Abstract
Many steps of viral replication are dependent on the interaction of viral proteins with host cell components. To identify rhinovirus proteins involved in such interactions, human rhinovirus 39 (HRV39), a virus unable to replicate in mouse cells, was adapted to efficient growth in mouse cells producing the viral receptor ICAM-1 (ICAM-L cells). Amino acid changes were identified in the 2B and 3A proteins of the adapted virus, RV39/L. Changes in 2B were sufficient to permit viral growth in mouse cells; however, changes in both 2B and 3A were required for maximal viral RNA synthesis in mouse cells. Examination of infected HeLa cells by electron microscopy demonstrated that human rhinoviruses induced the formation of cytoplasmic membranous vesicles, similar to those observed in cells infected with other picornaviruses. Vesicles were also observed in the cytoplasm of HRV39-infected mouse cells despite the absence of viral RNA replication. Synthesis of picornaviral nonstructural proteins 2C, 2BC, and 3A is known to be required for formation of membranous vesicles. We suggest that productive HRV39 infection is blocked in ICAM-L cells at a step posttranslation and prior to the formation of a functional replication complex. The observation that changes in HRV39 2B and 3A proteins lead to viral growth in mouse cells suggests that one or both of these proteins interact with host cell proteins to promote viral replication.
Picornaviruses are small RNA viruses with a single-stranded, positive-sense genome of approximately 7 to 8 kb enclosed in a nonenveloped, icosahedral capsid. These viruses are responsible for a wide range of diseases including foot-and-mouth disease, hepatitis, poliomyelitis, and the common cold. Following cell entry, the viral genome is translated as a single polyprotein that is co- and posttranslationally cleaved by viral proteases, yielding both precursor proteins and end products. Many of the precursor proteins are essential for virus replication, with functions distinct from the end products (29, 40, 42). Four structural proteins produced from the P1 region of the genome form the viral capsid. Nonstructural proteins of the P2 and P3 region of the genome encode all of the viral proteins responsible for genomic RNA replication.
Viral genome replication occurs on the surface of cytoplasmic vesicles induced by the synthesis of viral proteins 2C and 3A and the precursor 2BC (70). These vesicles are formed in conjunction with massive cell membrane proliferation (4, 7) and increased cellular synthesis of phospholipids (32). With the exception of the two viral proteases 2Apro and 3Cpro, all of the nonstructural proteins have been found on the surface of these vesicles and are thought to interact in the replication complex (26, 59, 61, 70). The vesicles assemble into a rosette, and the replication complex forms on the inner face of the rosette (13, 26). Salt-induced dissociation of the rosette demonstrated that the individual vesicles themselves can also support replication initiation and elongation (26). Neither the composition of the replication complex nor its mechanism of formation has been fully elucidated.
The 2B protein and its cleavage precursor, 2BC, bind membranes and are responsible for an increase in cell membrane permeability that occurs late in infection (3, 76). This increased permeability leads to calcium efflux from the endoplasmic reticulum (ER) and may contribute to release of virus particles from the cell (5). It is thought that 2B acts as a viroporin, oligomerizing and inserting into membranes via its two hydrophobic domains to form a pore (1, 31, 51, 74, 75). When synthesized in uninfected cells, 2B causes a block in protein secretion from the Golgi complex (24). Protein 2C is a multifunctional protein. Amphipathic α-helices at both the N and C termini are thought to mediate a peripheral association of 2C with cell membranes (25, 64) and contribute to its ability to induce vesicles in the cytoplasm (68). Protein 2C also has nucleoside triphosphate-binding domains (56), which are found in the nonstructural proteins of other similar positive-strand RNA viruses such as flaviviruses (28, 41). The flavivirus NS3 protein, which contains the nucleoside triphosphate-binding domain, possesses helicase activity; however, this activity has not been demonstrated for a picornaviral 2C protein (56). Protein 2C also has RNA-binding activity (6, 55, 56) and may have a role in encapsidation of viral RNA (73).
The precise function of 3A is not known; however, 3A has a strongly hydrophobic region near its C terminus that mediates association with membranes and inclusion in the replication complex (72). Amino acid changes in the 3A protein alter host range and tropism of picornaviruses, although how they do so has not been determined (9, 44, 52). Synthesis of 3A alone in uninfected cells leads to a block in protein secretion from the ER and massive ER swelling but not vesicle formation (23, 24). The 3AB protein, a precursor to 3A, may serve as an anchor for the replication complex by binding both viral RNA and membranes (11). Protein 3AB binds 3Dpol and its precursor, 3CDpro, and stimulates the activities of these proteins (10, 43, 53). In addition, it has been postulated that 3AB is the donor for 3B(VPg), the protein primer for positive-strand RNA synthesis. The arrangement of viral proteins in the replication complex is unknown, but it is suspected that many of the nonstructural proteins interact with each other. For example, it has been suggested that proteins 2B and 3A interact (71) and that 2C binds 2B and 3A in the replication complex (65).
Because of the limited coding capacity of viral genomes, many steps of viral replication are dependent on host cell components. Several cellular proteins that interact with the picornaviral genome have been found. Electrophoretic mobility shift assays identified poly(rC) binding protein 2 (14-16) and poly(A) binding protein 1 (62) as viral RNA-binding proteins. Nucleolin was shown to bind to the poliovirus 3′ noncoding region (78) and stimulate viral translation (38). In addition, unr, as well as a unr-interacting protein called unrip, interacts with polypyrimidine tract-binding protein to stimulate picornavirus internal ribosome entry site-mediated translation (17, 36). La autoantigen has been shown to have a role in the stimulation of translation of several picornaviruses (19, 47, 48). The only host cell proteins known to be involved in poliovirus RNA replication are poly(rC) binding protein 2 (79) and poly(A) binding protein 1 (34).
One way to identify viral proteins that interact with host cell proteins is to study viral variants with expanded host range. We have pursued this approach with two serotypes of rhinovirus. Wild-type human rhinovirus type 16 (HRV16) productively infects mouse ICAM-L cells (33), but viral yields are low and cytopathic effect is absent. Variants of HRV16 with improved growth in these cells were selected by alternately passaging the virus between HeLa and mouse L cells producing the viral receptor ICAM-1 (ICAM-L cells) (33). Amino acid changes in the 2C protein of HRV16 conferred an altered growth phenotype, with increased cytopathic effect, elevated levels of viral RNA production, and higher viral titers in mouse cells (33). However, attempts to identify host cell proteins that interact with viral protein 2C were unsuccessful. In contrast to HRV16, human rhinovirus 39 (HRV39) is unable to replicate in mouse cells. Here we describe the adaptation of HRV39 to growth in mouse cells. Changes in the mouse-adapted virus are located in viral proteins that are involved in the formation of the replication complex. The mechanisms of vesicle production and replication complex formation in picornavirus-infected cells have not been defined. The vesicles, which are induced by the synthesis of 2BC, 2C, and 3A proteins, may initially form as COPII vesicles (57), autophagic vesicles (59, 61), or COPI vesicles (30). Regardless of the origin of picornavirus-induced vesicles, some viral proteins must interact with host cell proteins to generate the membrane alterations that occur in the cell and lead to the formation of replication complexes. The adaptation of picornaviruses for growth in nonpermissive host cells can identify viral proteins involved in these processes and will facilitate identification of cellular proteins needed in viral replication.
MATERIALS AND METHODS
Viruses and cell lines.
Mouse L cells stably producing ICAM-1 (ICAM-L cells) were generated and maintained as described previously (33). HeLa R19 cells, which synthesize elevated levels of ICAM-1, were a kind gift from Bert Semler (University of California, Irvine). Plaque assays were performed as previously described (33) using HeLa R19 cells. HRV39 was received from the American Type Culture Collection (Manassas, VA). Virus was plaque purified three times on monolayers of HeLa R19 cells at 33°C before preparation of a stock in the same cells.
Cloning viruses.
Viral RNA was isolated from the supernatant of infected cells or from pelleted virus (40,000 rpm for 120 min in an SW41 rotor) using Trizol reagent (Gibco), according to the manufacturer's instructions. 5′ and 3′ rapid amplification of cDNA ends was performed using the 5′/3′ rapid amplification of cDNA ends kit from Roche (catalog no. 1734792) to determine the nucleotide sequence of the ends of HRV39 RNA. This information was used to design primers to amplify a DNA copy of the genome. The 5′ virus-specific primer includes a SacI site and a T7 RNA polymerase binding site. Reverse transcription was performed using Superscript II (Gibco), and the first-strand DNA was amplified as two half-genome fragments. The 5′ half of the genome (nucleotides 1 to 3800) was amplified with Expand Long Template DNA polymerase (Boehringer Mannheim), cloned into pCR-XL-TOPO (Invitrogen), and excised using EcoRV and HindIII. This fragment was then inserted into pACYC177 cleaved with HindIII/SmaI to generate pACYC177-5′39. The 3′ half of the genome (nucleotides 3400 to 7150) was amplified and inserted into pCR-XL-TOPO and excised with PpuMI/BciVI. This fragment was inserted into pACYC177-5′39 that had also been cleaved with PpuMI/BciVI to produce pACYC177-39. This procedure was also used to produce a DNA copy of the genome of RV39/L (pACYC177-39/L). The plasmids were linearized by cleavage with SphI, and infectious RNA was generated using in vitro transcription with T7 RNA polymerase (Promega). Multiple attempts to insert a full-length DNA copy of the viral genome into a higher-copy-number vector (pUC19) always resulted in deletions or alterations in the genome after bacterial propagation that rendered the viral transcripts noninfectious in mammalian cells.
Isolation of RV39/IGE and RV39/RM.
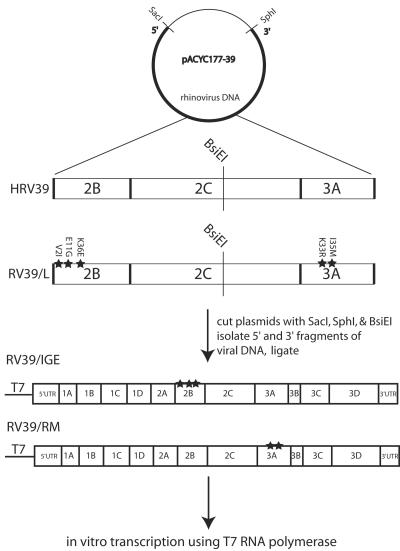
DNA from both pACYC177-39 and pACYC177-39/L was cut at the unique restriction site BsiEI, located between the 2B and 3A proteins (Fig. 3). The 5′ portion of HRV39 DNA was ligated to the 3′ portion of RV39/L DNA, and the 5′ portion of RV39/L DNA was ligated to the 3′ portion of HRV39 DNA. The resulting DNAs were transcribed in vitro. Infectious viruses containing either the 2B changes alone (RV39/IGE) or the 3A changes alone (RV39/RM) were isolated after transfection of viral RNA transcripts and plaque purification on HeLa R19 cells. Total RNA was extracted from the infected cell supernatant, and the 2BC3AB region was amplified using reverse transcription-PCR (RT-PCR). The nucleotide sequence of PCR products was determined to ensure that changes were present only in the 2B or the 3A coding region and that the sample was free of contaminating wild-type HRV39 or RV39/L.
FIG. 3.
Schematic for generation of viral transcripts containing mouse-adapted amino acid changes in either the 2B or 3A protein. A DNA copy of the HRV39 and RV39/L viral genomes was cloned into pACYC177. The plasmids were cleaved with BsiEI and SacI or BsiEI and SphI, and DNA fragments containing (i) the T7 promoter and the 5′ half of the viral cDNA or (ii) the 3′ half of the viral cDNA and the poly(A) sequence were purified. Ligations of the 5′ half of HRV39 DNA and the 3′ half of RV39/L DNA or the 5′ half of HRV39/L DNA and the 3′ half of HRV39 DNA resulted in full-length DNAs with changes in either 2B or 3A. RNA transcripts were generated by in vitro transcription, and the RNA was introduced into cells by transfection to produce virus.
Electron microscopy.
To prepare samples for electron microscopy, cells were either infected or mock infected, the infections were stopped at the indicated times, and cells were fixed with 6% glutaraldehyde (Ladd Research) for 30 min on ice. Cells were then pelleted, resuspended in 2% osmium tetroxide (Ladd Research) for 1 hour, and enrobed in 0.4% agar. An acetone dehydration series (30% to 100%) was used to dry the cell pellet, and the pellet was embedded in Epon resin (Energy Beam Sciences; no. EK-TT), following the manufacturer's directions for a medium hard block. Ultrathin sections, collected on uncoated 200-mesh copper grids, were obtained using a Porter-Blum MT-2 ultramicrotome fitted with a diamond knife. The sections were stained with Reynold's alkaline lead citrate and observed with a Philips TEM 201 electron microscope operated at an accelerating voltage of 60 kV. Images captured on black-and-white film were digitally scanned and assembled using Adobe Photoshop.
RNA analysis.
Dot blot assays were performed as described previously (33). Briefly, replica plates were infected at a multiplicity of 10 PFU per cell, and total cellular RNA was isolated with Trizol (Gibco) at 0, 6, and 12 h postinfection. The RNA was transferred onto a nitrocellulose membrane and cross-linked, and the blot was hybridized to a 32P-labeled RNA probe. The hybridization probe included nucleotides 1600 to 7150 of the HRV39 sequence and was complementary to positive-strand viral RNA. The blot was exposed to a PhosphorImager, and RNA levels were quantitated with ImageQuant software (Molecular Dynamics).
Viral growth curve analysis.
Growth curves were performed by infecting duplicate plates of 105 ICAM-L or HeLa cells at a multiplicity of 10 PFU/cell. Infections were stopped at various times postinfection, HEPES buffer (Gibco) was added to a final concentration of 50 mM, and the cells were frozen. After 48 h, all samples were frozen and thawed three times and centrifuged, and viral titers in the supernatants were determined by plaque assay. Plaque assays were performed as described previously (33).
Nucleotide sequence accession number.
The complete sequence of HRV39 has been deposited in NCBI (accession number AY751783).
RESULTS
Isolation of a rhinovirus type 39 mutant that replicates in mouse cells.
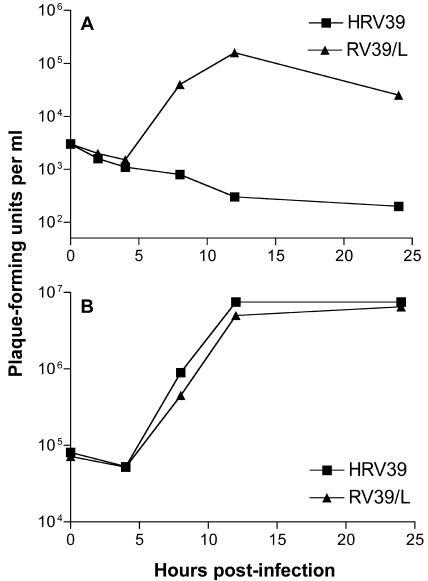
HRV39 replicates in human cells, but replication cannot be detected in mouse cells (Fig. 1) (33, 46). Selection of an HRV39 variant that replicates in ICAM-L cells was accomplished by alternately passaging the virus between HeLa and ICAM-L cells. A gradual increase in virus titers following each passage in ICAM-L cells was observed. The mouse-adapted strain of HRV39, called RV39/L, was isolated after approximately 15 alternate passages. Infection with RV39/L produced titers of 105 to 106 PFU/ml in ICAM-L cells, and in HeLa cells RV39/L replicated to similar titers as HRV39 (Fig. 1).
FIG. 1.
Growth of HRV39 and RV39/L in cultured cells. Wild-type HRV39 or RV39/L was used to infect (A) ICAM-L cells or (B) HeLa R19 cells at a multiplicity of infection of 10. Infections were halted at the indicated times postinfection, and viral titers were determined by plaque assay.
Changes in protein 2B partially mediate adaptation to mouse cells.
Full-length DNA copies of both HRV39 and the adapted RV39/L genomes were produced from viral RNA. HRV39 and RV39/L viral RNAs produced by in vitro transcription were infectious in HeLa R19 cells; however, only viral transcripts of RV39/L produced virus when transfected into ICAM-L cells (data not shown). Comparison of HRV39 and RV39/L sequences revealed five amino acid differences, located in the 2B and 3A proteins (Fig. 2). No other sequence changes were found in coding or noncoding regions.
FIG. 2.
Multiple alignment of picornaviral 2B and 3A proteins. The boxed regions in both proteins represent hydrophobic domains. Dashed boxes in 3A correspond to the 3A homodimer interface. Amino acid differences between HRV39 and RV39/L proteins are indicated by underlined residues in the HRV39 sequence. The amino acid changes in RV39/L are shown above the HRV39 residue.
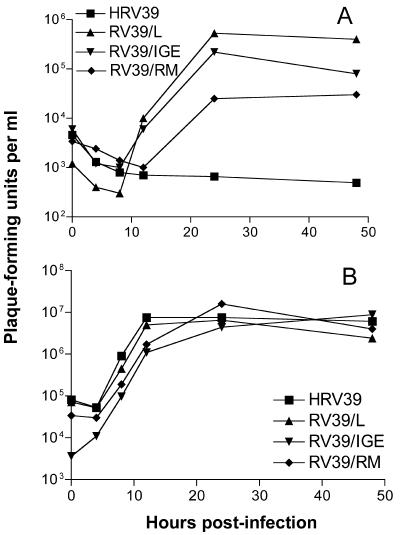
To determine if changes in 2B, 3A, or both were responsible for the adaptation to mouse cells, viruses were produced that contained only the alterations in either 2B or 3A (Fig. 3). Viruses containing either the 2B changes alone (RV39/IGE) or the 3A changes alone (RV39/RM) were isolated after transfection of viral RNA into HeLa R19 cells and plaque purification. The 2BC3AB region of each viral genome was amplified using RT-PCR and sequenced to ensure that the changes were present in only the 2B or the 3A protein and the virus stock was free of contaminating wild-type HRV39 or RV39/L. One-step growth curve analysis in ICAM-L cells indicated that RV39/IGE replicated with kinetics similar to those for RV39/L; however, final virus yields were slightly lower (Fig. 4A). RV39/RM also replicated in mouse cells, although to lower titers than RV39/L and RV39/IGE. However, sequence analysis of RV39/RM viral RNA at 24 h postinfection in ICAM-L cells revealed the presence of a mixed population of viruses (Table 1). Approximately 30% of the RT-PCR product amplified from RV39/RM viral RNA contained a K36N coding change in protein 2B. Amino acid 36 of 2B is also altered in RV39/L (Fig. 2). It is possible that viruses containing the K36N change contributed to the limited growth of RV39/RM observed in ICAM-L cells (see next paragraph and Discussion). None of the other viruses had coding changes at 24 h postinfection in ICAM-L cells (Table 1). The most robust growth in ICAM-L cells was observed during infection with RV39/L, which contains changes in both proteins. All four viruses replicated with similar kinetics in HeLa R19 cells, demonstrating that amino acid changes in 2B and 3A do not confer a general replication advantage (Fig. 4B).
FIG. 4.
Growth of HRV39, RV39/L, RV39/IGE, and RV39/RM in cultured cells. Infections were carried out in (A) ICAM-L cells or (B) HeLa R19 cells at a multiplicity of infection of 10. Infections were halted at the indicated times postinfection, and viral titers were determined by plaque assay.
TABLE 1.
Amino acid changes in 2B and 3A of HRV39 during growth in ICAM-L cells
| Virus | Viral protein | Amino acid
|
||
|---|---|---|---|---|
| Residue | 0 hpia | 24 hpi | ||
| HRV39 | 2B | 2 | V | V |
| 11 | E | E | ||
| 36 | K | K | ||
| 3A | 33 | K | K | |
| 35 | I | I | ||
| RV39/L | 2B | 2 | I | I |
| 11 | G | G | ||
| 36 | E | E | ||
| 3A | 33 | R | R | |
| 35 | M | M | ||
| RV39/IGE | 2B | 2 | I | I |
| 11 | G | G | ||
| 36 | E | E | ||
| 3A | 33 | K | K | |
| 35 | I | I | ||
| RV39/RM | 2B | 2 | V | V |
| 11 | E | E | ||
| 36 | K | 0.7 K/0.3 Nb | ||
| 3A | 33 | R | R | |
| 35 | M | 0.5 I/0.5 Mb | ||
hpi, hour(s) postinfection.
Predicted fraction of each amino acid determined by measuring the peak heights on traces derived from nucleotide sequence analysis of RT-PCR products.
Selective pressure for amino acid changes in 2B during infection of ICAM-L cells.
It has been suggested that changes in P2 proteins mediate adaptation of HRV39 to mouse cells and that these changes arise sporadically during viral passage in HeLa cells (46). Although our initial stock of HRV39 was unable to replicate in mouse cells, we subsequently observed low-level replication of HRV39 in ICAM-L cells with a different viral stock. Determination of the nucleotide sequence of the P2 and P3 regions of this virus 24 h after infection of ICAM-L cells revealed a single amino acid change at the K36 residue of 2B but no changes elsewhere. Approximately 50% of the RT-PCR product amplified from viral RNA contained a mutation predicted to change amino acid 36 from K to E, which is identical to the K36 change in protein 2B of RV39/L (Fig. 2). Although this coding change was not detected in viral RNA from the virus stock used to infect ICAM-L cells, it was likely present in a small fraction of the stock virus population and was selected during infection. These findings strongly suggest that the K36 residue of 2B is critical for the adaptation of HRV39 to mouse cells.
Replication of HRV39/L but not HRV39 viral RNA early in infection.
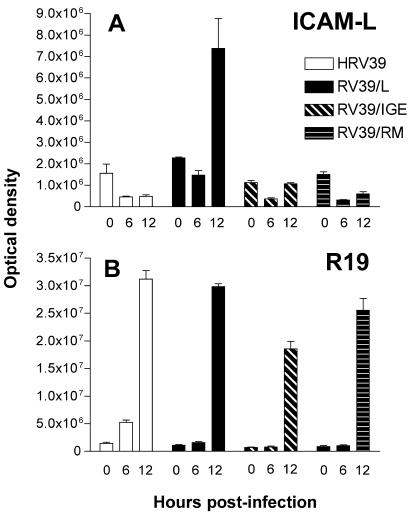
To identify the step at which HRV39 replication is blocked in ICAM-L cells, we examined positive-strand viral RNA synthesis in infected cells by dot blot hybridization analysis. While all viruses produced significant levels of viral RNA by 12 h postinfection in HeLa R19 cells, high levels of positive-strand RNA in ICAM-L cells were observed only during infection with RV39/L (Fig. 5A and B). Although both RV39/IGE and RV39/RM had begun producing positive-strand viral RNA at 12 h postinfection, neither virus produced as much viral RNA as RV39/L. These observations provide additional support for a role for both the 2B and the 3A proteins in adaptation of HRV39 to mouse cells. The results also demonstrate that HRV39 replication is blocked in ICAM-L cells at a step prior to positive-strand viral RNA production.
FIG. 5.
Positive-strand viral RNA production in infected cells. Infections were carried out in (A) ICAM-L cells or (B) HeLa R19 cells at a multiplicity of infection of 10. At 0, 6, and 12 h postinfection, total RNA was isolated from infected cells and transferred onto a nitrocellulose membrane. A radiolabeled RNA hybridization probe complementary to positive-strand viral RNA was used to measure levels of viral RNA present at various times postinfection. The amount of hybridized RNA probe was determined with a PhosphorImager, analyzed using ImageQuant software, and reported as optical density.
Both HRV39 and RV39/L induce membrane alterations in infected cells.
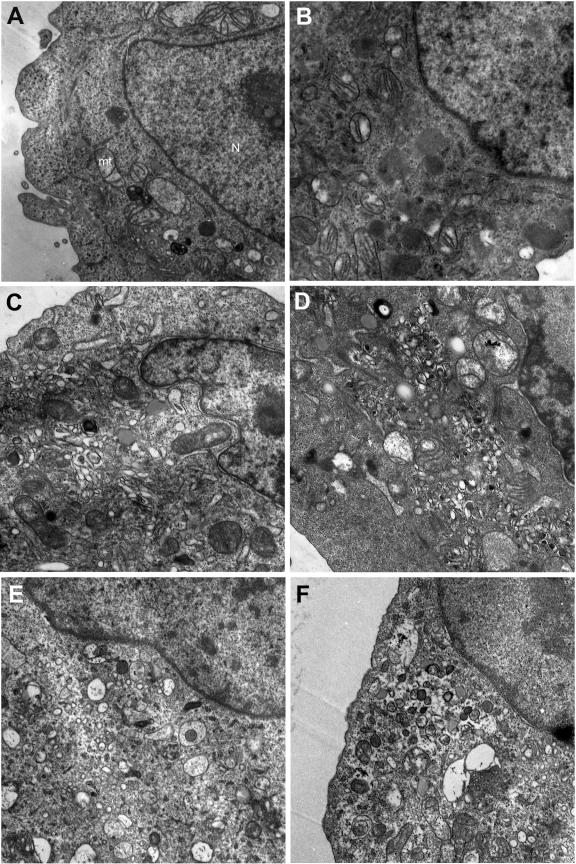
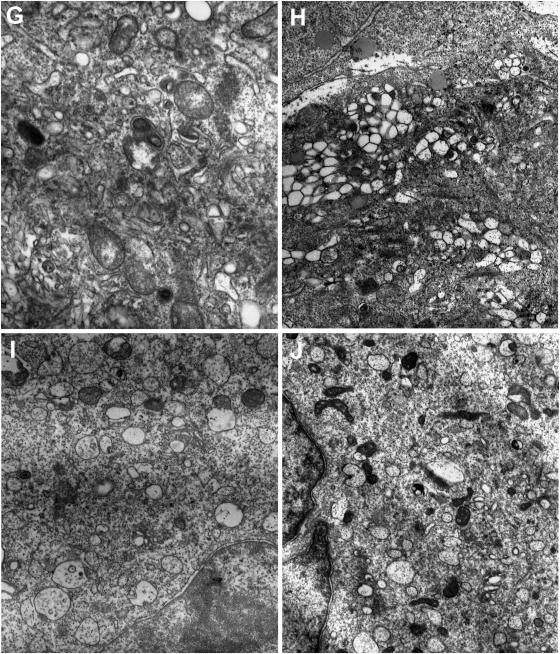
Cytoplasmic vesicles are induced by the synthesis of the 2BC, 2C, and 3A proteins of picornaviruses, and these vesicles are critical for viral replication (61, 66, 68). However, ultrastructural changes have not been described in rhinovirus-infected cells. To determine if these morphological changes occur during HRV39 infection, we examined infected HeLa R19 and ICAM-L cells by electron microscopy. Substantial vesicle formation and concomitant destruction of intracellular membranes were observed throughout the cytoplasm of HRV39- and RV39/L-infected HeLa R19 cells (Fig. 6C, D, G, and H; compare to Fig. 6A). Because HRV39 does not replicate in mouse cells, it was of interest to determine whether this virus could induce vesicle formation in ICAM-L cells. To our surprise, infection of ICAM-L cells with both HRV39 and RV39/L leads to vesicle formation (Fig. 6E, F, I, and J; compare to Fig. 6B). However, the vesicles formed in infected ICAM-L cells appear different from those in HeLa R19 cells. In ICAM-L cells (Fig. 6I and J), virus-induced vesicles are less clumped and more dispersed than those in HeLa R19 cells (Fig. 6G and H). These data demonstrate that, although RNA replication does not occur during HRV39 infection in ICAM-L cells, the 2BC and 3A proteins are competent to induce cytoplasmic membranous vesicles.
FIG. 6.
Formation of membranous vesicles in infected cells visualized by electron microscopy. HeLa R19 cells are (A) uninfected or infected with (C and G) HRV39 or (D and H) RV39/L. ICAM-L cells were (B) uninfected or infected with (E and I) HRV39 or (F and J) RV39/L.
DISCUSSION
The replication cycle of picornaviruses occurs entirely in the cytoplasm. Infection comprises many processes that are poorly understood, including the induction of increased phospholipid synthesis, a block in protein secretion from the ER and Golgi complex, the formation of cytoplasmic vesicles which harbor the viral replication complex, and alterations in intracellular calcium content in the cell (5, 23, 24, 32, 37, 76). While the viral proteins responsible for some of these changes have been identified, their mechanisms of action are not known. Many consequences of viral infection are likely to be effected by interactions between viral and host cell proteins. Despite this assumption, very few such interactions have been identified.
To identify viral proteins that might interact with cell proteins, we selected a variant of HRV39 that replicates in mouse cells. Although replication of this serotype cannot be detected in mouse cells, stocks of this virus may contain low levels of viral variants that are able to grow in mouse cells. These variants are presumably selected upon passage in ICAM-L cells. The genome of the mouse-adapted virus RV39/L encodes amino acid changes in both the 2B and the 3A proteins. Previously, human rhinovirus 2 (HRV2), a minor-group serotype of rhinovirus related to HRV39, was adapted to mouse cells (81). In infected cells, the adapted virus RV2/L produced P2 proteins that migrate with altered mobility on protein gels compared with P2 proteins of HRV2. Nucleotide sequence analysis of RNA encoding 2ABC3AB revealed 10 coding changes in the genome of RV2/L, three of which are identical to changes seen in RV39/L (V2I and K36E of 2B and I35M of 3A) (Fig. 2) (F. Yin, personal communication). RV2/L also exhibited altered sensitivity to inhibitors that specifically block viral RNA replication, leading the authors to suggest that the process of viral RNA replication was altered in an unknown manner. The influence of the individual amino acid changes of RV2/L in the adaptation to mouse cells has not been determined. The same group subsequently isolated mouse-adapted variants of HRV39 by transfection of viral RNA in L cells (46). Of three independent virus stocks used, only one produced infectious virus after RNA transfection. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis indicated that a P2 protein of the virus that replicated in mouse cells migrated differently from that of the other two virus stocks. Nucleotide sequence from the adapted virus was not obtained. The authors suggested that a P2 protein was an essential viral component that could function in mouse L cells only in an altered form and that genetic changes leading to this altered form arose spontaneously during viral growth in permissive human cells (46). A P2 protein was also implicated in the adaptation of HRV16 to growth in mouse cells (33). Amino acid changes needed for replication of HRV16 in mouse cells are located in protein 2BC and appear to cause conformational changes in the viral protein that may influence interaction with a host cell protein.
To identify the specific protein(s) involved in our adaptation of HRV39 to mouse cells, viruses with the amino acid changes of RV39/L in either the 2B or 3A protein were produced (Fig. 3). The results of one-step growth curve analysis in ICAM-L cells indicated that RV39/IGE replicated with kinetics similar to those of RV39/L (Fig. 4). While the titers of both RV39/IGE and RV39/L reached similar levels by 12 h postinfection, the titer of RV39/IGE did not exceed the starting titer, while RV39/L titers increased 10-fold. These differences were reflected in the results of RNA analysis: at 12 h postinfection, the amount of RV39/IGE RNA did not exceed starting levels, while the amount of RV39/L viral RNA increased (Fig. 5). However, despite the large difference in viral RNA levels at 12 h postinfection, titers of RV39/L and RV39/IGE were nearly identical. The reason for this observation is not known, but one possible explanation is that RNA packaging or assembly of RV39/L virions is affected by the amino acid changes in 2B and/or 3A. Overall, the growth curve and RNA analyses indicate that, while the amino acid changes in 2B contribute significantly to the adaptation to mouse cells, the additional changes in 3A are required for complete adaptation. In particular, changes in the K36 residue of 2B seem to be important in mouse cell adaptation of HRV39. The similar growth in human cells of viruses with amino acid changes in either the 2B or 3A protein confirmed that the adaptation was specific to mouse cells (Fig. 4B). Analysis of viral RNA production in mouse cells demonstrated that amino acid changes in both 2B and 3A were required to achieve comparable levels of viral positive-strand genome production to that seen in cells infected with RV39/L. RNA levels could not be determined at later times postinfection due to the presence of substantial cytopathic effect.
There are two hydrophobic regions in 2B that are both important in the functions of the protein (75) (Fig. 2). HR1, the more amino-terminal hydrophobic region, is a cationic amphipathic α-helix thought to span the membrane. The second hydrophobic region, HR2, is a transmembrane domain. Both HR2 and the hydrophilic region between HR1 and HR2 have been shown to be important in multimerization of coxsackievirus B3 2B (21). This viral protein localizes to the Golgi complex in COS-1 cells: deletion of HR1 changes the localization to the ER (22), although HR2 is also important in proper localization. Other studies have demonstrated that 2B localizes to the ER and the Golgi complex (58). The properties of 2B suggest that it functions as a viroporin, oligomerizing and inserting into membranes to create pores (31). The formation of these pores may explain the increase in intracellular calcium observed in the cytoplasm of infected cells by allowing the passage of calcium from either the ER lumen or the extracellular milieu (37, 76). Another known effect of 2B (and 2BC) synthesis is to block protein export from the Golgi complex (8, 24). At least the N-terminal 30 amino acids of 2BC are likely to be involved in this function (7). Alterations in the 2B protein that impair virus growth but do not affect the ability of the protein to permeabilize membranes or block protein secretion have been described, suggesting additional functions for 2B (77).
Three amino acid changes were identified in the 2B protein of our mouse-adapted RV39/L, including one amino acid change—K36E—in HR1. This residue is a glutamate in several of the picornaviruses shown in Fig. 2. Of the viruses with a glutamate residue at this position, all are able to grow in mouse cells (2, 50, 54, 82), suggesting that this residue may be critical in determining host range. This amino acid is also altered from a K to an E in mouse-adapted HRV2/L (F. Yin, personal communication). The other two amino acid changes in protein 2B of RV39/L, V2I and E11G, do not appear to exhibit any particular pattern in the other viruses that would suggest an obvious role in host range (Fig. 2). It seems unlikely that any amino acid change involved in host range expansion would affect a critical function such as multimerization, pore formation, or inhibition of protein secretion. However, these activities may require a host protein, and the K36 residue could be critical in the interaction between 2B and a host protein.
Considerable evidence implicates the 3A protein and its precursor, 3AB, in RNA replication (10, 35, 53, 69, 80). A strongly hydrophobic region in the C terminus of 3A is involved in membrane association and is likely to mediate inclusion of 3A in the replication complex (45, 72) (Fig. 2). Recently, the solution structure of the soluble N-terminal domain of poliovirus 3A was determined, showing that 3A forms homodimers with unstructured N- and C-terminal regions in each monomer (60). The G41 residue of poliovirus 3A, immediately following the last residue in the second of two amphipathic α-helices, corresponds to the K33 residue of rhinovirus 39 3A (Fig. 2). The W42 residue of poliovirus 3A, which acts in concert with a number of other N-terminal residues to “bury” the dimer interface, is directly adjacent to the rhinovirus I35 residue. While these regions are not directly involved in dimerization of poliovirus 3A, they do contribute to 3A structure. It was suggested that long-range contacts between G41 or its neighbor, W42, and N-terminal residues mediate the structure of poliovirus 3A. Given these data, it seems likely that the amino acid changes observed in RV39/L 3A cause conformational changes in the protein that may alter its interactions with host cell proteins or other viral proteins. Evidence for possible interaction between 2B and 3A comes from studies of chimeric viruses with the hydrophobic domain of poliovirus 3A exchanged for that of rhinovirus 3A. These chimeric viruses are viable but do not replicate as well as wild-type virus. Viruses with improved replication were isolated that contain compensatory changes in the 2B protein, suggesting that 2B and 3A may interact during infection (71). Recent data also suggest interactions between 2B, 2C, and 3A (65).
We observed selection for an amino acid change at residue K36 of 2B protein during growth of HRV39 in mouse cells (Table 1). However, changes in 2B do not fully account for the adaptation of RV39/L to mouse cells: RV39/IGE does not grow as well as RV39/L in mouse cells, nor does it produce the same amount of RNA as RV39/L by 12 h postinfection (Fig. 5). Although RV39/RM did not replicate as well as RV39/IGE, in the presence of the 2B changes, alterations in 3A clearly enhance viral replication in mouse cells. How do these changes facilitate adaptation to mouse cells? It is possible that the interaction of 2B (or a 2B precursor) with a host cell protein mediates growth in mouse cells. If contacts also occur between 2B and 3A, amino acid changes in 3A could facilitate improved interactions with 2B protein of mouse-adapted RV39/L. We were unable to demonstrate interaction between HRV39 2B and 3A using the yeast two-hybrid system (data not shown), but this result should not be considered definitive evidence of a lack of interaction during infection. Alternatively, 2B and 3A might interact with the same host cell protein, or with two different proteins. In either case, the interaction of 2B with its putative cellular partner would have a greater influence on growth in mouse cells than the interaction of 3A and its cellular partner.
Most of what is known about virus-induced alterations of cell membranes comes from studies of poliovirus, the prototype picornavirus. The most drastic structural change observed in poliovirus-infected cells is the loss of intact ER and Golgi complex and the accumulation of membranous vesicles in the cytoplasm (18). Most of the nonstructural proteins and some nonstructural protein precursors have been found on the surface of these vesicles (12, 26, 59, 61, 70). The synthesis of 2BC and 3A in uninfected cells leads to the formation of vesicles identical in buoyant density and appearance to those present during poliovirus infection (61). The vesicles include markers from different organelles, including the ER and the Golgi complex (23, 57, 58). The requirement for de novo phospholipid synthesis for poliovirus replication suggests that vesicle membranes may be newly synthesized rather than exclusively derived from preexisting membranes (32).
There is controversy over the origin of the vesicles induced by poliovirus infection. Many of the vesicles found in poliovirus-infected cells are double membraned, originate from multiple organelles, and have cytosolic content, suggesting a mechanism of formation similar to cellular autophagy (59, 61). However, recent evidence demonstrated that poliovirus-induced vesicles are morphologically similar to COPII vesicles, which are involved in anterograde transport (57). The COPII proteins Sec13/31 were shown to colocalize with the viral 2B protein on the surface of the vesicles. The authors found that viral vesicles bud from the ER but do not fuse with the ER/Golgi intermediate complex and suggested that the cellular COPII machinery participates in the induction of vesicles suitable for poliovirus replication. However, the inhibition of poliovirus and rhinovirus RNA replication by brefeldin A directly contradicts this hypothesis (20, 30). Brefeldin A inhibits formation of COPI but not COPII vesicles. In addition, overproduction of dominant-negative Sar1, a COPII component, was found to block anterograde transport but not poliovirus replication (63). These observations argue against a COPII origin for poliovirus vesicles.
The vesicles formed by HRV39 in ICAM-L and HeLa R19 cells appear different from each other when examined by electron microscopy. The vesicles in HeLa R19 cells are densely packed, similar to vesicles observed during infection with other picornaviruses (12, 27). The vesicle shape is elongated and irregular, and the vesicles appear to contain electron-dense material. Vesicles induced in ICAM-L cells are mostly round, larger, and more dispersed. Most of the rhinovirus-induced vesicles appear to have single membranes, in contrast to poliovirus-induced vesicles. However, the formation of predominantly single-membraned vesicles has been reported for other picornaviruses (49), and vesicles of this morphology have also been observed during poliovirus infection (61).
Despite the absence of detectable HRV39 RNA replication in mouse cells (Fig. 5), vesicles are formed during HRV39 infection (Fig. 6E and I). This observation suggests that the viral proteins translated from genomes that initially enter the cell are able to induce vesicles in mouse cells. Therefore, interactions of wild-type viral proteins with membranes or with host cell proteins required for vesicle formation are presumably not compromised in mouse cells. The absence of HRV39 viral replication in mouse cells demonstrates that HRV39 must be blocked at a step subsequent to vesicle formation but prior to the initiation of replication, such as replication complex assembly or RNA synthesis. The replication complex is believed to form in cis, coupling vesicle formation, genome translation, and viral RNA replication (67). Preformed vesicles are unable to support replication of superinfecting poliovirus (27), and trans-complementation with nonstructural viral proteins is extremely limited (39, 40, 70). Our data suggest that the vesicle-forming functions of the 2B and 3A proteins (or their precursors) are not linked to their ability to facilitate replication. The viral precursor proteins may recruit cellular proteins at a step prior to the formation of vesicles that either initiates or nucleates replication complex formation. Alternatively, replication complexes may form and subsequently require cell proteins to render them replication competent. As wild-type HRV39 is able to replicate efficiently in HeLa R19 cells, the block to HRV39 growth in mouse cells cannot be due to viral cis-acting factors. Host cell proteins provided in trans must play a role in the formation of a functional replication complex. Efforts to isolate these proteins are under way.
Acknowledgments
This work was supported by Public Health Service grant AI50754 from the National Institute of Allergy and Infectious Diseases.
We thank O. Roger Anderson, Lamont-Doherty Earth Observatory of Columbia University, for electron microscopy and helpful suggestions.
REFERENCES
- 1.Agirre, A., A. Barco, L. Carrasco, and J. L. Nieva. 2002. Viroporin-mediated membrane permeabilization. Pore formation by nonstructural poliovirus 2B protein. J. Biol. Chem. 277:40434-40441. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J., J. Choi, C. H. Joo, I. Seo, D. Kim, S. Y. Yoon, Y. K. Kim, and H. Lee. 2004. Susceptibility of mouse primary cortical neuronal cells to coxsackievirus B. J. Gen. Virol. 85:1555-1564. [DOI] [PubMed] [Google Scholar]
- 3.Aldabe, R., A. Barco, and L. Carrasco. 1996. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 271:23134-23137. [DOI] [PubMed] [Google Scholar]
- 4.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 5.Aldabe, R., A. Irurzun, and L. Carrasco. 1997. Poliovirus protein 2BC increases cytosolic free calcium concentrations. J. Virol. 71:6214-6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barco, A., and L. Carrasco. 1995. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 14:3349-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barco, A., and L. Carrasco. 1998. Identification of regions of poliovirus 2BC protein that are involved in cytotoxicity. J. Virol. 72:3560-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beard, C. W., and P. W. Mason. 2000. Genetic determinants of altered virulence of Taiwanese foot-and-mouth disease virus. J. Virol. 74:987-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beneduce, F., A. Ciervo, Y. Kusov, V. Gauss-Muller, and G. Morace. 1999. Mapping of protein domains of hepatitis A virus 3AB essential for interaction with 3CD and viral RNA. Virology 264:410-421. [DOI] [PubMed] [Google Scholar]
- 11.Beneduce, F., A. Ciervo, and G. Morace. 1997. Site-directed mutagenesis of hepatitis A virus protein 3A: effects on membrane interaction. Biochim. Biophys. Acta 1326:157-165. [DOI] [PubMed] [Google Scholar]
- 12.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 13.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blyn, L. B., R. Chen, B. L. Semler, and E. Ehrenfeld. 1995. Host cell proteins binding to domain IV of the 5′ noncoding region of poliovirus RNA. J. Virol. 69:4381-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boussadia, O., M. Niepmann, L. Creancier, A. C. Prats, F. Dautry, and H. Jacquemin-Sablon. 2003. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 77:3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 19.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 24:6861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuconati, A., A. Molla, and E. Wimmer. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jong, A. S., I. W. Schrama, P. H. Willems, J. M. Galama, W. J. Melchers, and F. J. van Kuppeveld. 2002. Multimerization reactions of coxsackievirus proteins 2B, 2C and 2BC: a mammalian two-hybrid analysis. J. Gen. Virol. 83:783-793. [DOI] [PubMed] [Google Scholar]
- 22.de Jong, A. S., E. Wessels, H. B. Dijkman, J. M. Galama, W. J. Melchers, P. H. Willems, and F. J. van Kuppeveld. 2003. Determinants for membrane association and permeabilization of the coxsackievirus 2B protein and the identification of the Golgi complex as the target organelle. J. Biol. Chem. 278:1012-1021. [DOI] [PubMed] [Google Scholar]
- 23.Doedens, J. R., T. H. Giddings, Jr., and K. Kirkegaard. 1997. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J. Virol. 71:9054-9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14:894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 26.Egger, D., L. Pasamontes, R. Bolten, V. Boyko, and K. Bienz. 1996. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J. Virol. 70:8675-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallivan, J. P., and M. J. McGarvey. 2003. The importance of the Q motif in the ATPase activity of a viral helicase. FEBS Lett. 554:485-488. [DOI] [PubMed] [Google Scholar]
- 29.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gazina, E. V., J. M. Mackenzie, R. J. Gorrell, and D. A. Anderson. 2002. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J. Virol. 76:11113-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez, M. E., and L. Carrasco. 2003. Viroporins. FEBS Lett. 552:28-34. [DOI] [PubMed] [Google Scholar]
- 32.Guinea, R., and L. Carrasco. 1990. Phospholipid biosynthesis and poliovirus genome replication, two coupled phenomena. EMBO J. 9:2011-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris, J. R., and V. R. Racaniello. 2003. Changes in rhinovirus protein 2C allow efficient replication in mouse cells. J. Virol. 77:4773-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus 3D polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunt, S. L., J. J. Hsuan, N. Totty, and R. J. Jackson. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irurzun, A., J. Arroyo, A. Alvarez, and L. Carrasco. 1995. Enhanced intracellular calcium concentration during poliovirus infection. J. Virol. 69:5142-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izumi, R. E., B. Valdez, R. Banerjee, M. Srivastava, and A. Dasgupta. 2001. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 76:17-29. [DOI] [PubMed] [Google Scholar]
- 39.Johnson, K. L., and P. Sarnow. 1991. Three poliovirus 2B mutants exhibit noncomplementable defects in viral RNA amplification and display dosage-dependent dominance over wild-type poliovirus. J. Virol. 65:4341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jurgens, C., and J. B. Flanegan. 2003. Initiation of poliovirus negative-strand RNA synthesis requires precursor forms of P2 proteins. J. Virol. 77:1075-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuang, W. F., Y. C. Lin, F. Jean, Y. W. Huang, C. L. Tai, D. S. Chen, P. J. Chen, and L. H. Hwang. 2004. Hepatitis C virus NS3 RNA helicase activity is modulated by the two domains of NS3 and NS4A. Biochem. Biophys. Res. Commun. 317:211-217. [DOI] [PubMed] [Google Scholar]
- 42.Kusov, Y., and V. Gauss-Muller. 1999. Improving proteolytic cleavage at the 3A/3B site of the hepatitis A virus polyprotein impairs processing and particle formation, and the impairment can be complemented in trans by 3AB and 3ABC. J. Virol. 73:9867-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lama, J., A. V. Paul, K. S. Harris, and E. Wimmer. 1994. Properties of purified recombinant poliovirus protein 3AB as substrate for viral proteinases and as co-factor for RNA polymerase 3Dpol. J. Biol. Chem. 269:66-70. [PubMed] [Google Scholar]
- 44.Lama, J., M. A. Sanz, and L. Carrasco. 1998. Genetic analysis of poliovirus protein 3A: characterization of a non-cytopathic mutant virus defective in killing Vero cells. J. Gen. Virol. 79:1911-1921. [DOI] [PubMed] [Google Scholar]
- 45.Liu, J., T. Wei, and J. Kwang. 2004. Membrane-association properties of avian encephalomyelitis virus protein 3A. Virology 321:297-306. [DOI] [PubMed] [Google Scholar]
- 46.Lomax, N. B., and F. H. Yin. 1989. Evidence for the role of the P2 protein of human rhinovirus in its host range change. J. Virol. 63:2396-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meerovitch, K., J. Pelletier, and N. Sonenberg. 1989. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 3:1026-1034. [DOI] [PubMed] [Google Scholar]
- 48.Meerovitch, K., Y. V. Svitkin, H. S. Lee, F. Lejbkowicz, D. J. Kenan, E. K. Chan, V. I. Agol, J. D. Keene, and N. Sonenberg. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J. Virol. 67:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monaghan, P., H. Cook, T. Jackson, M. Ryan, and T. Wileman. 2004. The ultrastructure of the developing replication site in foot-and-mouth disease virus-infected BHK-38 cells. J. Gen. Virol. 85:933-946. [DOI] [PubMed] [Google Scholar]
- 50.Nadkarni, S. S., and J. M. Deshpande. 2003. Recombinant murine L20B cell line supports multiplication of group A coxsackieviruses. J. Med. Virol. 70:81-85. [DOI] [PubMed] [Google Scholar]
- 51.Nieva, J. L., A. Agirre, S. Nir, and L. Carrasco. 2003. Mechanisms of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett. 552:68-73. [DOI] [PubMed] [Google Scholar]
- 52.Nunez, J. I., E. Baranowski, N. Molina, C. M. Ruiz-Jarabo, C. Sanchez, E. Domingo, and F. Sobrino. 2001. A single amino acid substitution in nonstructural protein 3A can mediate adaptation of foot-and-mouth disease virus to the guinea pig. J. Virol. 75:3977-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plotch, S. J., and O. Palant. 1995. Poliovirus protein 3AB forms a complex with and stimulates the activity of the viral RNA polymerase, 3Dpol. J. Virol. 69:7169-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Racaniello, V. R., R. Ren, and M. Bouchard. 1993. Poliovirus attenuation and pathogenesis in a transgenic mouse model for poliomyelitis. Dev. Biol. Stand. 78:109-116. [PubMed] [Google Scholar]
- 55.Rodriguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez, P. L., and L. Carrasco. 1993. Poliovirus protein 2C has ATPase and GTPase activities. J. Biol. Chem. 268:8105-8110. [PubMed] [Google Scholar]
- 57.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandoval, I. V., and L. Carrasco. 1997. Poliovirus infection and expression of the poliovirus protein 2B provoke the disassembly of the Golgi complex, the organelle target for the antipoliovirus drug Ro-090179. J. Virol. 71:4679-4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strauss, D. M., L. W. Glustrom, and D. S. Wuttke. 2003. Towards an understanding of the poliovirus replication complex: the solution structure of the soluble domain of the poliovirus 3A protein. J. Mol. Biol. 330:225-234. [DOI] [PubMed] [Google Scholar]
- 61.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Svitkin, Y. V., H. Imataka, K. Khaleghpour, A. Kahvejian, H. D. Liebig, and N. Sonenberg. 2001. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7:1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor, M. P., W. D. Heo, T. Meyer, and K. Kirkegaard. 2004. Presented at the Seventh International Symposium on Positive Strand RNA Viruses, San Francisco, CA, 27 May to 1 June 2004.
- 64.Tershak, D. R. 1984. Association of poliovirus proteins with the endoplasmic reticulum. J. Virol. 52:777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teterina, N., M. S. Rinaudo, A. E. Gorbalenya, K. Bienz, D. Egger, E. Levenson, and E. Ehrenfeld. 2004. Presented at the Seventh International Symposium on Positive Strand RNA Viruses, San Francisco, CA, 27 May to 1 June 2004.
- 66.Teterina, N. L., K. Bienz, D. Egger, A. E. Gorbalenya, and E. Ehrenfeld. 1997. Induction of intracellular membrane rearrangements by HAV proteins 2C and 2BC. Virology 237:66-77. [DOI] [PubMed] [Google Scholar]
- 67.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teterina, N. L., M. S. Rinaudo, and E. Ehrenfeld. 2003. Strand-specific RNA synthesis defects in a poliovirus with a mutation in protein 3A. J. Virol. 77:12679-12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teterina, N. L., W. D. Zhou, M. W. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Towner, J. S., D. M. Brown, J. H. Nguyen, and B. L. Semler. 2003. Functional conservation of the hydrophobic domain of polypeptide 3AB between human rhinovirus and poliovirus. Virology 314:432-442. [DOI] [PubMed] [Google Scholar]
- 72.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 73.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Kuppeveld, F. J., J. M. Galama, J. Zoll, and W. J. Melchers. 1995. Genetic analysis of a hydrophobic domain of coxsackie B3 virus protein 2B: a moderate degree of hydrophobicity is required for a cis-acting function in viral RNA synthesis. J. Virol. 69:7782-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van Kuppeveld, F. J., J. M. Galama, J. Zoll, P. J. van den Hurk, and W. J. Melchers. 1996. Coxsackie B3 virus protein 2B contains cationic amphipathic helix that is required for viral RNA replication. J. Virol. 70:3876-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Kuppeveld, F. J., J. G. Hoenderop, R. L. Smeets, P. H. Willems, H. B. Dijkman, J. M. Galama, and W. J. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 16:3519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Kuppeveld, F. J., W. J. Melchers, K. Kirkegaard, and J. R. Doedens. 1997. Structure-function analysis of coxsackie B3 virus protein 2B. Virology 227:111-118. [DOI] [PubMed] [Google Scholar]
- 78.Waggoner, S., and P. Sarnow. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xiang, W., A. Cuconati, A. V. Paul, X. Cao, and E. Wimmer. 1995. Molecular dissection of the multifunctional poliovirus RNA-binding protein 3AB. RNA 1:892-904. [PMC free article] [PubMed] [Google Scholar]
- 81.Yin, F. H., and N. B. Lomax. 1983. Host range mutants of human rhinovirus in which nonstructural proteins are altered. J. Virol. 48:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, S., and V. R. Racaniello. 1997. Persistent echovirus infection of mouse cells expressing the viral receptor VLA-2. Virology 235:293-301. [DOI] [PubMed] [Google Scholar]