Abstract
Significant challenges to poultry health are posed by chicken anemia virus (CAV), which induces immunosuppression and causes increased susceptibility to secondary infections. The effective management and containment of CAV within poultry stocks require precise and prompt diagnosis. However, a deficiency persists in the availability of low-cost, rapid, and portable CAV detection devices. In this study, an immunochromatographic lateral-flow test strip-based assay was developed for CAV detection using in-house generated monoclonal antibodies (MABs) against CAV viral protein 1 (VP1). The recombinant truncated VP1 protein (Δ60VP1), with amino acid residues 1 to 60 of the native protein deleted, was produced via a prokaryotic expression system and utilized for immunizing BALB/c mice. Subsequently, high-affinity MABs against Δ60VP1 were generated and screened using conventional hybridoma technology combined with serial dilution assays. Two MABs, MAB1, and MAB3, both binding to distinct epitopes of Δ60VP1, were selected for the development of a lateral-flow assay. Sensitivity analysis demonstrated that the Δ60VP1 antigen could be detected by our homemade lateral-flow assay at concentrations as low as 625 ng/mL, and this sensitivity was maintained for at least 6 mo. The assay exhibited high specificity, as evidenced by its lack of reactivity with surrogate recombinant proteins and the absence of cross-reactivity with other chicken viruses and viral antigens. Comparative analysis with quantitative PCR data demonstrated substantial agreement, with a Kappa coefficient of 0.66, utilizing a sample set comprising 305 clinical chicken serum samples. In conclusion, the first lateral-flow assay for CAV detection was developed in this study, utilizing 2 specific anti-VP1 MABs. It is characterized by simplicity, rapidity, sensitivity, and specificity.
Key words: chicken anemia virus, detection, lateral flow assay, monoclonal antibody, VP1
INTRODUCTION
Chicken anemia virus (CAV), an important global threat to poultry health, gravely impairs the avian immune system, culminating in diminished productivity (Schat, 2009; Fatoba and Adeleke, 2019). This viral pathogen indiscriminately afflicts both genders across all age groups of chickens. However, its primary impact is observed in chicks aged between 2 and 5 wk (Schat, 2009; Fatoba and Adeleke, 2019). CAV exhibits tropism for the hemocytoblasts in the bone marrow and precursor lymphocytes in the thymus, instigating anemia, thymic atrophy, and immunosuppression (Adair, 2000). Clinical manifestations in infected poultry include lethargy, stunted growth, pallor of combs, elevated mortality rates, and an escalated susceptibility to secondary pathogenic infections (Zhang et al., 2015). The dissemination of CAV predominantly occurs horizontally via the oral-fecal modality through excreta of the infected birds and has the potential for vertical transmission from infected hens to their progeny (Miller and Schat, 2004; Fatoba and Adeleke, 2019). The persistence of this virus in various environmental matrices and on contaminated surfaces accentuates its transmission within avian cohorts. The ensuing afflictions can cause a significant economic loss to the poultry sector.
Chicken anemia virus, classified under the Gyrovirus genus of the Anelloviridae family, is characterized as a nonenveloped virus with a diameter of approximately 25 nm (Rosario et al., 2017). The virus possesses a circular, single-stranded, negative-sense DNA genome spanning roughly 2.3 kb. Within its genome, the open reading frames exhibit partial overlap and encode 3 pivotal viral proteins (VPs) (Lacorte et al., 2007). Among these, the 51 kDa VP1, which is the virus's only structural protein, plays a vital role in inducing neutralizing antibodies against CAV. VP1 also hinders type I interferon production, notably by inhibiting TBK1 phosphorylation and disrupting the cGAS-STING signaling pathways (Chen et al., 2023). The nonstructural VP2 (28 kDa) serves a dual function: facilitating VP1 configuration (Noteborn et al., 1998) and executing phosphatase activities on serine/threonine and tyrosine residues (Peters et al., 2005). Additionally, VP3 (13 kDa), known as apoptin, not only triggers cellular apoptosis but also orchestrates various functions in the viral replication process (Feng et al., 2020; Zhang et al., 2021). Recent discoveries have revealed 2 novel amino acid mutations in VP1 and VP3, along with 3 nucleotide mutations in a noncoding region, in a highly pathogenic CAV strain that has been shown to cause disease in older chickens (Fang et al., 2023).
Rapid and precise diagnosis of CAV is pivotal in deploying pertinent countermeasures, encompassing vaccination protocols, biosecurity norms, and poultry management strategies. Serological evaluations, particularly the commercially endorsed CAV ELISA kit (Flockchek, CAV, IDEXX, Hoofddorp, North Holland) and the BioChek kit (Reeuwijk, the Netherlands), have been instrumental in detecting CAV antibodies (Hadimli et al., 2008; Kurian et al., 2012; Chansiripornchai, 2017; Van et al., 2021; Al-Masodi et al., 2023; Shao et al., 2023). The advent of peptide-based ELISA, exploiting a synthetic 21-amino acid sequence at VP1′s N-terminus, has showcased enhanced detection efficacy compared to its commercial counterparts (Shao et al., 2023).
In tandem with serodiagnostic advancements, nucleic acid-based diagnostic modalities, such as quantitative PCR (qPCR) assays and loop-mediated isothermal amplification, have been meticulously appraised and endorsed (Huang et al., 2010; Han et al., 2019; Techera et al., 2019; Kaffashi et al., 2021; Kannaki et al., 2021; Luan et al., 2022). Emerging techniques like droplet digital PCR and real-time recombinase-aided amplification assay offer promising detection avenues (Li et al., 2019; Wu et al., 2022). Nevertheless, these methodologies can be labor-intensive, and time-consuming, and necessitate specialized laboratory infrastructure and equipment.
Our consortium recently reported encouraging outcomes in CAV serodiagnosis utilizing a recombinant truncated VP1 protein (Δ60VP1) expressed in Escherichia coli (Wanganurakkul et al., 2020). Building upon this, the current study describes the formulation of a lateral flow diagnostic strip for CAV, employing monoclonal antibodies (MABs) specific to Δ60VP1. We rigorously scrutinized parameters influencing the diagnostic strip's fidelity and juxtaposed its performance with qPCR, affirming its potential as an efficient CAV diagnostic instrument. This innovation augments the poultry diagnostic arsenal, offering a swift, field-deployable solution.
MATERIALS AND METHODS
Ethical Statement
The handling and execution of all experimental procedures strictly adhered to the guidelines and recommendations established by the Institutional Animal Care and Use Committee (IACUC) of Chulalongkorn University, with approval number 1673004. Furthermore, all sample collection procedures were carried out in accordance with the standard diagnostic manual for livestock diseases in Thailand (NIAH, 1998).
Expression and Purification of Recombinant Δ60VP1, VP1-F1, and VP1-F2 of Chicken Anemia Virus
In this study, 3 versions of the CAV VP1 proteins were utilized. The Δ60VP1 (L61-P389) was used for mouse immunization and MAB generation. To verify that the 2 selected MABs subsequently bind to distinct epitopes, which is crucial for the assay development, 2 additional constructs, VP1-F1 (residues L61-M193) and VP1-F2 (residues G194-Y250), were employed to narrow down the region of interaction.
The expression of these proteins was achieved using the E. coli strain Rosetta-gami. This strain was transformed with one of the recombinant plasmids pET-28a(+) encoding either Δ60VP1 (Wanganurakkul et al., 2020), VP1-F1 (Sittidech and Assavalapsakul, 2018), or VP1-F2 (Sittidech and Assavalapsakul, 2018). Cultivation of the bacteria was performed on Luria-Bertani agar, supplemented with 34 μg/mL chloramphenicol (Biobasic, Canada) and 50 μg/mL kanamycin (T.P Drug Laboratories, Thailand), and incubated at 30°C for 12 to 16 h. A single colony of the recombinant bacteria was then inoculated into Luria-Bertani broth containing the same antibiotics and incubated at 25°C with shaking at 250 rpm. Once the optical density at 600 nm reached 0.4, protein expression was induced by adding isopropyl-β-D-1-thiogalactopyranoside (Himedia, India) to a final concentration of 0.2 mM. After 3 h of incubation, the cells were harvested by centrifugation, and the expressed proteins were analyzed using SDS-PAGE and Western blotting. Specifically, Δ60VP1 was resolved on a 13% Tris-Glycine-SDS gel, while recombinant VP1-F1 and VP1-F2 were resolved on a 16% Tricine-SDS gel. Western blot analysis utilized a primary antibody, His Tag Antibody (R&D Systems, Minneapolis, MN), at a dilution of 1:5,000, and a secondary antibody, Goat anti-Mouse IgG F(ab′)2-Peroxidase (Sigma Aldrich, St. Louis, MO).
For purification, the cell pellet was subjected to anionic denaturing detergent purification, following the method described by Schlager et al. (2012). Briefly, the cells were resuspended in a PCL lysis buffer at 0.1 volume of the bacterial culture and then lysed by sonication with 45 s pulse on, 5 s pulse off, and amplitude 40% for 4 min. The lysate was subsequently incubated on ice for an hour. After centrifugation at 15,557 × g at 4°C for 20 min, the supernatant was adjusted to a volume of 10 mL with PCW binding buffer and filtered through a 0.45 μm PVDF membrane syringe filter (AWLSCI Technologies, China). The protein of interest was then purified using a 5 mL HisTrap HP affinity chromatography column (Cytiva, Marlborough, MA). The column was washed with PCW binding buffer, and the bound protein was eluted using PCW elution buffer containing increasing concentrations of imidazole (40, 60, 100, 200, and 300 mM). The remaining proteins were eluted by washing the column with Stripping buffer. The selected protein fraction was exchanged with 1 × PBS using an Amicon Ultra Centrifugal Filter (Merck Millipore, Ireland), and the protein concentration was determined using the Bradford assay (Bio-Rad, Hercules, MA) at 595 nm with a BSA standard (Sigma Aldrich, St. Louis, MO).
Immunization and MAB Generation
Four female BALB/c mice at 8 wk of age were utilized for the immunization process. The purified recombinant Δ60VP1 was mixed with complete Freund's adjuvant (Sigma Aldrich, St. Louis, MO) at a 1:1 volume ratio. Each mouse received 30 µg of protein via intraperitoneal injection. Subsequently, 3 more injections of the same amount of protein were administered at 2-wk intervals, this time mixed with incomplete Freund's adjuvant (Sigma Aldrich, St. Louis, MO).
Sera were collected from the mice 1 wk before the third injection by extracting blood via mandibular puncture. The antisera were then centrifuged at 9,838 × g for 10 min at 4°C. Prior to hybridoma production, the levels of serum antibodies to the Δ60VP1 antigen were quantified using indirect ELISA.
After humane euthanization, spleen cells were fused with a murine myeloma cell (P3 × 63/Ag8.653) using 50% (wt/vol) polyethylene glycol (MW 1,500 Da; Sigma Aldrich, St. Louis, MO) at a ratio 2:1. The resulting fusion products were plated onto 96-well plates. Positive hybridoma clones producing MABs were selected through an initial screening using indirect ELISA. Subsequently, positive clones were recloned using the limiting dilution method and preserved in liquid nitrogen for future use. The isotype of MABs from hybridoma clones was determined using the Mouse Typer Isotyping Panel (Bio-Rad, Hercules, CA).
Preparation and Purification of MABs Specific to Recombinant Δ60VP1
The hybridoma cells, designated as 6-5B-5B-12F-10A (23) (clone No. 1) and 3-F1-H3 (clone No. 3), producing MABs specific to recombinant Δ60VP1 were transitioned to a serum-free culture medium (ISF-1 medium; Biochrom, Germany). Subsequently, the cells were transferred to a 1-L container and cultured for 10 d. The cell residue was eliminated, and the resulting culture medium was collected by centrifugation at 1,510 × g for 5 min. To purify the MAB, the culture medium was filtered through a 0.45 μm filter membrane and subjected to purification using a 5 mL HiTrap Protein G HP antibody purification column (Cytiva, Marlborough, MA) on an ÄKTA start protein purification system (Cytiva, Marlborough, MA). Briefly, the column was pre-equilibrated with binding buffer (20 mM sodium phosphate, pH 7.2). Then, 500 mL of the filtered hybridoma supernatant was loaded onto the column at a flow rate of 1 mL/min. The column was washed with 50 mL of binding buffer to remove unbound proteins. The MAB was subsequently eluted from the protein G using 100 mM glycine-HCl, pH 2.7 and 30 fractions (1 mL) collected. The pH of each fraction was adjusted by the addition of 40 µL of 1 M Tris-HCl buffer, pH 9.0 to each fraction and all fractions were combined. The mixture containing MAB from hybridoma clone No. 1 (MAB1) underwent dialysis against 1 × PBS for its application on the test line of the lateral flow assay. The second purified MAB from hybridoma clone No. 3 (MAB3) underwent dialysis with 2 mM sodium borate buffer, pH 8.6 for coating on colloidal gold. The amount of MAB each was determined using the BCA assay (Thermo Scientific, Waltham, MA), and Western blotting was performed using each purified MAB as a primary antibody at a dilution of 1:12,000 to assess its specificity toward recombinant Δ60VP1. In addition, the affinity of MAB1 and MAB3 was determined with VP1-F1 or VP1-F2 polypeptide by Western blotting.
Labeling of Purified MABs With Colloidal Gold
To determine the optimal ratio of MAB to colloidal gold for labeling, a range of MAB concentrations (0–200 μg/mL) was prepared by diluting the MAB3 with 2 mM sodium borate buffer, pH 8.6. Each 20 μL concentration of MAB3 was added to a well of a 96-well plate, followed by the addition of 200 μL of colloidal gold solution (40 nm diameter, OD522nm = 1.164, pH 8.2; K.Bio Sciences, Thailand) to each well. After incubating the plate at ambient temperature with continuous shaking for 1 h, 10% (wt/vol) NaCl (80 μL) was added, and the absorbance at 520 nm was measured.
For MAB-colloidal gold coating, colloidal gold solution (10 mL, pH 8.2) was mixed with MAB3 (1 mL) at a ratio of 10:1 and incubated at ambient temperature with shaking for 1 h. Next, 5% (wt/vol) BSA was added, and the shaking continued for an additional hour. The mixture underwent centrifugation at 25,000 × g for 30 min at 4°C, and the supernatant was discarded. The MAB3-colloidal gold pellet was resuspended in 1 mL of sodium borate buffer, followed by 3 repeated cycles of centrifugation and resuspension to remove unbound protein. Finally, the MAB3-colloidal gold was resuspended in 1 mL of 2% (wt/vol) sucrose in sodium borate buffer and stored at 4°C until required.
Preparation and Interpretation of the Colloidal Gold Immunochromatographic Lateral Flow Assay
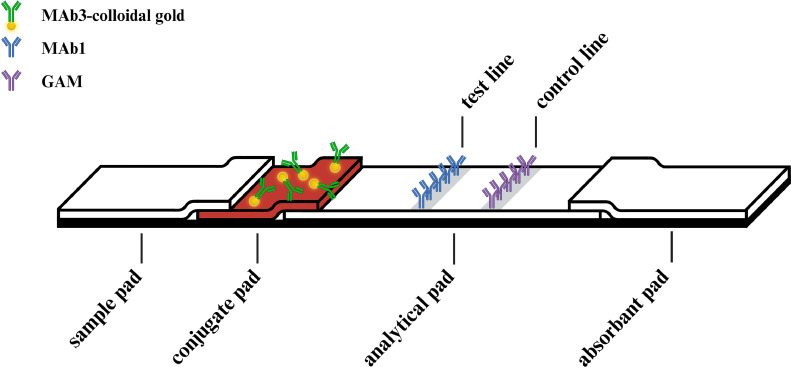
The sandwich lateral flow assay consists of 4 major connecting parts: the sample pad, the conjugate pad, the analytical pad, and the absorbent pad (Figure 1). The sample pad (Standard 17, Whatman, UK) was prepared by soaking in Tris buffer saline, pH 7.5 (TBS) containing 1% (wt/vol) BSA and 0.1% (vol/vol) Tween 20 for 30 min, followed by pat drying and incubation at 37°C for 1 h. Similarly, the conjugate pad (GF33; Whatman Schleicher & Schuell, Germany) was submerged in PB buffer [1% (wt/vol) BSA, 2% (wt/vol) sucrose, 0.2% (vol/vol) Tween 20] for 30 min, pat dried, and incubated at 37°C for 1 h. The conjugate pad was sprayed with the MAB3-colloidal gold solution at a rate of 10 μL/cm and dried at 37°C for 30 min. The FF170 analytical pad (Whatman, UK) was joined to the plastic backing card and incubated at 37°C for 15 min. Subsequently, the control line on the analytical pad was sprayed with Goat anti-Mouse IgG (GAM; Lampire Biological Laboratories, Pipersville, PA) (1 mg/mL), while the test line was sprayed with MAB1 (2 mg/mL) at a rate of 1 μL/cm to act as the capture antibody. The absorbent pad was incubated at 37°C for 1 h. Finally, all components were assembled as shown in Figure 1. The test strip was cut using a Guillotine Cutter (BioDot, Irvine, CA) with a size of 4 mm × 25 mm and inserted into a prefabricated cassette (K.Bio Sciences, Thailand).
Figure 1.
Schematic of the components of a sandwich lateral flow assay cassette. The figure illustrates the various components of a sandwich lateral flow assay cassette. These components include the sample pad, the conjugate pad, the analytical pad, and the absorbent pad. The sample pad is where the sample solution is applied, which then flows through the conjugate pad and then reaches the analytical pad. The analytical pad contains immobilized monoclonal antibodies specific to the target antigen, forming the test line and control line positions. Finally, the absorbent pad facilitates the flow of liquid and maintains the integrity of the assay.
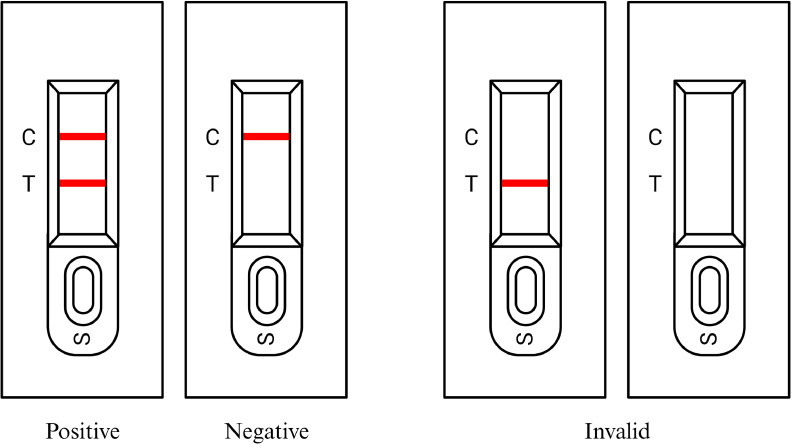
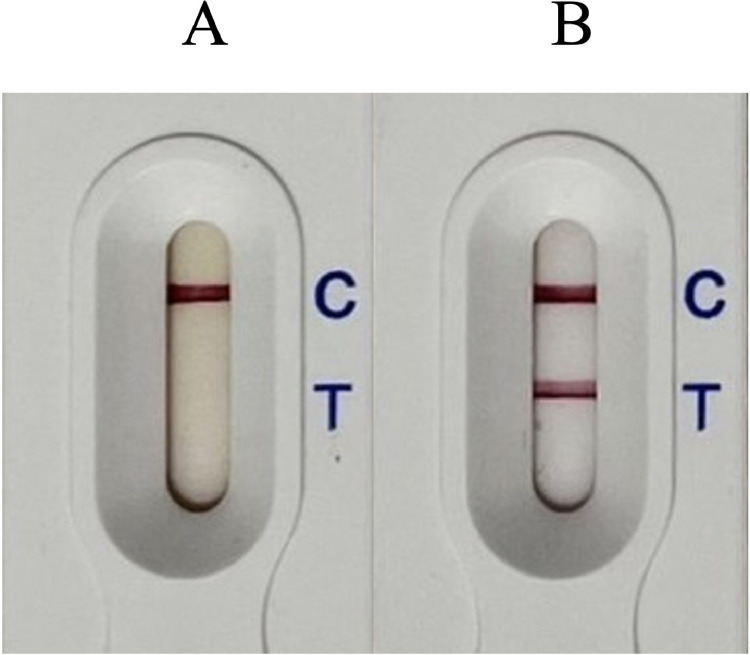
Interpretation of the test outcome was done visually without the aid of specialized equipment. After dispensing 100 µL of the test sample into the sample dropper, the test was incubated at room temperature for 15 min. The results of the test were recorded at 30 min following sample application. A positive result is indicated when both the control line and test line on the analytical pad show colored bands, indicating the presence of the tested antigen in the sample solution. Conversely, a negative result is indicated by the presence of a single band on the control line, signifying the absence of the tested antigen in the sample solution. If the lateral flow assay shows only 1-color band at the test line position or does not show any bands at all, it indicates a problem with the lateral flow assay. In such cases, the result is invalid as the assay cannot be accurately interpreted (Figure 2).
Figure 2.
Examples of test methods and interpretations of sandwich lateral flow assay. The figure showcases examples of different outcomes of the test methods and their corresponding interpretations using sandwich lateral flow assay. (A) An example of a positive test scenario where both the test line and control line show colored bands, indicating the presence of the target antigen in the sample. (B) An example of a negative test scenario where only the control line displays a colored band, suggesting the absence of the target antigen in the sample. However, the control band is observed, confirming a valid test outcome. (C) and (D) Examples of invalid test outcomes. If no bands appear or if only the test line shows a colored band, the result of the assay is considered invalid, indicating a potential issue with the lateral flow assay or sample preparation.
Performance of Prepared Lateral Flow Assay
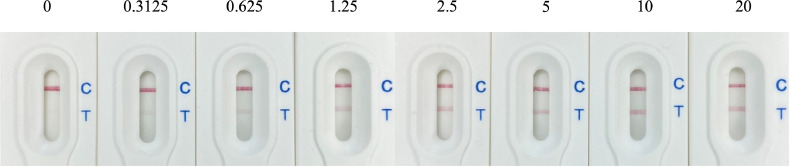
Sensitivity of the Lateral Flow Assay. To examine the sensitivity of the lateral flow assay, they were prepared as described above. The sample solutions included commercial chicken serum prepared with TBS containing 1% (vol/vol) Tween 20 and 0.1% (wt/vol) BSA (TBST) at a ratio of 1:1, supplemented with recombinant Δ60VP1 at concentrations of 0, 0.3125, 0.625, 1.25, 2.50, 5, 10, and 20 μg/mL After applying the solution onto the lateral flow assay, the changes in color intensity at the control line and test line positions of the analytical pad were observed and the sensitivity of the test band was determined.
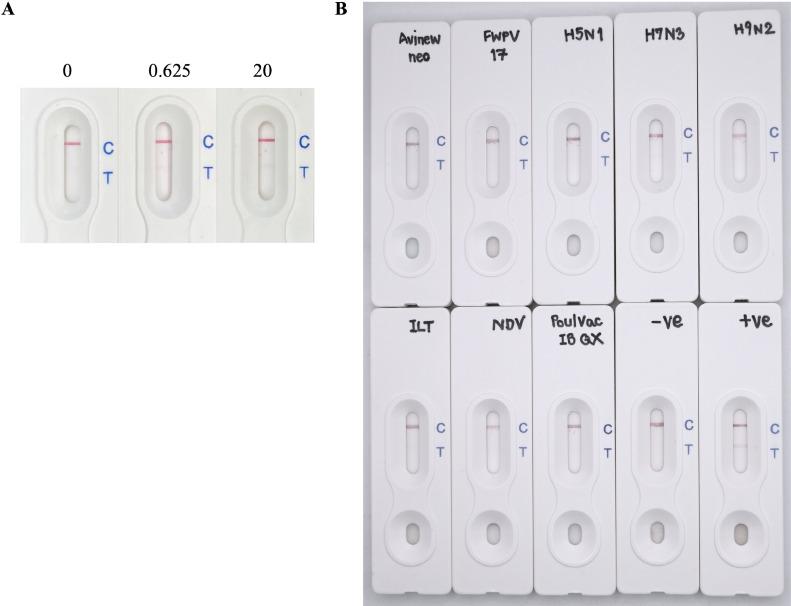
Specificity of the Lateral Flow Assay. To examine the specificity of the lateral flow assay, the sample solution consisted of TBST, supplemented with another recombinant protein tagged with polyhistidine, like the recombinant protein Δ60VP1. This recombinant protein was derived from the capsid protein of the Nervous necrosis virus (NNV) and was used at concentrations of 0, 0.625, and 20 µg/mL. Cross-reactivity examinations were conducted with other chicken viruses and viral antigens, half-diluted with TBST at predetermined titers or concentrations. The tested chicken viruses included the Newcastle disease virus (VG/GA-AVINEW strain, Merial Avinew NeO Vaccine, Boehringer Ingelheim Animal Health, UK), Fowl pox virus (Poxine), Avian Infectious Bronchitis virus (Poulvac IB QX, Lot no. 272234, Zoetis UK Limited), and Infectious Laryngotracheitis virus (ILT). Concurrently, antigen from different strains of Avian Influenza virus (H5N1 Lot no. 110166 S, H7N3 Lot no. 170266 S, and H9N2 Lot no. 140266 S) and that of New Castle disease virus (NDV Lot 220865 S) obtained from the National Institute of Animal Health (NIAH), Thailand, were produced from embryonic chicken eggs and inactivated for examination. To ensure accuracy, CAV and CAV-free chicken serums were employed as positive and negative controls, respectively. After applying the sample solution onto the lateral flow assay, the recorded color intensity results at both the control and test line positions were utilized in evaluating the assay's specificity against the test sample.
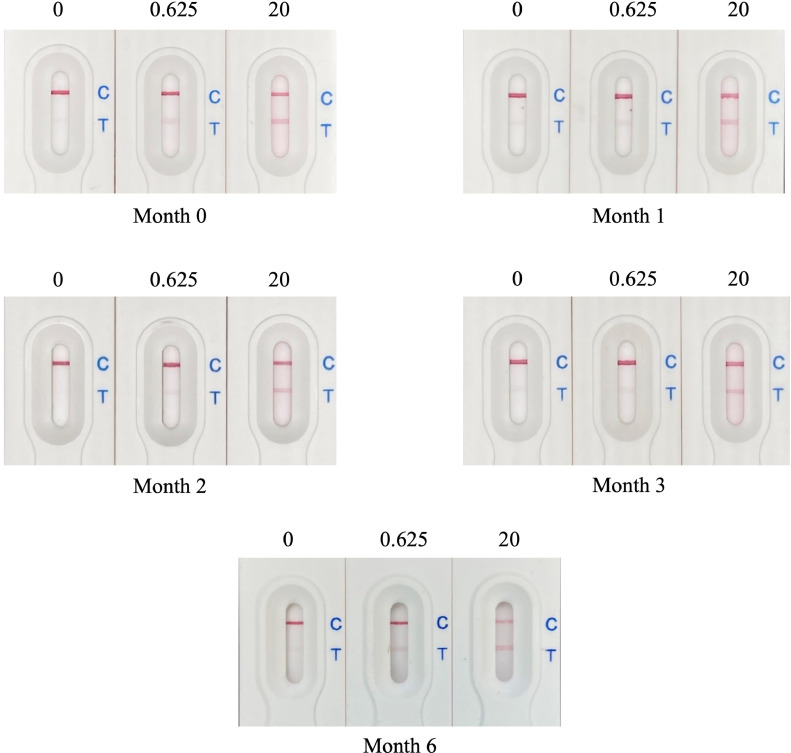
Shelf Life of the Lateral Flow Assay. To examine the stability of the lateral flow assay, they were stored at room temperature (25°C) for a period of 6 mo. The stability was tested at mo 0, 1, 2, 3, and 6 using the sample solution consisting of TBST, supplemented with recombinant Δ60VP1 at concentrations of 0, 0.625, and 20 µg/mL. The sample solution was dropped onto the assay, and the color intensity results at the control and test lines were recorded. The stability of the lateral flow assay was evaluated at each time point.
Examination of Clinical Samples and Comparison of Lateral Flow Assay Performance With qPCR
A collection of serum specimens was acquired from a diverse population of laying hens, spanning different breeds and age groups. A total of 305 samples were sourced from poultry farms situated in the eastern region of Thailand, facilitated by the Veterinary Research and Development Center, Department of Livestock Development, Chonburi Province, spanning the period from 2010 to 2022. In the context of the lateral flow assay, the collected sera were diluted at a ratio of 1:1 using TBST, and 100 µL was added to the sample pad. The subsequent interpretations of the assay results were made in accordance with previously established and defined criteria.
To evaluate the relative performance of the lateral flow assay for CAV detection, the qPCR assay was used to re-test the chicken serum samples described above. DNA was extracted from 100 µL each of the 305 serum specimens utilizing the Quick-DNA Miniprep Kit (Zymo Research, Irvine, CA), strictly adhering to the protocol described by the manufacturer. Postextraction, the DNA was eluted from the column in 50 µL and preserved at −20°C until further analysis. The qPCR assays were conducted by deploying specific primers and probes that target the conserved domains of the CAV, as described by Li et al. (2019). Each PCR reaction, with a total volume of 20 μL, comprised of 1 × FastStart TaqMan Probe Master (Roche, Germany), 250 nM of both CIAV-F (5ʹGCAGGGGCAAGTAATTTCAA 3ʹ) and CIAV-R primers (5ʹGCCACACAGCGATAGAGTGA 3ʹ), 1 μM of the probe (FAM-5ʹACTGCAGAGAGATCCGGATTGGTATCG-3ʹ BHQ), and 8.2 µL of the DNA template. The amplification process was undertaken on a qPCR LightCycler 480 system (Roche, Germany) with the following cycling parameters: initial denaturation at 95°C for 10 s, succeeded by 45 cycles of denaturation at 95°C for 10 s, and annealing at 60°C for 1 min. The fluorescence signals were then cataloged within the 465 to 510 nm spectrum. Samples registering a cycle threshold (CT) value ≤40 were designated as positive for CAV. Samples with CT values >40 were designated as negative for CAV.
Statistical Analysis
The collected data were analyzed to determine the diagnostic sensitivity, specificity, and accuracy of the test results obtained from the 305 serum samples using the lateral flow assay and the qPCR analysis. The following formulas were employed to calculate the diagnostic sensitivity, diagnostic specificity, and accuracy:
For the purposes of this study, true positives were defined as samples where the CT value from the qPCR assay was ≤40, while true negatives were defined as samples with a CT value >40.
Additionally, the level of agreement between the lateral flow assay results and qPCR was assessed by calculating kappa (κ) using a 2 × 2 contingency table in GraphPad Prism version 10.0.0 for Windows (GraphPad Software, Boston, MA). The degree of concordance between the lateral flow assay results and qPCR was then evaluated based on the criteria established by Landis and Koch (1977) (Table 1).
Table 1.
The degree of concordance of test results assessed using the kappa (κ) value and Landis and Koch criteria.
| Kappa (κ) | Degree of concordance |
|---|---|
| <0.00 | Poor |
| 0.00–0.20 | Slight |
| 0.21–0.40 | Fair |
| 0.41–0.60 | Moderate |
| 0.61–0.80 | Substantial |
| 0.81–1.00 | Almost perfect |
RESULTS
Expression and Purification of Recombinant Δ60VP1, VP1-F1, and VP1-F2 of Chicken Anemia Virus
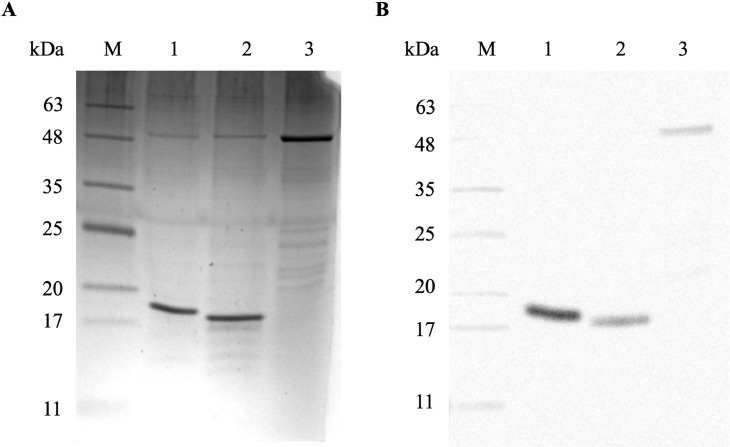
The expression and purification of recombinant Δ60VP1, VP1-F1, and VP1-F2 of CAV were performed successfully. The recombinant Δ60VP1 had an estimated molecular weight 45 kDa and encompasses the VP1-F1 and VP1-F2 polypeptide proteins as subunits, with molecular weights of 17 kDa and 16 kDa, respectively. The purification of the polypeptides was carried out using a HisTrap HP chromatography columns with an anionic denaturing detergent. The purified recombinant proteins were eluted in PCW fractions at imidazole concentrations of 60, 100, 200, and 300 mM (Supplementary Figures S1–S3). The purity and size of the proteins were confirmed through SDS-PAGE and Western blotting, using His Tag Antibody as the primary antibody. The SDS-PAGE (Figure 3A) and Western blot analysis (Figure 3B) showed bands at approximately 45 kDa for recombinant Δ60VP1, 17 kDa for VP1-F1 polypeptide, and 16 kDa for VP1-F2 polypeptide, respectively. These results indicate the successful expression and purification of the target polypeptides.
Figure 3.
Analysis of purified recombinant polypeptides Δ60VP1, VP1-F1, and VP1-F2, utilized in this study. The purified recombinant F1, F2, and Δ60VP1 proteins were subjected to analysis by SDS-PAGE (A) and Western blotting using His Tag Antibody as the primary antibody (B). The gel lanes are labeled as follows: Lane M: CozyHi Prestained Protein Ladder (highQu, Germany); Lane 1: Recombinant VP1-F1 (17 kDa); Lane 2: VP1-F2 (16 kDa); Lane 3: Δ60VP1 (45 kDa).
Production and Purification of MAB1 and MAB3 Specific to Recombinant Δ60VP1 and Its Counterparts
To generate MABs, mice were immunized with recombinant Δ60VP1. Two MABs, MAB1, and MAB3, specific to Δ60VP1 protein determined to be IgG kappa (IgG2a and IgG2b) were selected: MAB1, targeting the recombinant VP1-F1 polypeptide, and MAB3, targeting the recombinant VP1-F2 polypeptide (Sittidech, 2017). Therefore, these MABs were utilized in the preparation of the prototype test strip based on the sandwich lateral flow assay principle. Noticeably, the alignment of amino acid sequences between VP-F1 and VP-F2 of Δ60VP1 and corresponding sequences from other isolates including those from the USA (ABJ90436.1), Japan (BAA90491.1), Taiwan (AVO63704.1), Turkey (QVL22486.1), Iran (ANC50810.1), India (AXF92409.1), and various areas of China (QQN72596.1, FF176599.1, WLD15706.1, WIM51797.1, WFK20752.1, UBT23944.1) reveals a high sequence similarity (Supplementary Figure S4).
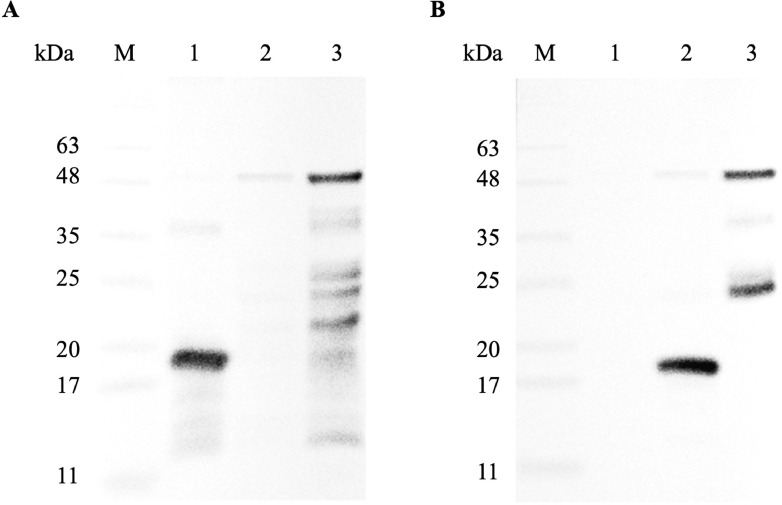
The MABs were subsequently purified using protein G affinity chromatography columns. The specificity of the purified MABs against recombinant Δ60VP1 and either VP1-F1 or VP1-F2 polypeptides was confirmed through Western blot analysis. The results demonstrated that a band corresponding to the recombinant VP1-F1 polypeptide at approximately 17 kDa and a band corresponding to the recombinant Δ60VP1 at approximately 45 kDa were observed when MAB1 was used as the primary antibody (Figure 4A). No reactivity was observed for MAB1 to the VP1-F2 polypeptide. When MAB3 was used as the primary antibody, a band against the recombinant VP1-F2 polypeptide at approximately 16 kDa and a band against the recombinant Δ60VP1 at approximately 45 kDa were detected (Figure 4B). No reactivity was observed for MAB3 to the VP1-F1. It is noted that degradation of the recombinant Δ60VP1 was observed with both MABs. Overall, these Western blotting analyses confirmed the specificity of the monoclonal antibodies against their respective target polypeptide.
Figure 4.
Specificity of monoclonal antibodies to purified recombinant polypeptides VP1-F1, VP1-F2, and Δ60VP1. The specificity of monoclonal antibodies (MABs) against purified recombinant polypeptides VP1-F1, VP1-F2, and Δ60VP1 was confirmed through Western blot analysis using primary antibodies as MAB1 (A), and MAB3 (B). Lane M: CozyHi Prestained Protein Ladder (highQu, Germany); Lane 1: Recombinant VP1-F1 (17 kDa); Lane 2: VP1-F2 (16 kDa); Lane 3: Δ60VP1 (45 kDa).
The optimal ratio of MAB3 for labeling with gold particles was evaluated within the range of 0 to 200 µg/mL of the antibody. A concentration of 160 µg/mL of MAB3 was required to ensure stable labeling with the gold particles for use in the production of lateral flow assay.
Performance Assessment of Lateral Flow Assay
Sensitivity Evaluation. The sensitivity of the lateral flow assay was evaluated by applying commercial chicken serum prepared with TBST at a ratio of 1:1, supplemented with recombinant Δ60VP1 at concentrations ranging from 0 to 20 µg/mL (100 µL). It was observed that the color intensity of the test line increased in direct proportion to the concentration of antigen applied. The lowest concentration at which a discernible color was visible on the test line of the assay was observed for Δ60VP1 was at a concentration of 0.625 µg/mL (Figure 5). This concentration equates to a limit of detection of 62.5 ng of the Δ60VP1 antigen.
Figure 5.
Evaluation of the lateral flow assay at various recombinant protein Δ60VP1 concentrations. The lateral flow assay was assessed using a sample solution containing different concentrations of the recombinant protein Δ60VP1. The concentrations ranged from 0 to 20 µg/mL, with incremental concentrations of 0.3125, 0.625, 1.25, 2.50, 5, 10, and 20 µg/mL, respectively.
Stability Evaluation. The stability of the lateral flow assay was evaluated at monthly intervals for 6 mo by monitoring changes in color intensity at both control and test line locations at ambient temperature. When assayed with a TBST-only solution (devoid of recombinant protein), the control line maintained consistent color intensity, at all time points evaluated, confirming the stability of this component of the assay. However, the test line exhibited a nonspecific background signal across the 0-, 1-, 3-, and 6-mo time points, rendering the interpretation of results ambiguous. To further scrutinize stability, a solution with recombinant Δ60VP1 at 0.625 µg/mL was applied. Throughout the observation months, both control and test lines consistently retained their color intensity. Furthermore, using a higher concentration of 20 µg/mL Δ60VP1 rendered the test line noticeably darker, yet the color stability remained consistent (Figure 6). These results clearly demonstrated the lateral flow assay remained stable and maintained its sensitivity of antigen detection over a 6-mo period when stored at room temperature.
Figure 6.
Stability evaluation of the lateral flow assay over time. The stability of the lateral flow assay was evaluated by testing sample solutions containing recombinant protein Δ60VP1 at concentrations of 0, 0.625, and 20 µg/mL at mo 0, 1, 2, 3, and 6, respectively.
Specificity Evaluation. To evaluate the lateral flow assay's specificity, potential cross-reactivity was investigated using the recombinant capsid of NNV as a surrogate antigen. When 100 µL of recombinant NNV capsid (20 µg/mL) was applied to the lateral flow assay, color development was observed on the control line, signifying a valid test outcome. In contrast, the test line of the assay remained devoid of coloration (Figure 7A). In addition, examination with other chicken viruses and viral antigens gave similar results (Figure 7B). This observation underscored the lateral flow assay's specificity as it only reacted to its target recombinant protein CAV VP1 and did not react with a recombinant protein from an unrelated virus.
Figure 7.
Specificity evaluation of the lateral flow assay. The lateral flow assay was evaluated for specificity using sample solutions containing recombinant NNV capsid protein at concentrations of 0, 0.625, and 20 µg/mL, respectively (A) and other chicken viruses and viral antigens (B). Avinew neo, Newcastle disease virus; FWPV17, Fowl pox virus; H5N1, H7N3, H9N2, antigen from different strains of Avian Influenza virus; ILT, Infectious Laryngotracheitis virus; NDV, antigen from New Castle disease virus; Poulvac IB QX, Avian Infectious Bronchitis virus; −ve, CAV negative serum; +ve, CAV positive serum.
Integrated analyses suggest the lateral flow assay had the required stability, sensitivity, and specificity, to warrant testing its capacity to detect CAV in field samples.
Test Performance Comparison: Lateral Flow Assay and qPCR Analysis of Chicken Serum Samples
The performance of the lateral flow assay was evaluated by comparing lateral flow assay results with qPCR analysis using primers specific to CAV. First, when the assays were used to test chicken serum samples without the presence of CAV, the results were negative according to the qPCR analysis. Only a single red bar appeared at the control line, confirming a valid test (Figure 8A). Subsequently, the assays were tested on chicken serum samples infected with the CAV, which yielded positive results through the qPCR analysis. In this case, 2 red bars appeared at both the test line and control line (Figure 8B).
Figure 8.
Lateral flow assay evaluation with real chicken serum samples. The lateral flow assay was used to test chicken serum samples for the presence of chicken anemia virus. In panel A, the assay result was negative when testing chicken serum samples without the anemia virus. In panel B, the assay result was positive when testing chicken serum samples infected with the chicken anemia virus.
In total, 305 chicken serum samples were examined using the lateral flow assay to detect the presence of CAV, and the results were compared to those obtained by qPCR analysis, which served as the reference test in this study. Examples of the lateral flow assay results using serum samples are depicted in Supplementary Figure S5. The lateral flow assay produced positive results for 45 samples and negative results for 260 samples. On the other hand, the qPCR analysis yielded positive results for 32 samples and negative results for 273 samples (Supplementary Table S1). Based on the test results, the lateral flow assay demonstrated a diagnostic sensitivity of 84.38%, diagnostic specificity of 93.41%, and overall accuracy of 92.46%. Moreover, the agreement between the lateral flow assay and the qPCR analysis was substantial, as indicated by a κ-value of 0.66 (Table 2).
Table 2.
Comparison of lateral flow assay results with chicken serum samples to qPCR.
| Test | qPCR |
||
|---|---|---|---|
| Positive | Negative | Total | |
| Lateral flow assay | |||
| Positive | 27 | 18 | 45 |
| Negative | 5 | 255 | 260 |
| Total | 32 | 273 | 305 |
| Diagnostic sensitivity | 27/32 = 84.38% | ||
| Diagnostic specificity | 255/273 = 93.41% | ||
| Accuracy | (27 + 255)/305 = 92.46% | ||
| Kappa (κ) | 0.66 (Substantial agreement) | ||
DISCUSSION
CAV poses a significant threat to young chickens, with infection resulting in anemia, reduced weight gain, immunosuppression, and elevated mortality. Immunocompromised chickens are at increased risk of secondary infections of increased severity. With no specific treatments currently available, early detection combined with effective control measures is essential to limit the spread of the disease and minimize economic losses. While qPCR stands out as the most robust method for the sensitive detection of viral nucleic acids (Techera et al., 2019; Kaffashi et al., 2021; Kannaki et al., 2021; Luan et al., 2022), the recent emergence of the real-time recombinase-aided amplification assay shows promise in CAV detection (Wu et al., 2022). However, the field application of these technologies often demands the availability of specialized, battery-supported, equipment and skilled personnel to undertake the analysis accurately. Thus, there is a critical need for intuitive, accurate, and rapid detection techniques to support clinical endeavors.
The current study has addressed this need by developing and validating a lateral flow assay designed to enable the rapid, sensitive, and simplified CAV detection at poultry farms, to facilitate the timely implementation of interventions as a component of regular farm management. Development of the assays was enabled by using a recombinant form of CAV VP1 with an N-terminal truncation for reduced toxicity and efficient synthesis in E. coli cells (Lee et al., 2009). In a previous study, the capacity of Δ60VP1 was extended by demonstrating its application in CAV serodiagnosis via indirect ELISA (Wanganurakkul et al., 2020). The Δ60VP1 specific monoclonals, MAB1 and MAB3 generated in this study, were specific to the different epitopes on VP1-F1 and VP1-F2 polypeptides that enable the development of a sandwich lateral flow assay for detecting CAV in chicken serum samples. Further, amino acid sequence comparisons from different countries demonstrated a high degree of sequence similarity for CAV VP1. This implies that MAB1 and MAB3, which have an affinity for VP1-F1 and VP1-F2 respectively on the lateral flow assay, could have a universal applicability in other regions with varying strains of CAV. Importantly there was strong agreement between the results of the nominated gold standard assay, qPCR, and the lateral flow assay.
In our study, we utilized the recombinant capsid of NNV, an infectious piscine agent, distinguished by its N-terminal His Tag (MGSSHHHHHHSSGLVPRGSH), to evaluate the specificity of the lateral flow assay. The outcome, marked by the absence of color intensity at the test line, strongly validates the efficacy of the lateral flow assay. This result suggests that neither MAB3-conjugated colloidal gold nor MAB1 interacted with the N-terminal His-Tag region. Similar observations were noted when testing other chicken viruses and viral antigens, reaffirming the lateral flow assay's specificity for the CAV capsid protein.
In our study, the sensitivity threshold of the lateral flow assay was determined to be 0.625 µg/mL for the Δ60VP1 antigen. This sensitivity is lower than the immunochromatography strip developed by Lee et al. (2021). They utilized paired polyclonal antibodies and MAB specific to VP1 and VP2 of the Sacbrood virus, an insect virus. As with our approach, their recombinant VPs were produced from E. coli cells. Lee et al. employed polyclonal antibodies conjugated with colloidal gold, which potentially provides a higher chance of viral capture compared to our use of MAB. Nevertheless, as noted by Ince and Sezgintürk (2022), the sensitivity of lateral flow assays can vary depending on the virus in question. To enhance sensitivity, we plan to further explore MAB1, which exhibited greater sensitivity than MAB3 (Sittidech, 2017), in the development of more sensitive tools like immunosensors.
Importantly, the lateral flow assay also demonstrated an excellent shelf life, maintaining its effectiveness for at least 6 mo under ambient temperature. Interestingly, we observed that the 2-mo-old lateral flow assay exhibited optimal performance, displaying no noticeable background reactivity at the test line in the absence of the recombinant Δ60VP1 CAV. However, it is worth acknowledging that the detectable background of the lateral flow assay might arise due to the in-house preparation process, particularly during the application of MAB1 and MAB3-colloidal gold onto the pads in the general laboratory setting. Therefore, to ensure consistency and accuracy, production of the lateral flow assay would ideally be conducted within an ISO-17025-certified laboratory. This recommendation underpins our confidence in the potential of the lateral flow assay to demonstrate extended storage and field use.
This study reports a novel lateral flow assay with the capacity to improve the management of CAV. When comparing lateral flow assay results against those generated by qPCR of the sample pool strong agreement was detected. The assay had excellent diagnostic sensitivity, specificity, and accuracy in CAV detection from the same serum samples. This lends support to the lateral flow assay as a credible diagnostic tool, especially crucial in agricultural contexts demanding rapid, user-friendly solutions for detecting the disease of interest. When combined with its cost-effectiveness, rapid result turnaround, and user-friendliness, even for nonexperts, its potential is further underscored. This is the first report of an immunochromatographic assay for CAV detection. With further development and validation, this could be readily adopted for practical application in the field.
CONCLUSIONS
In summary, this study made several significant findings regarding the detection and diagnosis of CAV. The study successfully expressed and purified the recombinant Δ60VP1 protein, an important antigenic component of CAV, and generated specific monoclonal antibodies that recognized distinct epitopes of the polypeptide. The MABs enabled a sandwich lateral flow assay to be developed, demonstrating high sensitivity to low concentrations of Δ60VP1 protein. The stability of the performance of the assay was maintained over a 6-mo period, yielding consistent results. Importantly, the lateral flow assay showed specificity, with no cross-reactivity observed. Comparative analysis with qPCR revealed good diagnostic sensitivity, specificity, and accuracy of the lateral flow assay. There was a substantial level of agreement between the lateral flow assay and qPCR results further supported its reliability. Overall, these findings first establish the effectiveness of the developed lateral flow assay as a rapid alternative for diagnosing CAV infection, facilitating the timely implementation of control measures and disease management in poultry populations.
DECLARATION OF AI AND AI-ASSISTED TECHNOLOGIES IN THE WRITING PROCESS
During the preparation of this work, the authors used ChatGTP in order to enhance grammar, refine the manuscript's flow, and improve readability. After using this tool/service, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
ACKNOWLEDGMENTS
This research was generously supported by the Thailand Science Research and Innovation Fund at Chulalongkorn University (Grant No. CUFRB65_food(8)_116_23_46) and The 90th Anniversary of Chulalongkorn University Scholarship (Ratchadapisek Somphot Endowment Fund) (Grant No. GCUGR1125601042M). We express our sincere gratitude for their financial contributions, which made this study possible. We thank Klairoong Thonsranoi, the Veterinarian, Professional Level at the National Institute of Animal Health (NIAH), The Department of Livestock Development, Ministry of Agriculture and Cooperatives, Thailand, for providing the other chicken viruses and viral antigens used in this study.
DISCLOSURES
The authors declare no conflicts of interest. The funders of this work had no role in study design, data collection and analysis, decision to publish, or the preparation and revision of the manuscript. A patent application under the title “Lateral flow test strip for Chicken anemia virus detection” (No. 2303000210) was submitted to the Department of Intellectual Property, Ministry of Commerce, Thailand, on February 28, 2023.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.psj.2024.103432.
Appendix. Supplementary materials
REFERENCES
- Adair B.M. Immunopathogenesis of chicken anemia virus infection. Dev. Comp. Immunol. 2000;24:247–255. doi: 10.1016/s0145-305x(99)00076-2. [DOI] [PubMed] [Google Scholar]
- Al-Masodi E.T., Al-Azzawi A.K., Al-Ajeeli K., Bande F., Sheikh M.O.B. Serological and molecular detection of chicken anemia virus in some flocks of broiler chickens in Diyala Providence/Iraq. AIP Conf. Proc. 2023;2475 [Google Scholar]
- Chansiripornchai N. Field study of seroconversion of three different commercial vaccines of chicken infectious anemia virus in Thailand. Thai J. Vet. Med. 2017;46:699–704. [Google Scholar]
- Chen J., Yuan X., Ma Z., Wang G., Wang Y., Cao H., Li X., Zheng S.J., Gao L. Chicken infectious anemia virus (CIAV) VP1 antagonizes type I interferon (IFN-I) production by inhibiting TBK1 phosphorylation. Virus Res. 2023;327 doi: 10.1016/j.virusres.2023.199077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Jia H., Hu Y., Wang Y., Cui Z., Qi L., Zhao P. Molecular characterization and pathogenicity study of a highly pathogenic strain of chicken anemia virus that emerged in China. Front. Cell. Infect. Microbiol. 2023;13 doi: 10.3389/fcimb.2023.1171622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatoba A.J., Adeleke M.A. Chicken anemia virus: a deadly pathogen of poultry. Acta Virol. 2019;63:19–25. doi: 10.4149/av_2019_110. [DOI] [PubMed] [Google Scholar]
- Feng C., Liang Y., Teodoro J.G. The role of apoptin in chicken anemia virus replication. Pathogens. 2020;9:294. doi: 10.3390/pathogens9040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadimli H.H., Erganiş O., Güler L., Uçan U.S. Investigation of chicken infectious anemia virus infection by PCR and ELISA in chicken flocks. Turk. J. Vet. Anim. Sci. 2008;32:79–84. [Google Scholar]
- Han C., Niu X., Liu L., Li J., Li J., Yao S., Song X., Gao H., Gao Y., Qi X., Zeng X., Wang Y., Wang X. Development of a loop-mediated isothermal amplification assay for the detection of chicken anemia virus. Poult. Sci. 2019;98:1176–1180. doi: 10.3382/ps/pey495. [DOI] [PubMed] [Google Scholar]
- Huang C.-H., Lai G.-H., Lee M.-S., Lin W.-H., Lien Y.-Y., Hsueh S.-C., Kao J.-Y., Chang W.-T., Lu T.-C., Lin W.-N., Chen H.-J. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of chicken anaemia virus. J. Appl. Microbiol. 2010;108:917–924. doi: 10.1111/j.1365-2672.2009.04481.x. [DOI] [PubMed] [Google Scholar]
- Ince B., Sezgintürk M.K. Lateral flow assays for viruses diagnosis: up-to-date technology and future prospects. TrAC - Trends Anal. Chem. 2022;157 doi: 10.1016/j.trac.2022.116725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffashi A., Mahmoudzadeh M., Kachooei S.Ataei. Development of taqman real-time polymerase chain reaction assay for detection of chicken anemia virus in Newcastle disease vaccines. Arch. Razi Inst. 2021;76:421–427. doi: 10.22092/ari.2020.342677.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannaki T.R., Priyanka E., Subbiah M., Haunshi S. Development and validation of high throughput real-time polymerase chain reaction assay for quantitative detection of chicken infectious anemia virus. Virus Dis. 2021;32:343–346. doi: 10.1007/s13337-020-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian A., Neumann E.J., Hall W.F., Marks D. Effects of blood sample mishandling on ELISA results for infectious bronchitis virus, avian encephalomyelitis virus and chicken anaemia virus. Vet. J. 2012;192:378–381. doi: 10.1016/j.tvjl.2011.08.028. [DOI] [PubMed] [Google Scholar]
- Lacorte C., Lohuis H., Goldbach R., Prins M. Assessing the expression of chicken anemia virus proteins in plants. Virus Res. 2007;129:80–86. doi: 10.1016/j.virusres.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Lee M.-S., Lien Y.-Y., Feng S.-H., Huang R.-L., Tsai M.-C., Chang W.-T., Chen H.-J. Production of chicken anemia virus (CAV) VP1 and VP2 protein expressed by recombinant Escherichia coli. Process. Biochem. 2009;44:390–395. [Google Scholar]
- Lee S.H., Oh T.-K., Oh S., Kim S., Noh H.B., Vinod N., Lee J.Y., Moon E.S., Choi C.W. Development of a kit for rapid immunochromatographic detection of Sacbrood virus infecting Apis cerana (AcSBV) based on polyclonal and monoclonal antibodies raised against recombinant VP1 and VP2 expressed in Escherichia coli. Viruses. 2021;13:2439. doi: 10.3390/v13122439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhang Y., Meng F., Jiang H., Xu G., Ding J., Zhang Y., Dong G., Tian S., Chang S., Zhao P. A new strategy for the detection of chicken infectious anemia virus contamination in attenuated live vaccine by droplet digital PCR. Biomed. Res. Int. 2019;2019 doi: 10.1155/2019/2750472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan Q., Jiang Z., Wang D., Wang S., Yin Y., Wang J. A sensitive triple nanoparticle-assisted PCR assay for detection of fowl adenovirus, infectious bursal disease virus and chicken anemia virus. J. Virol. Methods. 2022;303 doi: 10.1016/j.jviromet.2022.114499. [DOI] [PubMed] [Google Scholar]
- Miller M.M., Schat K.A. Chicken infectious anemia virus: an example of the ultimate host-parasite relationship. Avian Dis. 2004;48:734–745. doi: 10.1637/7271-090304R. [DOI] [PubMed] [Google Scholar]
- NIAH . National Institute of Animal Health; Bangkok, Thailand: 1998. Pages 10–11 in Collection of Samples. Standard Diagnostic Manual for Livestock Diseases Disease in Thailand. [Google Scholar]
- Noteborn M.H.M., Verschueren C.A.J., Koch G., Van Der Eb A.J. Simultaneous expression of recombinant baculovirus-encoded chicken anaemia virus (CAV) proteins VP1 and VP2 is required for formation of the CAV-specific neutralizing epitope. J. Gen. Virol. 1998;79:3073–3077. doi: 10.1099/0022-1317-79-12-3073. [DOI] [PubMed] [Google Scholar]
- Peters M.A., Jackson D.C., Crabb B.S., Browning G.F. Mutation of chicken anemia virus VP2 differentially affects serine/threonine and tyrosine protein phosphatase activities. J. Gen. Virol. 2005;86:623–630. doi: 10.1099/vir.0.80197-0. [DOI] [PubMed] [Google Scholar]
- Rosario K., Breitbart M., Harrach B., Segalés J., Delwart E., Biagini P., Varsani A. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- Schat K.A. In: Pages 151–183 in TT Viruses: The Still Elusive Human Pathogens. de Villiers E.M., zur Hausen H., editors. Springer; Heidelberg, Berlin, Germany: 2009. Chicken anemia virus. [Google Scholar]
- Schlager B., Straessle A., Hafen E. Use of anionic denaturing detergents to purify insoluble proteins after overexpression. BMC Biotechnol. 2012;12:95. doi: 10.1186/1472-6750-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H., Li J., Zhang J., Zhang Q., Ma L., Lu J., Li T., Xie Q., Wan Z., Qin A., Ye J. Research Note: A novel peptide-based ELISA for efficient detection of antibody against chicken infectious anemia virus. Poult. Sci. 2023;102 doi: 10.1016/j.psj.2022.102284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittidech, A. 2017. Monoclonal antibody against VP1 protein of chicken anemia virus and epitope mapping. M.Sc. Diss. Chulalongkorn Univ., Thailand. (In Thai)
- Sittidech A., Assavalapsakul W. Molecular cloning and expression of four regions of VP1 protein from chicken anemia virus. Proc. Int. Conf. Agric. Nat. Res. 2018;2018:257–260. [Google Scholar]
- Techera C., Tomás G., Panzera Y., Banda A., Perbolianachis P., Pérez R., Marandino A. Development of real-time PCR assays for single and simultaneous detection of infectious bursal disease virus and chicken anemia virus. Mol. Cell. Probes. 2019;43:58–63. doi: 10.1016/j.mcp.2018.11.004. [DOI] [PubMed] [Google Scholar]
- Van N.T.B., Cuong N.V., Nhung N.T., Yen N.T.P., Vy L., Kiet B.T., Hoang N.V., Hien V.B., Thu H.T.V., Chansiripornchai N., Thwaites G.E., Choisy M., Carrique-Mas J. A seroepidemiological investigation on major viral and bacterial pathogens in small-scale chicken flocks in the Mekong delta region of Vietnam. Thai J. Vet. Med. 2021;51:729–733. [Google Scholar]
- Wanganurakkul S., Smith D.R., Chintapitaksakul L., Assavalapsakul W. Effective production of recombinant Δ60VP1 chicken anemia virus protein in Escherichia coli and its application to a serodiagnostic indirect ELISA. J. Virol. Methods. 2020;282 doi: 10.1016/j.jviromet.2020.113887. [DOI] [PubMed] [Google Scholar]
- Wu X., Kong J., Yao Z., Sun H., Liu Y., Wu Z., Liu J., Zhang H., Huang H., Wang J., Chen M., Zeng Y., Huang Y., Chen F., Xie Q., Zhang X. A new rapid and sensitive method for detecting chicken infectious anemia virus. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.994651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu B., Liu Y., Chen W., Dai Z., Bi Y., Xie Q. Assessing the efficacy of an inactivated chicken anemia virus vaccine. Vaccine. 2015;33:1916–1922. doi: 10.1016/j.vaccine.2015.02.066. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhang X., Cheng A., Wang M., Yin Z., Huang J., Jia R. Apoptosis triggered by ORF3 proteins of the Circoviridae family. Front. Cell. Infect. Microbiol. 2021;10 doi: 10.3389/fcimb.2020.609071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.