Abstract
The human papillomavirus type 16 (HPV-16) E5 protein is a small, hydrophobic polypeptide that is expressed in virus-infected keratinocytes and alters receptor signaling pathways, apoptotic responses, and endosomal pH. Despite its ability to inhibit endosomal acidification, the HPV-16 E5 protein is found predominantly in the endoplasmic reticulum (ER), suggesting that its effect may be indirect and perhaps global. To determine whether E5 alters the pHs of additional intracellular compartments, we transduced human keratinocytes with a codon-optimized E5 vector and then quantified endosomal and trans-Golgi pHs using sensitive, compartment-specific, ratiometric pHluorin constructs. E5 protein increased endosomal pH from 5.9 to 6.9 but did not affect the normal trans-Golgi pH of 6.3. Confirming the lack of alteration in trans-Golgi pH, we observed no alterations in the acidification-dependent processing of the proH3 protein. C-terminal deletions of E5, which retained normal expression and localization in the ER, were defective for endosomal alkalization. Thus, E5 does not uniformly alkalinize intracellular compartments, and its C-terminal 10 amino acids appear to mediate interactions with critical ER targets that modulate proton pump function and/or localization.
Infection by the high-risk human papillomaviruses (HPVs) is a critical step in the genesis of cervical cancer, and nearly 99% of all cervical cancers contain and express HPV genomes (65). Two of the HPV early genes, E6 and E7, are responsible for inhibiting host cell differentiation as well as for facilitating cellular immortalization (reviewed in references 18, 34, 36, 40, 50, and 59). The E5 protein can augment cellular immortalization by E6/E7 via mechanisms that are not clearly defined (51). E5 expression appears to induce a plethora of cellular changes, including enhanced growth factor signaling (12, 13, 53, 58, 64), the activation of mitogen-activated protein kinase pathways (12, 14, 25), and the alkalization of endosomes (52), which may contribute to alterations in endosomal trafficking (56). The HPV type 16 (HPV-16) E5 protein can also induce the anchorage-independent growth of immortalized fibroblasts, although this activity in primary keratinocytes is not observed (32, 41, 53).
Despite the above myriad of activities, the precise role of HPV E5 protein in the viral life cycle is unclear. Recently, two reports demonstrated that the E5 gene in the intact genomes of HPV-16 and HPV-31 could be mutated with only minimal effects on host cell and viral DNA synthesis or viral late gene expression (17, 20). These two studies, using a raft culture system that mimics some of the events during natural infection, indicate that the effect of E5 is subtle yet potentially necessary for the in vivo maintenance of the host cells' undifferentiated phenotype and ability to synthesize DNA. In vivo, in situ hybridization of numerous low-grade cervical intraepithelial neoplasias and premalignant cervical lesions has shown that mRNA species capable of expressing the E5 open reading frame are expressed in the basal cells (11, 16, 49, 60) and are the most abundant transcripts within these cells (19, 49). As the cells migrate upward from the basement membrane, the level of E5 transcripts declines but is still detectable, whereas the level of E6 and E7 transcripts becomes predominant (49). Using E5-specific antibodies, the E5 protein itself has been detected in the lower one-third of the epidermis in low-grade lesions and throughout the epidermis in higher-grade lesions (7, 31), indicating that the undifferentiated, basal-like cells are the site of E5 synthesis. Thus, in vitro and in vivo data suggest that E5 is expressed predominantly in the least differentiated cells of the stratified squamous epithelium, a finding which is compatible with experiments demonstrating that it has little influence on viral DNA amplification which occurs in the more differentiated layers of the epithelium.
One of the more reproducible activities of E5 is its ability to augment epidermal growth factor (EGF) receptor signaling (13, 41, 53, 64), a pathway that inhibits keratinocyte differentiation (58). One hypothesis for this enhanced EGF receptor signaling is that E5, which is known to interact with the 16K subunit of the vacuolar H+-ATPase (V-ATPase) (10), directly inhibits the proton pump, thereby leading to the alkalinization of endosomes and consequent recycling of the endosome and receptor (52, 53). Defining a specific interaction between the E5 and 16K proteins, however, has been complicated by their extreme hydrophobicity (and tendency to associate with other hydrophobic proteins) as well as the need to overexpress these proteins to observe binding. As a consequence, studies evaluating the functional interaction of these proteins often yield contradictory information. For example, the HPV-16 E5 protein has been shown to both bind an epitope-tagged version of the bovine 16K subunit and functionally inhibit the yeast V-ATPase (1). However, in another study, E5 was shown to bind the 16K subunit without altering the function of the yeast V-ATPase (2). In addition, the 16K binding domain in E5 has been shown to reside within amino acid residues 41 to 54 in one study (1) and within amino acids 54 to 78 in another study (43). Clearly, the interactions between HPV E5 and 16K proteins are not as well defined as those between the BPV-1 E5 protein and 16K, where a single amino acid change in E5 can abrogate both 16K binding and effects on the proton pump (21, 22, 46).
In a previous study, we localized the HPV-16 E5 protein to the endoplasmic reticulum of foreskin keratinocytes (HFK) (15). Despite previous reports that HPV-16 E5 can alkalinize endosomes (52, 56), we were unable to find significant amounts of E5 associated with endosomes, suggesting that its effect on endosomal pH might be indirect. To address this issue, we first performed experiments with newer and more quantitative techniques to validate that HPV-16 E5 protein alkalinized endosomes. Assuming that we could verify that E5 could alkalinize a cellular compartment in which we could not detect its presence, we also examined whether E5 might be acting indirectly and perhaps generically to inhibit the acidification of other compartments. Specifically, we examined whether E5 could alter trans-Golgi pH, another cellular compartment in which we could find little E5. Finally, we have also examined the biological activity of C-terminal deletions of E5 that have previously been shown to bind the 16K subunit of the V-ATPase but were unable to augment enhancement of EGF receptor activation (43). In brief, our experiments verify that the HPV-16 E5 protein does indeed alkalinize endosomes, but that this activity is compartment specific since the acidic trans-Golgi is unaffected. More importantly, the predominant localization of E5 to the endoplasmic reticulum (ER) suggests that endosome alkalinization by E5 is not due to direct E5/16K binding within this compartment. We hypothesize that E5, via its C terminus, alters endosomal pH indirectly, and we discuss several models which could explain this activity.
Wild-type and mutant HPV-16 E5 proteins are expressed at similar levels and localize to the endoplasmic reticulum.
Epitope-tagged forms of wild-type and mutant E5 proteins were generated by PCR using the codon-optimized version of the HPV-16 E5 gene as a template (15). Two deletion mutants which lacked the C-terminal 10 and 20 amino acids were constructed. Deletion of the hydrophilic, C-terminal amino acids has already been shown to abrogate the ability of E5 to augment EGF receptor activation but not to interfere with its ability to associate with the 16K pore-forming subunit of the V-ATPase (43). The E5 constructs were subcloned into the LXSN retroviral vector, and retroviruses were produced as previously described (15). Primary human foreskin keratinocytes were grown in KGM medium (Invitrogen) and infected with the appropriate retroviruses. Following selection in G418 (100 μg/ml for 10 days), the cells were screened by immunoprecipitation (IP) and Western blotting (IB) for expression of the indicated E5 proteins using the AU1 antibody (Covance). Figure 1A shows that the two E5 mutant proteins, missing 10 or 20 amino acids at the C terminus, are both expressed at levels similar to the full-length E5 protein. To ensure that the deletions did not interfere with the targeting of E5 to the ER, we also examined their localization in HFK cells. Antibodies against calnexin, an endoplasmic reticulum protein, were used to determine if the E5 mutant proteins were normally localized to the ER. As shown in Fig. 1B, both wild-type and mutant E5 proteins (red) colocalize with calnexin (green), producing the dominant yellow merged image. In some cells, there are small amounts of E5 and calnexin which are not coincident. Thus, the deletions made in E5 do not perturb its expression or localization.
FIG. 1.
Expression and localization of full-length E5 protein and two deletion mutants in HFK cells. Panel A shows the expression of E5* and deletion mutants missing 10 or 20 amino acids from the carboxy terminus. LXSN denotes the empty retrovirus-infected HFK cells. Equal amounts of proteins were IP and IB with the AU1 antibody. Panel B shows the localization of E5* and the deletion mutants in the same HFK cells. For colocalization, the cellular endoplasmic reticulum protein calnexin (1:100; Santa Cruz) was used as a marker. E5 was detected in the tetramethyl rhodamine isocyanate channel with a goat anti-mouse secondary antibody conjugated to Texas Red. Calnexin was detected in the fluorescein isothiocyanate (FITC) channel with goat anti-rabbit secondary antibodies conjugated to FITC. Images were then merged, and colocalization is indicated in yellow.
The M2 protein of influenza virus is present in the Golgi apparatus in HFK cells.
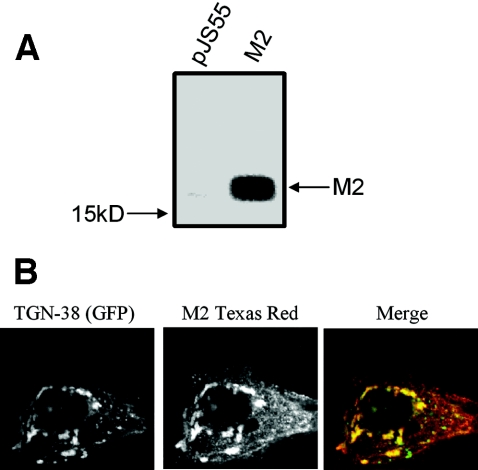
The M2 protein of influenza virus is an acid-activated, H+-selective, pore-forming protein (reviewed in reference 42) that has previously been shown to inhibit acidification of the Golgi apparatus in epithelial cells (8, 28-30, 44). We used this protein as a positive control for our experiments evaluating alterations in Golgi pH. Initial experiments were carried out to ensure that the M2 protein was localized to the Golgi apparatus in primary human keratinocytes. The protein was epitope tagged with AU1 and its expression evaluated by IP/Western blotting and immunofluorescence techniques. HFK (LXSN) cells (106) were plated in 100-mm-diameter tissue culture dishes and transiently transfected 24 h later with empty vector, pJS55 (48), or pJS55-M2 (M2 construct) using Fugene 6 (Roche Biomedical). Twenty-four hours later, the cells were lysed in radioimmunoprecipitation assay buffer, and equal amounts of protein were immunoprecipitated with AU1 antibody. The immunoprecipitated proteins were then transferred to a polyvinylidene difluoride membrane (ImmunoBlot), reacted with AU1 monoclonal antibody, and visualized by chemiluminescence. As shown in Fig. 2A, the M2 protein was highly expressed in HFK cells. To verify that M2 was in the Golgi as previously reported (28), we evaluated whether it colocalized with the fluorescent Golgi-specific protein, trans-Golgi network (TGN)-38GFP (39, 45). HFK cells were plated in two-chamber slides (Labtek) and transiently cotransfected with the TGN-38GFP vector and the M2 vector (pJS55-M2). One day later, the cells were fixed and M2 protein visualized with AU1 antibody and rhodamine-conjugated secondary antibody. Figure 2B shows one of several experiments showing that the M2 protein (red) colocalized predominantly with TGN-38GFP (green) in the Golgi (see yellow signal in the merged image). Consistent with previous studies (30), the merged image in Fig. 2 also indicates that there is additional M2 protein (red) in membrane compartments other than the trans-Golgi.
FIG. 2.
Expression and localization of M2 in LXSN-infected HFK cells. Panel A shows and IP/IB using the AU1 antibody. pJS55 is the empty vector, and the M2 lane is pJS55-M2 when transiently transfected into HFK (LXSN) cells. Panel B shows the localization of M2 using TGN-38GFP as the marker protein. TGN-38GFP images were captured in the FITC channel and the M2 images were captured in the tetramethyl rhodamine isocyanate channel with the use of a goat anti-mouse secondary conjugated to Texas Red. The images were then merged, and colocalization is shown in yellow.
Calibration curve for measuring pH with ratiometric pHluorin constructs.
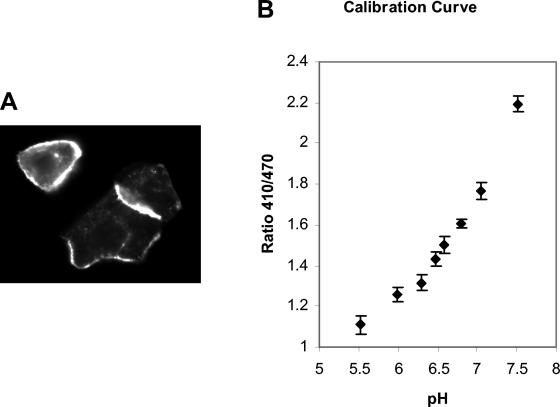
In order to measure Golgi and endosomal pH within the cells, we utilized compartment-specific, ratiometric pHluorin constructs (39, 45). Three constructs were utilized: glycosylphosphatidylinositol-green fluorescent protein (GPI-GFP), which anchors the pHluorin on the outside of the plasma membrane (6); cellubrevin-GFP, which places the pHluorin on the lumenal side of early endosomes (37); and TGN-38GFP, which targets the pHluorin to the lumen of the trans-Golgi network (33). These constructs have been used to measure organelle pH in various cells (38, 39, 45, 55) and have generated results that agree with previously published pH values for those organelles. Early endosomes exhibit an average pH of 6.1 and the trans-Golgi apparatus an average pH of 6.3 (reviewed in reference 23). A calibration curve for the pH dependence of GFP fluorescence was established by transfecting the GPI-anchored pHluorin (GPI-GFP) construct into keratinocytes, where the expressed protein was found on the cell surface as predicted (Fig. 3A). Due to the surface location of the expressed protein, a calibration curve was generated by simply exposing cells to buffers with different pHs. The images were captured using a Photometrics Cool Snap HQ camera mounted on an Axiovert 35 microscope at ×100 power outfitted with a dichromatic mirror (500DCXR) and bandpass filter (HQ535/50), both from Chroma Technologies. Briefly, cells were transfected with the GPI-GFP construct in two-chamber slides and analyzed 24 h later. For the calibration curve, the cells were rinsed in and then analyzed in prewarmed buffers at the following pHs: 7.5, 7.2, 7.0, 6.7, 6.5, 6.3, 6.0, and 5.5 (39, 46). The images were captured at wavelengths of 410 nm and 470 nm consecutively, and a ratio of 410/470 was determined. For each pH point, a minimum of 20 cells were analyzed. The data were imported into Microsoft Excel for further analysis, and the resulting calibration curve is shown in Fig. 3B. All experimental results were within this pH range, and values were determined by interpolation.
FIG. 3.
Calibration curve for pH measurements using the ratiometric pHluorins. Panel A shows the expression and localization of the GPI-GFP ratiometric pHluorin transfected into LXSN-infected HFK cells at a wavelength of 410 nm. Panel B shows the results of the calibration curve when the images were captured at 410 and 470 nm consecutively and then a ratio was determined. The error bars indicate the standard deviations for at least 20 samples for each point.
Endosomal and Golgi pH measurements.
Once a calibration curve had been established, the ratiometric pHluorin constructs fused to cellubrevin (for endosomal localization) and TGN-38 (for Golgi localization) were used to determine the pHs of these two organelles in control and E5-expressing cells (Fig. 4).
FIG. 4.
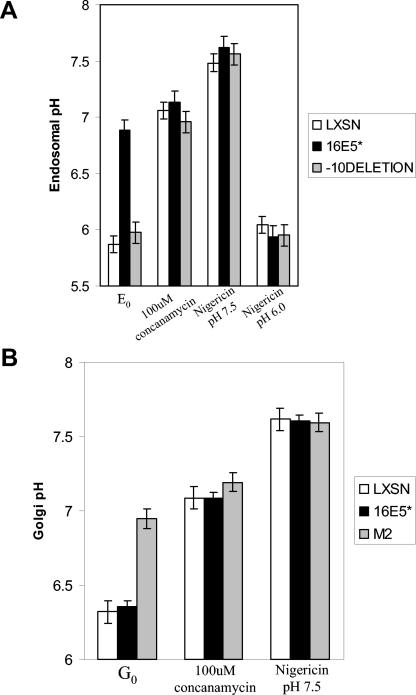
Endosomal and trans-Golgi pH measurements using the ratiometric pHluorins. Panel A shows the results of the endosomal pH measurements in LXSN, E5*, and the −10 deletion mutant of E5-expressing cells transfected with the cellubrevin-GFP pHluorin. E0 stands for the initial measurements of endosomal pH in the various cells with no treatments. The second set of bars shows the resulting pH when the cells were sequentially treated with 100 μM concanamycin. Finally, the last two sets of bars show the resulting pH when the cells were treated with 5 μM nigericin and a buffered solution with pH 7.5 and 6.0. The cells were treated sequentially while on the heated stage. The error bars indicate standard deviations for at least 20 cells for each line. Panel B shows the results of the Golgi pH measurements in LXSN-, E5*-, and M2-expressing cells transfected with the TGN-38GFP pHluorin. The procedure followed was the same as for panel A. G0 represents the initial Golgi pH measurements before the cells were treated. The additional bar sets represent measurements after the listed chemical treatments. Error bars are the same as for panel A.
The first goal was to use this more quantitative technology to verify that HPV E5 did indeed alkalinize endosomes (Fig. 4A). HFK cells expressing LXSN (control vector), HPV-16 E5 protein, or the −10 deletion E5 mutant protein were transfected with the cellubrevin-GFP pHluorin construct. Control cells exhibited an endosomal pH of 5.9 (E0), consistent with previous reports (39, 55). However, E5-expressing cells had an endosomal pH of 6.9. Cells expressing the −10 E5 deletion mutant exhibited an endosomal pH of 6.0, as did the −20 mutant (data not shown). Thus, in agreement with previously published data (52, 56), HPV E5 elevated endosomal pH. It is also clear that the C-terminal 10 amino acids of E5 are critical for its biological activity.
In addition, we evaluated the response of control and E5-expressing cells to concanamycin and nigericin treatments. Concanamycin is a potent and specific inhibitor of the H+-ATPase (46) and it raised endosomal pH to 7.0 to 7.2 in all cell lines, demonstrating the role of the proton pump in maintaining the acidic environment. Finally, the cells were treated with 5 μM of the proton ionophore nigericin (Molecular Probes), and then equilibrated in calibration buffer at either pH 6.0 or pH 7.5. All cell types were altered in their endosomal pH according to the corresponding buffer, indicating that the pHluorin responded appropriately in all cell lines. The more acidic control was added in this case to ensure that E5 was not directly affecting the response of the pHluorin to an acidic environment.
Another pHluorin reagent, TGN-38GFP, was used to measure Golgi pH in control HFK cells or cells expressing HPV-16 E5 protein or M2 protein (Fig. 4B). The Golgi pH (G0) of E5-expressing cells was calculated to be 6.35 to 6.4, nearly identical to the Golgi pH of the control cells (6.3 to 6.4). In contrast, the Golgi pH of M2-transfected cells was elevated to 6.9, consistent with previous studies and the known ability of M2 to dissipate proton gradients (28, 30, 44). As with the above endosomal pH measurements, we verified the response of the various cell lines to concanamycin and nigericin treatments. Concanamycin raised the Golgi pHs to 7.0 to 7.2 in all cell lines. Finally, when the cells were treated with 5 μM of the proton ionophore, nigericin, and then equilibrated in pH 7.5 calibration buffer, we documented that all cells responded by increasing their Golgi pHs to 7.5.
E5 does not affect pH-sensitive processing of cellular proteins in the Golgi.
We also employed a biological assay to verify the above fluorescence analysis of Golgi pH. The cellular protein proH3 undergoes autocatalytic cleavage in the Golgi at an acidic pH (57), and this cleavage can be used to evaluate the acid environment of the Golgi. ProH3 is synthesized as a 105-kDa proprotein, which is cleaved in acidic environments near the C terminus to yield the H3 protein (75 kDa) and a 30-kDa fragment. Both the full-length (ProH3) and cleaved (H3) forms are secreted from the cell. However, if the Golgi is alkalinized, the cleavage does not occur and only the 105-kDa form is secreted. HFK cells expressing LXSN (control vector) or HPV-16 E5 vector were transfected with the proH3 construct, and 24 h later they were labeled with 35S-labeled mix (Amersham) for 3 h. Some cultures were supplemented with 100 μM concanamycin for the duration of the labeling. The supernatant was harvested and immunoprecipitated with proH3 antiserum, and the products were separated electrophoretically on polyacrylamide gels and detected by autoradiography. Figure 5 shows that control and E5-expressing cells secreted both the 105-kDa ProH3 protein and the 75-kDa H3 protein, indicating that the Golgi was acidic and that the normal cleavage of ProH3 was occurring. When the Golgi proton pump was inactivated with concanamycin, both control cells and E5 cells secreted only the proH3 protein, confirming that acidification was essential for proper protein processing. Thus, both fluorometric and biological data indicate that the HPV-16 E5 protein does not inhibit Golgi acidification.
FIG. 5.
Cleavage and secretion of the proH3 protein in LXSN or E5* cells. Cells transfected with the proH3 construct were metabolically labeled and treated without concanamycin (lanes 1 and 3) or with 100 μM concanamycin (lanes 2 and 4). ProH3 or the cleavage product H3 was immunoprecipitated from the supernatant. The blot shows the cleaved form of proH3, H3, and the full-length form of proH3.
The data presented in this study verify that the HPV-16 E5 protein alkalinizes early endosomes, an effect that can have profound effects on cellular physiology. For example, endosome alkalinization can alter growth factor receptor downregulation, receptor recycling, and receptor/ligand interactions (3; reviewed in references 4, 5, 9, 54, 61, and 62), thereby potentially contributing to the observed effects of E5 on EGF receptor (EGFR) signaling (12, 13, 43, 53, 64). In addition, the alkalinization of endosomes can interfere with viral antigen presentation (47, 63), and such effects might contribute significantly to the ability of HPV-infected cells to escape immune surveillance.
While we and others have speculated that the effects of E5 are due primarily to altered endosomal pH, a recent study using LysoTracker Red and pH probes delivered by endocytosis has suggested that E5 blocks early-to-late endosome trafficking and that late endosome/lysosome pH is unaltered in E5-expressing cells (56). In the current study, we have demonstrated that HPV E5 can raise the pH of the early endosome compartment and that this alkalinization displays site specificity since the trans-Golgi was not affected. By using the pHluorin methodology to measure the pH of early endosomes and the trans-Golgi, we have avoided artifacts associated with the endosome-mediated uptake of pH probes. In addition, the pHluorin constructs have provided us with a more sensitive and site-specific analysis of compartment pH than can be obtained with LysoTracker Red. Our results do not necessarily conflict with those of Thomsen et al. Our study demonstrated that the pH of early endosomes was perturbed, and it is possible that the pHs of late endosomes/lysosomes might be unaltered, as reported. The conclusion by Thomsen et al. that early-to-late endosome trafficking is affected by E5 is also compatible with our results. An acidic lumenal pH is essential for directing endosome trafficking as well the recruitment of accessory signaling/trafficking molecules, such as ARNO and Arf1, from the cytoplasm to the endosomal compartment (24, 35) Thus, it is possible that alterations in early endosomal pH alter trafficking along the endocytic pathway, preventing fusion with late endosomes and, ultimately, lysosomes.
Since E5 is found predominantly in the ER, it is perplexing how it mediates pH changes specifically in endosomes. Our data suggest that this is not a direct effect of E5 on endosomal proton pumps, since very little E5 is found in this compartment. However, we cannot rule out the possibility that the level of E5 observed in endosomes by immunofluorescence is artifactually low. That is, the amount of E5 in endosomes might be underestimated due to antigen masking by endosomal proteins or lipids. While this is possible, the immunofluorescence signal for E5 in ER membranes is very strong, suggesting that the simple embedding of E5 into membranes does not interfere with antibody reactivity. The use of an AU1 epitope on E5 would also be anticipated to help identify E5 immunologically. Lastly, if E5 levels in endosomes are truly higher than anticipated from the immunofluorescence data, then we should also observe pH changes in the trans-Golgi, where there is similar, minimal expression of E5.
Another explanation for the alkalinizing activity of E5 is that, rather than directly inactivating the V-ATPase in endosomes, it functions in the ER to sequester a critical component/regulator of the V-ATPase. For example, studies in yeast indicate that V-ATPase assembly occurs in the ER. If E5 complexed with 16K in the ER, it could potentially interfere with the transfer of the proton pump into endosomes. It is also possible that there are additional E5 targets in the ER which participate in the transfer of the proton pump to endosomes. However, either of these models would require that the activity in the ER would preferentially inactivate proton pumps in endosomes rather than the trans-Golgi. Some insight into this site specificity has been observed with the influenza M2 protein which, under very high levels of expression, can selectively inhibit the acidification of early endosomes but not affect the lysosomal pH (30).
Finally, although the activation of EGFR signaling by E5 can be explained by interference with the vacuolar proton pump, it cannot be ruled out that pH changes and receptor signaling are independent activities of E5. Recent studies have shown that activated receptors, which are internalized via the endosomal pathway, transit to the endoplasmic reticulum (ER), where they are desensitized by the protein tyrosine phosphatase 1B (PTP1B) before fusion with lysosomal bodies (27). Interestingly, PTP1B knockout cells exhibit enhancements in EGF receptor phosphorylation without a corresponding significant increase in mitogen-activated protein kinase phosphorylation (26), a phenotype which closely mimics that observed for E5-expressing cells (13, 64).
In brief, the HPV-16 E5 protein specifically alkalinizes early endosomes, and the C terminus of E5 appears to play a critical role in this activity. It is anticipated that the E5 C-terminal deletion mutant will be helpful in identifying targets for full-length E5, which may explain alterations in the endosomal pH.
Acknowledgments
We thank James Rothman for the kind gift of the ratiometric pHluorin constructs and Maria Thuveson for the proH3 construct and antisera.
This work was supported by a grant to R.S. (RO1 CA53371) from the National Cancer Institute.
REFERENCES
- 1.Adam, J. L., M. W. Briggs, and D. J. McCance. 2000. A mutagenic analysis of the E5 protein of human papillomavirus type 16 reveals that E5 binding to the vacuolar H+-ATPase is not sufficient for biological activity, using mammalian and yeast expression systems. Virology 272:315-325. [DOI] [PubMed] [Google Scholar]
- 2.Ashby, A. D., L. Meagher, M. S. Campo, and M. E. Finbow. 2001. E5 transforming proteins of papillomaviruses do not disturb the activity of the vacuolar H+-ATPase. J. Gen. Virol. 82:2353-2362. [DOI] [PubMed] [Google Scholar]
- 3.Balbis, A., G. Baquiran, V. Dumas, and B. I. Posner. 2004. Effect of inhibiting vacuolar acidification on insulin signaling in hepatocytes. J. Biol. Chem. 279:12777-12785. [DOI] [PubMed] [Google Scholar]
- 4.Benmerah, A. 2004. Endocytosis: signaling from endocytic membranes to the nucleus. Curr. Biol. 14:R314-R316. [DOI] [PubMed] [Google Scholar]
- 5.Burke, P., K. Schooler, and H. S. Wiley. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell 12:1897-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caras, I. W., G. N. Weddell, M. A. Davitz, V. Nussenzweig, and D. W. Martin, Jr. 1987. Signal for attachment of a phospholipid membrane anchor in decay accelerating factor. Science 238:1280-1283. [DOI] [PubMed] [Google Scholar]
- 7.Chang, J. L., Y. P. Tsao, D. W. Liu, S. J. Huang, W. H. Lee, and S. L. Chen. 2001. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J. Biomed. Sci. 8:206-213. [DOI] [PubMed] [Google Scholar]
- 8.Ciampor, F., C. A. Thompson, S. Grambas, and A. J. Hay. 1992. Regulation of pH by the M2 protein of influenza A viruses. Virus Res. 22:247-258. [DOI] [PubMed] [Google Scholar]
- 9.Clague, M. J., and S. Urbe. 2001. The interface of receptor trafficking and signalling. J. Cell Sci. 114:3075-3081. [DOI] [PubMed] [Google Scholar]
- 10.Conrad, M., V. J. Bubb, and R. Schlegel. 1993. The human papillomavirus type 6 and 16 E5 proteins are membrane-associated proteins which associate with the 16-kilodalton pore-forming protein. J. Virol. 67:6170-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crum, C. P., M. Symbula, and B. E. Ward. 1989. Topography of early HPV 16 transcription in high-grade genital precancers. Am. J. Pathol. 134:1183-1188. [PMC free article] [PubMed] [Google Scholar]
- 12.Crusius, K., E. Auvinen, and A. Alonso. 1997. Enhancement of EGF- and PMA-mediated MAP kinase activation in cells expressing the human papillomavirus type 16 E5 protein. Oncogene 15:1437-1444. [DOI] [PubMed] [Google Scholar]
- 13.Crusius, K., E. Auvinen, B. Steuer, H. Gaissert, and A. Alonso. 1998. The human papillomavirus type 16 E5-protein modulates ligand-dependent activation of the EGF receptor family in the human epithelial cell line HaCaT. Exp. Cell Res. 241:76-83. [DOI] [PubMed] [Google Scholar]
- 14.Crusius, K., I. Rodriguez, and A. Alonso. 2000. The human papillomavirus type 16 E5 protein modulates ERK1/2 and p38 MAP kinase activation by an EGFR-independent process in stressed human keratinocytes. Virus Genes 20:65-69. [DOI] [PubMed] [Google Scholar]
- 15.Disbrow, G. L., I. Sunitha, C. C. Baker, J. Hanover, and R. Schlegel. 2003. Codon optimization of the HPV-16 E5 gene enhances protein expression. Virology 311:105-114. [DOI] [PubMed] [Google Scholar]
- 16.Durst, M., D. Glitz, A. Schneider, and H. zur Hausen. 1992. Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology 189:132-140. [DOI] [PubMed] [Google Scholar]
- 17.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fehrmann, F., and L. A. Laimins. 2003. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene 22:5201-5207. [DOI] [PubMed] [Google Scholar]
- 19.Forslund, O., P. Lindqvist, K. Haadem, J. Czegledy, and B. G. Hansson. 1997. HPV 16 DNA and mRNA in cervical brush samples quantified by PCR and microwell hybridization. J. Virol. Methods 69:209-222. [DOI] [PubMed] [Google Scholar]
- 20.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldstein, D. J., R. Kulke, D. Dimaio, and R. Schlegel. 1992. A glutamine residue in the membrane-associating domain of the bovine papillomavirus type 1 E5 oncoprotein mediates its binding to a transmembrane component of the vacuolar H+-ATPase. J. Virol. 66:405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, D. J., W. Li, L.-M. Wang, M. A. Heidaran, S. Aaronson, R. Shinn, R. Schlegel, and J. H. Pierce. 1994. The bovine papillomavirus type 1 E5 transforming protein specifically binds and activates the β-type receptor for the platelet-derived growth factor but not other related tyrosine kinase-containing receptors to induce cellular transformation. J. Virol. 68:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabe, M., and G. Oster. 2001. Regulation of organelle acidity. J. Gen. Physiol. 117:329-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu, F., and J. Gruenberg. 2000. ARF1 regulates pH-dependent COP functions in the early endocytic pathway. J. Biol. Chem. 275:8154-8160. [DOI] [PubMed] [Google Scholar]
- 25.Gu, Z., and G. Matlashewski. 1995. Effect of human papillomavirus type 16 oncogenes on MAP kinase activity. J. Virol. 69:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haj, F. G., B. Markova, L. D. Klaman, F. D. Bohmer, and B. G. Neel. 2003. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J. Biol. Chem. 278:739-744. [DOI] [PubMed] [Google Scholar]
- 27.Haj, F. G., P. J. Verveer, A. Squire, B. G. Neel, and P. I. Bastiaens. 2002. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science 295:1708-1711. [DOI] [PubMed] [Google Scholar]
- 28.Henkel, J. R., G. Apodaca, Y. Altschuler, S. Hardy, and O. A. Weisz. 1998. Selective perturbation of apical membrane traffic by expression of influenza M2, an acid-activated ion channel, in polarized Madin-Darby canine kidney cells. Mol. Biol. Cell 9:2477-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henkel, J. R., G. A. Gibson, P. A. Poland, M. A. Ellis, R. P. Hughey, and O. A. Weisz. 2000. Influenza M2 proton channel activity selectively inhibits trans-Golgi network release of apical membrane and secreted proteins in polarized Madin-Darby canine kidney cells. J. Cell Biol. 148:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henkel, J. R., J. L. Popovich, G. A. Gibson, S. C. Watkins, and O. A. Weisz. 1999. Selective perturbation of early endosome and/or trans-Golgi network pH but not lysosome pH by dose-dependent expression of influenza M2 protein. J. Biol. Chem. 274:9854-9860. [DOI] [PubMed] [Google Scholar]
- 31.Kell, B., R. J. Jewers, J. Cason, F. Pakarian, J. N. Kaye, and J. M. Best. 1994. Detection of E5 oncoprotein in human papillomavirus type 16-positive cervical scrapes using antibodies raised to synthetic peptides. J. Gen. Virol. 75:2451-2456. [DOI] [PubMed] [Google Scholar]
- 32.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 33.Luzio, J. P., B. Brake, G. Banting, K. E. Howell, P. Braghetta, and K. K. Stanley. 1990. Identification, sequencing and expression of an integral membrane protein of the trans-Golgi network (TGN38). Biochem. J. 270:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansur, C. P., and E. J. Androphy. 1993. Cellular transformation by papillomavirus oncoproteins. Biochim. Biophys. Acta 1155:323-345. [DOI] [PubMed] [Google Scholar]
- 35.Maranda, B., D. Brown, S. Bourgoin, J. E. Casanova, P. Vinay, D. A. Ausiello, and V. Marshansky. 2001. Intra-endosomal pH-sensitive recruitment of the Arf-nucleotide exchange factor ARNO and Arf6 from cytoplasm to proximal tubule endosomes. J. Biol. Chem. 276:18540-18550. [DOI] [PubMed] [Google Scholar]
- 36.McCance, D. J. 1998. Human papillomaviruses and cervical cancer. J. Med. Microbiol. 47:371-373. [DOI] [PubMed] [Google Scholar]
- 37.McMahon, H. T., Y. A. Ushkaryov, L. Edelmann, E. Link, T. Binz, H. Niemann, R. Jahn, and T. C. Sudhof. 1993. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature 364:346-349. [DOI] [PubMed] [Google Scholar]
- 38.Mellman, I., R. Fuchs, and A. Helenius. 1986. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 55:663-700. [DOI] [PubMed] [Google Scholar]
- 39.Miesenbock, G., D. A. De Angelis, and J. E. Rothman. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192-195. [DOI] [PubMed] [Google Scholar]
- 40.Nead, M. A., and D. J. McCance. 1998. Activities of the transforming proteins of human papillomaviruses, p. 225-251. In D. J. McCance (ed.), Human tumor viruses. ASM Press, Washington, D.C.
- 41.Pim, D., M. Collins, and L. Banks. 1992. Human papillomavirus type 16 E5 gene stimulates the transforming activity of the epidermal growth factor receptor. Oncogene 7:27-32. [PubMed] [Google Scholar]
- 42.Pinto, L. H., and R. A. Lamb. 2004. Viral ion channels as models for ion transport and targets for antiviral drug action. FEBS Lett. 560:1-2. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez, M. I., M. E. Finbow, and A. Alonso. 2000. Binding of human papillomavirus 16 E5 to the 16 kDa subunit c (proteolipid) of the vacuolar H+-ATPase can be dissociated from the E5-mediated epidermal growth factor receptor overactivation. Oncogene 19:3727-3732. [DOI] [PubMed] [Google Scholar]
- 44.Sakaguchi, T., Q. Tu, L. H. Pinto, and R. A. Lamb. 1997. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc. Natl. Acad. Sci. USA 94:5000-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sankaranarayanan, S., D. De Angelis, J. E. Rothman, and T. A. Ryan. 2000. The use of pHluorins for optical measurements of presynaptic activity. Biophys. J. 79:2199-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schapiro, F., J. Sparkowski, A. Adduci, F. Suprynowicz, R. Schlegel, and S. Grinstein. 2000. Golgi alkalinization by the papillomavirus E5 oncoprotein. J. Cell Biol. 148:305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sinnathamby, G., and L. C. Eisenlohr. 2003. Presentation by recycling MHC class II molecules of an influenza hemagglutinin-derived epitope that is revealed in the early endosome by acidification. J. Immunol. 170:3504-3513. [DOI] [PubMed] [Google Scholar]
- 48.Sparkowski, J., J. Anders, and R. Schlegel. 1994. Mutation of the bovine papillomavirus E5 oncoprotein at amino acid 17 generates both high- and low-transforming variants. J. Virol. 68:6120-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoler, M. H., C. R. Rhodes, A. Whitbeck, S. M. Wolinsky, L. T. Chow, and T. R. Broker. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 23:117-128. [DOI] [PubMed] [Google Scholar]
- 50.Stoppler, H., M. C. Stoppler, and R. Schlegel. 1994. Transforming proteins of the papillomaviruses. Intervirology 37:168-179. [DOI] [PubMed] [Google Scholar]
- 51.Stoppler, M. C., S. W. Straight, G. Tsao, R. Schlegel, and D. J. McCance. 1996. The E5 gene of HPV-16 enhances keratinocyte immortalization by full-length DNA. Virology 223:251-254. [DOI] [PubMed] [Google Scholar]
- 52.Straight, S. W., B. Herman, and D. J. McCance. 1995. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J. Virol. 69:3185-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Straight, S. W., P. M. Hinkle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 67:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szymkiewicz, I., O. Shupliakov, and I. Dikic. 2004. Cargo- and compartment-selective endocytic scaffold proteins. Biochem J. 383:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teter, K., G. Chandy, B. Quinones, K. Pereyra, T. Machen, and H. P. Moore. 1998. Cellubrevin-targeted fluorescence uncovers heterogeneity in the recycling endosomes. J. Biol. Chem. 273:19625-19633. [DOI] [PubMed] [Google Scholar]
- 56.Thomsen, P., B. van Deurs, B. Norrild, and L. Kayser. 2000. The HPV16 E5 oncogene inhibits endocytic trafficking. Oncogene 19:6023-6032. [DOI] [PubMed] [Google Scholar]
- 57.Thuveson, M., and E. Fries. 2000. The low pH in trans-Golgi triggers autocatalytic cleavage of pre-α-inhibitor heavy chain precursor. J. Biol. Chem. 275:30996-31000. [DOI] [PubMed] [Google Scholar]
- 58.Tomakidi, P., H. Cheng, A. Kohl, G. Komposch, and A. Alonso. 2000. Modulation of the epidermal growth factor receptor by the human papillomavirus type 16 E5 protein in raft cultures of human keratinocytes. Eur. J. Cell Biol. 79:407-412. [DOI] [PubMed] [Google Scholar]
- 59.Villa, L. L. 1997. Human papillomaviruses and cervical cancer. Adv. Cancer Res. 71:321-341. [DOI] [PubMed] [Google Scholar]
- 60.Vormwald-Dogan, V., B. Fischer, H. Bludau, U. K. Freese, L. Gissmann, D. Glitz, E. Schwartz, and M. Durst. 1992. Sense and antisense transcripts of human papillomavirus type 16 in cervical cancers. J. Gen. Virol. 73(Pt. 7):1833-1838. [DOI] [PubMed] [Google Scholar]
- 61.Wiley, H. S., and P. M. Burke. 2001. Regulation of receptor tyrosine kinase signaling by endocytic trafficking. Traffic 2:12-18. [DOI] [PubMed] [Google Scholar]
- 62.Yarden, Y. 2001. The EGFR family and its ligands in human cancer. Signalling mechanisms and therapeutic opportunities. Eur. J. Cancer 37(Suppl. 4):S3-S8. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, B., P. Li, E. Wang, Z. Brahmi, K. W. Dunn, J. S. Blum, and A. Roman. 2003. The E5 protein of human papillomavirus type 16 perturbs MHC class II antigen maturation in human foreskin keratinocytes treated with interferon-gamma. Virology 310:100-108. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, B., D. F. Spandau, and A. Roman. 2002. E5 protein of human papillomavirus type 16 protects human foreskin keratinocytes from UV B-irradiation-induced apoptosis. J. Virol. 76:220-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]