transduce these signals (Wu et al. 2017; Jubic et al. 2019; van Wersch et al. 2020; Gong et al. 2023).
Characterized RNLs include ACTIVATED DISEASE RESISTANCE 1 (ADR1) and N Required Gene 1 (NRG1) (Grant et al. 2003; Peart et al. 2005; Adachi et al. 2019; Jubic et al. 2019; Feehan et al. 2020). They are considered to be central immune receptors that function together with the lipase-like immune proteins ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) and its homologs PHYTOALEXIN DEFICIENT 4 (PAD4) and SENESCENCE-ASSOCIATED GENE 101 (SAG101) as central signaling hubs (Sun et al. 2021; Gong et al. 2023).
In Arabidopsis (Arabidopsis thaliana), AtNRG1s, AtEDS1, and AtSAG101 form a module that mediates various TNL-triggered ETI responses, while AtADR1s cooperate with AtEDS1 and AtPAD4 in both TNL- and CNL-triggered ETI responses and in PRR-mediated PTI responses in the leaf apoplast (Lapin et al. 2019; Wu et al. 2019; Saile et al. 2020; Pruitt et al. 2021; Sun et al. 2021; Tian et al. 2021). The TIR domains of TNLs cleave NAD+ to produce pRib-AMP/ADP and ADPr-ATP, which induce the formation of AtADR1-L1-AtEDS1-AtPAD4 and AtNRG1.1-AtEDS1-AtSAG101 heterotrimers, respectively (Horsefield et al. 2019; Wan et al. 2019; Essuman et al. 2022; Huang et al. 2022; Jia et al. 2022). Activated AtNRG1s and AtADR1s may oligomerize and function as calcium influx channels at the plasma membrane, triggering cell death (Jacob et al. 2021; Wu et al. 2021; Wang et al. 2023).
In Nicotiana benthamiana, infection by the bacterial pathogens Pseudomonas syringae pv. tomato (Pst) DC3000 and Xanthomonas euvesicatoria (Xe) 85-10 causes ETI responses because they contain the homologous effectors HR and pathogenicity (Hrp) outer protein (hop) Q (HopQ1) and Xanthomonas outer protein Q (XopQ), respectively (Wei et al. 2007; Adlung and Bonas 2017). The TNL Recognition of XopQ 1 (NbROQ1) in N. benthamiana recognizes HopQ1 and XopQ directly, and it acts through NbNRG1 and NbEDS1 to trigger ETI responses, including the HR and resistance to Pst DC3000 and Xe 85-10 (Schultink et al. 2017; Qi et al. 2018; Martin et al. 2020). In addition, NbNRG1, NbEDS1, or NbSAG101 can mediate various other TNL-mediated ETI responses, including TNLN, recognition of Peronospora parasitica 1 (RPP1), and resistance to P. syringae 4 (RPS4)-triggered HRs, and TNLN-mediated resistance to tobacco mosaic virus (Peart et al. 2005; Qi et al. 2018; Castel et al. 2019; Gantner et al. 2019; Wu et al. 2019). On the other hand, how the RNL NbADR1 and its putative immune partner NbPAD4 function in plant immunity is unknown.
Here, we show that NbADR1, together with NbEDS1 and NbPAD4, controls bacteria- and flg22-induced stomatal closure via SA and ABA pathways and that it confers resistance to bacterial pathogens introduced by spray inoculation. NbADR1 interacts with the NbEDS1-NbPAD4 complex upon inoculation with a bacterial pathogen. The transcription factor NbWRKY40e associates with NbEDS1 and NbPAD4 and acts as a key positive regulator of stomatal immunity by modulating the SA and ABA pathways. Furthermore, we demonstrate the conserved roles of ADR1-EDS1-PAD4 in stomatal immunity in Arabidopsis. Our findings highlight the essential role of the intracellular immune receptor complex ADR1-EDS1-PAD4 in stomatal immunity.
Results
NbADR1 promotes Pst DC3000 resistance in spray-inoculated plants
To explore the immune functions of NbADR1, we generated or obtained the following mutants: N. benthamiana adr1 (Supplemental Fig. S1A), nrg1 (a control; Qi et al. 2018), adr1 nrg1, and the presumed partner mutant eds1 (Schultink et al. 2017). We then examined their immune responses following spray inoculation with Pst DC3000, which triggers various immune reactions, including stomatal immunity, apoplast PTI responses, and ETI responses (via recognition of HopQ1).
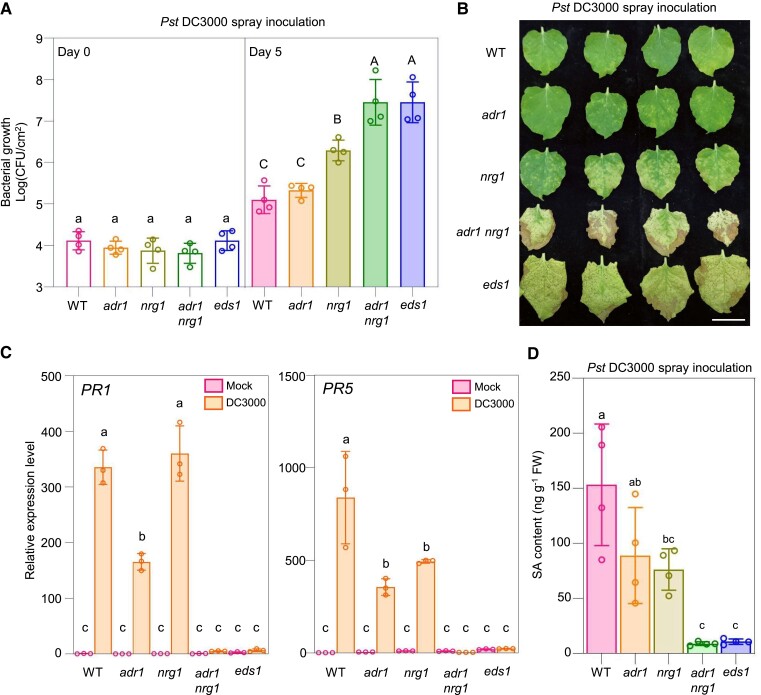
The adr1 mutant harbored a bacterial population that was similar in size to that of wild type, while nrg1 had a larger bacterial population (Fig. 1A). Compared with nrg1, both the adr1 nrg1 and eds1 mutants possessed significantly greater numbers of pathogenic bacteria. The adr1 mutant exhibited a similar resistance phenotype to wild type, while nrg1 displayed a disease phenotype and the adr1 nrg1 and eds1 mutants had the most severe disease phenotypes (Fig. 1B). Furthermore, the expression levels of the representative defense-related genes PATHOGENESIS-RELATED GENE 1 (PR1) and PR5 (Qi et al. 2018) in wild type were upregulated upon spray inoculation with Pst DC3000, whereas their expression levels following Pst DC3000 treatment were lower in adr1 (for PR1/5) or nrg1 (for PR1) compared with wild type and were the lowest in the adr1 nrg1 and eds1 mutants (Fig. 1C).
Figure 1.
NbADR1 plays a role in immune responses under Pst DC3000 spray inoculation. A, B) Bacterial populations A) and disease symptoms B) in leaves of Nicotiana benthamiana wild-type (WT) and the mutants adr1, nrg1, adr1 nrg1, and eds1 at 0 and 5 d A), or 10 d B) postspray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 (OD600 = 0.4), respectively. Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). The experiments were repeated 3 times with similar results. Scale bars represent 3 cm. C) Quantitative real-time PCR analysis of PR1 and PR5 in N. benthamiana wild-type (WT) and the indicated mutants at 20 h after spray inoculation with mock (10 mM MgCl2) or Pst DC3000 (OD600 = 0.4). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). The experiments were repeated 3 times with similar results. D) SA contents in leaves of N. benthamiana WT and the indicated mutants at 24 h postspray inoculation with Pst DC3000 (OD600 = 0.4). Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). FW, fresh weight; CFU, for colony-forming unit.
SA is a central defense phytohormone in the response to bacterial pathogens (Peng et al. 2021). We measured the SA contents in the aforementioned plants after Pst DC3000 spray inoculation (Fig. 1D; Supplemental Fig. S2). Compared with that in wild type, the SA contents were lower in adr1 and nrg1, and they were the lowest in adr1 nrg1 and eds1. Taken together, these results suggest that NbADR1 plays a role in resistance to Pst DC3000 following spray inoculation.
NbADR1, NbPAD4, and NbEDS1 control stomatal immunity
Given that spray inoculation with Pst DC3000 triggered ETI activation, PTI activation in the apoplast, and stomatal immunity, we dissected the immune function of NbADR1. First, we examined ETI activation in plants based on TNL-triggered HRs and resistance to syringe inoculation with Pst DC3000 and Xe 85-10 harboring the homologs HopQ1 or XopQ (Supplemental Fig. S3).
Consistent with previous findings (Qi et al. 2018), transient expression of XopQ, HopQ1, and the effector/TNL pairs A. thaliana recognized 1 (ATR1)/RPP1 or p50/N triggered the HR in leaves of wild-type plants, but not in nrg1 or eds1 mutant leaves; ATR1/RPP1 and p50/N triggered the HR in roq1 plants, whereas XopQ or HopQ1 did not. Here, we found that transient expression of XopQ, HopQ1, ATR1/RPP1, or p50/N triggered the HR in adr1, but not in adr1 nrg1 plants (Supplemental Fig. S3A). Bacterial infection assays with syringe inoculation showed that nrg1, eds1, and roq1 were susceptible to Pst DC3000 and Xe 85-10 (Qi et al. 2018), whereas adr1 exhibited similar resistance to wild type, and adr1 nrg1 displayed similar susceptibility to nrg1, eds1, and roq1 (Supplemental Fig. S3, B and C). These results suggest that NbADR1 is not involved in TNL-mediated ETI responses, including the HR and resistance to bacterial pathogens.
We also generated adr1, nrg1, and adr1 nrg1 mutant plants carrying a transgene for the expression of bacterial spot resistance gene Bs2 from pepper (Capsicum annuum), which recognizes the effector AvrBs2 from Xanthomonas (Leister et al. 2005), and we tested their phenotypes in terms of CNLBs2-mediated HRs and resistance (Supplemental Fig. S4). The adr1, nrg1, and adr1 nrg1 mutations did not affect AvrBs2-triggered Bs2-mediated HRs or Bs2-mediated resistance to the bacterial pathogen Xe 85-10 ΔXopQ (containing AvrBs2), indicating that NbADR1 is not required for CNLBs2-mediated ETI responses.
Next, we explored whether NbADR1 functions in PTI activation in the apoplast of leaves. We infiltrated the virulent strain Pst DC3000 ΔHopQ1 or Xe 85-10 ΔXopQ, which cannot trigger ETI responses, into adr1, nrg1, adr1 nrg1, and eds1 mutant plants and into their presumed partner mutants pad4 and sag101 (Fig. 2A; Supplemental Figs. S1B, S1C, and S5). Disease symptoms and bacterial growth assays showed that the mutants all exhibited similar susceptibility to wild type upon syringe inoculation, suggesting that NbADR1, NbNRG1, NbEDS1, NbPAD4, and NbSAG101 are not involved in PTI activation in the leaf apoplast.
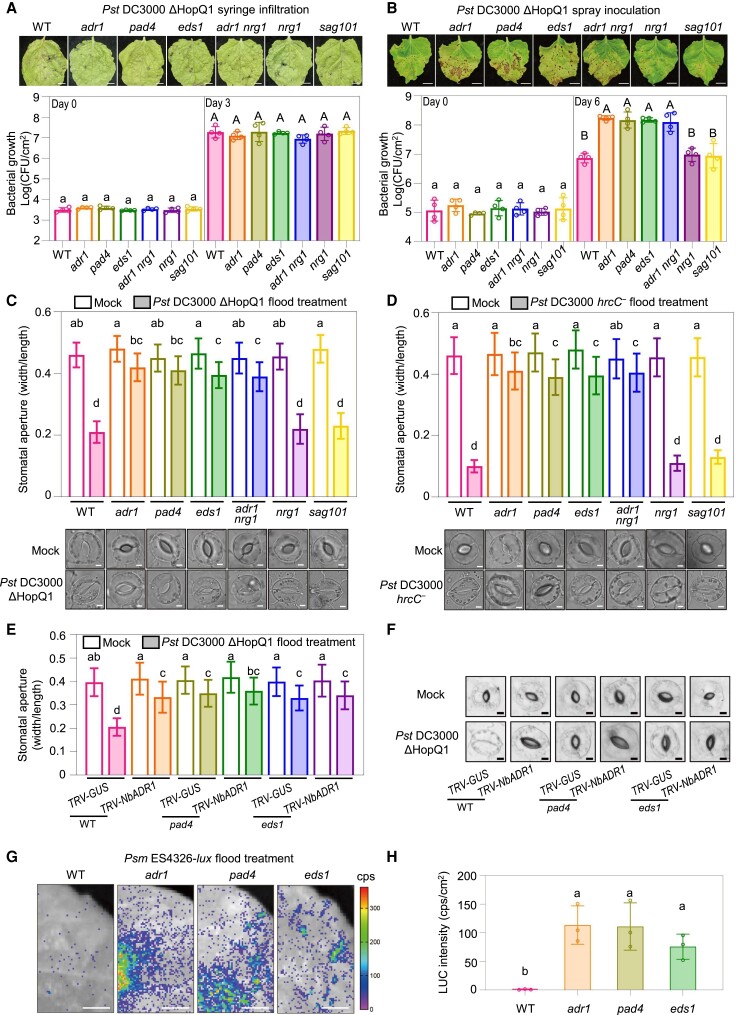
Figure 2.
NbADR1, NbPAD4, and NbEDS1 mediate bacterial pathogen-induced stomatal closure. A, B) Disease symptoms and bacterial populations in leaves of the Nicotiana benthamiana wild-type (WT), adr1, pad4, eds1, adr1 nrg1, nrg1, and sag101 at 4 d (A, upper panel), or at 0 and 3 d (A, lower panel) postsyringe infiltration, or at 10 d (B, upper panel), or 0 and 6 d (B, lower panel) postspray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 ΔHopQ1 (OD600 = 0.0001 for syringe, OD600 = 0.4 for spray), respectively. Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). The experiments were repeated 3 times with similar results. Scale bars represent 1 cm. C, D) Stomatal apertures and images of stomata in leaves of N. benthamiana WT and the indicated mutants after 1 h of flood treatment with mock (10 mM MgCl2) or Pst DC3000 ΔHopQ1 (OD600 = 0.4) (C), and Pst DC3000 hrcC− (OD600 = 0.4). D) Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. E, F) Stomatal apertures E) and images of stomata F) in leaves of N. benthamiana WT and the indicated mutants with TRV-GUS or TRV-NbADR1, after 1 h of flood treatment with mock (10 mM MgCl2) or Pst DC3000 ΔHopQ1 (OD600 = 0.4). Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. G, H) Bacterial pathogen entry assay in leaves of N. benthamiana WT and the indicated mutants. The leaves were photographed and the luciferase (LUC) intensities were quantified by a CCD imaging system after 1 h of P. syringae pv. maculicola (Psm) ES4326-lux flood treatment (OD600 = 0.5). Data are means (± Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. The experiments were repeated 3 times with similar results. CFU, colony-forming unit; cps, counts per second.
To test whether NbADR1 regulates stomatal immunity, we performed spray inoculation with Pst DC3000 ΔHopQ1 using adr1, nrg1, adr1 nrg1, eds1, pad4, and sag101 mutant plants (Fig. 2B). The adr1, adr1 nrg1, pad4, and eds1 mutants were more susceptible to the pathogen than wild type, whereas nrg1 and sag101 exhibited a similar disease phenotype to wild type. These results imply that NbADR1, NbPAD4, and NbEDS1, but not NbNRG1 or NbSAG101, play a role in stomatal immunity.
To further confirm the involvement of NbADR1, NbPAD4, and NbEDS1 in stomatal immunity, we measured the stomatal apertures in leaves of the mutants exposed to mock treatment, Pst DC3000 ΔHopQ1 or Pst DC3000 hrcC−, which is defective in type III secretion (Melotto et al. 2006) (Fig. 2, C and D). The bacteria induced stomatal closure in wild-type, nrg1, and sag101 plants at similar levels, whereas the stomatal closure response was compromised in adr1, pad4, eds1, and adr1 nrg1 to a similar degree, demonstrating that NbADR1, NbPAD4, and NbEDS1, but not NbNRG1 or NbSAG101, mediate pathogen-induced stomatal closure.
Silencing NbADR1 in pad4 and eds1 by virus-induced gene silencing did not enhance their defects in stomatal closure upon treatment with Pst DC3000 ΔHopQ1 (Fig. 2, E and F; Supplemental Fig. S6A), suggesting that NbADR1, NbPAD4, and NbEDS1 function in the same pathway to mediate stomatal closure.
Next, bacterial pathogen entry assays were conducted using luxCDABE-tagged P. syringae pv. maculicola (Psm) ES4326 (Fan et al. 2008; Su et al. 2017). Approximately, 40- to 60-fold more bacteria entered the leaf apoplast in eds1, adr1, and pad4 compared with wild type (Fig. 2, G and H), further supporting the essential roles of NbADR1, NbEDS1, and NbPAD4 in stomatal immunity.
Taken together, our results demonstrate that the immune receptor NbADR1 and its presumed partners NbPAD4 and NbEDS1 function in the same pathway and mediate stomatal immunity, whereas NbNRG1 and NbSAG101 do not affect stomatal immunity.
NbADR1, NbPAD4, and NbEDS1 are required for flg22-induced stomatal immunity
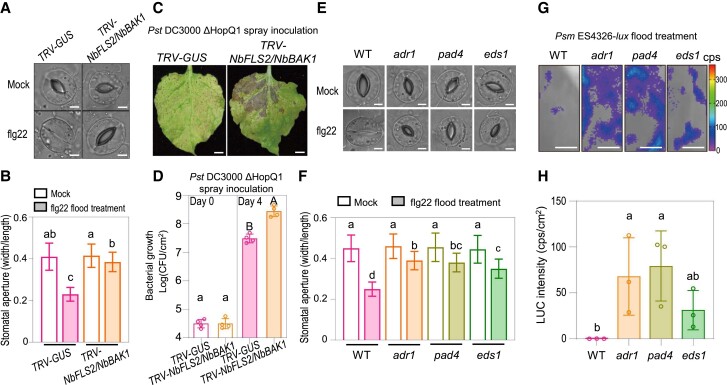
In Arabidopsis, the PAMP flg22, which is perceived by the plasma membrane receptor AtFLS2 and its coreceptor AtBAK1, induces stomatal closure (Chinchilla et al. 2007; Zeng and He 2010). We found that NbFLS2 and NbBAK1 were required for flg22-induced stomatal closure and for resistance to spray inoculation with Pst DC3000 ΔHopQ1 (Fig. 3, A to D; Supplemental Fig. S6, B and C). To explore the potential involvement of the NbADR1-NbPAD4-NbEDS1 module in flg22-regulated stomatal immunity, we examined the response to flg22 in wild-type, adr1, pad4, and eds1 plants. As shown in Fig. 3, E and F, flg22-induced stomatal closure was compromised in these mutants. Accordingly, with flg22 pretreatment, the bacterial populations that entered the leaf apoplast in adr1, pad4, and eds1 were larger than those in wild type (Fig. 3, G and H). Thus, NbADR1, NbPAD4, and NbEDS1 may mediate flg22-NbFLS2 and NbBAK1-induced stomatal immunity in N. benthamiana.
Figure 3.
NbADR1, NbPAD4, and NbEDS1 mediate flg22-induced stomatal immunity. A, B) Images of stomata A) and stomatal apertures B) in leaves of Nicotiana benthamiana TRV-GUS and TRV-NbFLS2/NbBAK1 after 1 h of flood treatment with mock (ddH2O) or 100 nM flg22. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. C, D) Disease symptoms C) and bacterial populations D) in leaves of N. benthamiana TRV-GUS and TRV-NbFLS2/NbBAK1 at 7 d C), or at 0 and 4 d D) postspray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 ΔHopQ1 (OD600 = 0.4). Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). The experiments were repeated 3 times with similar results. CFU stands for colony-forming unit. Scale bars represent 1 cm. E, F) Images of stomata E) and stomatal apertures F) in leaves of N. benthamiana wild-type (WT) and the indicated mutants after 1 h of flood treatment with mock (ddH2O) or 100 nM flg22. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. G, H) Bacterial pathogen entry assay in leaves of N. benthamiana WT and the indicated mutants. The leaves were pretreated with 100 nM flg22 for 1 h before bacteria treatment. The leaves were photographed and quantified by a CCD imaging system after 1 h of P. syringae pv. maculicola ES4326-lux flood treatment (OD600 = 0.5). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. cps, counts per second.
NbADR1, NbPAD4, and NbEDS1 mediate DC3000 hrcC− spray infection-induced expression of SA or ABA biosynthesis or response-related genes for stomatal immunity
Our results demonstrated that NbADR1 contributes to SA content in plants spray inoculated with Pst DC3000 (Fig. 1D), and it is known that the phytohormones SA and ABA play crucial roles in stomatal immunity (Melotto et al. 2006; Zheng et al. 2012; Montillet and Hirt 2013; Melotto et al. 2017; Roussin-Léveillée et al. 2022). Therefore, we investigated whether NbADR1, NbPAD4, and NbEDS1 exert their effects on pathogen-induced stomatal closure via SA and ABA pathways.
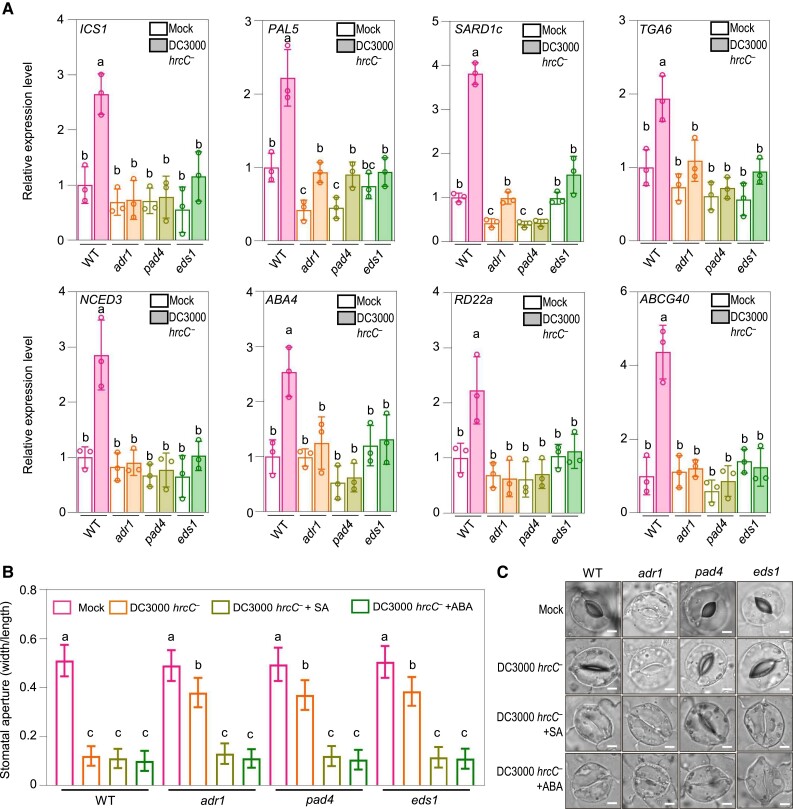
Reverse transcription quantitative PCR (RT-qPCR) analysis showed that the expression of the SA biosynthesis or response-related genes ISOCHORISMATE SYNTHASE 1 (ICS1), PHENYLALANINE AMMONIA-LYASE 5 (PAL5), SAR DEFICIENT 1c (SARD1c), and TGACG motif-binding factor 6 (TGA6) (Sun et al. 2015; Zheng et al. 2015; Medina-Puche et al. 2020) and of the ABA biosynthesis or response-related genes NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3), ABA DEFICIENT 4 (ABA4), RESPONSIVE TO DESICCATION 22a (RD22a), and ATP-BINDING CASSETTE G40 (ABCG40) (Dall’Osto et al. 2007; Sato et al. 2018; Roussin-Léveillée et al. 2022) was upregulated in wild-type plants spray inoculated with Pst DC3000 hrcC− (Fig. 4A). However, after Pst DC3000 hrcC− treatment, the expression levels of these genes were lower in adr1, pad4, and eds1 than in wild type.
Figure 4.
NbADR1, NbEDS1, and NbPAD4 function upstream of SA and ABA pathways in stomatal closure. A) Quantitative reverse transcription PCR analysis of the representative SA pathway genes ICS1, PAL5, SARD1c, and TGA6, and the representative ABA pathway genes NCED3, ABA4, RD22a and ABCG40 in Nicotiana benthamiana wild-type (WT), adr1, pad4, and eds1 at 2 h postspray inoculation with mock (10 mM MgCl2) or Pseudomonas syringae pv. tomato (Pst) DC3000 hrcC− (OD600 = 0.4). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). B, C) Stomatal apertures B) and images of stomata C) in leaves of N. benthamiana WT and the indicated mutants after 1 h of flood treatment with mock (10 mM MgCl2), Pst DC3000 hrcC− (OD600 = 0.4), or Pst DC3000 hrcC−(OD600 = 0.4) plus SA (the form of sodium salicylate) or ABA, respectively. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results.
We next investigated whether exogenous application of SA and ABA could rescue the stomatal closure phenotype in adr1, pad4, and eds1 plants treated with Pst DC3000 hrcC−. As shown in Fig. 4, B and C, treatment with SA or ABA restored stomatal closure in adr1, pad4, and eds1 plants treated with Pst DC3000 hrcC−. These data indicate that NbADR1, NbEDS1, and NbPAD4 are essential for the DC3000 hrcC− spray infection-induced expression of SA or ABA biosynthesis or response-related genes during stomatal immunity.
NbADR1 recruits the NbPAD4-NbEDS1 complex during Pst DC3000 ΔHopQ1-induced stomatal immunity
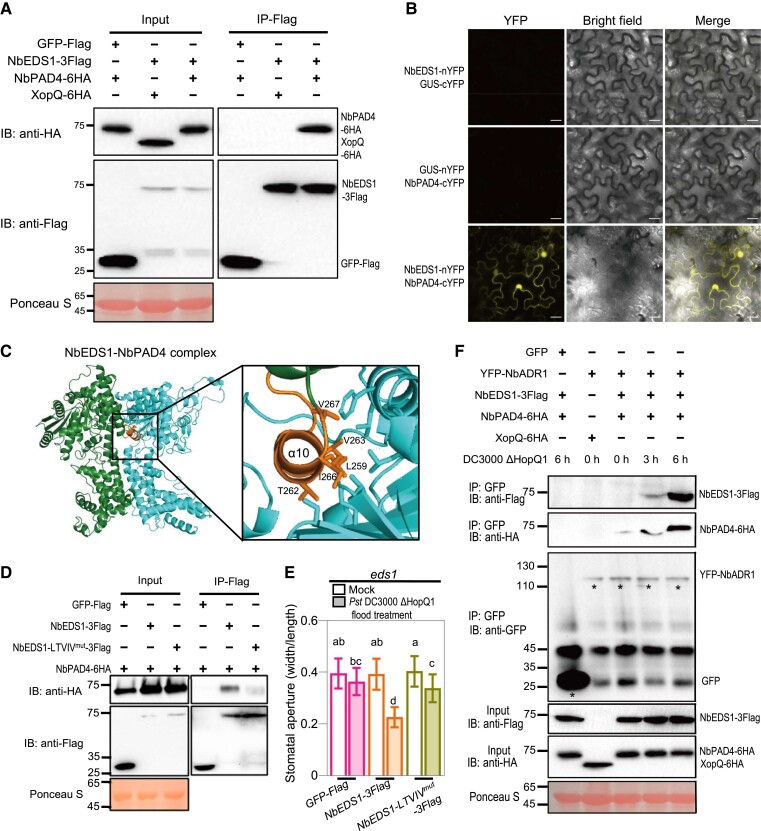
Having shown that NbADR1, NbEDS1, and NbPAD4 mediate stomatal immunity, we investigated whether they interact with each other and form a complex. Coimmunoprecipitation (IP) showed that NbEDS1 interacted with NbPAD4 under basal conditions (Fig. 5A). Additionally, bimolecular fluorescence complementation (BiFC) assays demonstrated their interactions in both the epidermal and guard cells of N. benthamiana leaves, while the negative controls did not show any interaction (Fig. 5B; Supplemental Fig. S7A).
Figure 5.
NbADR1 physically associates with the preformed NbPAD4-NbEDS1 complex upon Pst DC3000 ΔHopQ1 infection. A) Co-IP assay showed the interaction of NbEDS1 with NbPAD4. The NbPAD4-6HA or XopQ-6HA (as a negative control) was transiently coexpressed with NbEDS1-3flag or the GFP-flag control in Nicotiana benthamiana leaves. The OD600 for each Agrobacterium was adjusted to 0.3. The total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. B) BiFC assay showed the interactions of NbEDS1 with NbPAD4 in epidermal cells of N. benthamiana leaves. The N-terminal fragment of yellow fluorescent protein (nYFP)-fused NbEDS1, the C-terminal fragment of YFP (cYFP)-fused NbPAD4, and the controls β-glucuronidase (GUS)-nYFP and GUS-cYFP were transiently coexpressed in N. benthamiana leaves, respectively. The OD600 for each Agrobacterium was adjusted to 0.3. YFP fluorescence was detected under the confocal microscope with identical gain settings (laser, 514 nm, 6%; collection bandwidth, 526 to 588 nm; pinhole, 1.20 AU; master gain, 660.0) at 30 h after coexpression. Scale bars represent 20 μm. C) The homology model of the NbEDS1-NbPAD4 protein complex based on the structure of the Arabidopsis AtEDS1-AtPAD4 complex. The key residues predicted for NbEDS1-NbPAD4 interface were marked with orange in NbEDS1. D) Co-IP assay showed that the interaction of NbEDS1 with NbPAD4 is largely disrupted by the “LTVIV” mutation (L259E, T262F, V263E, I266E, and V267E) within NbEDS1. The OD600 for each Agrobacterium was adjusted to 0.3. The total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. E) Stomatal apertures in leaves of N. benthamiana eds1 with transient expression of GFP-Flag, NbEDS1-3Flag, or NbEDS1-LTVIV-3Flag, after 1 h of flood treatment with mock (10 mM MgCl2) or Pseudomonas syringae pv. tomato (Pst) DC3000 ΔHopQ1 (OD600 = 0.4). The OD600 for each Agrobacterium was adjusted to 0.5. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). F) Co-IP assay showed that NbADR1 recruits NbPAD4 and NbEDS1 upon Pst DC3000 ΔHopQ1 spray infection. The YFP-NbADR1 and GFP were transiently coexpressed with XopQ-6HA, NbEDS1-3Flag, or NbPAD4-6HA variants in N. benthamiana leaves, respectively, and the leaves were subsequently inoculated with Pst DC3000 ΔHopQ1 (OD600 = 0.4). The OD600 for each Agrobacterium was adjusted to 0.3 (except that for YFP-NbADR1 was 0.6). The total proteins were extracted from the leaves at 0, 3, or 6 h after treatment and were immunoprecipitated with the anti-GFP agarose beads. The IP product proteins were detected by immunoblotting using the anti-Flag or anti-HA antibody. Ponceau-S staining of Rubisco served as a loading control. Asterisks indicate the bands of GFP or YFP-ADR1. All these experiments were repeated 3 times with similar results. IB, immunoblotting; IP, immunoprecipitation.
Using the cryo-EM structure of the AtEDS1-AtPAD4 heterodimer from Arabidopsis as a reference (Huang et al. 2022), we modeled NbEDS1 and NbPAD4, and we identified a hydrophobic pocket on NbPAD4 that presumably contacts the α10 helix of NbEDS1 (Fig. 5C). To validate this interaction interface, we mutated the predicted NbPAD4-contacting residues in NbEDS1 (L259E, T262F, V263E, I266E, and V267E to produce NbEDS1-LTVIVmut) and found that these mutations largely compromised the interaction between NbEDS1 and NbPAD4 (Fig. 5D). Compared with wild-type NbEDS1, transient expression of NbEDS-LTVIVmut could not rescue pathogen-induced stomatal closure in eds1 (Fig. 5E). These findings suggest that the NbEDS1–NbPAD4 interaction is crucial for stomatal immunity.
Co-IP further showed that NbADR1 hardly interacted with NbEDS1 and NbPAD4 under basal conditions, while spray inoculation with Pst DC3000 ΔHopQ1 induced its interaction with NbEDS1 and NbPAD4 (Fig. 5F). Consistent with this result, a BiFC assay showed that the NbADR1 LRR domain interacted with NbPAD4 in the presence of NbEDS1 in guard cells upon infection with Pst DC3000 ΔHopQ1 (Supplemental Fig. S7B). Together, these results suggest that NbADR1, NbEDS1, and NbPAD4 function as a complex in stomatal immunity.
NbWRKY40e associates with NbEDS1 and NbPAD4 and mediates stomatal immunity
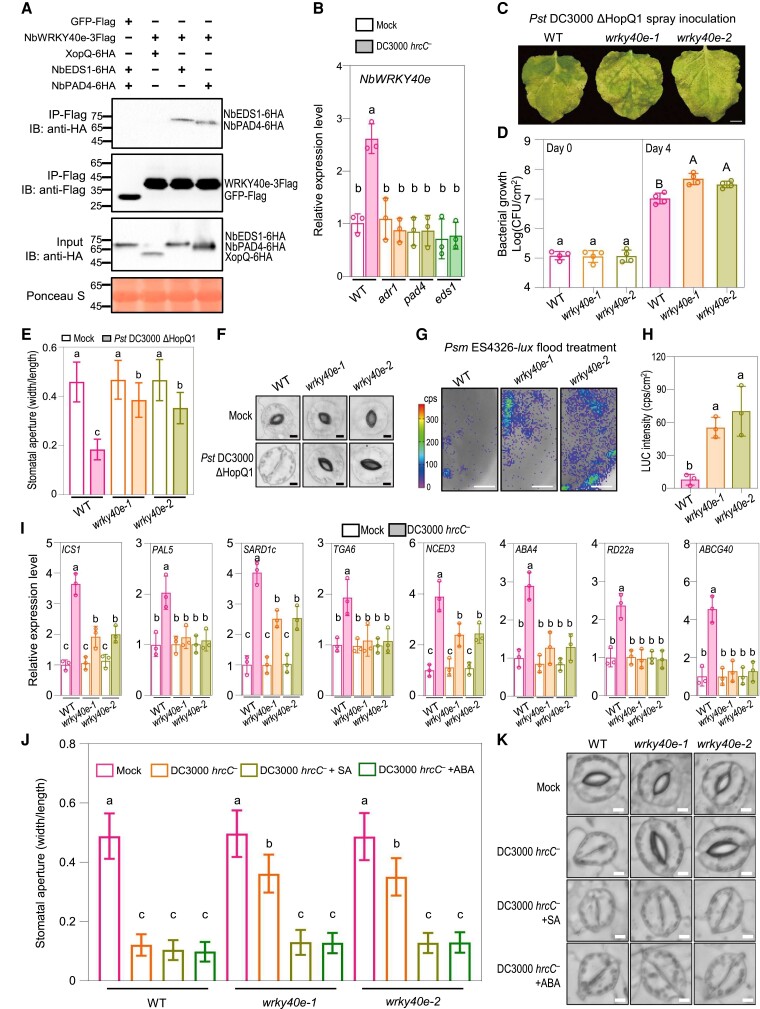
To dissect the molecular activity of the NbADR1-NbEDS1-NbPAD4 module in stomatal immunity, we employed the TurboID proximity labeling strategy to screen for NbEDS1-interacting proteins. NbWRKY40e was identified as a candidate. Co-IP showed that NbEDS1 and NbPAD4, but not XopQ (the control), were coimmunoprecipitated with NbWRKY40e (Fig. 6A). RT-qPCR analysis revealed that NbWRKY40e expression was induced by DC3000 hrcC− infection in wild type, while this induction was compromised in adr1, eds1, and pad4 plants (Fig. 6B). These results imply that NbWRKY40e participates in NbEDS1 and NbPAD4-mediated stomatal immunity.
Figure 6.
NbWRKY40e associates with NbEDS1 and NbPAD4 and mediates stomatal immunity. A) Co-IP assay showed that NbWRKY40e associates with NbEDS1 and NbPAD4. The NbEDS1-6HA, NbPAD4-6HA, or XopQ-6HA was transiently coexpressed with NbWRKY40e-3flag or the GFP-flag control in Nicotiana benthamiana leaves, respectively. The OD600 for each Agrobacterium was adjusted to 0.3. Total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. B) Quantitative real-time PCR analysis of NbWRKY40e in N. benthamiana wild-type (WT) and the indicated mutants at 2 h postspray inoculation with mock (10 mM MgCl2) or Pseudomonas syringae pv. tomato (Pst) DC3000 hrcC− (OD600 = 0.4). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). C, D) Disease symptoms C) and bacterial populations D) in leaves of indicated plants at 6 d C), or 0 and 4 d D) postspray inoculation with Pst DC3000 ΔHopQ1 (OD600 = 0.4). Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. E, F) Stomatal apertures E) and images of stomata F) in leaves of N. benthamiana WT, wrky40e-1 and wrky40e-2 after 1 h of flood treatment with mock (10 mM MgCl2) or Pst DC3000 ΔHopQ1 (OD600 = 0.4). Data are means ± Sd; n = 50. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. G, H) Bacterial pathogen entry assay in leaves of N. benthamiana WT and the indicated wrky40e mutants. The leaves were photographed and quantified by a CCD imaging system after 1 h of Psm ES4326-lux flood treatment (OD600 = 0.5). Data are means (± Sd) of 3 biological replicates from 3 independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. I) Quantitative real-time PCR analysis of the representative SA and ABA pathway genes in N. benthamiana WT and the indicated wrky40e mutants at 2 h postspray inoculation with mock (10 mM MgCl2) or Pst DC3000 hrcC− (OD600 = 0.4). Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by one-way ANOVA analysis (Tukey's post hoc test, P < 0.05). J, K) Stomatal apertures J) and images of stomata K) in leaves of N. benthamiana WT and the indicated wrky40e mutants after 1 h of flood treatment with mock (10 mM MgCl2), Pst DC3000 hrcC− (OD600 = 0.4), or Pst DC3000 hrcC−(OD600 = 0.4) plus SA (the form of sodium salicylate) or ABA, respectively. Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. IB, immunoblotting; IP, immunoprecipitation; CFU, colony-forming unit; cps, counts per second.
We next silenced NbWRKY40e and tested the stomatal immune response (Supplemental Fig. S8). In comparison with the control, plants in which NbWRKY40e was silenced contained larger bacterial populations after spray inoculation with DC3000 ΔHopQ1 (Supplemental Fig. S8, A and B). Consistent with this finding, bacteria-induced stomatal closure was compromised in NbWRKY40e-silenced plants (Supplemental Fig. S8, C and D).
We next generated 2 NbWRKY40e knockout mutants, wrky40e-1 and wrky40e-2, using CRISPR-Cas9 technology (Supplemental Fig. S1D). Compared with wild type, the wrky40e mutants were more susceptible to spray inoculation with DC3000 ΔHopQ1, and they displayed compromised stomatal closure upon flood treatment with DC3000 ΔHopQ1 (Fig. 6, C to F). Additionally, more bacteria entered the leaf apoplast in the wrky40e mutants compared with wild type (Fig. 6, G and H). After spray inoculation with Pst DC3000 hrcC−, the expression levels of SA- and ABA-related genes were lower in the wrky40e mutants than in wild type, and supplementation with SA or ABA rescued stomatal closure in the wrky40e mutants following Pst DC3000 hrcC− treatment (Fig. 6, I to K).
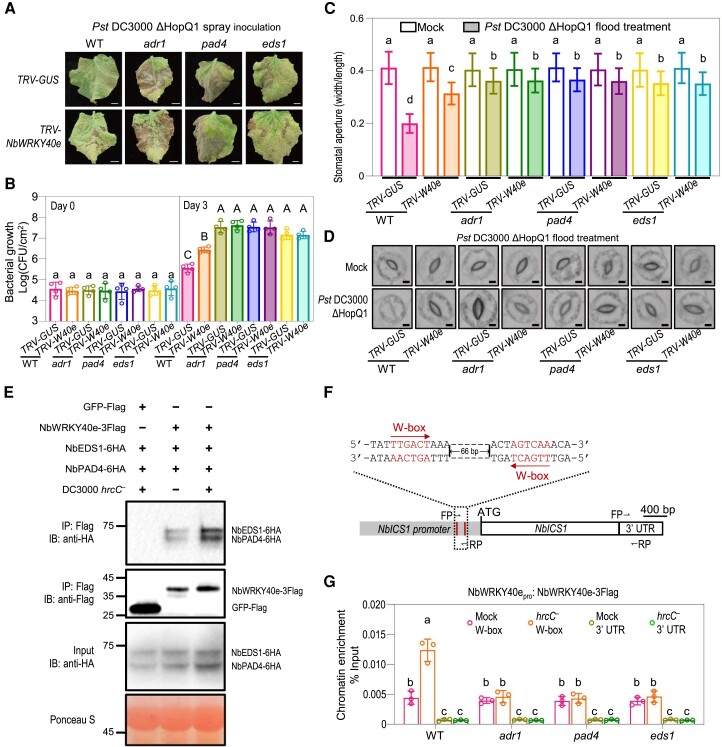
To confirm the involvement of NbWRKY40e in NbADR1, NbEDS1, and NbPAD4-mediated stomatal immunity, we silenced NbWRKY40e in 4 backgrounds (wild type, adr1, eds1, and pad4) and examined their stomatal immune responses. Silencing NbWRKY40e did not enhance the defects in bacterium-induced stomatal closure or plant susceptibility to spray inoculation with DC3000 ΔHopQ1, in adr1, eds1, and pad4 (Supplemental Fig. S8E; Fig. 7, A to D), suggesting that NbWRKY40e functions in the same pathway as NbADR1, NbEDS1, and NbPAD4 to mediate stomatal immunity.
Figure 7.
NbWRKY40e participates in NbADR1-, NbEDS1-, and NbPAD4-mediated stomatal immunity. A, B) Disease symptoms A) and bacterial populations B) in leaves of Nicotiana benthamiana wild-type (WT), adr1, pad4, and eds1 with TRV-GUS or TRV-NbWRKY40e at 7 d (A), or 0 and 3 d (B) postspray inoculation with Pseudomonas syringae pv. tomato (Pst) DC3000 ΔHopQ1 (OD600 = 0.4). Data are means (±Sd) of 4 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 1 cm. C, D) Stomatal apertures C) and images of stomata D) in leaves of N. benthamiana WT, adr1, pad4, and eds1 with TRV-GUS or TRV-NbWRKY40e after 1 h of flood treatment with mock (10 mM MgCl2) or Pst DC3000 ΔHopQ1 (OD600 = 0.4). Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. All these experiments were repeated 3 times with similar results. E) Co-IP assay showed that the association between NbWRKY40e and NbEDS1-NbPAD4 was significantly induced by Pst DC3000 hrcC− (OD600 = 0.4) inoculation at 3 hpi. NbEDS1-6HA and NbPAD4-6HA were transiently coexpressed with NbWRKY40e-3flag or the GFP-flag control in N. benthamiana leaves, respectively. The OD600 for each Agrobacterium was adjusted to 0.3. Total proteins were immunoprecipitated with the anti-Flag agarose beads, and the IP product proteins were detected by immunoblotting using the anti-flag or anti-HA antibody. Ponceau-S staining of Rubisco was used as a loading control. F) Schematic diagrams of the NbICS1 promoter. The W-box motifs were shown in red, with the red arrows indicating their directions. The FP and RP represented the primers for the chromatin immunoprecipitation-quantitative PCR assay. G) Chromatin immunoprecipitation-quantitative PCR analysis of the NbWRKY40-3Flag binding affinity to the NbICS1 promoter in WT, adr1, pad4, and eds1 at 2 h postinoculation with mock (10 mM MgCl2) or Pst DC3000 hrcC− (OD600 = 0.4). The NbWRKY40e promoter-driven NbWRKY40e-3Flag was transient expressed in WT, adr1, pad4, and eds1. The chromatin extracted from N. benthamiana leaves was immunoprecipitated with anti-Flag antibody. Data are means (±Sd) of 3 biological replicates from independent plants. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). IB, immunoblotting; IP, immunoprecipitation; CFU, colony-forming unit; FP, forward primer; RP, reverse primer; UTR, untranslated region.
We next explored how NbADR1, NbEDS1, and NbPAD4 affect the function of NbWRKY40e. Co-IP assays revealed that the interaction of NbEDS1 or NbPAD4 with NbWRKY40e was enhanced by spray inoculation with Pst DC3000 hrcC− (Fig. 7E). Chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) analysis showed that spray inoculation with Pst DC3000 hrcC− enhanced the binding of NbWRKY40e, but not of the negative control GFP, to the W-box motif region of the ICS1 promoter (Fig. 7, F and G; Supplemental Fig. S9). However, the enhanced binding of NbWRKY40e to the ICS1 promoter in response to Pst DC3000 hrcC− treatment was attenuated in adr1, eds1, and pad4 (Fig. 7, F and G). Thus, NbADR1, NbEDS1, and NbPAD4 are required for the enhanced binding of NbWRKY40e to the ICS1 promoter upon Pst DC3000 hrcC− treatment.
Taken together, these results suggest that NbWRKY40e associates with NbEDS1 and NbPAD4 and participates in NbADR1, NbEDS1, and NbPAD4-mediated stomatal immunity.
The Arabidopsis ADR1-EDS1-PAD4 module plays a conserved role in stomatal immunity
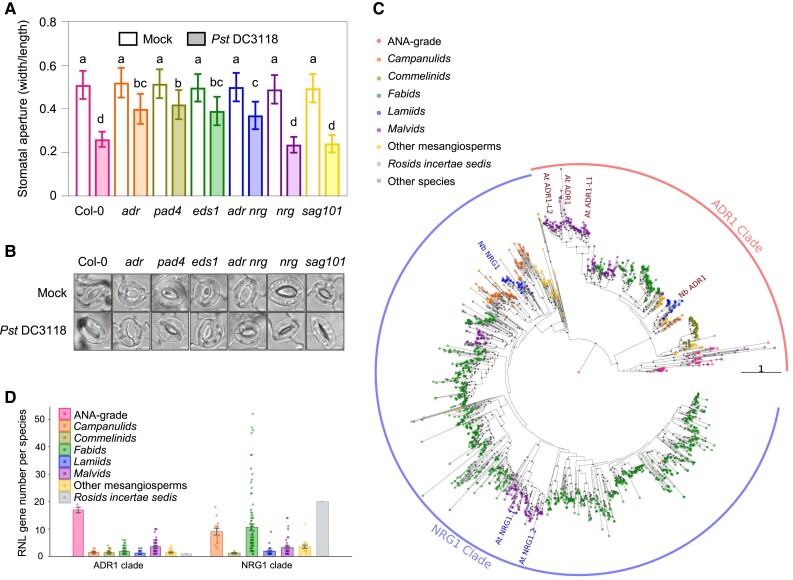
To investigate whether the function of ADR1-EDS1-PAD4 in stomatal immunity is conserved, we measured the stomatal apertures in wild-type and mutant Arabidopsis plants (wild-type Col-0, adr1 adr1-l1 adr1-l2, eds1a eds1b, pad4, nrg1.1 nrg1.2, adr1 adr1-l1 adr1-l2 nrg1.1 nrg1.2, and sag101) following flood treatment with a mock solution or the coronatine-defective strain Pst DC3118 (Fig. 8, A and B; Supplemental Fig. S1, E and F). Pst DC3118 induced stomata closure significantly in wild type, nrg1.1/1.2, and sag101, whereas stomata closure was compromised in adr1 adr1-l1/l2, adr1 adr1-l1/l2 nrg1.1/1.2, eds1a/1b, and pad4, indicating that the Arabidopsis AtADR1s-AtEDS1-AtPAD4 module, but not AtNRG1s or AtSAG101, mediates stomatal closure in response to bacterial infection. These data suggest a conserved role for ADR1-EDS1-PAD4 in stomatal immunity.
Figure 8.
The ADR1-EDS1-PAD4 module plays a conserved role in stomatal immunity. A, B) Stomatal apertures A) and images of stomata B) in leaves of the Arabidopsis Col-0 wild-type, adr1 adr1-l1 adr1-l2 (adr), pad4, nrg1.1 nrg1.2 (nrg), sag101, adr1 adr1-l1 adr1-l2 nrg1.1 nrg1.2 (adr nrg), and eds1 after 1 h of flood treatment with mock (10 mM MgCl2) or Pseudomonas syringae pv. tomato (Pst) DC3118 (OD600 = 0.2). Data are means ± Sd; n = 50 stomata. Letters indicate significant differences by 1-way ANOVA analysis (Tukey's post hoc test, P < 0.05). Scale bars represent 5 μm. The experiments were repeated 3 times with similar results. C) A maximum likelihood tree based on NB-ARC sequences of 1,979 RNL proteins predicted from 277 flowering plants. Eighty percent or better bootstrap values are shown as gray dots on branches. Branch length indicates the number of substitutions per site. The tree was rooted with the single RNL from Amborella trichopoda. All RNL proteins falls into 2 clades: ADR1 and NRG1 clades. Branch length indicates the number of substitutions per site. D) Numbers of predicted ADR1 and NRG1 genes per species. Error bars represent mean of standard error (SEM).
To explore the phylogeny and diversity of ADR1- and NRG1-type RNLs in flowering plants, we conducted a phylogenetic analysis using 1,979 RNL sequences from 277 angiosperm species (Fig. 8C). Amborella trichopoda, the sister lineage to all other flowering plants, was used as the root of the phylogenetic tree. The RNLs were divided into 2 groups, the ADR1-clade and NRG1-clade, corresponding to ADR1s and NRG1s, respectively. Interestingly, ADR1-clade proteins were found to be widely distributed across almost all angiosperm lineages, indicating their ancient and ancestral nature in angiosperms. In contrast, NRG1-clade RNLs were absent from the ANA grade angiosperms (Amborellales, Nymphaeales, and Austrobaileyales) and monocots. Notably, the ADR1-type RNLs exhibited considerable expansion in 3 the ANA grade angiosperm species, while the NRG1-type RNLs underwent a massive duplication in eudicots, particularly in Campanulids and Fabids (Fig. 8D). These findings suggest that ADR1- and NRG1-type RNLs have followed different evolutionary trajectories in flowering plants, which may account for their functional differences in stomatal immunity. Despite their phylogenetic distance, both AtADR1s and NbADR1, placed in the Malvids and Fabids clades, respectively, are involved in stomatal immunity, suggesting that the function of ADR1s in stomatal immunity is conserved across many plant species.
Discussion
The role and underlying mechanism of the intracellular RNL immune receptor NbADR1 and its putative immune partner NbPAD4 in innate immunity have remained enigmatic. In this study, we found that the Nbadr1 mutation did not affect the TNL- or CNL-triggered HR and resistance to bacterial pathogens or PTI activation in the leaf apoplast (Fig. 2A; Supplemental Figs. S3 and S4), consistent with a study showing that NbADR1 is not required for PRRCf4, FLS2-mediated PTI activation in the leaf apoplast (Znnchen et al. 2022). However, we unexpectedly discovered that NbADR1, NbPAD4, and NbEDS1 were essential for resistance to spray inoculation with bacterial pathogens and played a profound role in stomatal closure and immunity via SA and ABA pathways (Figs. 1 to 4).
During ETI signaling, the TIR domains-produced molecules pRib-AMP/ADP and ADPr-ATP enhance AtADR1-L1-AtEDS1-AtPAD4 and AtNRG1.1-AtEDS1-AtSAG101 complex formation, respectively (Horsefield et al. 2019; Wan et al. 2019; Huang et al. 2022; Jia et al. 2022). Meanwhile, during PTI-stomatal defense signaling, pathogen infection induced interactions of NbADR1 with NbPAD4 and NbEDS1 in N. benthamiana guard cells at 1 h following pathogen infection (Fig. 5; Supplemental Fig. S7B). The molecular signals and how these members including NbADR1 perceive the signals and subsequently form stomatal immunity-related complexes remain to be elucidated.
Compared with NbADR1, the RNL NbNRG1 mediates TNL-triggered ETI responses (Qi et al. 2018; Supplemental Fig. S3), but not leaf apoplast PTI responses or stomatal immunity (Fig. 2, A to D). As stomatal immunity and ETI responses were activated by Pst DC3000 spray inoculation in N. benthamiana, the disease phenotypes observed in nrg1, adr1, and adr1 nrg1 (Fig. 1) were caused by ETI response disruption and stomatal immunity activation in nrg1, ETI response activation and stomatal immunity disruption in adr1, and simultaneous ETI response and stomatal immunity disruption in adr1 nrg1. Thus, NbADR1 and NbNRG1 seem to play redundant or additive roles in plant resistance to spray inoculation with Pst DC3000. Following toothpick inoculation with Pst DC3000 ΔHopQ1 in N. benthamiana, distant colonization in the xylem (a spreading symptom) occurred and HopQ1 triggered an ETI response to block distant colonization (Misas-Villamil et al. 2011). It will be interesting to explore whether NbADR1 affects distant colonization by Pst DC3000 ΔHopQ1.
It is worth noting that bacteria-induced stomatal closure was not completely disrupted in adr1, pad4, and eds1 (Fig. 2, C to F), suggesting the existence of NbADR1, NbEDS1, or NbPAD4-independent pathways in regulating stomatal immunity. In addition, NbADR1 was expressed not only in the guard cells but also in the epidermal cells of N. benthamiana leaves (Supplemental Fig. S7, C and D). Meanwhile, a recent study showed that NbADR1 is involved in stromule formation in leaf epidermal cells (Prautsch et al. 2023). It will be interesting to explore the functions of NbADR1 beyond those in guard cells.
In this study, NbWRKY40e was characterized as NbEDS1- and NbPAD4-interacting transcription factors that function in the same pathway as NbADR1, NbEDS1, and NbPAD4 to mediate stomatal immunity by regulating SA and ABA pathways (Figs. 6 and 7). However, no interaction was detected between NbADR1 and NbWRKY40e, even in the presence of NbEDS1/NbPAD4 and pathogen infection (Supplemental Fig. S10), implying that NbADR1-NbEDS1-NbPAD4 and NbWRKY40e mediate stomatal immunity not by forming a 4-membered complex. As outlined in Fig. 9, we speculate that in response to pathogen invasion, the intracellular immune receptor NbADR1 and its immune partners NbPAD4/NbEDS1 perceive immune signals from flg22-NbFLS2-NbBAK1 and that they form NbADR1-NbPAD4-NbEDS1 complexes. Subsequently, NbEDS1 and NbPAD4 interact incrementally with NbWRKY40e, enhance its function, and activate SA and ABA pathways, thereby leading to stomatal closure for preventing pathogen entry.
Figure 9.
A simplified model for ADR1-EDS1-PAD4 in stomatal immunity. Upon bacterial pathogen invasion, the intracellular immune receptor NbADR1 and its immune partners NbPAD4 and NbEDS1 sense the immune signals from flg22-NbFLS2-NbBAK1 and form the NbADR1-NbPAD4-NbEDS1 complexes; NbEDS1 and NbPAD4 interact with NbWRKY40e and affect gene expression (e.g. ICS1); they regulate SA and ABA pathways, control stomatal aperture, and mediate stomatal immunity to prevent pathogen entry.
It is widely accepted that intracellular NLR receptors trigger hypersensitive cell death to inhibit pathogen growth during ETI responses (Zhang et al. 2004; Peart et al. 2005; Rairdan et al. 2008). This study uncovers a pathway in which the intracellular RNL receptor module NbADR1-NbEDS1-NbPAD4 functions with NbWRKY40e to mediate stomatal immunity (Fig. 9). Thus, our work provides valuable insight into the role of RNL receptors in innate immunity. It is noteworthy that the molecular details of NbADR1 recruiting NbEDS1 and NbPAD4 and its regulation in guard cells remain unclear and require future investigation. There might be PRR signaling-activated molecules that could be perceived by NbADR1 and enhance NbADR1-NbPAD4-NbEDS1 complex formation, and after some yet unknown signaling transduction events (e.g. configuration change), NbEDS1 and NbPAD4 would be released to control NbWRKY40e for stomatal immunity. With the development of technical systems for studying guard cells, it would be helpful to obtain images of guard cell-specific NbADR1-mediated stomatal immune signaling, such as signaling molecules, the formation and dynamic actions of NbADR1-NbPAD4-NbEDS1 and NbEDS1-NbPAD4-NbWRKY40e complexes, and subsequent transcriptional regulation in guard cells.
Both N. benthamiana and Arabidopsis ADR1s and their immune partners PAD4s and EDS1s are essential for stomatal immunity to bacterial pathogens (Figs. 2 to 5 and 8), suggesting that the ADR1-PAD4-EDS1 module mediates stomatal immunity in a wide range of plant species. It would be significant to test this speculation and to dissect the common and unique characteristics of ADR1-EDS1-PAD4 module-based immune signaling in various plants to further our understanding of plant innate immune systems.
Materials and methods
Plant materials, growth conditions, and vector construction
The N. benthamiana mutants nrg1, eds1, adr1, nrg1 adr1, sag101, pad4, wrky40e-1, wrky40e-2, roq1, and the Arabidopsis (A. thaliana) mutants adr1 (sail_842_b05), adr1-l1 (sail_302_c06), adr1-l2 (salk_076159), eds1a eds1b, pad4 (salk_089936), sag101 (salk_022911), nrg1.1 nrg1.2 (salk_020974), and adr1 adr1-l1 adr1-l2 nrg1.1 nrg1.2 were described previously, obtained from NASC or ABRC, or generated by the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene editing method and crosses (Supplemental Fig. S1; Yan et al. 2015; Ge et al. 2017; Qi et al. 2018). The N. benthamiana and Arabidopsis plants were grown under a light intensity of 2,000 to 4,000 lux provided by full-spectrum fluorescent bulbs (TL5 Essential 28W/865; PHILIPS) and under a 16 h-light/8 h-dark photoperiod (24 to 26 °C) or a 12 h-light/12 h-dark photoperiod (18 to 23 °C), respectively. For vector construction, the coding sequences of ADR1s, EDS1, SAG101, PAD4, WRKY40e, and their derivatives were constructed into the pE1776 or the modified pCAMBIA1300 or pEarlyGate100 vectors (Leister et al. 2005). The primers for vector construction and genotyping are listed in Supplemental Data Set 1.
Agrobacterium-mediated transient protein expression
The Agrobacterium (Agrobacterium tumefaciens) (GV3101) bacterial strains containing the expression vector were cultivated overnight, pelleted, and resuspended in infiltration buffer (10 mM 2-[N-morpholino] ethanesulfonic acid [MES], 10 mM MgCl2, 200 μM acetosyringone, pH 5.6), kept under dark at room temperature for approximately 2 h, and subsequently infiltrated into leaves of 4-wk-old N. benthamiana plants. The inoculum concentration for each Agrobacterium strain was adjusted to OD600 = 0.1 to 0.6. For Co-IP assays, bacterial pathogen strains (OD600 = 0.4) or mock were inoculated at the indicated time before harvest.
For observation of HR, the Agrobacterium strains containing the constructs expressing XopQ, HopQ1, ATR1, RPP1, p50, N, or AvrBs2 (Qi et al. 2018) were resuspended in the infiltration buffer (10 mM MES, 10 mM MgCl2, and 0.2 mM acetosyringone) and were coinfiltrated into leaves of 4-wk-old N. benthamiana using a needleless syringe. The infiltrated leaves were wrapped up with aluminum foil and were imaged at 3 d post infiltration.
For transient expression assays in guard cells, leaves from 4-wk-old N. benthamiana were flooded in infiltration buffer containing 0.01% (v/v) Tween-20 and the indicated Agrobacterium strains with their abaxial surfaces facing upwards for 24 h in the dark. Subsequently, bacterial pathogen strains (OD600 = 0.4) or mock were inoculated at the indicated time before confocal imaging or stomatal aperture quantification.
Bacterial growth assay
P. syringae pv. tomato (Pst) DC3000, Pst DC3000 ΔHopQ1, X. euvesicatoria (Xe) 85-10, and Xe 85-10 ΔXopQ strains (Qi et al. 2018) were cultivated overnight, pelleted, and resuspended in autoclaved 10 mM MgCl2. Four-week-old (Fig. 1A, 1B, 2A, 2B, 6C, and 6D; Supplemental Figs. S3B, S3C, S4B, and S5) or 5-wk-old (Fig. 3D, 7A, and 7B; Supplemental Fig. S8B) N. benthamiana plants were syringe-infiltrated or spray inoculated with the bacterial inocula (OD600 = 0.0001) or the bacteria solution (OD600 = 0.4) in 0.02% (v/v) Silwet-77, respectively. Leaf samples were collected at the indicated time points after infection, homogenized, and subjected to gradient dilutions as described previously (Qi et al. 2018).
RT-qPCR analysis
Leaves from 4-wk-old N. benthamiana were spray inoculated with mock or Pst DC3000 (OD600 = 0.4) and were harvested 24 h after inoculation (Fig. 1C). Leaves from 4-wk-old N. benthamiana were spray inoculated with mock or Pst DC3000 hrcC− (OD600 = 0.4) and were harvested at 2 h after inoculation (Fig. 4A, 6B, and 6I). Three-week-old N. benthamiana were infiltrated with Agrobacterium bacteria containing TRV vectors for gene silencing, and leaves from the plants were harvested at 2 wk postinfiltration (Supplemental Fig. S6, S8A, and S8E). Following the standard plant RT-qPCR protocols (Udvardi et al. 2008; Remans et al. 2014), total RNA was isolated from N. benthamiana leaves using the RNA-easy Isolation Reagent (Vazyme, R701-01). Reverse transcription was performed using All-in-One 5×RT MasterMix (abm, no. G592), and RT-qPCR was conducted with BlasTaq 2×qPCR MasterMix (abm, no. G891) on a BIO-RAD CFX96 real-time system. NbActin was used as the reference gene, as previously described (Qi et al. 2018). The relative expression levels of different genes were calculated using the “2-ΔΔCt” method (Livak and Schmittgen 2001). The primers used for RT-qPCR are listed in Supplemental Data Set 1.
Extraction and determination of SA
Leaves from 4-wk-old N. benthamiana were spray inoculated with Pst DC3000 (OD600 = 0.4) and were harvested 24 h after inoculation. For each sample, 100 mg of leaf material was homogenized in liquid nitrogen and transferred into a 2 mL centrifuge tube. To extract the SA, 1.5 mL extraction buffer (isopropanol: formic acid = 99.5:0.5, v/v, with a final concentration of 200 ng/L of d4-SA as internal standard) was added, followed by vortexing and a 30-min ultrasonic treatment. After centrifugation at 14,000 g for 15 min, 1.4 mL of the supernatant was transferred and dried using a LABCONCO CentriVap vacuum centrifugal concentrator and resuspended in 1 mL methanol solvent (85:15, v/v).
For sample purification, a Waters Sep-pak C18 SPE tube was utilized, and a total of 2 mL eluent was collected. The eluent was subsequently dried and resuspended in 200 µL methanol solvent (20:80, v/v). The SA content was detected by a triple quadrupole mass spectrometer and calculated with calculation curve made by standards.
Negative ionization mode data were acquired using an ACQUITY UPLC I-Class (Waters) coupled to an AB Sciex 4,500 QTRAP triple quadrupole mass spectrometer (AB SCIEX) equipped with a 50 × 2.1 mm, 1.7 μm ACQUITY UPLC BEH C18 column (Waters). Ten microliter of samples were loaded each time and then eluted at a flow rate of 200 μL/min with initial conditions of 20% (v/v) mobile phase A (0.1% [v/v] formic acid in acetonitrile) and 80% (v/v) mobile phase B (0.1% [v/v] formic acid in water). The gradients of mobile phase were as follows: 0 min, 80% (v/v) B; 0 to 3 min, 60% (v/v) B; 3 to 5 min, 40% (v/v) B; 5 to 7 min, 0% (v/v) B; 7 to 9 min, 0% (v/v) B; 9 to 9.1 min, 80% (v/v) B; and 9.1 to 12 min, 80% (v/v) B. The auto-sampler was set at 10 °C.
Mass spectrometry is operated separately in negative electrospray ionization mode. The [M - H] of analyte was selected as the precursor ion. The quantification mode was multiple reaction monitoring (MRM) mode using the mass transitions (precursor ions/product ions). Temperature of ESI ion source was set at 500 °C. Curtain gas flow was set as 25 psi, collisionally activated dissociation (CAD) gas was set as medium, and the ion spray voltage was (−) 4500 V for negative ionization mode, with ion gas 1 and 2 set as 50 psi. The collision energies for different MRM pairs were shown as follows for SA (precursor ion, 137.00; Product ion, 93.10; DP [volts], −75; CE [volts], −23) and d4-SA (precursor ion, 142.00; Product ion, 66.00; DP [volts], −190; CE [volts], −21). Data acquisition and processing were performed using AB SCIEX analyst 1.6.3 software (Applied Biosystems).
Stomatal aperture quantification
Leaves from 4-wk-old (Figs. 2C, 2D, 3E, 3F, 4B, 4C, 5E, 6E, 6F, 6J, and 6K) and 5-wk-old (Figs. 2E, 2F, 3A, 3B, 7C, and 7D; Supplemental Fig. S8, C and D) N. benthamiana were harvested and flooded in approximately 20 mL of freshly prepared MES/KCl buffer (50 mM KCl, 0.1 mM CaCl2, 10 mM MES-KOH, pH 6.15) with the abaxial surfaces facing the light for 2 to 3 h. Subsequently, the leaves were treated with mock, 100 nM flg22, or the suspension (OD600 = 0.4) of Pst DC3000 ΔHopQ1 or Pst DC3000 hrcC− (Melotto et al. 2006), or the Pst DC3000 hrcC− suspension (OD600 = 0.4) containing mock, sodium salicylate (5 mM), or ABA (10 μM). After 1 h of treatment, leaf abaxial epidermal tissue was peeled off with tweezers, and a total of 50 stomata for each genotype and treatment were observed and photographed by a Nikon Ti2-U microscope with a PCO Panda 4.2 camera.
For Arabidopsis, abaxial epidermal peels from fully expanded leaves of 4-wk-old plants were flooded in a stomatal opening buffer solution (50 mM KCl in 10 mM MES buffer, pH 6.15 [KOH]). The peels were incubated under light for 2 h to promote stomatal opening before stomatal aperture measurement. Mock or Pst DC3118 (Melotto et al. 2006) suspension (OD600 = 0.2) was added to the stomata opening buffer. The peels were mounted on slides, and microscopic images of stomata were captured after 1 h of inoculation. Peels from at least 3 plants and a minimum of 50 stomata were examined for each sample. Stomata aperture equaled the width/length ratio of the stomata opening.
Pathogen entry assay
Leaves from 4-wk-old (Fig. 2G, 2H, 3G, 3H, 6G, and 6H) N. benthamiana were flooded in approximately 20 mL of freshly prepared MES/KCl buffer and placed under light for 4 h. After that, the leaves were exposed to the luxCDABE-tagged P. syringae pv. maculicola (Psm) ES4326 strain (Fan et al. 2008) (OD600 = 0.5) for 1 h. The leaves were gently stirred and then washed in 0.02% (v/v) Silwet-77 for about 10 s. The intensity of luciferase (LUC) emitted by the entered pathogens was imaged and quantified by a CCD imaging system (Berthold Technologies, NightSHADE LB 985).
Virus induced gene silencing
The VIGS plants were generated with the TRV-based method (Liu et al. 2002). The Agrobacterium bacteria (OD600 = 0.2 for each strain) containing TRV1 and TRV2-NbFLS2/NbBAK1, TRV2-NbWRKY40e, or TRV2-GUS were coinfiltrated into leaves of 4-wk-old plants, and the leaves from the plants were used for pathogen infection at 2 wk after coinfiltration of the TRV vectors. The primers used for constructing TRV2-NbFLS2/NbBAK1 and TRV2-NbWRKY40e vectors are listed in Supplemental Data Set 1.
BiFC assay
Agrobacterium strains with N-terminal fragment of yellow fluorescent protein (nYFP) or C-terminal fragment of YFP (cYFP)-fused NbEDS1, NbPAD4, NbADR1-LRR, and GUS constructs were mixed and introduced into leaves from 4-wk-old N. benthamiana by Agrobacterium-mediated transformation. YFP signal in the epidermal guard cells and pavement cells of the leaves were detected with a Zeiss laser scanning confocal microscope (LSM 880).
Co-IP assay
The leaves from 4-wk-old N. benthamiana were infiltrated with the Agrobacterium (GV3101) strains with the indicated vectors, subsequently infected with either mock, Pst DC3000 ΔHopQ1 (OD600 = 0.4), or Pst DC3000 hrcC− (OD600 = 0.4), and were collected and homogenized in liquid nitrogen before being suspended in protein extraction buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.5% [v/v] Nonidet P-40, 5% [v/v] glycerol, 10 mM DTT, 6 mM ß-mercaptoethanol, 1×Protease Inhibitor cocktail). The samples were centrifuged at 12,000 g for 10 min at 4 °C, and this step was repeated twice. The supernatant was transferred to a tube containing anti-Flag beads (A2220, Sigma) or anti-GFP beads (SA070001, Smart-Lifesciences), followed by incubation for 3 h at 4 °C. The beads were subsequently washed with IP buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.2% [v/v] nonidet P-40, 1% [v/v] glycerol, 1 mM DTT) for 4 times. The bound proteins were eluted by resuspending the beads in 3× Laemmli buffer, boiling for 5 min, and centrifuging at 2,000 g for 5 min. The eluted proteins were further analyzed by immunoblotting with anti-flag (BE2005, Easybio, rabbit, 1:5,000 dilution), anti-HA (11867423001, Roche, rat, 1:5,000 dilution), or anti-GFP antibodies (BE2002, Easybio, rabbit, 1:5,000 dilution).
ChIP-qPCR analysis
The ChIP-qPCR was conducted with minor modifications based on a previously published research article (Adachi et al. 2015; Ranawaka et al. 2020). Leaves from 4-wk-old N. benthamiana were infiltrated with the indicated Agrobacterium (GV3101) strains and were spray inoculated with either mock or Pst DC3000 hrcC− (OD600 = 0.4) at 28 h after infiltration, and the leaves were collected at 2 h postspray inoculation. After crosslinking in 1 (v/v) % formaldehyde for 20 min, the leaves were washed 3 times with ddH2O and homogenized. The chromatin was isolated and sonicated 3 times for 5 min each (total of 15 min). The fragmented chromatin was incubated with 50 μL anti-Flag beads (A2220, Sigma) overnight at 4 °C. The beads were washed for 4 times, and the chromatin was eluted and decrosslinked. The chromatin DNA was purified using the ChIP-DNA Clean & Concentrator kit (D5201, ZYMO) and subsequently analyzed by qPCR assays. The enrichment of the promoter DNA was calculated from the qPCR results using the % input method as published (Adachi et al. 2015). Negative controls, including the amplification of the NbICS1 3′UTR fragment and infiltration of GFP-Flag, were included. The primers used for ChIP-qPCR were listed in Supplemental Data Set 1.
RNLs protein phylogenetic analysis
The sequences of 1,979 RESISTANCE TO POWDERY MILDEW8-type NLR receptors proteins were obtained from the ANNA database (https://biobigdata.nju.edu.cn/ANNA/), which includes NLR proteins from more than 300 angiosperm genomes. The classification of the species involved was acquired from the Angiosperm Phylogeny Group (APG) IV (THE ANGIOSPERM PHYLOGENY GROUP 2009). HMMER3.0 was used to conduct HMM search for the NB-ARC domain (Pfam: PF00931) within the RNL proteins. The NB-ARC domain sequences were extracted using an in-house script calling Biopython module (Cock et al. 2009). All NB-ARC domain sequences were aligned using MAFFT v7.471 and manually corrected using MEGA7. A maximum likelihood (ML) tree was constructed using IQ-TREE v2.1.2 with the following command: “iqtree -s input.aa.fa in –prefix RNL -m MFP -B 1000 -alrt 1000 -T AUTO.” The ML tree was visualized in R v4.1.2 using packages ggplot2, ggtree, and treeio. The sequence alignments and machine-readable tree files have been provided as Supplemental Files S1 and S2, respectively.
Statistical analysis
The statistical analysis was performed using 1-way ANOVA followed by Tukey's post hoc test with significance level of 0.05. Letters indicate significant differences. The sample size was presented in each figure legend. All the ANOVA tables are provided in Supplemental Data Set 2.
Accession numbers
The N. benthamiana and Arabidopsis Genome Initiative accession numbers for the genes mentioned in this study are as follows: NbABA4 (Niben101Scf10375g01002.1), NbABCG40 (Niben101Scf04958g00015.1), NbADR1 (Niben101Scf02422g02015.1), NbBAK1 (Niben101Scf00279g02022.1), NbEDS1 (Niben101Scf06720g01024.1), NbFLS2 (Niben101Scf01785g10011), NbICS1 (Niben101Scf05166g06006.1), NbNCED3 (Niben101Scf04174g06004.1), NbNRG1 (Niben101Scf02118g00018.1), NbPAD4 (Niben101Scf02544g01012.1), NbPAL5 (Niben101Scf04652g00007.1), NbPR1 (Niben101Scf00107g03008.1), NbPR5 (Niben101Scf04053g02006.1), NbRD22a (Niben101Scf04764g05003.1), NbSAG101b1 (Niben101Scf01300g01009.1), NbSARD1c (Niben101Scf11071g04005.1), NbTGA6 (Niben101Scf00596g01005.1), NbWRKY40e (Niben101Scf04944g05002.1), AtADR1 (AT1G33560), AtADR1-L1 (AT4G33300), AtADR1-L2 (AT5G04720), AtEDS1a (AT3G48090), AtEDS1b (AT3G48080), AtNRG1.1 (AT5G66900), AtNRG1.2 (AT5G66910), AtPAD4 (AT3G52430), and AtSAG101 (AT5G14930).
Supplementary Material
Acknowledgments
We thank Myeong-Je Cho, Yilong Gong, Bei Liu, and Jingzhi Ma for their technical assistance or materials.
Contributor Information
Hanling Wang, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Susheng Song, College of Life Sciences, Capital Normal University, Beijing 100048, China.
Shang Gao, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Qiangsheng Yu, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Haibo Zhang, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Xiulin Cui, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Jun Fan, MOA Key Lab of Pest Monitoring and Green Management, College of Plant Protection, China Agricultural University, Beijing 100193, China.
Xiufang Xin, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Yule Liu, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Brian Staskawicz, Department of Plant and Microbial Biology, University of California, Berkeley, CA 94720, USA; Innovative Genomics Institute, University of California, Berkeley, CA 94720, USA.
Tiancong Qi, Center for Plant Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
Author contributions
T.Q. conceived and designed the project; H.W., T.Q., Q.Y., H.Z., and X.C. performed experiments; T.Q., H.W., S.G., S.S., J.F., X.X., Y.L., and B.S. analyzed data; T.Q. and S.S. wrote the manuscript with assistance from H.W., S.G., and Q.Y. All authors contributed to project discussion and manuscript preparation.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene editing mutants used in this study.
Supplemental Figure S2. SA intensity of Pst DC3000 spray-inoculated N. benthamiana WT, adr1, nrg1, adr1 nrg1, and eds1 leaves detected by ACQUITY UPLC.
Supplemental Figure S3. The Nbadr1 mutation cannot affect TNL-mediated HR and ROQ1-mediated resistance to syringe infection of bacterial pathogens in N. benthamiana.
Supplemental Figure S4. NbADR1 and NbNRG1 do not regulate CNLBs2-mediated ETI in N. benthamiana.
Supplemental Figure S5. NbADR1, NbNRG1, NbEDS1, NbPAD4, and NbSAG101 are not required for plant resistance to syringe-infiltrated Xe 85-10 ΔXopQ.
Supplemental Figure S6. Analysis of gene expression levels in virus-induced gene silencing plants.
Supplemental Figure S7. NbADR1 interacts with NbPAD4-NbEDS1 in guard cells of N. benthamiana upon Pst DC3000 ΔHopQ1 infection.
Supplemental Figure S8. Silencing NbWRKY40e compromises stomatal immunity in N. benthamiana.
Supplemental Figure S9. The GFP-Flag negative control for the ChIP-qPCR analysis.
Supplemental Figure S10. The interaction between NbWRKY40e and NbADR1 was not detected in N. benthamiana.
Supplemental Data Set 1. Primer sequences used in this study.
Supplemental Data Set 2. Statistical analysis results.
Supplemental File 1. Sequence alignments.
Supplemental File 2. Machine-readable tree files.
Funding
We thank the Ministry of Science and Technology-Key Research and Development Program (2021YFA1301800), the National Natural Science Foundation of China (32370756 and 32270765), and the Tsinghua-Peking Center for Life Sciences for funding support.
Data availability
All data to support the conclusions of this manuscript are provided in the main figures and supplementary information.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Adachi H, Derevnina L, Kamoun S. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr Opin Plant Biol. 2019:50:121–131. 10.1016/j.pbi.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Adachi H, Nakano T, Miyagawa N, Ishihama N, Yoshioka M, Katou Y, Yaeno T, Shirasu K, Yoshioka H. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell. 2015:27(9):2645–2663. 10.1105/tpc.15.00213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlung N, Bonas U. Dissecting virulence function from recognition: cell death suppression in Nicotiana benthamiana by XopQ/HopQ1-family effectors relies on EDS1-dependent immunity. Plant J. 2017:91(3):430–442. 10.1111/tpj.13578 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A. 2011:108(39):16463–16468. 10.1073/pnas.1113726108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundalovic-Torma C, Lonjon F, Desveaux D, Guttman DS. Diversity, evolution, and function of Pseudomonas syringae effectoromes. Annu Rev Phytopathol. 2022:60(1):211–236. 10.1146/annurev-phyto-021621-121935 [DOI] [PubMed] [Google Scholar]
- Castel B, Ngou PM, Cevik V, Redkar A, Kim DS, Yang Y, Ding P, Jones JDG. Diverse NLR immune receptors activate defence via the RPW8-NLR NRG1. New Phytol. 2019:222(2):966–980. 10.1111/nph.15659 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007:448(7152):497–500. 10.1038/nature05999 [DOI] [PubMed] [Google Scholar]
- Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009:25(11):1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact. 2011:24(8):918–931. 10.1094/MPMI-03-11-0050 [DOI] [PubMed] [Google Scholar]
- Dall’Osto L, Cazzaniga S, North H, Marion-Poll A, Bassi R. The Arabidopsis aba4-1 mutant reveals a specific function for neoxanthin in protection against photooxidative stress. Plant Cell. 2007:19(3):1048–1064. 10.1105/tpc.106.049114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Ma W. How to win a tug-of-war: the adaptive evolution of Phytophthora effectors. Curr Opin Plant Biol. 2021:62:102027. 10.1016/j.pbi.2021.102027 [DOI] [PubMed] [Google Scholar]
- Essuman K, Milbrandt J, Dangl JL, Nishimura MT. Shared TIR enzymatic functions regulate cell death and immunity across the tree of life. Science. 2022:377(6605):eabo0001. 10.1126/science.abo0001 [DOI] [PubMed] [Google Scholar]
- Fan J, Crooks C, Lamb C. High-throughput quantitative luminescence assay of the growth in planta of Pseudomonas syringae chromosomally tagged with Photorhabdus luminescens luxCDABE. Plant J. 2008:53(2):393–399. 10.1111/j.1365-313X.2007.03303.x [DOI] [PubMed] [Google Scholar]
- Feehan JM, Castel B, Bentham AR, Jones JDG. Plant NLRs get by with a little help from their friends. Curr Opin Plant Biol. 2020:56:99–108. 10.1016/j.pbi.2020.04.006 [DOI] [PubMed] [Google Scholar]
- Freh M, Gao J, Petersen M, Panstruga R. Plant autoimmunity-fresh insights into an old phenomenon. Plant Physiol. 2022:188(3):1419–1434. 10.1093/plphys/kiab590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner J, Ordon J, Kretschmer C, Guerois R, Stuttmann J. An EDS1-SAG101 complex is essential for TNL-mediated immunity in Nicotiana benthamiana. Plant Cell. 2019:31(10):2456–2474. 10.1105/tpc.19.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Bergonci T, Zhao Y, Zou Y, Du S, Liu MC, Luo X, Ruan H, Garcia-Valencia LE, Zhong S, et al. Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science. 2017:358(6370):1596–1600. 10.1126/science.aao3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Tian L, Kontos I, Li J, Li X. Plant immune signaling network mediated by helper NLRs. Curr Opin Plant Biol. 2023:73:102354. 10.1016/j.pbi.2023.102354 [DOI] [PubMed] [Google Scholar]
- Grant JJ, Chini A, Basu D, Loake GJ. Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol Plant Microbe Interact. 2003:16(8):669–680. 10.1094/MPMI.2003.16.8.669 [DOI] [PubMed] [Google Scholar]
- Horsefield S, Burdett H, Zhang X, Manik MK, Shi Y, Chen J, Qi T, Gilley J, Lai JS, Rank MX, et al. NAD(+) cleavage activity by animal and plant TIR domains in cell death pathways. Science. 2019:365(6455):793–799. 10.1126/science.aax1911 [DOI] [PubMed] [Google Scholar]
- Hu Y, Ding Y, Cai B, Qin X, Wu J, Yuan M, Wan S, Zhao Y, Xin XF. Bacterial effectors manipulate plant abscisic acid signaling for creation of an aqueous apoplast. Cell Host Microbe. 2022:30(4):518–529 e6. 10.1016/j.chom.2022.02.002 [DOI] [PubMed] [Google Scholar]
- Huang S, Jia A, Song W, Hessler G, Meng Y, Sun Y, Xu L, Laessle H, Jirschitzka J, Ma S, et al. Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity. Science. 2022:377(6605):eabq3297. 10.1126/science.abq3297 [DOI] [PubMed] [Google Scholar]
- Jacob P, Kim NH, Wu F, El-Kasmi F, Chi Y, Walton WG, Furzer OJ, Lietzan AD, Sunil S, Kempthorn K, et al. Plant “helper” immune receptors are ca(2+)-permeable nonselective cation channels. Science. 2021:373(6553):420–425. 10.1126/science.abg7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia A, Huang S, Song W, Wang J, Meng Y, Sun Y, Xu L, Laessle H, Jirschitzka J, Hou J, et al. TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity. Science. 2022:377(6605):eabq8180. 10.1126/science.abq8180 [DOI] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL. Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol. 2019:50:82–94. 10.1016/j.pbi.2019.03.013 [DOI] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari D, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell. 2019:31(10):2430–2455. 10.1105/tpc.19.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ. Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell. 2005:17(4):1268–1278. 10.1105/tpc.104.029637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002:30(4):415–429. 10.1046/j.1365-313X.2002.01297.x [DOI] [PubMed] [Google Scholar]
- Liu Z, Hou S, Rodrigues O, Wang P, Luo D, Munemasa S, Lei J, Liu J, Ortiz-Morea FA, Wang X, et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature. 2022:605(7909):332–339. 10.1038/s41586-022-04684-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001:25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, et al. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science. 2020:369:370–371. 10.1126/science.abc9520 [DOI] [PubMed] [Google Scholar]
- Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science. 2020:370(6521):eabd9993. 10.1126/science.abd9993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Puche L, Tan H, Dogra V, Wu M, Rosas-Diaz T, Wang L, Ding X, Zhang D, Fu X, Kim C, et al. A defense pathway linking plasma membrane and chloroplasts and co-opted by pathogens. Cell. 2020:182(5):1109–1124 e1125. 10.1016/j.cell.2020.07.020 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006:126(5):969–980. 10.1016/j.cell.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY. Stomatal defense a decade later. Plant Physiol. 2017:174(2):561–571. 10.1104/pp.16.01853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misas-Villamil JC, Kolodziejek I, van der Hoorn RA. Pseudomonas syringae colonizes distant tissues in Nicotiana benthamiana through xylem vessels. Plant J. 2011:67(5):774–782. 10.1111/j.1365-313X.2011.04632.x [DOI] [PubMed] [Google Scholar]
- Montillet JL, Hirt H. New checkpoints in stomatal defense. Trends Plant Sci. 2013:18(6):295–297. 10.1016/j.tplants.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Murata Y, Mori IC, Munemasa S. Diverse stomatal signaling and the signal integration mechanism. Annu Rev Plant Biol. 2015:66(1):369–392. 10.1146/annurev-arplant-043014-114707 [DOI] [PubMed] [Google Scholar]
- Ngou BPM, Jones JDG, Ding P. Plant immune networks. Trends Plant Sci. 2022:27(3):255–273. 10.1016/j.tplants.2021.08.012 [DOI] [PubMed] [Google Scholar]
- Outram MA, Figueroa M, Sperschneider J, Williams SJ, Dodds PN. Seeing is believing: exploiting advances in structural biology to understand and engineer plant immunity. Curr Opin Plant Biol. 2022:67:102210. 10.1016/j.pbi.2022.102210 [DOI] [PubMed] [Google Scholar]
- Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, A CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol. 2005:15(10):968–973. 10.1016/j.cub.2005.04.053 [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang J, Li X, Zhang Y. Salicylic acid: biosynthesis and signaling. Annu Rev Plant Biol. 2021:72(1):761–791. 10.1146/annurev-arplant-081320-092855 [DOI] [PubMed] [Google Scholar]
- Prautsch J, Erickson JL, Özyurek S, Gormanns R, Franke L, Lu Y, Marx J, Niemeyer F, Parker JE, Stuttmann J, et al. Effector XopQ-induced stromule formation in Nicotiana benthamiana depends on ETI signaling components ADR1 and NRG1. Plant Physiol. 2023:191(1):161–176. 10.1093/plphys/kiac481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, Karelina D, Hua C, Frohlich K, Wan WL, et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature. 2021:598(7881):495–499. 10.1038/s41586-021-03829-0 [DOI] [PubMed] [Google Scholar]
- Qi D, Innes RW. Recent advances in plant NLR structure, function, localization, and signaling. Front Immunol. 2013:4:348. 10.3389/fimmu.2013.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho MJ, Schultink A, Staskawicz BJ. NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci U S A. 2018:115(46):E10979–E10987. 10.1073/pnas.1814856115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008:20(3):739–751. 10.1105/tpc.107.056036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranawaka B, Tanurdzic M, Waterhouse P, Naim F. An optimised chromatin immunoprecipitation (ChIP) method for starchy leaves of Nicotiana benthamiana to study histone modifications of an allotetraploid plant. Mol Biol Rep. 2020:47(12):9499–9509. 10.1007/s11033-020-06013-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Keunen E, Bex GJ, Smeets K, Vangronsveld J, Cuypers A. Reliable gene expression analysis by reverse transcription-quantitative PCR: reporting and minimizing the uncertainty in data accuracy. Plant Cell. 2014:26(10):3829–3837. 10.1105/tpc.114.130641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussin-Léveillée C, Lajeunesse G, St-Amand M, Veerapen VP, Silva-Martins G, Nomura K, Brassard S, Bolaji A, He SY, Moffett P. Evolutionarily conserved bacterial effectors hijack abscisic acid signaling to induce an aqueous environment in the apoplast. Cell Host Microbe. 2022:30(4):489–501 e484. 10.1016/j.chom.2022.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile SC, Jacob P, Castel B, Jubic LM, Salas-Gonzales I, Bäcker M, Jones JDG, Dangl JL, El Kasmi F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 2020:18(9):e3000783. 10.1371/journal.pbio.3000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Takasaki H, Takahashi F, Suzuki T, Iuchi S, Mitsuda N, Ohme-Takagi M, Ikeda M, Seo M, Yamaguchi-Shinozaki K, et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc Natl Acad Sci U S A. 2018:115(47):E11178–E11187. 10.1073/pnas.1811491115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017:92(5):787–795. 10.1111/tpj.13715 [DOI] [PubMed] [Google Scholar]
- Su J, Zhang M, Zhang L, Sun T, Liu Y, Lukowitz W, Xu J, Zhang S. Regulation of stomatal immunity by interdependent functions of a pathogen-responsive MPK3/MPK6 cascade and abscisic acid. Plant Cell. 2017:29(3):526–542. 10.1105/tpc.16.00577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Zhang Y, Li Y, Zhang Q, Ding Y, Zhang Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat Commun. 2015:6(1):10159. 10.1038/ncomms10159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Lapin D, Feehan JM, Stolze SC, Kramer K, Dongus JA, Rzemieniewski J, Blanvillain-Baufume S, Harzen A, Bautor J, et al. Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat Commun. 2021:12(1):3335. 10.1038/s41467-021-23614-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- THE ANGIOSPERM PHYLOGENY GROUP . An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009:161(2):105–121. 10.1111/j.1095-8339.2009.00996.x [DOI] [Google Scholar]
- Tian H, Wu Z, Chen S, Ao K, Huang W, Yaghmaiean H, Sun T, Xu F, Zhang Y, Wang S, et al. Activation of TIR signalling boosts pattern-triggered immunity. Nature. 2021:598(7881):500–503. 10.1038/s41586-021-03987-1 [DOI] [PubMed] [Google Scholar]
- Udvardi MK, Czechowski T, Scheible WR. Eleven golden rules of quantitative RT-PCR. Plant Cell. 2008:20(7):1736–1737. 10.1105/tpc.108.061143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Weyer AL, Monteiro F, Furzer OJ, Nishimura MT, Cevik V, Witek K, Jones JDG, Dangl JL, Weigel D, Bemm F. A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell. 2019:178(5):1260–1272 e1214. 10.1016/j.cell.2019.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch S, Tian L, Hoy R, Li X. Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 2020:1(1):100016. 10.1016/j.xplc.2019.100016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung EH, Osborne Nishimura E, DiAntonio A, Milbrandt J, Dangl JL, et al. TIR domains of plant immune receptors are NAD(+)-cleaving enzymes that promote cell death. Science. 2019:365(6455):799–803. 10.1126/science.aax1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. 2019a:364(6435):eaav5870. 10.1126/science.aav5870 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang HW, et al. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science. 2019b:364:6435. 10.1126/science.aav5868 [DOI] [PubMed] [Google Scholar]
- Wang S, Li S, Wang J, Li Q, Xin XF, Zhou S, Wang Y, Li D, Xu J, Luo ZQ, et al. A bacterial kinase phosphorylates OSK1 to suppress stomatal immunity in rice. Nat Commun. 2021:12(1):5479. 10.1038/s41467-021-25748-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pruitt RN, Nürnberger T, Wang Y. Evasion of plant immunity by microbial pathogens. Nat Rev Microbiol. 2022:20(8):449–464. 10.1038/s41579-022-00710-3 [DOI] [PubMed] [Google Scholar]
- Wang Z, Liu X, Yu J, Yin S, Cai W, Kim NH, El Kasmi F, Dangl JL, Wan L. Plasma membrane association and resistosome formation of plant helper immune receptors. Proc Natal Acad Sci U S A. 2023:120(32):e2222036120. 10.1073/pnas.2222036120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007:51(1):32–46. 10.1111/j.1365-313X.2007.03126.x [DOI] [PubMed] [Google Scholar]
- Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, Kamoun S. NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci U S A. 2017:114(30):8113–8118. 10.1073/pnas.1702041114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Tian L, Liu X, Zhang Y, Li X. TIR signal promotes interactions between lipase-like proteins and ADR1-L1 receptor and ADR1-L1 oligomerization. Plant Physiol. 2021:187(2):681–686. 10.1093/plphys/kiab305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Li M, Dong OX, Xia S, Liang W, Bao Y, Wasteneys G, Li X. Differential regulation of TNL-mediated immune signaling by redundant helper CNLs. New Phytol. 2019:222(2):938–953. 10.1111/nph.15665 [DOI] [PubMed] [Google Scholar]
- Yan L, Wei S, Wu Y, Hu R, Li H, Yang W, Xie Q. High-efficiency genome editing in Arabidopsis using YAO promoter-driven CRISPR/cas9 system. Mol Plant. 2015:8(12):1820–1823. 10.1016/j.molp.2015.10.004 [DOI] [PubMed] [Google Scholar]
- Ye W, Munemasa S, Shinya T, Wu W, Ma T, Lu J, Kinoshita T, Kaku H, Shibuya N, Murata Y. Stomatal immunity against fungal invasion comprises not only chitin-induced stomatal closure but also chitosan-induced guard cell death. Proc Natl Acad Sci U S A. 2020:117(34):20932–20942. 10.1073/pnas.1922319117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010:153(3):1188–1198. 10.1104/pp.110.157016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Melotto M, He SY. Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr Opin Biotechnol. 2010:21(5):599–603. 10.1016/j.copbio.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Coaker G, Zhou JM, Dong X. Plant immune mechanisms: from reductionistic to holistic points of view. Mol Plant. 2020:13(10):1358–1378. 10.1016/j.molp.2020.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dorey S, Swiderski M, Jones JD. Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 2004:40(2):213–224. 10.1111/j.1365-313X.2004.02201.x [DOI] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012:11(6):587–596. 10.1016/j.chom.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Zhou M, Yoo H, Pruneda-Paz JL, Spivey NW, Kay SA, Dong X. Spatial and temporal regulation of biosynthesis of the plant immune signal salicylic acid. Proc Natl Acad Sci U S A. 2015:112(30):9166–9173. 10.1073/pnas.1511182112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zönnchen J, Gantner J, Lapin D, Barthel K, Eschen-Lippold L, Erickson JL, Villanueva SL, Zantop S, Kretschmer C, Joosten M, et al. EDS1 complexes are not required for PRR responses and execute TNL-ETI from the nucleus in Nicotiana benthamiana. New Phytol. 2022:236(6):2249–2264. 10.1111/nph.18511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to support the conclusions of this manuscript are provided in the main figures and supplementary information.