Abstract
The outcome of feline leukemia virus (FeLV) infection in nature is variable, including malignant, proliferative, and degenerative disorders. The determinants of disease outcome are not well understood but are thought to include viral, host, and environmental factors. In particular, genetic variations in the FeLV long terminal repeat (LTR) and SU gene have been linked to disease outcome. FeLV-945 was previously identified as a natural isolate predominant in non-T-cell neoplastic and nonneoplastic diseases in a geographic cohort. The FeLV-945 LTR was shown to contain unique repeat elements, including a 21-bp triplication downstream of the enhancer. The FeLV-945 SU gene was shown to encode mutational changes in functional domains of the protein. The present study details the outcomes of infection with recombinant FeLVs in which the LTR and envelope (env) gene of FeLV-945, or the LTR only, was substituted for homologous sequences in a horizontally transmissible prototype isolate, FeLV-A/61E. The results showed that the FeLV-945 LTR determined the kinetics of disease. Substitution of the FeLV-945 LTR into FeLV-A/61E resulted in a significantly more rapid disease onset but did not alter the tumorigenic spectrum. In contrast, substitution of both the FeLV-945 LTR and env gene changed the disease outcome entirely. Further, the impact of FeLV-945 env on the disease outcome was dependent on the route of inoculation. Since the TM genes of FeLV-945 and FeLV-A/61E are nearly identical but the SU genes differ significantly, FeLV-945 SU is implicated in the outcome. These findings identify the FeLV-945 LTR and SU gene as determinants of disease.
Feline leukemia virus (FeLV) is a naturally occurring gammaretrovirus of the domestic cat. FeLV is endemic in free-roaming urban domestic cats, serological survey of which shows that at least 50% of adult animals have been infected (27). The disease outcome of natural FeLV infection is variable and rather unpredictable. Among persistently infected animals, the majority succumb to degenerative diseases, including anemia or immunodeficiency; however, a substantial minority develop neoplastic or proliferative diseases, including lymphoma, leukemia, or myeloproliferative disorder (18, 26). The determinants of disease outcome in natural FeLV infection have not been clearly defined but probably involve a combination of host, viral, and environmental factors. While there is little doubt that the genetic heterogeneity of the outbreeding mammalian host exerts an influence on disease outcome, the genetic heterogeneity of FeLV in nature clearly has an impact as well. Like other natural retrovirus populations, FeLV is not a single genomic species but represents a family of closely related viruses. Four natural subgroups of FeLV (A, B, C, and T) have been described on the basis of sequence differences in the surface glycoprotein (SU) and on receptor interactions required for entry (20). Subgroup A FeLV (FeLV-A) includes the ecotropic, weakly pathogenic viruses that are horizontally transmitted in nature. Infection with FeLV-A is associated with prolonged, asymptomatic persistent infection that may lead to malignant lymphoma, typically of T-cell origin. For example, experimental infection with FeLV-A/61E in several studies induced thymic lymphoma in some animals after prolonged latency for up to 2 years (22, 25, 28), but other animals remained healthy for even longer periods of observation (25). FeLV-A is present in all natural infections and gives rise to the other subgroups by envelope (env) gene mutation, insertion, or recombination events de novo (19, 20, 34). FeLV-B is a polytropic virus that arises by recombination with endogenous FeLV-related sequences (34, 35). The disease association of FeLV-B infection remains unclear; however, FeLV-B is unusually common in animals with lymphoid malignancy and thus may be linked to the induction of that disease (14, 36). FeLV-C is also a polytropic virus that arises by mutation in the SU gene. FeLV-C is strongly associated with aplastic anemia in infected animals (19). FeLV-T has recently been classified and includes T-cell-tropic cytopathic viruses that cause lymphoid depletion and fatal immunodeficiency disease in infected cats (20, 21, 29). FeLV-T evolves from FeLV-A by mutation and insertion in the SU gene (9, 12). The association of particular outcomes with FeLV subgroups as described above suggests that the nature of the virus isolate is the major disease determinant in FeLV infection. In fact, in the case of anemia or immunodeficiency induced by FeLV-C or FeLV-T, the genetic regions responsible for directing disease outcome have been localized to mutations or insertions in the FeLV SU gene (9, 12, 13, 19). By comparison, the viral determinants of neoplastic disease have not been as clearly defined.
We recently examined FeLV genetic variation in diseased tissues from naturally infected cats in a geographic and temporal cohort (6). The cohort included animals diagnosed upon necropsy with thymic lymphoma, multicentric lymphoma, myeloproliferative disorder, mast cell leukemia, or anemia. Thymic lymphomas from the cohort were shown to contain proviral DNA with tandemly repeated transcriptional enhancer sequences in the long terminal repeat (LTR), as described by others from similar tumors (17, 19, 30). The lengths and termini of LTR enhancer duplications were observed to vary among the natural tumors while uniformly conserving LVb/Ets and CORE binding sites. Regardless of length, enhancer duplications in the LTR conferred little transcriptional advantage as measured in reporter gene assays (6). In non-T-cell malignant, proliferative, and degenerative diseases, the predominant LTR species contained a single transcriptional enhancer element followed downstream by a 21-bp sequence triplicated in tandem (6). This unique LTR, characteristic of a prototype isolate from the cohort previously designated FeLV-945 (2, 16), was shown to confer a replicative advantage on the virus in a manner dependent on the 21-bp triplication (24). Recent analysis revealed that the 21-bp triplication encodes binding sites across the repeat junctions for the transcription factor c-Myb, that the triplication-containing LTR is responsive to c-Myb, and further, that c-Myb in complex with the 21-bp triplication recruits the transcriptional coactivator, CBP, to the FeLV-945 LTR (10). The unique transcription factor complex that forms on the FeLV-945 LTR may be responsible for the utilization of a novel set of common insertion sites in the induction of lymphoma (2, 15, 16). Analysis of the FeLV-945 SU gene also demonstrated it to be unique in sequence; specifically, FeLV-945 SU was shown to be most closely related to FeLV-A, but it also contained several point mutations in the functional domains VRA, VRB, and PRR. Despite the sequence differences, measurements of receptor utilization allowed the assignment of FeLV-945 to subgroup A (5). Initial studies of the impact of the FeLV-945 elements on pathogenesis in cats demonstrated that substitution of the FeLV-945 LTR into prototype FeLV-A/61E did not alter the tumorigenic spectrum of that virus; however, substitution of the FeLV-945 LTR and env gene into FeLV-A/61E altered the disease spectrum entirely from thymic lymphoma of T-cell origin to a non-T-cell multicentric lymphoma (5). The present study details the pathogenesis and disease outcome following infection with those recombinant viruses compared to contemporary controls infected with FeLV-A/61E. The results show that the kinetics of tumor induction is determined by the unique FeLV-945 LTR while the tumor spectrum is determined by the FeLV-945 SU protein.
MATERIALS AND METHODS
Preparation of virus stocks and in vivo challenge.
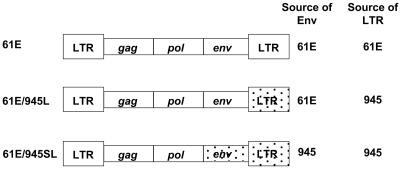
Recombinant infectious FeLV proviruses in which the FeLV-945 LTR and env gene, or the FeLV-945 LTR only, was substituted for homologous sequences in FeLV-A/61E were constructed as previously described (5). The recombinants were designated 61E/945SL and 61E/945L, respectively (Fig. 1). To prepare infectious viral stocks, plasmid DNA containing the proviral genome of FeLV-A/61E, 61E/945SL, or 61E/945L was introduced by transfection into feline embryonic fibroblasts. Three weeks later, culture supernatants were harvested, passaged through a 0.22-μm-pore-size filter, and concentrated 16-fold using Centriprep centrifugal filter units (Millipore Corp., Billerica, Mass.). The titer of each virus stock was determined by quantifying the 50% tissue culture infectious dose (TCID50). For this purpose, feline embryonic fibroblasts seeded at a density of 5 × 103 per well in 24-well culture dishes were challenged with threefold serial dilutions of each stock in quadruplicate wells in the presence of hexadimethrine bromide (Sigma-Aldrich, St. Louis, Mo.) at 8 μg/ml. After 48 h, the cells were washed twice with Hanks' balanced salt solution and were maintained for 1 month. At that time, 50 μl of supernatant from each well was assayed by antigen capture enzyme-linked immunosorbent assay (ELISA) (Synbiotics Corp., San Diego, Calif.) for the presence of FeLV p27Gag antigen. The TCID50 was defined as the lowest dilution of the virus stock that resulted in infection in 50% of the wells.
FIG. 1.
Diagram of recombinant FeLV proviruses constructed by substituting the envelope gene and/or the LTR of FeLV-945 for homologous sequences in FeLV-A/61E as previously described (5).
Specific-pathogen-free pregnant dams were obtained from Liberty Labs, Inc., New Jersey. Within the first 24 h postpartum, neonatal kittens were inoculated intraperitoneally (i.p.) with infectious virus particles or intradermally (i.d.) with plasmid DNA. For intraperitoneal inoculation, kittens were injected with 5 × 105 TCID50 of FeLV-A/61E, 61E/945SL, or 61E/945L in a volume of 0.5 ml. For intradermal inoculation, kittens were inoculated with 50 μg of plasmid DNA encoding the 61E/945SL provirus combined with 0.40 mg of a cationic lipid compound (DOTAP; Roche Applied Science, Indianapolis, Ind.) in a final volume of 0.6 ml of HEPES-buffered saline. The DNA-DOTAP mixture was inoculated intradermally into five sites over the dorsal thorax and thighs as described by others (22).
Longitudinal monitoring of infection and disease progression.
Infected animals were observed by daily monitoring and biweekly physical examinations for evidence of disease, including progressive weight loss, anorexia, diarrhea, dehydration, pallor, inactivity, or debilitation. One animal served as an age-matched uninoculated control and was housed in a separate room. Peripheral blood was collected from each animal every 2 weeks for the first year and every 8 weeks thereafter and was submitted for complete blood count (Anilytics, Inc., Gaithersburg, Md.). Upon evidence of disease, animals were euthanized by intravenous barbiturate overdose (Beuthanasia-D; Schering Plough Animal Health, Union, N.J.) and submitted for complete necropsy and histopathological examination. At necropsy, diseased and normal tissues were collected for formalin fixation, for freezing at −80°C, and for the long-term storage of viable tumor cells at −80°C in 90% fetal bovine serum (FBS) with 10% dimethyl sulfoxide.
FeLV viremia was detected in blood samples by ELISA (Synbiotics Corp., San Diego, Calif.) for the detection of p27Gag antigen. For precise quantitation of p27Gag antigen in serum, an ELISA protocol was developed by Custom Monoclonals International (West Sacramento, Calif.) as follows. Immulon 2HB microtiter plates (Thermo Electron Corp., Milford, Mass.) were coated overnight at room temperature with anti-FeLV p27 capture antibody CM2 (Custom Monoclonals International, West Sacramento, Calif.) in phosphate-buffered saline (PBS) at 0.5 μg/well and were then washed twice in PBS with 0.5% Tween 20. Serum samples were added to each well at doubling dilutions of 1:2 to 1:128 in B3T buffer (0.88% [wt/vol] NaCl, 0.79% [wt/vol] Tris-HCl, 0.03% [wt/vol] EDTA, 2% [wt/vol] bovine serum albumin, 3.3% fetal bovine serum, 0.07% Tween 20, and 1% thimerosal [2% solution]). Purified FeLV p27 antigen (Custom Monoclonals International, West Sacramento, Calif.) was used as a standard on each microtiter plate by adding it to duplicate wells in doubling dilutions between 250 ng/ml and 2 ng/ml. After incubation for 45 min at room temperature, the wells were washed three times in PBS with 0.5% Tween 20. The biotin-tagged anti-FeLV p27 probe antibody, CM1 (Custom Monoclonals International, West Sacramento, Calif.) was then added at 0.5 μg/well. After incubation for 45 min at room temperature, the wells were washed three times in PBS with 0.5% Tween 20, and streptavidin-horseradish peroxidase (BD Biosciences, San Jose, Calif.) was added at a dilution of 1:1,000 in B3T buffer. After incubation for 45 min at room temperature, the wells were washed three times in PBS with 0.5% Tween 20, and a chromogenic substrate solution was added containing 0.4 mg/ml of o-phenylenediamine dihydrochloride (Sigma-Aldrich, St. Louis, Mo.) in 0.05 M phosphate citrate buffer (pH 5.0). After a 6-minute incubation, the reaction was stopped with 3 N HCl and analyzed by quantifying the optical density at 492 nm using an automated ELISA plate reader. FeLV p27Gag antigen in each serum sample was thereby quantified by comparison to a standard curve generated in parallel on each microtiter plate.
Southern blot analysis of genomic DNAs from tumor samples.
High-molecular-weight DNA was isolated from tumors, and Southern blot analysis was performed as previously described (2). For analysis of the beta chain gene of the murine T-cell receptor (TCR-β) locus, 8 μg of DNA was digested with HincII and hybridized to probe 86T5, a 600-bp EcoRI fragment from the murine TCR-β cDNA. For analysis of recombinant FeLV-B proviruses, 8 μg of DNA was digested with KpnI and hybridized to probe B/S, a Sau3A fragment from the env gene of FeLV-B/Gardner-Arnstein (35).
Immunophenotypic analysis of tumor cells.
For analysis of surface phenotype by flow cytometry, tumor cells (1 × 105 to 5 × 105 per assay) were suspended in 200 μl of ice-cold standard azide buffer (SAB) with FBS (1% [wt/vol] FA Bacto Buffer, 0.1% [wt/vol] NaN3, 1% heat-inactivated fetal bovine serum). Primary antibody (1 μg per assay) was added to the cells and incubated on ice for 60 min. When secondary antibody was required, cells were washed and resuspended in SAB with FBS, followed by addition of secondary antibody (1 μg per assay) and incubation for 30 min on ice. The cells were then washed twice and resuspended in 500 μl of SAB without serum for analysis. The antibodies used were as follows: fluorescein isothiocyanate-conjugated mouse anti-feline CD4 monoclonal antibody (3-4F; Southern Biotechnology, Birmingham, Ala.), phycoerythrin-conjugated mouse anti-feline CD8 monoclonal antibody (fCD8; Southern Biotechnology, Birmingham, Ala.), phycoerythrin-Cy5-conjugated rat anti-mouse CD45R/B220 monoclonal antibody (RA3-6B2; BD Biosciences, San Jose, Calif.), mouse anti-FeLV monoclonal antibody (C11D8; Custom Monoclonal Antibodies International, West Sacramento, Calif.), and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) polyclonal secondary antibody (Southern Biotechnology, Birmingham, Ala.). Mouse IgG1(κ) and rat IgG2a(κ) isotype control antibodies (Southern Biotechnology, Birmingham, Ala.) were also used. The cells were analyzed on a Becton Dickinson FACSCalibur flow cytometer and interpreted with BD Biosciences CELLQuest Pro Software. Forward and side scatter characteristics were used to measure cell size and density, respectively, for the purpose of gating out cell debris. FeLV-infected feline 3201 T-lymphoid cells (CD4+ CD8+ FeLV+) were used for electronic compensation for spectral overlap of the fluorochromes.
Formalin-fixed paraffin-embedded sections (5 μm thick) from lymphomas were also examined by immunohistochemical staining using the following mouse monoclonal antibodies for the primary stain: (i) clone CM7 directed against feline IgM (Custom Monoclonal Antibodies International, West Sacramento, Calif.), (ii) clone JCB117 directed against human CD79a (DakoCytomation, Carpinteria, Calif.), and (iii) clone C11D8 directed against FeLV SU protein (Custom Monoclonal Antibodies International, West Sacramento, Calif.). Reactivity to primary antibodies was detected using an avidin-biotin complex enzyme technique (Vectastain ABC kit; Vector Laboratories, Burlingame, Calif.), followed by the addition of diaminobenzidine as a peroxidase substrate. Sections were counterstained with Mayer's hematoxylin for visualization. As controls, duplicate sections were processed without primary antibody.
PCR amplification of FeLV env gene and LTR sequences from tumor DNA.
FeLV env gene and LTR sequences were amplified from tumor DNA by PCR using primers H18 (5′ ACA TAT CGT CCT CCT GAC CAC 3′) and H20 (5′ GAA GGT CGA ACT CTG GTC AAC T 3′), which recognize, respectively, sequences in the pol gene upstream of the env gene start codon and sequences in the U3 region of the 3′ LTR that are conserved among exogenous FeLVs (7). To ensure high fidelity of amplification products, PCR was performed using Pfu DNA polymerase-based enzyme mixes (Expand High Fidelity PCR System [Roche Applied Science, Indianapolis, Ind.] and Herculase Hotstart DNA polymerase [Stratagene, La Jolla, Calif.]). Multiple amplifications were performed from each DNA sample, and the predominant products were cloned into TA cloning vectors, pCR2.1 (Invitrogen Corp., Carlsbad, Calif.) or pGEM-T Easy (Promega Corp., Madison, Wis.).
Nucleotide sequence accession numbers.
Three to five clones from each DNA sample were submitted for automated nucleotide sequence analysis. The results were submitted to GenBank (accession numbers AY706341 to AY706357 for SU sequences and AY706360 to AY706380 for LTR sequences).
RESULTS AND DISCUSSION
To study the influence of the distinctive sequence elements of FeLV-945 on pathogenesis, recombinant infectious FeLV proviruses were constructed in which the envelope gene (env) and LTR of FeLV-945 were substituted for homologous sequences in FeLV-A/61E or in which only the FeLV-945 LTR was substituted. These recombinant FeLVs were designated 61E/945SL and 61E/945L, respectively (Fig. 1). Four neonatal kittens born to an FeLV-free dam were inoculated intraperitoneally with 5 × 105 particles (TCID50) of FeLV-61E/945SL within the first 24 h after birth. FeLV-free litters of five or four neonatal animals were similarly inoculated with FeLV-61E/945L or FeLV-A/61E, respectively (Table 1). At regular intervals thereafter, viremia was measured by antigen capture ELISA for the major capsid protein p27Gag in peripheral blood. By this measure, persistent viremia was detected in all animals beginning at 2 to 4 weeks postinoculation. Since the kittens were housed together with the dam until weaning at 8 weeks of age, the possibility of horizontal transmission to the dam was also examined by ELISA. Viremia was never detected in the dam of the litter infected with FeLV-A/61E, but a self-limiting viremia was observed in the dam of kittens inoculated with 61E/945L beginning at week 5 and lasting for 1 month. In the dam of animals inoculated with 61E/945SL, viremia was first detected beginning at week 6 and has persisted throughout the course of the study, now ongoing for >82 weeks.
TABLE 1.
Summary of inocula, routes of infection, survival times, and disease outcomes in cats inoculated with FeLV or infectious recombinant viruses
| Animal | Inoculuma | Routeb | Initial detection of viremiac (weeks p.i.) | Survival time (weeks p.i.) | Disease outcome |
|---|---|---|---|---|---|
| O3 | FeLV-A/61E | i.p. | 2 | >82 | No disease symptoms |
| O4 | FeLV-A/61E | i.p. | 4 | 68 | Thymic lymphoma |
| O5 | FeLV-A/61E | i.p. | 4 | >82 | No disease symptoms |
| O6 | FeLV-A/61E | i.p. | 2 | >82 | No disease symptoms |
| O23 | 61E/945L | i.p. | 4 | 51 | Thymic lymphoma |
| O24 | 61E/945L | i.p. | 2 | 55 | Thymic lymphoma |
| O25 | 61E/945L | i.p. | 2 | 57 | Thymic lymphoma |
| O26 | 61E/945L | i.p. | 2 | 48 | Thymic lymphoma |
| O27 | 61E/945L | i.p. | 2 | 26 | Thymic lymphoma |
| N89 | 61E/945SL | i.p. | 2 | 72 | Uncharacterized |
| N90 | 61E/945SL | i.p. | 2 | 42 | Multicentric lymphoma |
| N91 | 61E/945SL | i.p. | 2 | 38 | Multicentric lymphoma |
| N92 | 61E/945SL | i.p. | 4 | 49 | Multicentric lymphoma |
| N65 | 61E/945SL | i.d.d | 6 | 49 | Thymic lymphoma |
| N67 | 61E/945SL | i.d. | 8 | 33 | Thymic lymphoma |
Neonatal animals were inoculated with FeLV-A/61E or with recombinant virus in which the FeLV-945 LTR, or the FeLV-945 LTR and env gene, was substituted for homologous sequences in FeLV-A/61E. The recombinant viruses were designated 61E/945L and 61E/945SL, respectively.
Animals were inoculated i.p. with a whole-virus inoculum or i.d. with plasmid DNA encoding infectious provirus.
FeLV viremia was determined by ELISA from blood samples collected every 2 weeks after inoculation.
Five additional littermates were inoculated intradermally with 61E/945SL but did not become viremic. These animals were euthanized at 12 weeks postinoculation and are not included in Table 1.
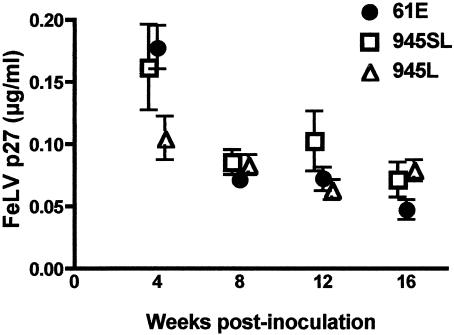
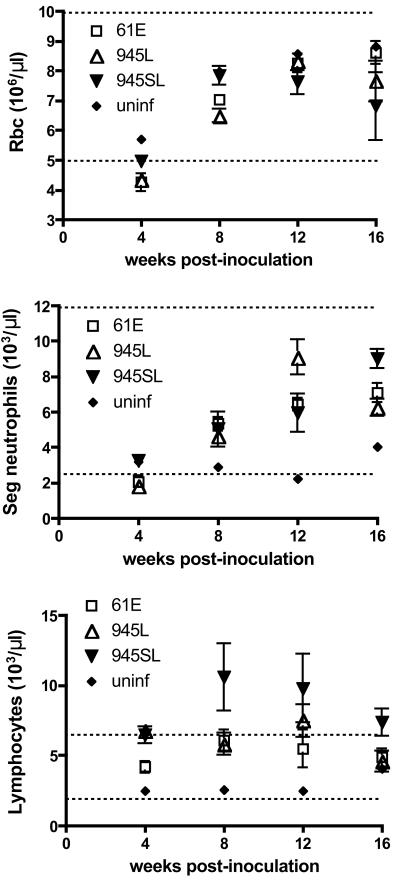
Previous studies had shown that the FeLV-945 LTR confers a replicative advantage on the virus in cultured cells (6, 24). To test the prediction that the FeLV-945 LTR may similarly confer a replicative advantage in vivo, FeLV p27Gag was quantified by ELISA in serum samples collected serially during the first 16 weeks of infection. The results showed that mean serum antigen levels for all challenge groups were highest during the first 4 weeks of infection and then declined. Through 16 weeks of infection, measurements of antigenemia in peripheral blood were statistically indistinguishable among the challenge groups (Fig. 2). Thus, if the FeLV-945 LTR confers a replicative advantage in vivo, it is not apparent as elevated virus load in peripheral blood. A possibility as yet untested is that differences in FeLV antigen levels among the challenge groups may be evident in target tissues for transformation or in other infected tissues. Peripheral blood collected from infected animals at regular intervals during the course of disease was also submitted for complete blood count. Examination of hemograms revealed a significant depression in the number of circulating red blood cells and segmented neutrophils during the first 4 weeks postinoculation, particularly in animals infected with FeLV-A/61E or 61E/945L. By 8 weeks postinoculation, the numbers of these cells in peripheral blood had recovered to normal levels (Fig. 3). In animals infected with 61E/945SL, a significant lymphocytosis was observed in peripheral blood early in infection beginning at 8 weeks postinoculation and persisting for at least 1 month (Fig. 3). Other studies of naturally and experimentally infected cats have also associated the acute stages of FeLV infection with a transient anemia, lymphopenia, or atypical lymphocytosis and neutropenia (31-33).
FIG. 2.
Quantitation of FeLV p27Gag antigen in peripheral blood of animals infected with FeLV-A/61E (61E), 61E/945L (945L), or 61E/945SL (945SL). Peripheral blood was collected at 4, 8, 12, and 16 weeks postinoculation. For each sample, the amount of FeLV p27Gag antigen was determined in quadruplicate by ELISA. Shown are the mean and standard error at each timed collection.
FIG. 3.
Cell counts in peripheral blood of animals infected with FeLV-A/61E (61E), 61E/945L (945L), or 61E/945SL (945SL) and in an age-matched uninfected control (uninf). Peripheral blood was collected at 4, 8, 12, and 16 weeks postinoculation. Shown are the mean and standard error of red blood cell (Rbc), segmented (Seg) neutrophil, and lymphocyte counts at each timed collection. Dashed horizontal lines demarcate the normal levels of each cell count in the peripheral blood of cats (Anilytics, Inc., Gaithersburg, Md).
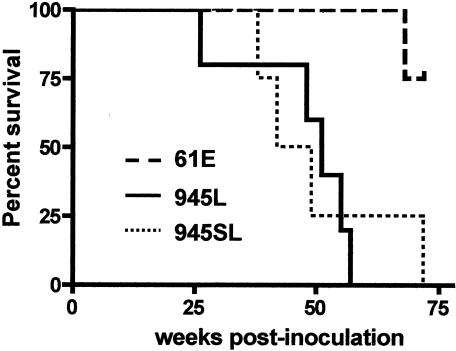
The kinetics of disease induction were distinct among the challenge groups. Kaplan-Meier survival estimates and analysis of the data by the log-rank test demonstrated that 61E/945L and 61E/945SL induced diseases with indistinguishable kinetics but that both induced disease significantly more rapidly than did FeLV-A/61E (P = 0.03) (Fig. 4). Of four animals infected with FeLV-A/61E, one succumbed to lymphoma at 68 weeks postinoculation. The three remaining animals have demonstrated persistent viremia as measured by ELISA for p27Gag in peripheral blood (data not shown) but have shown no signs of disease throughout the course of the study, ongoing now for >82 weeks (Table 1). These observations are consistent with previous studies demonstrating FeLV-A/61E to be a weakly pathogenic virus. Experimental infection with FeLV-A/61E has been associated with the induction of thymic lymphoma in some animals after prolonged latency for up to 2 years (22, 25, 28), but other infected animals have remained healthy in studies that continued for as long as 812 days (25). By comparison, animals infected with 61E/945L succumbed to disease between 26 and 57 weeks postinoculation (average, 47 weeks), and those infected with 61E/945SL succumbed to disease between 38 and 72 weeks postinoculation (average, 50 weeks) (Table 1). 61E/945L and 61E/945SL have in common the FeLV-945 LTR (Fig. 1), thus implicating its unique 21-bp triplication as a potential disease determinant. Previous studies have shown that the unique 21-bp triplication in the FeLV-945 LTR provides transcriptional enhancer function to the LTR, modulates LTR transcriptional activity, and confers a replicative advantage in a cell-type-specific manner (3, 6, 24). These observations suggest that the FeLV-945 LTR may act in pathogenesis by increasing virus replication in target tissues for transformation and/or by more efficiently activating expression of cellular oncogenes relevant to disease induction.
FIG. 4.
Kaplan-Meier survival estimates for animals inoculated intraperitoneally with FeLV-A/61E (61E), 61E/945L (945L), or 61E/945SL (945SL). The log-rank test showed a significant difference between survival rates of FeLV-A/61E-infected cats and those infected with 61E/945L or 61E/945SL (P = 0.03).
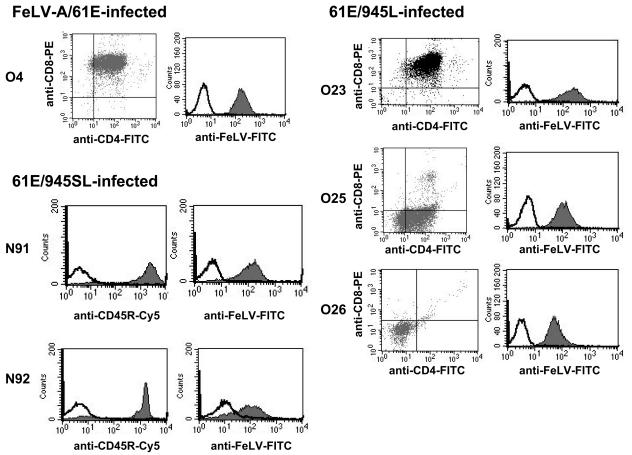
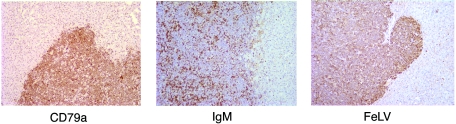
The disease outcomes among the challenge groups were also distinct (Table 1). In the single animal to have succumbed to FeLV-A/61E infection to date, and in all animals infected with 61E/945L, necropsy revealed a large thymic tumor that occupied most of the chest, compressed the lungs, and extended through the thoracic inlet, causing compression on the esophagus and surrounding tissues. Southern blot analysis of tumor DNA from each case demonstrated clonal rearrangement of the TCR-β locus; thus, all tumors induced by FeLV-A/61E or 61E/945L were identified as lymphomas of T-cell origin (representative data are shown in Fig. 5). Flow cytometric analysis of tumor cells demonstrated surface phenotypes characteristic of immature thymocytes, e.g., CD4+ CD8+ (cats O4 and O23) or CD4− CD8− (cat O26). One case (cat O27) was characterized as predominantly CD4+ CD8−, and another (cat O25) was of mixed phenotype, including CD4+ CD8− (65%), CD4− CD8− (23%), and CD4+ CD8+ (12%) (representative data are shown in Fig. 6). In all cases, tumor cells were shown to express FeLV SU protein on the surface as measured by reactivity to the anti-SU monoclonal antibody, C11D8 (Fig. 6). These findings are consistent with other studies showing that FeLV-A/61E typically induces thymic lymphoma of T-cell origin after prolonged latency and that tumor cells exhibit the surface phenotype of immature thymocytes (22, 28). The findings further demonstrate that substitution of the FeLV-945 LTR into FeLV-A/61E did not alter the tumorigenic spectrum. In contrast, 61E/945SL-infected animals succumbed to disease with relatively rapid kinetics but in no case developed T-cell lymphoma of the thymus. Rather, as recently reported (5), three 61E/945SL-infected animals succumbed between 38 and 49 weeks postinoculation to a multicentric lymphoma that involved multiple organs, including liver, kidney, and lungs, but excluded the thymus (Table 1). Southern blot analysis of DNAs from those tumors revealed the TCR-β locus to be in germ line configuration, indicative of non-T-cell origin (Fig. 5). Consistent with these findings, flow cytometric analysis demonstrated the absence of CD4 or CD8 expression on the surfaces of tumor cells (data not shown). Rather, flow cytometry demonstrated high levels of expression of CD45R/B220, indicative of B-cell origin of the tumors. Surface expression of FeLV SU protein was also demonstrated (Fig. 6). The origin of the lymphoma from cat N91 was confirmed by immunohistochemical analysis demonstrating abundant expression of a second B-cell marker, CD79a. Surface IgM expression was also apparent on a fraction of tumor cells (Fig. 7). In addition, immunohistochemical analysis confirmed the expression of FeLV SU protein by the majority of tumor cells (Fig. 7). FeLV-induced B-cell lymphomas are relatively uncommon; however, a previous analysis demonstrated FeLV expression in only one of six B-cell tumors examined (26). Thus, the lymphomas induced by 61E/945SL are unusual with respect to the B-cell origin and the abundant expression of FeLV. One 61E/945SL-infected animal, cat N89, survived significantly longer than its littermates but succumbed to disease at 72 weeks postinoculation (Table 1). Cat N89 showed severe lymphocytopenia beginning from week 41 postinoculation and throughout the remainder of the disease course. During this period, the lymphocyte count varied between 0.65 × 103 and 1.3 × 103 per μl compared to normal limits of 1.5 × 103 to 7.0 × 103 per μl. Necropsy revealed normal thymus, liver, and spleen. The lungs appeared generally healthy, but small nodular lesions were observed. A mesenteric lymph node was significantly enlarged, but other lymph nodes appeared normal. While the disease presentation was clearly distinct from 61E/945L-induced thymic lymphoma, the disorder present in cat N89 remains to be characterized in detail. Overall, the distinct disease outcome in cats infected with 61E/945SL compared to 61E/945L implicates the FeLV-945 env gene as a determinant of the disease spectrum. Recent analysis of the FeLV-945 env gene demonstrated that the SU protein differs significantly from FeLV-A/61E across functional domains, although the TM genes are nearly identical (5). These observations specifically implicate FeLV-945 SU as a determinant of disease in 61E/945SL-infected cats.
FIG. 5.
Southern blot analysis of the TCR-β locus in genomic DNA from tumors induced by infection with FeLV-A/61E, 61E/945L, or 61E/945SL introduced i.p. or with 61E/945SL introduced i.d. DNA samples were digested with HincII and hybridized to a probe representing the beta chain gene of the murine T-cell receptor (TCR-β). Indicated in each set are the identifying animal number and a control sample from the liver of an uninfected cat (C). Indicated are the 8.4-kb and 2.9-kb fragments definitive of germ line organization of the feline TCR-β locus (closed circles) and a polymorphism occasionally detected in the TCR-β locus (arrowhead). A portion of these data was reported previously (5).
FIG. 6.
Surface phenotypes of tumor-derived cells from animals inoculated intraperitoneally with FeLV-A/61E, 61E/945L, or 61E/945SL. The surface expression of CD4, CD8, CD45R/B220, and FeLV SU on tumor cells was quantified by flow cytometry.
FIG. 7.
Immunohistochemical analysis of the multicentric lymphoma from cat N91 invading the liver. Shown is immunohistochemical staining for expression of (left) B-cell marker CD79a, (right) FeLV SU protein, and (center) IgM.
In parallel studies, 61E/945SL was introduced into FeLV-free neonatal cats by intradermal inoculation of plasmid DNA encoding the infectious viral genome. Plasmid DNA (50 μg) was mixed with a cationic lipid compound and injected intradermally into five sites over the dorsal thoraxes and thighs of seven neonatal littermates as described by others (7, 22). In studies by others, FeLV-A/61E plasmid DNA introduced intradermally was shown to establish persistent infection within 4 weeks, leading to a typical thymic lymphoma of T-cell origin in three of four animals by 66 weeks postinoculation (22). In the present study, 61E/945SL plasmid DNA was introduced intradermally, and viremia was followed by ELISA for p27Gag in peripheral blood collected at biweekly intervals thereafter. The results demonstrated viremia in only two of seven animals beginning at 6 and 8 weeks postinoculation (Table 1). The level of viremia was comparable to that in animals inoculated intraperitoneally with 61E/945SL (data not shown) but was delayed in appearance (6 to 8 weeks following intradermal inoculation versus 2 to 4 weeks following intraperitoneal inoculation). Viremia was not detected in the remaining animals up to 12 weeks postinoculation, and they were euthanized at that time without evidence of infection or disease. Thus, it appears that direct intradermal inoculation of proviral DNA was inefficient in establishing infection by 61E/945SL, perhaps an indication that this route did not offer access to the optimal target cells. The two animals that became persistently infected after intradermal inoculation succumbed to disease at 33 and 49 weeks postinoculation, a latency period comparable to that in animals inoculated intraperitoneally. The disease outcome, however, was entirely distinct. Animals inoculated intradermally presented at necropsy with large thymic tumors typical of infection with FeLV-A/61E. Southern blot analysis of tumor DNA demonstrated clonal somatic rearrangement of the TCR-β locus, thus identifying the tumors as lymphomas of T-cell origin (Fig. 5). These findings demonstrated that 61E/945SL, containing the env gene and LTR of FeLV-945, is capable of inducing either T-cell lymphoma or non-T-cell disease, depending on the route of inoculation. By comparison, it is noteworthy that FeLV-945 was not identified in T-cell tumors in the natural cohort from which it was originally isolated. Rather, it was identified in non-T-cell multicentric lymphomas, as well as in proliferative and degenerative diseases of non-T-cell origin (2, 6). The most frequent route of exposure to FeLV in nature is thought to be oronasal contact with infectious saliva; however, it has been speculated that the parenteral routes used experimentally (e.g., intraperitoneal and intravenous) may mimic the natural introduction of FeLV in the course of cat bites during fights or copulation (33). In the present studies, it is remarkable that intraperitoneal inoculation recapitulated the disease spectrum of FeLV-945 in nature while intradermal inoculation did not.
Studies were next performed to determine whether the uniquefeatures of FeLV-945 SU and LTR were conserved in end stage disease from experimentally infected animals. The 3′ end of proviral DNA was amplified by PCR from tumor samples using oligonucleotide primers H18 and H20, which recognize, respectively, sequences in the pol gene upstream of the env gene start codon and sequences in the U3 region of the 3′ LTR that are conserved among exogenous FeLVs (7). DNA samples representative of each tumor type were subjected to multiple independent amplifications, and the predominant products of each reaction were cloned and sequenced. Amplification products from a thymic lymphoma induced by 61E/945L (cat O27) contained an SU gene identical to that of FeLV-A/61E across the VRA, VRB, and PRR coding domains, with five predicted amino acid substitutions outside those domains. One of those mutations replaced a histidine residue at position 6 with proline within an SPHQ motif known to be critical for fusion upon virus entry (Fig. 8A) (4, 37). The identical H6P mutation in FeLV SU has been reported previously from viruses in natural and experimentally induced lymphomas (28). These viruses, like FeLV-T isolates containing the H6 mutation, may require trans-activation by a soluble cofactor for entry (1, 12). SU genes isolated from 61E/945SL-induced thymic lymphomas (cats N65 and N67) were identical to FeLV-945 across the VRA, VRB, and PRR coding domains and contained four predicted amino acid substitutions outside those domains. The SU gene from N65 also contained a 12-nucleotide insertion in the same region as that previously reported from T-cell cytopathic FeLV isolates 81T and 61C (Fig. 8B) (21, 28, 29). The insertions in 81T and 61C SU proteins were previously shown to confer T-cell cytopathic properties on the virus, but introduction of the 81T insertion into FeLV-A/61E rendered the virus defective for replication. Tissue culture-adapted variants competent to replicate in feline cells were shown to contain a compensatory glutamine-to-proline substitution at position 7 within the critical SPHQ motif (12). It is intriguing that N65 SU also contained the Q7P substitution (Fig. 8A), perhaps acquired as a compensatory mutation for the insertion in SU. SU genes isolated from 61E/945SL-induced multicentric lymphomas (cats N90, N91, and N92) were identical in sequence to FeLV-945 except for the predicted substitution of three amino acids in the PRR of N90 SU (Fig. 8C). It is not known what effect this substitution may have on the PRR-mediated modulation of SU conformation.
FIG. 8.
Comparison of the predicted amino acid sequences of portions of the SU protein of FeLV-A/61E (8), FeLV-945 (2), the immunodeficiency-inducing FeLV isolates 61C and 81T (21, 28), and proviral sequences from tumors induced by infection with 61E/945L (cat O27) or 61E/945SL (cats N65 and N90). The numbers above the sequence correspond to the amino acid positions of the mature FeLV-A/61E Env protein (8). Asterisks indicate amino acid identity. Dashes are included to optimize the sequence alignment, allowing for amino acid insertions. Shading demarcates an SPHQ motif previously shown to be critical for fusion events upon virus entry (4, 37).
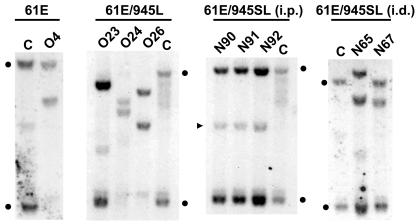
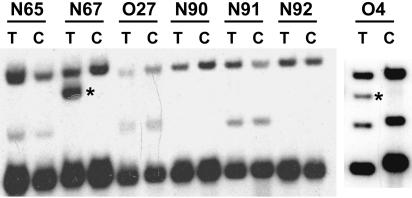
Another determinant implicated in FeLV-mediated lymphomagenesis is the emergence of FeLV subgroup B (FeLV-B)viruses that arise de novo during the course of infection. FeLV-B viruses contain a novel envelope gene derived by recombination between exogenous FeLV-A and endogenous FeLV-related sequences in a manner analogous to the generation of mink cell focus-forming recombinant viruses during murine leukemia virus infection. In nature, FeLV-B is overrepresented in the diseased tissues of animals with lymphoma compared to asymptomatic FeLV-infected cats. For these reasons, the emergence of FeLV-B recombinants has been linked to the induction of malignant disease in infected animals (14, 19, 34). In the present study, the PCR amplification of provirus sequences from tumor DNA samples yielded only a single FeLV-B amplification product that was represented in only one of four clones sequenced from tumor N90 (data not shown). This observation implies that the generation of FeLV-B recombinants was a relatively rare event in tumors induced by 61E/945L or 61E/945SL. It is possible, however, that some FeLV-B recombinant env genes would not have been successfully retrieved using our PCR approach because the binding site for the upstream primer, H18, may have been altered during the recombination event (21, 28). To evaluate the frequency with which FeLV-B viruses arose in tumors in the present study, a Southern blotting approach was used in which tumor DNA was digested with KpnI and hybridized to a probe (B/S) specific for the major classes of endogenous FeLV that serve as substrates for recombination (35). Using this approach, FeLV-B recombinant proviral DNA can be visualized in genomic DNA as a novel hybridizing fragment of ∼3.6 kb, distinct in size from the related endogenous sequences (28, 36). The results of this analysis demonstrated no evidence of FeLV-B recombinants in T-cell lymphomas induced by 61E/945L or in multicentric lymphomas induced by 61E/945SL. In contrast, FeLV-B was readily detected in one of two T-cell lymphomas induced by intradermal inoculation of 61E/945SL and in the single T-cell lymphoma resulting to date from infection with FeLV-A/61E (Fig. 9). Thus, while FeLV-B recombinants can be detected by this approach, they were apparently involved infrequently in the generation of lymphomas induced by virus containing FeLV-945 sequence elements.
FIG. 9.
Southern blot analysis of FeLV-B proviral DNAs in tumors induced by intradermal inoculation with 61E/945SL (cats N65 and N67) or intraperitoneal inoculation with 61E/945L (cat O27), 61E/945SL (cats N90, N91, and N92), or FeLV-A/61E (cat O4). DNA samples from the tumor (lanes T) or control uninvolved tissue from the same animal (lanes C) were digested with KpnI and hybridized to a Sau3A probe (B/S) derived from the env gene of FeLV-B/Gardner-Arnstein (35). The presence of recombinant FeLV-B proviral DNA in tumors from cats N67 and O4 is evident as a novel hybridizing fragment of ∼3.6 kb that is distinct from endogenous FeLV-related sequences (asterisks).
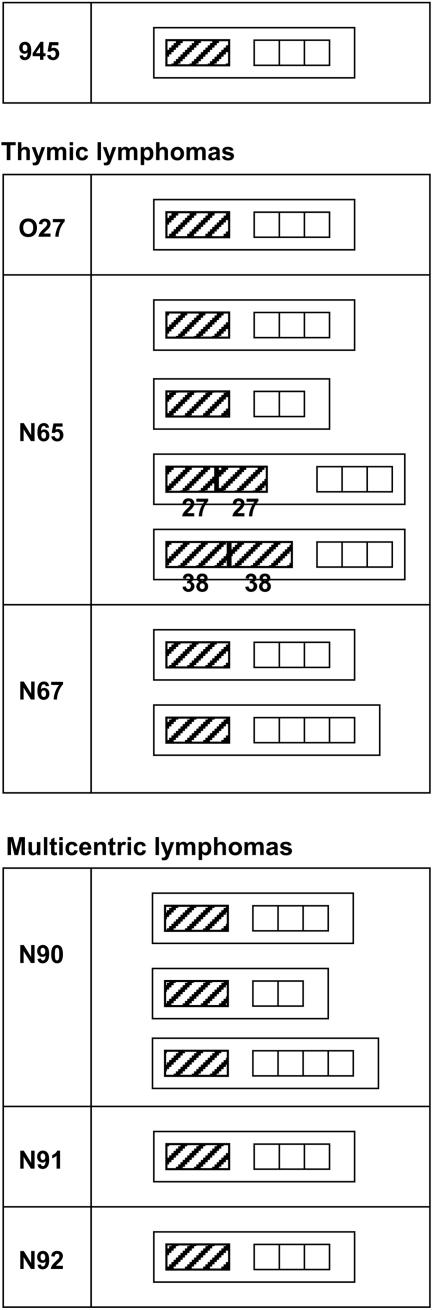
LTR sequences were also analyzed in amplification products from tumors induced by 61E/945L and 61E/945SL. The results demonstrated that the 21-bp triplication-containing FeLV-945 LTR was conserved in all tumors examined, although several tumors also contained LTR variants (Fig. 10). For example, multiple LTR species were detected in the T-cell lymphoma from animal N65: (i) LTRs with a single enhancer followed by three copies of the 21-bp sequence, (ii) LTRs with a single enhancer followed by two copies of the 21-bp sequence, and (iii) LTRs with duplicated enhancers of various repeat lengths followed by three copies of the 21-bp sequence. Duplication of enhancers is typical of the FeLV LTR in lymphomas of T-cell origin (6, 17, 19), and the generation of enhancer repeats de novo during the course of disease has been previously reported (30). Enhancer duplications were 27 or 38 bp in length, in both of which the LVb and CORE binding sites were conserved. These findings are consistent with previous reports showing that LVb and CORE sites, but not necessarily NF-1 or GRE, are typically duplicated in FeLV LTRs in T-cell lymphomas (6, 11, 23). In contrast, duplication of enhancers was not detected in T-cell lymphomas from animals O27 and N67; rather, these tumors contained LTRs with a single enhancer followed by triplication or quadruplication of the 21-bp repeat element. These observations are consistent with previous reports that the 21-bp triplication confers a strong replicative advantage on the virus in feline T cells (6). Thus, the FeLV-945 LTR may potentiate induction of T-cell disease even in the absence of a duplicated enhancer because of the influence of the 21-bp triplication. Duplication of enhancers was not detected in any of the multicentric lymphomas induced by 61E/945SL. Rather, those lymphomas contained LTRs with a single copy of the enhancer followed by two, three, or four copies of the 21-bp repeat element. Similar variation in copy numbers of the 21-bp element was previously reported in diseased tissues of the natural cohort from which FeLV-945 was originally identified. In that study, Southern blot analysis was used to demonstrate that LTRs with variable numbers of 21-bp elements were not artifacts of PCR amplification but were detectable in genomic DNA (6).
FIG. 10.
Diagrammatic representation of the LTR U3 region of FeLV-945 and of LTR sequences amplified from experimentally induced tumors. Represented are the predominant amplification products from thymic lymphomas induced by infection with 61E/945L (cat O27) or 61E/945SL (cats N65 and N67) and from multicentric lymphomas induced by infection with 61E/945SL (cats N90, N91, and N92). Indicated is the organization of repeat elements in the U3 region, including enhancer (hatched boxes) and the 21-bp repeat unit (open boxes). In some cases, multiple amplification products were obtained from the same tissue. When the LTR contained enhancer duplications, the lengths of the enhancer duplications (in bp) are indicated.
In summary, the present study examined the influence of the unique LTR and SU gene of FeLV-945 on pathogenesis in infected animals. The 21-bp triplication-containing LTR of FeLV-945 was previously shown to confer a replicative advantage in vitro (6, 24) and was shown in this study to determine the kinetics of disease induction. Specifically, substitution of the FeLV-945 LTR for homologous sequences in FeLV-A/61E resulted in a significantly more rapid induction of disease (Fig. 4). Substitution of the FeLV-945 LTR did not alter the tumorigenic spectrum of FeLV-A/61E, but substitution of both the FeLV-945 LTR and env gene changed the disease outcome entirely (Table 1). The 61E/945SL recombinant virus contained the entire env gene of FeLV-945; however, the env genes of FeLV-945 and FeLV-A/61E differ significantly only in the SU coding sequence (5). Thus, FeLV-945 SU is implicated in the outcome of 61E/945SL infection. Multicentric lymphomas induced by 61E/945SL were shown to express B-cell markers, as well as FeLV SU (Fig. 6 and 7). While the variable disease outcome of natural FeLV infection is likely due to both virus- and host-derived factors, these findings clearly implicate the FeLV-945 LTR and SU gene as determinants of disease outcome in infected cats.
Acknowledgments
This work was supported by NIH grant CA83823 from the National Cancer Institute and by Development Funds of the Tulane Cancer Center. I.P. was supported in part by NIH grant P51 RR000164 from the National Center for Research Resources. C.C. was supported in part by a grant from the Cancer Association of Greater New Orleans.
We gratefully acknowledge Xavier Alvarez for assistance with electronic imaging.
REFERENCES
- 1.Anderson, M. M., A. S. Lauring, C. C. Burns, and J. Overbaugh. 2000. Identification of a cellular cofactor required for infection by feline leukemia virus. Science 287:1828-1830. [DOI] [PubMed] [Google Scholar]
- 2.Athas, G. B., B. Choi, S. Prabhu, P. A. Lobelle-Rich, and L. S. Levy. 1995. Genetic determinants of feline leukemia virus-induced multicentric lymphomas. Virology 214:431-438. [DOI] [PubMed] [Google Scholar]
- 3.Athas, G. B., P. Lobelle-Rich, and L. S. Levy. 1995. Function of a unique sequence motif in the long terminal repeat of feline leukemia virus isolated from an unusual set of naturally occurring tumors. J. Virol. 69:3324-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bae, Y., S. M. Kingsman, and A. J. Kingsman. 1997. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J. Virol. 71:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandhasin, C., P. N. Coan, and L. S. Levy. 2005. Subtle mutational changes in the SU protein of a natural FeLV-A isolate alter disease spectrum. J. Virol. 79:1351-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandhasin, C., P. A. Lobelle-Rich, and L. S. Levy. 2004. Feline leukemia virus LTR variation and disease association in a geographic and temporal cluster. J. Gen. Virol. 85:2937-2942. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., M. K. Bechtel, Y. Shi, A. Phipps, L. E. Mathes, K. A. Hayes, and P. Roy-Burman. 1998. Pathogenicity induced by feline leukemia virus, Rickard strain, subgroup A plasmid DNA (pFRA). J. Virol. 72:7048-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donahue, P. R., E. A. Hoover, G. A. Beltz, N. Riedel, V. M. Hirsch, J. Overbaugh, and J. I. Mullins. 1988. Strong sequence conservation among horizontally transmissible, minimally pathogenic feline leukemia viruses. J. Virol. 62:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donahue, P. R., S. L. Quackenbush, M. V. Gallo, C. M. deNoronha, J. Overbaugh, E. A. Hoover, and J. I. Mullins. 1991. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J. Virol. 65:4461-4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finstad, S. L., S. Prabhu, K. R. Rulli, and L. S. Levy. 2004. Regulation of FeLV-945 by c-Myb binding and CBP recruitment to the LTR. Virology J. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulton, R., M. Plumb, L. Shield, and J. C. Neil. 1990. Structural diversity and nuclear protein binding sites in the long terminal repeats of feline leukemia virus. J. Virol. 64:1675-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gwynn, S. R., F. C. Hankenson, A. S. Lauring, J. L. Rohn, and J. Overbaugh. 2000. Feline leukemia virus envelope sequences that affect T-cell tropism and syncytium formation are not part of known receptor-binding domains. J. Virol. 74:5754-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoover, E. A., and J. I. Mullins. 1991. Feline leukemia virus infection and diseases. J. Am Vet. Med. Assoc. 199:1287-1297. [PubMed] [Google Scholar]
- 14.Jarrett, O. 1980. Natural occurrence of subgroups of feline leukemia virus, p. 603-611. In M. Essex, G. Todaro, and H. zur Hausen (ed.), Viruses in naturally occurring cancers. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Johnson, C., P. A. Lobelle-Rich, A. Puetter, and L. S. Levy. 2005. Substitution of feline leukemia virus long terminal repeat sequences into murine leukemia virus alters the pattern of insertional activation and identifies new common insertion sites. J. Virol. 79:57-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levesque, K. S., L. Bonham, and L. S. Levy. 1990. flvi-1, a common integration domain of feline leukemia virus in naturally occurring lymphomas of a particular type. J. Virol. 64:3455-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto, Y., Y. Momoi, T. Watari, R. Goitsuka, H. Tsujimoto, and A. Hasegawa. 1992. Detection of enhancer repeats in the long terminal repeats of feline leukemia viruses from cats with spontaneous neoplastic and nonneoplastic diseases. Virology 189:745-749. [DOI] [PubMed] [Google Scholar]
- 18.Mullins, J. I., and E. A. Hoover. 1990. Molecular aspects of feline leukemia virus pathogenesis, p. 87-116. In R. C. Gallo and F. Wong-Staal (ed.), Retrovirus biology and human disease. Marcel Dekker, Inc., New York, N.Y.
- 19.Neil, J. C., R. Fulton, M. Rigby, and M. Stewart. 1991. Feline leukaemia virus: generation of pathogenic and oncogenic variants. Curr. Top. Microbiol. Immunol. 171:67-93. [DOI] [PubMed] [Google Scholar]
- 20.Overbaugh, J., and C. R. Bangham. 2001. Selection forces and constraints on retroviral sequence variation. Science 292:1106-1109. [DOI] [PubMed] [Google Scholar]
- 21.Overbaugh, J., P. R. Donahue, S. L. Quackenbush, E. A. Hoover, and J. I. Mullins. 1988. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science 239:906-910. [DOI] [PubMed] [Google Scholar]
- 22.Phipps, A. J., H. Chen, K. A. Hayes, P. Roy-Burman, and L. E. Mathes. 2000. Differential pathogenicity of two feline leukemia virus subgroup A molecular clones, pFRA and pF6A. J. Virol. 74:5796-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plumb, M., R. Fulton, L. Breimer, M. Stewart, K. Willison, and J. C. Neil. 1991. Nuclear factor 1 activates the feline leukemia virus long terminal repeat but is posttranscriptionally down-regulated in leukemia cell lines. J. Virol. 65:1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prabhu, S., P. A. Lobelle-Rich, and L. S. Levy. 1999. The FeLV-945 LTR confers a replicative advantage dependent on the presence of a tandem triplication. Virology 263:460-470. [DOI] [PubMed] [Google Scholar]
- 25.Quackenbush, S. L., P. R. Donahue, G. A. Dean, M. H. Myles, C. D. Ackley, M. D. Cooper, J. I. Mullins, and E. A. Hoover. 1990. Lymphocyte subset alterations and viral determinants of immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J. Virol. 64:5465-5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezanka, L. J., J. L. Rojko, and J. C. Neil. 1992. Feline leukemia virus: pathogenesis of neoplastic disease. Cancer Investig. 10:371-389. [DOI] [PubMed] [Google Scholar]
- 27.Rogerson, P., W. Jarrett, and L. Mackey. 1975. Epidemiological studies on feline leukaemia virus infection. I. A serological survey in urban cats. Int. J. Cancer 15:781-785. [DOI] [PubMed] [Google Scholar]
- 28.Rohn, J. L., M. L. Linenberger, E. A. Hoover, and J. Overbaugh. 1994. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J. Virol. 68:2458-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohn, J. L., M. S. Moser, S. R. Gwynn, D. N. Baldwin, and J. Overbaugh. 1998. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J. Virol. 72:2686-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohn, J. L., and J. Overbaugh. 1995. In vivo selection of long terminal repeat alterations in feline leukemia virus-induced thymic lymphomas. Virology 206:661-665. [DOI] [PubMed] [Google Scholar]
- 31.Rojko, J. L., C. M. Cheney, P. W. Gasper, K. L. Hamilton, E. A. Hoover, L. E. Mathes, and G. J. Kociba. 1986. Infectious feline leukaemia virus is erythrosuppressive in vitro. Leukoc. Res. 10:1193-1199. [DOI] [PubMed] [Google Scholar]
- 32.Rojko, J. L., E. A. Hoover, L. E. Mathes, R. G. Olsen, and J. P. Schaller. 1979. Pathogenesis of experimental feline leukemia virus infection. J. Natl. Cancer Inst. 63:759-768. [DOI] [PubMed] [Google Scholar]
- 33.Rojko, J. L., and G. J. Kociba. 1991. Pathogenesis of infection by the feline leukemia virus. J. Am. Vet. Med. Assoc. 199:1305-1310. [PubMed] [Google Scholar]
- 34.Roy-Burman, P. 1996. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes 11:147-161. [DOI] [PubMed] [Google Scholar]
- 35.Stewart, M. A., M. Warnock, A. Wheeler, N. Wilkie, J. I. Mullins, D. E. Onions, and J. C. Neil. 1986. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J. Virol. 58:825-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsatsanis, C., R. Fulton, K. Nishigaki, H. Tsujimoto, L. Levy, A. Terry, D. Spandidos, D. Onions, and J. C. Neil. 1994. Genetic determinants of feline leukemia virus-induced lymphoid tumors: patterns of proviral insertion and gene rearrangement. J. Virol. 68:8296-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zavorotinskaya, T., Z. Qian, J. Franks, and L. M. Albritton. 2004. A point mutation in the binding subunit of a retroviral envelope protein arrests virus entry at hemifusion. J. Virol. 78:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]