Abstract
Background
Ovarian cancer is the leading cause of death from gynecological malignancies. Investigating the HRR-related gene status, notably BRCA1/2 in different regions and populations is of great significance for formulating accurate target therapy.
Methods
We collected 124 ovarian cancer cases from the Affiliated Hospital of.
Qingdao University, detected the genomic alteration of 32 genes by NGS, including.
19 HRR-related genes, 9 proto-oncogenes and 4 tumor suppressor genes. Clinicopathological characteristics, variants, clinical significance, and correlation with prognosis were analyzed.
Results
The incidence of HRR-related gene mutation was 59.68 % and no statistical significance was found with multiple clinicopathological characteristics. BRCA1/2 (27.42 %) were the most frequent mutated HRR genes. 23 (18.55 %) cases harbored gBRCA1/2 mutation, with all BRCA1 mutations were pathogenic/likely pathogenic and 2 cases of BRCA2 mutation was variant of uncertain significance. Somatic BRCA1/2 mutations were found in 12 (9.68 %) cases, and sBRCA1/2 had a higher frequency in less common ovarian cancer than high-grade serous carcinoma. HRR-related gene mutation status was associated with better prognosis than HRR wild-type.
Conclusions
Somatic BRCA1/2 mutation has higher incidence in less common ovarian cancer. HRR gene mutation status is an independent prognosis factor in ovarian cancer. Clarifying the HRR gene status is important for the selection of target therapy as well as the evaluation of prognosis.
Keywords: Homologous recombination repair, BRCA1/2 mutation, Next, Generation sequencing, Ovarian cancer, Prognosis
1. Introduction
Ovarian cancer is the third most common cancer type and the second leading cause of cancer-related death in female reproductive system worldwide [1]. In China, ovarian cancer is the second leading cause of incidence and mortality in gynecologic cancer, there were 36,900 new cases and 27,200 deaths from this malignant tumor according to the latest statistics data [2]. Ovarian cancer consists of several histologic subtypes, with epithelial ovarian cancer accounts for 90 % of this disease [3,4], which contains five principal pathological subtypes, including high-grade serous ovarian cancer (HGSOC, 70 %), low-grade serous ovarian cancer (LGSOC, <5 %), endometrioid carcinoma (EC, 10 %), clear cell carcinoma (CCC, 10 %) and mucinous carcinoma (MC, 3 %) [5,6], the biological properties and molecular characters of high-grade tumors are distinct from other subtypes [5,7]. Non-epithelial tumor inlcudes germ cell tumor such as dysgerminoma, immature teratoma, and malignant sex cord-stromal tumor like Sertoli-Leydig cell tumor [3]. Despite the advances in the early diagnosis, surgery and treatment therapy in cancer, survival rate of ovarian tumor has remained modest in decades [7,8].
Primary treatment of ovarian cancer including appropriate debulking surgery with or without systemic platinum-based chemotherapy [9]. Recently, as the advanced development of next-generation sequencing (NGS), the management of OC has entered the era of precision medicine. Homologous recombination repair (HRR) is an important double-stranded DNA (dsDNA) damage repair mechanism, it uses sister chromatids as a template to repair dsDNA damage by recruiting BRCA1, BRCA2 and PARP (Poly ADP-ribosepolymerase) proteins in the signaling pathway. Failure of HRR could be caused by the loss of function mutations such as BRCA1, BRCA2, RAD51C, and RAD51D, which will lead to the failure of dsDNA damage repair and make the tumor more sensitive to platinum-based chemotherapy and PARP inhibitors (PARPi) [[10], [11], [12]].
PARP protein plays a key role in the repair of single-stranded DNA (ssDNA) damage, PARPi could prevent this process by capturing PARP protein at the site of single-stranded DNA damage, leading to the accumulation of damaged DNA and eventually cause the death of cell on the basis of HRR gene mutation [13]. As the most well-studied HRR-related gene, several clinical trails and studies have showed that patient with germline and/or somatic BRCA1/2 mutation is the greatest beneficiary of PARPi, with significantly improved PFS [[14], [15], [16]]. Based on these studies, national comprehensive cancer network (NCCN) has recommended PARPi a maintenance option in patients with germline and/or somatic BRCA1 and BRCA2 status [17].
The advent of next-generation sequencing (NGS) technology gives the possibility to better understand the genomic landscape of ovarian cancer and offers the guidance to precision treatment. Compared with other prior sequencing method, NGS provides an affordable, high throughput, high resolution, and more comprehensive genomic testing in multiple cancer types, as well as a reliable method for discovering novel genetic mutations to help understand the pathogenesis of disease [18,19]. The current NCCN guideline recommends molecular testing both in the upfront setting and upon recurrence in ovarian cancer. For upfront setting, to determine the molecular alterations including BRCA1/2 status, loss of heterozygosity (LOH), homologous recombination gene status in the absence of germline BRCA mutation to help the selection of interventions with well-known benefits such as PARPi. Other tumor specific markers including (but not limited to) MSI (microsatellite instability), TMB (tumor mutational burden), BRAF, and NTRK were suggest to be detected in the relapse setting to provide possible treatment selection [17].
In this study, we collected 124 ovarian cancer cases and performed a comprehensive analysis including clinicopathological characteristics, genome landscape, treatments and prognosis. Our study provides valuable guidance and reference for the precision medical treatment of ovarian cancer patient in China.
2. Materials and methods
2.1. Patients
We collected retrospective data from 129 cases of various ovarian cancer histological types that were diagnosed and received systemic treatment in the Affiliated Hospital of Qingdao University from January 2019 to February 2023. Patients who refused to enroll in this research (4 cases) and developed more than one malignant tumor (1 case) were excluded. All hematoxylin-eosin (HE) stained slides of the specimen were cross-checked by two pathologists. All patients provided informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (Project identification code: QYFY WZLL 27873).
2.2. Sample preparation
For formalin-fixed paraffin-embedded (FFPE) samples, tissues were sectioned in 5 μm and enriched as previous described [20]. Genomic DNA was extracted using a Tiangen paraffin-embedded tissue DNA extraction kit (Tiangen Biotech, Beijing, China) according to the manufacturer's instructions. Genomic DNA of blood sample was extracted by a Fresh blood/marrow DNA extraction kit following the manufacturer's instructions (Amoy Diagnostics, Xiamen, China). DNA library was prepared by the HRR gene combination detection library preparation kit (Amoy Diagnostics, Xiamen, China) and applied to an HRR 32 gene panel detection, including HRR-related genes (ATM, ATR, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, CHEK2, FANCA, FANCL, MRE11A, NBN, PALB2, PPP2R2A, RAD51B, RAD51C, RAD51D, RAD54L), proto-oncogenes (AR, BRAF, ERBB2, ESR1, HDAC2, HOXB13, KRAS, NRAS, PIK3CA), and tumor suppressor genes (CDH1,PTEN, STK11, TP53).
2.3. Next-generation sequencing
DNA sequencing was performed on the NextSeq CN500 Illumina platform (Illumina, San Diego, CA, USA) using a NextSeq Mid Output Reagent Cartridge (Illumina) following the manufacturer's instructions. The average sequencing depth was 1,000X and the effective sequencing depth was over 300X, variants with variant allele fraction (VAF) greater than 3 % were retained. Variant types including single nucleotide polymorphism (SNP), indels, hot spot mutation were analyzed. Patients with BRCA1/2 mutations were further checked with blood sample to confirm germline or somatic mutations.
2.4. Statistical analysis
All data was processed with SPSS 26.0.0 statistical analysis software (IBM Corp., Armonk, NY, USA) and GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, USA). Statistical significance was defined at P < 0.05. Correlation factors of ovarian cancer prognosis were analyzed by the multivariate logistic regression and cox regression models with 95 % confidence intervals (CI). Disease free survival (DFS) was confirmed as the interval between the date of diagnosis and the date of recurrence/metastasis or last follow-up. Survival analysis was performed by the Kaplan–Meier survival curves using log-rank test.
3. Results
3.1. Clinical pathological characteristics of ovarian cancer patients
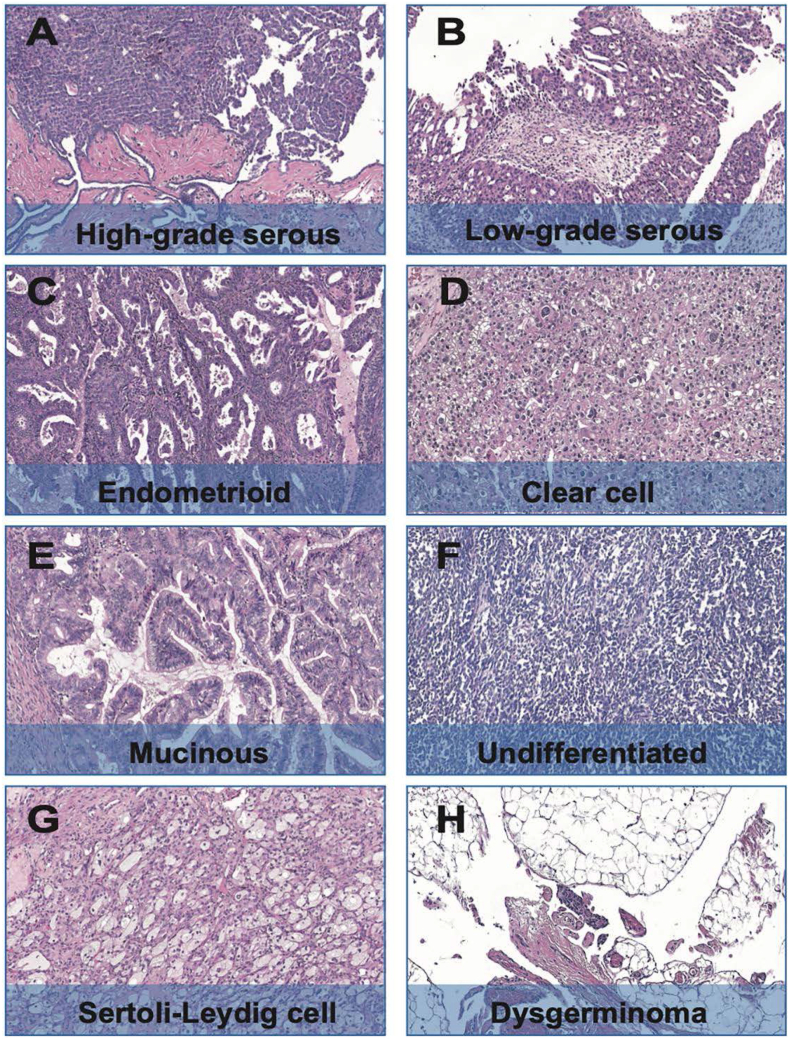
All the clinical pathological characteristics of 124 OC patients were summarized in Table 1. The median age of 124 OC patients was 57 years old (range from 25 to 79), 116 (93.54 %) of the patients undergo debulking surgery and the average tumor size was 6.3 cm. Serous carcinoma present a majority of pathological subtype (94/124, 75.81 %), other subtypes including clear cell carcinoma (11/124, 8.87 %), endometrioid carcinoma (10/124, 8.05 %), mucinous carcinoma (6/124, 4.84 %), dysgerminoma (1/124, 0.81 %), Sertoli-Leydig cell tumor (1/124, 0.81 %) and undifferentiated carcinoma (1/124, 0.81 %), the HE stained pathological images of each subtype were shown in Fig. 1. Over half of the patients (65/124, 52.42 %) were in FIGO (International Federation of Obstetricians and Gynecologists, FIGO) III stage at diagnosis, others were in FIGO I (23/124, 18.54 %), FIGO II (18/124, 14.52 %), FIGO IV (18/124, 14.52 %). 49 (39.52 %) patients received neoadjuvant chemotherapy before debulking surgery, with 117 (94.35 %) patients underwent systemic chemotherapy after surgery. Moreover, nearly half (62/123, 50.41 %) of the patients received postoperative targeted therapy including PARP inhibitors, Bevacizumab, and Anlotinib. The pathological characteristics listed in Table 1 showed no statistical significance with HRR-related gene mutation status.
Table 1.
Clinicopathological characteristics and HRR gene status of ovarian cancer.
| Clinicopathological characteristics | Overall (N = 124, %) | HRR-m (N = 74, %) | HRR-w (N = 50, %) | P value |
|---|---|---|---|---|
| Age/years | ||||
| Median/range | 57/25-79 | 57/25-79 | 58/28-79 | 0.910a |
| Mean ± SD | 55.6 ± 11.6 | 55.5 ± 1.4 | 55.7 ± 11.9 | |
| Tumor size/cm | ||||
| Median/range | 6.3/0.5–22 | 6/0.5–18 | 6.5/1.1–22 | 0.851a |
| Mean ± SD | 6.7 ± 4.2 | 6.7 ± 4.0 | 6.8 ± 4.4 | |
| Unknown | 13 | 9 | 4 | |
| Pathological type | ||||
| Serous carcinoma | 94 | 55 | 39 | 0.261a |
| Clear cell carcinoma | 11 | 9 | 2 | |
| Other | 19 | 10 | 9 | |
| FIGO | ||||
| I | 23 | 16 | 7 | 0.298a |
| II | 18 | 8 | 10 | |
| III | 65 | 41 | 24 | |
| IV | 18 | 9 | 9 | |
| Lymph node status | ||||
| Positive | 4 | 3 | 1 | 0.223a |
| Negative | 13 | 5 | 8 | |
| Unknown | 107 | 66 | 41 | |
| LVSI | ||||
| Positive | 7 | 5 | 2 | 0.798a |
| Negative | 117 | 69 | 48 | |
| Hormone receptor | ||||
| Positive | 89 | 50 | 39 | 0.168a |
| Negative | 19 | 11 | 8 | |
| Unknown | 16 | 13 | 3 | |
| PD-L1 | ||||
| Positive | 54 | 30 | 24 | 0.611a |
| Negative | 24 | 14 | 10 | |
| Unknown | 46 | 30 | 16 | |
| Ki67 | ||||
| Median/range | 45.0/3–90 | 50/3–90 | 37.5/5–80 | 0.332a |
| Mean ± SD | 45.1 ± 22.7 | 47 ± 22.4 | 42.6 ± 23.1 | |
| Unknown | 22 | 16 | 6 | |
| Type of surgery | ||||
| Neoadjuvant chemotherapy surgery | +49 | 25 | 24 | 0.170a |
| Radical surgery | 73 | 47 | 26 | |
| Other | 2 | 2 | 0 | |
| Postoperative | ||||
| chemotherapy Yes | 117 | 69 | 48 | 0.798a |
| No | 7 | 5 | 2 | |
| Postoperative targeted therapy | ||||
| Yes | 62 | 34 | 28 | 0.227a |
| No | 61 | 40 | 21 | |
| Unknown | 1 | 0 | 1 | |
Student's t-test; a Pearson's chi-squared test; LVSI, Lymphovascular space invasion; m, mutant; w, wildtype; Hormone receptor including estrogen receptor and progesterone receptor.
Fig. 1.
Photomicrographs (original magnification, 200×; hematoxylin-eosin stain) of the pathological subtypes of ovarian cancer in this study. (A) High-grade serous ovarian cancer (HGSOC); (B) Low-grade serous ovarian cancer (LGSOC); (C) Endometrioid carcinoma (EC); (D) Clear cell carcinoma (CCC); (E) Mucinous carcinoma; (F) Undifferentiated carcinoma; (G) Sertoli-Leydig cell carcinoma; (H) Dysgerminoma.
3.2. Variants of HRR-related genes and other genes
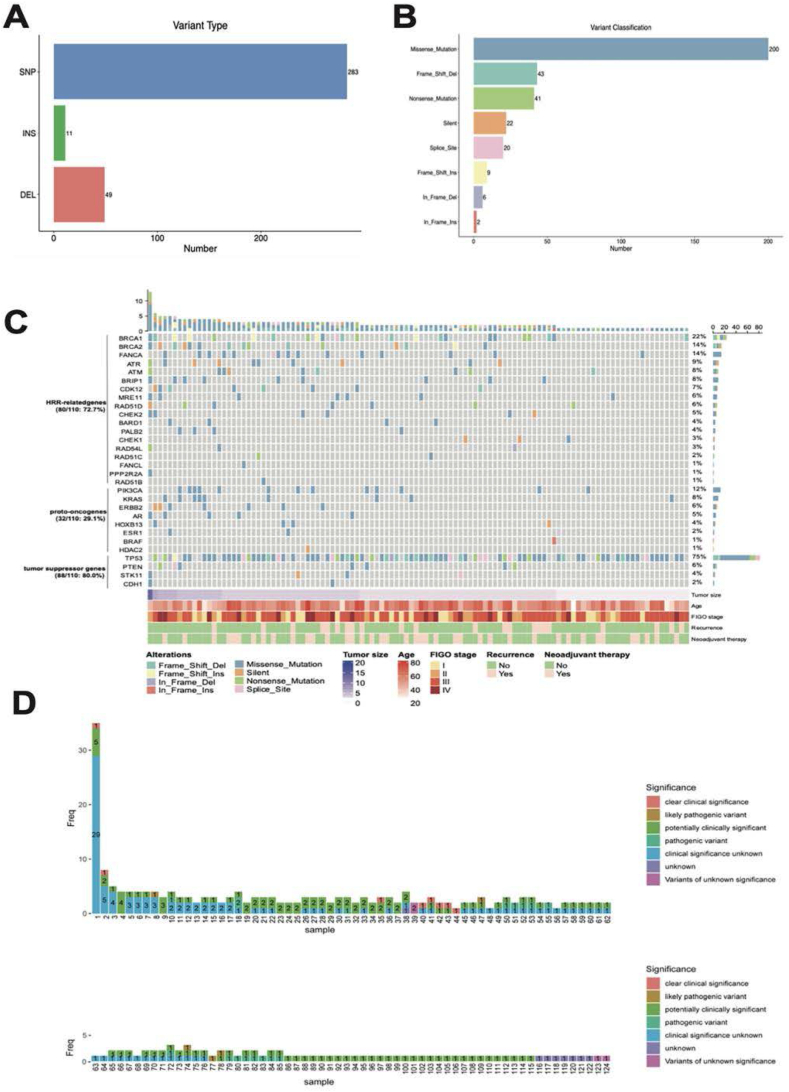
All the 124 OC cases were tested the genomic alterations by NGS and the variant files were summarized in Fig. 2. For all of the variants, SNP was the most common type (Fig. 2 A) and missense mutation was the leading subtype of variant classfication (Fig. 2 B). HRR-related gene variants were found in 74 (59.68 %) cases, with BRCA1 (19.35 %), BRCA2 (11.29 %) as the most frequent mutated genes, other genes including FANCA (14, 11.29 %), ATM (9, 7.26 %), ATR (7, 5.65 %), CDK12 (7, 5.65 %), MRE11 (6, 4.84 %), RAD51D (6, 4.84 %), BARD1 (4, 3.23 %), RAD54L (4, 3.23 %), RAD51C (4, 3.23 %), BRIP1 (3, 2.42 %), CHEK2 (3, 2.42 %), PALB2 (2, 1.61 %), FANCL (2, 1.61 %), PPP2R2A (2, 1.61 %), CHEK1 (1, 0.81 %) and RAD51B (1, 0.81 %). This NGS panel also detected the alterations of proto-oncogene including PIK3CA, KRAS, ERBB2, AR, HOXB13, ESR1, BRAF and HADC2, and 29 (23.39 %) cases were found with proto-oncogene variants. Furthermore, variants of tumor suppressor gene including TP53, PTEN, STK11, CDH1 were found in 89 (71.77 %) patients, with TP53 (83/89, 93.26 %) the most frequent mutated gene. To better represent the genomic variability in these ovarian cancer patients, we removed the genomic polymorphism alterations in all samples, as well as the germline mutations in blood samples, retained variants specified in the Clinvar database, and generated a waterfall plot of the mutations in 110 patents (Fig. 2 C). In addition, we reported the clinical significance of each mutation in all cases (Fig. 2 D).
Fig. 2.
Variants of the HRR-related gene, proto-oncogene and tumor suppressor gene in 124 cases of ovarian cancer. (A) The number of mutations in different variant type; (B) The number of mutations in different variant classifications; (C) The Oncoplot of multiple gene variants in 110 ovarian cases. Each row represents one gene and each column indicates one patient. The right panel shows the mutation frequeny of the individual gene. Classification of the alteration was indicated by different colors. Clinical pathological characteriatics including age, tumor size, FIGO stage, recurrence and neoadjuvant therapy were listed under the plot; (D) The clinical significance and frequency of each mutation in 124 ovarian cancer patients were delineated, significance including both germline and somatic mutations were indicated by different colors. SNP: single nucleotide polymorphism; INS: insertion; DEL: deletion; Freq: frequency.
3.3. Pathogenic/likely pathogenic (P/LP) alterations and variants of uncertain significance (VUS) in BRCA1/2 genes
In 124 ovarian cancer patients, 8 cases were tested with blood, 116 cases used paraffin tissues for sequencing, and only 20 cases using paraffin tissues with BRCA gene alteration were further confirmed for germline mutation. HGSOC accounted for most of the gBRCA1/2 mutations (95.65 %). For BRCA1/2 gene, 23 out of 124 (18.55 %) cases were found with germline mutations, including 14 (11.29 %) BRCA1 mutation, 8 (6.45 %) BRCA2 mutation, and 1 (0.81 %) case with both BRCA1/2 P/LP mutation. All BRCA1 mutations were P/LP and 2 cases of BRCA2 mutations were VUS. For 8 cases using blood samples, 32 genes listed in panel were all analyzed for germline mutation, and 2 cases were totally negative. P/LP mutations were found in 3 cases, 2 cases were BRCA1 and 1 case was RAD51D, 4 VUS were found in 3 cases, including BRIP1, FANCA, and RAD51C. All of the germline variants of 26 cases were concluded in Table 2. Classification of germline mutation was determined on the guidelines of ACMG/AMP [21].
Table 2.
Germline mutations of BRCA1/2 gene in ovarian cancer.
| Case Histologic NO. type | Tissue type | Variant | P/LP | VUS | ||
|---|---|---|---|---|---|---|
| 1 | HGSOC | P | BRCA1 | exon11 c.3841C > T p.Gln 1281a | Y | |
| 2 | HGSOC | P | BRCA1 | intron 21 c.5332+1 del | Y | |
| 3 | HGSOC | P | BRCA1 | exon11 c.3770_3771 del p.Glu1257Glyfsa9 | Y | |
| 4 | HGSOC | P | BRCA1 | exon11 c.2138C > G p.Ser 713a | Y | |
| 5 | HGSOC | P | BRCA1 | exon11 c.3748G > T p.Glu 1250a | Y | |
| 6 | HGSOC | P | BRCA1 | exon11 c.3352C > T p.Gln 1118a | Y | |
| 7 | HGSOC | P | BRCA1 | exon 24 c.5470_5477del p.Ile1824Aspfsa3 | Y | |
| 8 | HGSOC | P | BRCA2 | exon11 c.5645C > A p.Ser 1882a | Y | |
| 9 | HGSOC | P | BRCA1 | exon11 c.1961 dup p.Tyr655Valfsa18 | Y | |
| 10 | HGSOC | P | BRCA2 | exon11 c.6447_6448dup p.Lys2150Ilefsa19 | Y | |
| 11 | HGSOC | P | BRCA1 | exon 2 c.66 dup p.Glu23ArgfsTer18 | Y | |
| 12 | HGSOC | P | BRCA2 | exon19 c.8481T > G p.Pro 2827 = | Y | |
| 13 | HGSOC | P | BRCA2 | exon10 c.1905T > A p.Asp635Glu | Y | |
| 14 | HGSOC | P | BRCA1 | exon 22 c.5386 dup p.Ser1796PhefsTer34(S1796Ffsa34) | Y | |
| 15 | EC | P | BRCA2 | exon11 c.6235_6245del p.Val2079IlefsTer2(V2079Ifsa2) | Y | |
| 16 | HGSOC | P | BRCA2 | exon11 c.4415_4418del p.Lys1472ThrfsTer6(K1472Tfsa6) | Y | |
| 17 | HGSOC | P | BRCA2 | exon11 c.5789 del p.Leu1930TyrfsTer33 (L1930Yfsa33) | Y | |
| 18 | HGSOC | P | BRCA1 | exon11 c.2788_2795del p.Pro930TrpfsTer5(P930Wfsa5) | Y | |
| 19 | HGSOC | P | BRCA1 | exon11 c.1016 dup p.Val340GlyfsTer6(V340Gfsa6) | Y | |
| HGSOC | P | BRCA2 | exon8 c.658_659del p.Val220IlefsTer4(V220Ifsa4) | Y | ||
| 20 | HGSOC | P | BRCA1 | exon19 c.5162A > C p.Gln1721Pro(Q1721P) | Y | |
| 21 | HGSOC | P | BRCA1 | exon11 c.2747 dup p.Asn916LysfsTer9(N916Kfsa9) | Y | |
| 22 | HGSOC | B | RAD51D | exon 6 c.562C > T p.Arg 188a | Y | |
| 23 | HGSOC | B | BRIP1 | exon 16 c.2324A > G p. (Asn775Ser) | Y | |
| 24 | HGSOC | B | BRCA1 | exon 24 c.5521 del p.Ser1841Valfsa2 | Y | |
| 25 | HGSOC | B | FANCA | exon 42 c.4225C > T p.Arg1409Trp | Y | |
| B | RAD51C | exon 4 c.664_681 del p.Gln222_Pro227del | Y | |||
| 26 | HGSOC | B | BRCA1 | exon11 c.1016 del p.Lys339Argfsa2 | Y | |
| 27 | MC | B | FANCA | exon 42 c.4225C > T p.Arg1409Trp | Y | |
HGSOC: High-grade serous ovarian cancer; EC: Endometrioid carcinoma; MC: Mucinous carcinoma; P: Paraffin-embedded; B: Blood; Y:Yes.
3.4. Somatic mutations in BRCA genes
In 124 ovarian patients, we found 12 (9.68 %) cases harbored 23 BRCA1/2 gene alterations, including 9 (39.13 %) BRCA1 mutation and 14 (60.87 %) BRCA2 mutation, 1 case harbored both gBRCA1 and sBRCA2 mutation. Of the 12 cases with sBRCA1/2 mutations, HGSOC accounted for 58.33 % and LCOC had a proportion of 41.67 %. 73.91 % of the mutations were tier III (variants of unknown clinical significance), others were tier I (variants of strong clinical significance). Most of the variants were SNPs (Single Nucleotide Polymorphism, SNP), the others were introns. All of the somatic variants of 12 cases were summarized in Table 3. Clinical significance was identified according to the AMP/ASCO/CAP 2017 guidelines [22].
Table 3.
Somatic mutations of BRCA1/2 gene in ovarian cancer.
| Case NO. | Histologic Tissue Variant type type | Tier | |||
|---|---|---|---|---|---|
| 1 | CCC | P | BRCA2 | exon11 c.4189G > T p.Glu 1397a | I |
| BRCA1 | exon11 c.1740C > A p.F580L | III | |||
| BRCA1 | exon11 c.1576C > A p.Q526K | III | |||
| BRCA2 | exon10 c.953A > C p.K318T | III | |||
| BRCA2 | exon11 c.3069C > A p.N1023K | III | |||
| BRCA2 | exon11 c.3341T > G p.L1114R | III | |||
| BRCA2 | exon11 c.3800A > G p.D1267G | III | |||
| BRCA2 | exon11 c.4477G > A p.E1493K | III | |||
| BRCA2 | exon11 c.5705A > C p.D1902A | III | |||
| BRCA2 | exon11 c.6115T > G p.L2039V | III | |||
| BRCA2 | exon11 c.6772G > A p.E2258K | III | |||
| 2 | HGSOC | P | BRCA1 | exon11 c.3756_3759del p.Ser1253Argfsa10 I | |
| 3 | EC | P | BRCA2 | exon 24 c.9127G > T p.Glu 3043a I | |
| BRCA1 | intron 2 c.80 + 15G > T III | ||||
| 4 | EC | P | BRCA1 | exon 17 c.5066T > C p.Met1689Thr III | |
| 5 | HGSOC | P | BRCA1 | exon8 c.446A > C p.Glu149Ala III | |
| 6 | HGSOC | P | BRCA1 | exon11 c.1687C > T p.Gln563Ter I | |
| 7 | HGSOC | P | BRCA2 | intron 5 c.475+5G > C III | |
| 8 | MC | P | BRCA2 | exon11 c.5683G > A p.Glu1895Lys (E1895K) III | |
| 9 | HGSOC | P | BRCA2 | exon11 c.6591_6592del p.Glu2198AsnfsTer4 (E2198Nfsa4) I | |
| 10 | UC | P | BRCA1 | exon 7 c.343C > A p.Pro115Thr (P115T) III | |
| 11 | HGSOC | P | BRCA1 | exon11 c.2129 del p.Thr710IlefsTer26 (T710Ifsa26) I | |
| 12 | HGSOC | P | BRCA2 | exon 3 c.266C > T p.Pro89Leu (P89L) III | |
HGSOC: High-grade serous ovarian cancer; EC: Endometrioid carcinoma; MC: Mucinous carcinoma; UC: Undifferentiated carcinoma; P: Paraffin-embedded.
3.5. BRCA1/2 variants in high-grade serous ovarian cancer (HGSOC)
In our cohort, 92 (74.19 %) patients were diagnosed with HGSOC. 22 (23.91 %) were detected with gBRCA1/2 mutations, including 15 (16.30 %) with gBRCA1, 6 (6.52 %) with gBRCA2 and 1 (1.09 %) with both gBRCA1/2. 7 (7.61 %) cases were with sBRCA1/2 mutations, including 4 (4.35 %) with sBRCA1, 2 (2.17 %) with sBRCA2 and 1 (1.09 %) with both sBRCA1 and 2. TP53 mutation was found in 74 (80.43 %) patients and 23 (31.08 %) of them were combined with BRCA1/2 mutations.
3.6. BRCA1/2 variants in less common ovarian cancer (LCOC)
In our study, we investigated the BRCA1/2 variants in less common ovarian cancer, including low-grade serous ovarian cancer, clear cell carcinoma, mucinous carcinoma, dysgerminoma, Sertoli-Leydig cell tumor and undifferentiated carcinoma [23]. 32 out of 124 OC cases were identified with LCOC. BRCA2 germline mutation was found in 1 case of endometrioid carcinoma, and 23 sBRCA1/2 mutations were detected in 5 cases, including 2 cases of endometrioid carcinoma, 1 case of clear cell carcinoma, 1 case of mucinous carcinoma and 1 case of undifferentiated carcinoma. LCOC seemed to habor more somatic mutations than HGSOC and over half of the somatic mutations were variants of unknown clinical significance (17/23, 73.91 %). Detailed mutations were summarized in tables 2 and 3.
3.7. Genomic alterations and prognosis
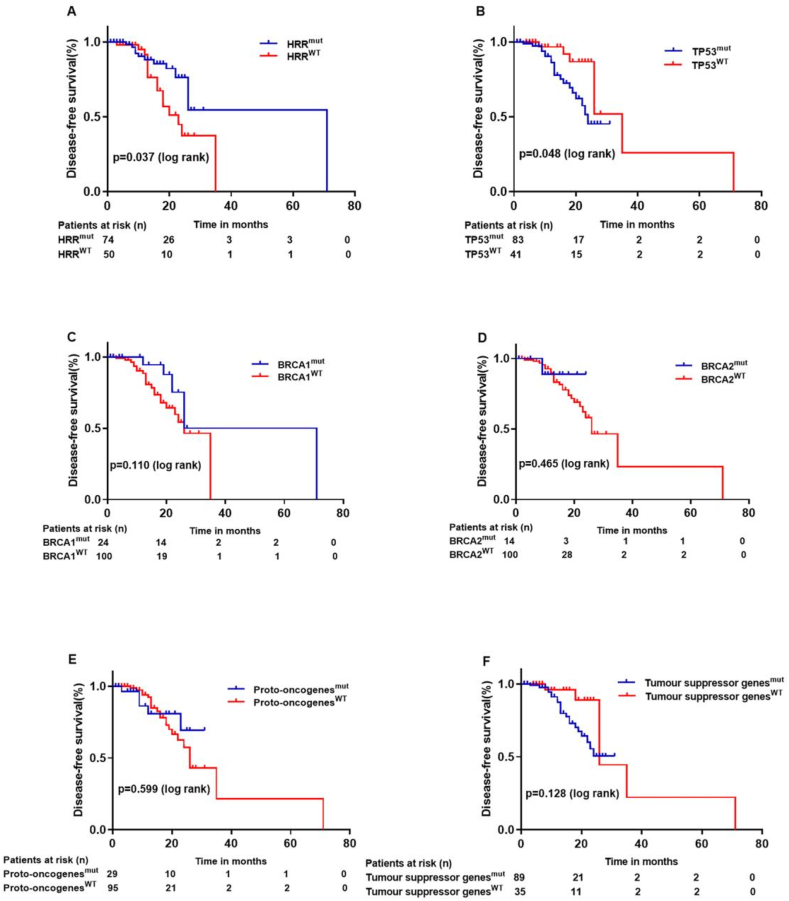
We followed up the patients from 1 month to 98 months, and the median follow-up time was 14 months. 27 cases were found with recurrence or new distant metastases, 1 case was dead of disease, and the median DFS was 12.5 months. Survival analysis indicated that patients with HRR-related gene mutations (P = 0.037) or TP53 mutations (P = 0.048) had better prognosis than HRR or TP53 wild-type patients (Fig. 3). Univariate cox analysis showed that patient age (P = 0.008), tumor size (P = 0.032), pathological subtype (P = 0.035), HRR-related gene status (P = 0.045), lymphovascular space invasion (LVSI, P = 0.020) and postoperative targeted therapy (P = 0.005) were related with prognosis of ovarian cancer. In addition, multivariate cox analysis showed that HRR-related gene status (P = 0.043) and postoperative targeted therapy (P = 0.020) were independent prognosis factors of ovarian cancer (Table 4).
Fig. 3.
Kaplan–Meier survival curves based on different gene mutations. (A) Patients with HRR-related gene mutation status had better DFS (P = 0.037); (B) Patients with TP53 gene mutation status had poor DFS (P = 0.048). (C–F) The gene status of (C) BRCA1 (P = 0.110), (D) BRCA2 (P = 0.465); (E) Proto-oncogenes (P = 0.599) and (F) Tumour suppressor genes (P = 0.128) had no statistical relationship with DFS. P values are calculated using the log-rank test.
Table 4.
Cox regression analysis for the identification of factors associated with the prognosis of ovarian cancer.
| Univariate HR (95%CI) | Multivariate P value HR (95%CI) P value | |||
|---|---|---|---|---|
| Age/years (25–79) | 1.053 (1.013–1.094) | 0.008 | ns | ns |
| Tumor size/cm (0.5–22) | 0.869 (0.765–0.988) | 0.032 | ns | ns |
| Pathological type (Serous vs other) | 0.116 (0.016–0.856) | 0.035 | ns | Ns |
| Tumor size/cm (0.5–22) | 0.869 (0.765–0.988) | 0.032 | ns | Ns |
| HRR gene status (m vs w) | 2.230 (1.019–4.880) | 0.045 | 2.979 (1.037–8.561) | 0.043 |
| LVSI (- vs +) | 3.702 (1.230–11.136) | 0.020 | ns 4.085 | ns |
| Postoperative targeted therapy (- vs +) | 2.406 (1.306–4.433) | 0.005 | 0.020 | (1.249–13.361) |
* LVSI, Lymphovascular space invasion; m, mutant; w, wildtype.
4. Discussion
Ovarian cancer is composed of several malignant tumors and has been a deadly gynecologic tumor for decades. 70 % patients were in advanced stage at first diagnosed [24] and about 70–80 % advanced cases were found relapsed in 5 years and developed platinum resistance, which indicated worse prognosis and higher mortality [25]. Earlier diagnosis and more precise targeted treatment can help patients with ovarian cancer to achieve longer survival. In recent years, several PARP inhibitors such as Olaparib were proved to be effective in recurrence ovarian cancer and have been approved by FDA for multiple indications, especially for those with BRCA gene mutations or homologous recombination deficiency (HRD) [[26], [27], [28]]. Therefore, identifying the genomic landscape of ovarian cancer patients, notably HRR-related genes such as BRCA1/2, is extremely critical for the selection of treatment and targeted drug.
In this retrospective study, we collected a cohort that were consist of 124 ovarian cancer patients with several histologic subtypes, grouped them into HRR-mutant and HRR-wildtype. Analysis of the correlation between HRR gene status and clinicopathological characteristics including age, tumor size, FIGO stage, treatments and so on, showed no statistical significance. Next, we analyzed the variants in HRR-related genes in our cohort, about 59.68 % patients were found with multiple types of mutations. Excluding the high mutation frequency in BRCA1/2, we also found several non-BRCA gene alterations, including FANCA (11.29 %), ATM (7.26 %), ATR (5.65 %), CDK12 (5.65 %), MRE11 (4.84 %), RAD51D (4.84 %), BARD1 (3.23 %), RAD54L (3.23 %), RAD51C (3.23 %), BRIP1 (2.42 %), CHEK2 (2.42 %), PALB2 (1.61 %), FANCL (1.61 %), PPP2R2A (1.61 %), CHEK1 (0.81 %) and RAD51B (0.81 %), the frequencies of these gene variants were varied from previous studies [29]. Although current treatment options were mostly focused on BRCA gene status, studies showed that non-BRCA1/2 specific HRR gene variants might represent a large number of HRD tumors, as well as a great number of patients that might benefit from targeted treatment [12,30]. Our study indicated that over half of the Chinese patients might be the potential beneficiary from targeted therapy such as PARP inhibitors.
BRCA1/2 gene status is critical for the selection of maintenance therapy in stage II-IV ovarian cancer after primary chemotherapy. Moreover, figuring out the alterations of BRCA1/2, especially germline mutation, is an effective way to identify the inherited risk of cancer and take preventive measures. Previous studies showed that about 8.6–24.5 % patients harbored gBRCA1/2 mutations, while 3.7–7.1 % patients presented sBRCA1/2 mutations [10,[31], [32], [33], [34], [35]]. Our study excluded the benign mutations and found the frequencies of g/s BRCA1/2 mutation were 18.55 % and 9.68 %. All the gBRCA1 mutations were P/LP, while 2 out of 8 gBRCA2 mutations were VUS, these results might be related to our small sample size, studies with a large number of samples are needed.
One interesting finding in our study was that sBRCA1/2 presented much higher frequency in LCOC patients than in HGSOC (41.67 % VS 4.35 %), and a majority (73.91 %) of these mutations were tier III (variants of unknown clinical significance). It is well known that the different histologic subtypes of OC had diversified clinical manifestation, outcomes, and responses to treatments, which mainly due to the diverse intrinsic tumor biology. Currently, whether to conduct comprehensive molecular testing for ovarian cancer remains controversial, but our results indicated that more complete molecular analysis might be particularly important for the uncommon histologic subtypes that had limited approved treatment options. Besides, those tier III mutations offer great possibilities for application of target therapies that might be useful.
With the widely application of PARPi maintenance therapy, treatment of ovarian cancer has gradually developed into a "surgery + chemotherapy + maintenance treatment" standard model. PARPi plays its anti-tumor role through the synthetic lethality effect, several clinical trails had proved that OC patients with gBRCA1/2 mutation had received significant PFS benefit from PARPi maintenance therpay [[36], [37], [38], [39]]. To date, the FDA has approved three PARPi drugs for the maintenance treatment of ovarian cancer, which are Olaparib, Rucaparib and Niraparib [[40], [41], [42]]. Furthermore, germline or somatic mutation in HRR-related genes is usually associated with better prognosis [43]. Beyond PARPi, the monotherpay or combined application of angiogenesis drug Bevacizumab, platinum-based chemotherapty, as well as Pembrolizumab in certain circumstances are also recommended by NCCN [17].
Here in our study, we investigated the correlations between multiple clinical features and prognosis, including the status of HRR-related gene, proto-oncogene, age, tumor size, and the application of postoperative targeted therapy. The postoperative targeted therapy in our study refers to the simultaneous or metachronous use of antiangiogenic therapy, PARPi therapy, and Anlotinib during the patient's care. Univariate cox analysis showed that age, tumor size, histologic type, HRR-related gene status, LVSI and postoperative targeted therapy were associated with OC patient prognosis, while multivariate cox analysis showed that HRR-related gene status and postoperative targeted therapy were independent prognosis factors of ovarian cancer. Survival analysis represented the same conclusion. Our results indicated that the evaluation of HRR-related genes showed clinical significance in the prognosis of ovarian cancer, consistent with previous conclusions [43,44].
There were several limitations to our study: (1) a single medical center study; (2) a small size of samples; (3) limited analysis of germline variants in non-BRCA1/2 HRR genes; (4) a relatively short following-up period and (5) no explicit classification by the therapeutic regimen of patient.
5. Conclusions
This study reported the accurate incidence of HRR-related genes, particularly BRCA1/2 in eastern China patients and found that BRCA1/2 somatic mutation showed a higher incidence in LCOC than HGSOC, HRR-related gene mutation status was significantly associated with the prognosis of ovarian cancer patients. This comprehensive analyze of HRR gene study provides a valuable reference for clinicians to select appropriate treatment plan for OC patient, as well as a better understanding of this malignant tumor.
Funding
This research was funded by the National Natural Science Foundation of China, grant number 81972329 to XM.X; the Science and Technology Project of the South District of Qingdao, grant number 2022-2-003-YY to XM.X; the Postdoctoral Innovation Project of Qingdao, grant number QDBSH20220201055 to YL.S.
Institutional review board statement
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the affliated hospital of Qingdao University (Project identification code: QYFY WZLL 27873).
Informed consent statement
All patients provided informed consent for inclusion before they participated in the study.
Data availability statement
The data presented in this study are available on request from the corresponding author.
CRediT authorship contribution statement
Yaolin Song: Writing - original draft, Investigation, Funding acquisition, Formal analysis, Conceptualization. Wenwen Ran: Methodology. Huiqing Jia: Formal analysis. Qin Yao: Formal analysis. Guangqi Li: Methodology. Yang Chen: Formal analysis. Xiaonan Wang: Methodology. Yujing Xiao: Methodology. Mengqi Sun: Formal analysis. Xiao Lu: Formal analysis. Xiaoming Xing: Writing - review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The manuscript titled “Next-Generation Sequencing-based Analysis of Homologous Recombination Repair Gene Variant in Ovarian Cancer” has not been published or presented elsewhere in part or in entirety and is not under consideration by another journal. We have read and understood your journal's policies, and we believe that neither the manuscript nor the study violates any of these. All authors declare no conflicts of interest.
Acknowledgments
We would like to thank all the patients and their families.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Zheng R.S., Zhang S.W., Sun K.X., Chen R., Wang S.M., Li L., Zeng H.M., Wei W.W., He J. [Cancer statistics in China, 2016] Zhonghua Zhongliu Zazhi. 2023;45:212–220. doi: 10.3760/cma.j.cn112152-20220922-00647. [DOI] [PubMed] [Google Scholar]
- 3.Park K.J., Selinger C.I., Alvarado-Cabrero I., Duggan M.A., Kiyokawa T., Mills A.M., Ordi J., Otis C.N., Plante M., Stolnicu S., et al. Dataset for the reporting of carcinoma of the cervix: recommendations from the international collaboration on cancer reporting (ICCR) Int. J. Gynecol. Pathol. 2022;41:S64–S89. doi: 10.1097/PGP.0000000000000909. [DOI] [PubMed] [Google Scholar]
- 4.Prat J. New insights into ovarian cancer pathology. Ann. Oncol. 2012;23(Suppl 10):x111–x117. doi: 10.1093/annonc/mds300. [DOI] [PubMed] [Google Scholar]
- 5.Kobel M., Kang E.Y. The evolution of ovarian carcinoma subclassification. Cancers. 2022;14 doi: 10.3390/cancers14020416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prat J. Pathology of borderline and invasive cancers. Best Pract. Res. Clin. Obstet. Gynaecol. 2017;41:15–30. doi: 10.1016/j.bpobgyn.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Lheureux S., Braunstein M., Oza A.M. Epithelial ovarian cancer: evolution of management in the era of precision medicine. CA Cancer J Clin. 2019;69:280–304. doi: 10.3322/caac.21559. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics. CA Cancer J Clin. 2022;72:7–33. doi: 10.3322/caac.21708. 2022. [DOI] [PubMed] [Google Scholar]
- 9.O'Malley D.M. New therapies for ovarian cancer. J Natl Compr Canc Netw. 2019;17:619–621. doi: 10.6004/jnccn.2019.5018. [DOI] [PubMed] [Google Scholar]
- 10.Miller R.E., Leary A., Scott C.L., Serra V., Lord C.J., Bowtell D., Chang D.K., Garsed D.W., Jonkers J., Ledermann J.A., et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020;31:1606–1622. doi: 10.1016/j.annonc.2020.08.2102. [DOI] [PubMed] [Google Scholar]
- 11.Harbin L.M., Gallion H.H., Allison D.B., Kolesar J.M. Next generation sequencing and molecular biomarkers in ovarian cancer-an opportunity for targeted therapy. Diagnostics. 2022;12 doi: 10.3390/diagnostics12040842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stover E.H., Fuh K., Konstantinopoulos P.A., Matulonis U.A., Liu J.F. Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol. Oncol. 2020;159:887–898. doi: 10.1016/j.ygyno.2020.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Yap T.A., Sandhu S.K., Carden C.P., de Bono J.S. Poly(ADP-ribose) polymerase (PARP) inhibitors: exploiting a synthetic lethal strategy in the clinic. CA Cancer J Clin. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- 14.Moore K., Colombo N., Scambia G., Kim B.G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 15.Ray-Coquard I., Leary A., Pignata S., Cropet C., Gonzalez-Martin A., Marth C., Nagao S., Vergote I., Colombo N., Maenpaa J., et al. Olaparib plus bevacizumab first-line maintenance in ovarian cancer: final overall survival results from the PAOLA-1/ENGOT-ov25 trial. Ann. Oncol. 2023 doi: 10.1016/j.annonc.2023.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Martin A., Pothuri B., Vergote I., DePont Christensen R., Graybill W., Mirza M.R., McCormick C., Lorusso D., Hoskins P., Freyer G., et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2019;381:2391–2402. doi: 10.1056/NEJMoa1910962. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong D.K., Alvarez R.D., Backes F.J., Bakkum-Gamez J.N., Barroilhet L., Behbakht K., Berchuck A., Chen L.M., Chitiyo V.C., Cristea M., et al. NCCN guidelines(R) insights: ovarian cancer, version 3.2022. J Natl Compr Canc Netw. 2022;20:972–980. doi: 10.6004/jnccn.2022.0047. [DOI] [PubMed] [Google Scholar]
- 18.Meldrum C., Doyle M.A., Tothill R.W. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin. Biochem. Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Dean D.C., Hornicek F.J., Shi H., Duan Z. RNA sequencing (RNA-Seq) and its application in ovarian cancer. Gynecol. Oncol. 2019;152:194–201. doi: 10.1016/j.ygyno.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Song Y., Wang L., Ran W., Li G., Xiao Y., Wang X., Zhang L., Xing X. Effect of tumor location on clinicopathological and molecular markers in colorectal cancer in eastern China patients: an analysis of 2,356 cases. Front. Genet. 2020;11:96. doi: 10.3389/fgene.2020.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the association for molecular pathology, American society of clinical oncology, and college of American pathologists. J. Mol. Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.In Ovarian Cancers: Evolving Paradigms in Research and Care. 2016. Washington (DC) [PubMed] [Google Scholar]
- 24.Roett M.A., Evans P. Ovarian cancer: an overview. Am. Fam. Physician. 2009;80:609–616. [PubMed] [Google Scholar]
- 25.Ryu J., Thomas S.N. Quantitative mass spectrometry-based proteomics for biomarker development in ovarian cancer. Molecules. 2021;26 doi: 10.3390/molecules26092674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gelmon K.A., Tischkowitz M., Mackay H., Swenerton K., Robidoux A., Tonkin K., Hirte H., Huntsman D., Clemons M., Gilks B., et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- 27.Fong P.C., Yap T.A., Boss D.S., Carden C.P., Mergui-Roelvink M., Gourley C., De Greve J., Lubinski J., Shanley S., Messiou C., et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J. Clin. Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 28.Friedlander M., Matulonis U., Gourley C., du Bois A., Vergote I., Rustin G., Scott C., Meier W., Shapira-Frommer R., Safra T., et al. Long-term efficacy, tolerability and overall survival in patients with platinum-sensitive, recurrent high-grade serous ovarian cancer treated with maintenance olaparib capsules following response to chemotherapy. Br. J. Cancer. 2018;119:1075–1085. doi: 10.1038/s41416-018-0271-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadler Z.K., Maio A., Chakravarty D., Kemel Y., Sheehan M., Salo-Mullen E., Tkachuk K., Fong C.J., Nguyen B., Erakky A., et al. Therapeutic implications of germline testing in patients with advanced cancers. J. Clin. Oncol. 2021;39:2698–2709. doi: 10.1200/JCO.20.03661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ledermann J.A., Drew Y., Kristeleit R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer. 2016;60:49–58. doi: 10.1016/j.ejca.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Kwong A., Ho C.Y.S., Shin V.Y., Au C.H., Luk W.P., Fung L.H., Chan T.L., Chan K.K.L., Ngan H.Y.S., Ma E.S.K. Germline mutations in Chinese ovarian cancer with or without breast cancer. Mol Genet Genomic Med. 2022;10 doi: 10.1002/mgg3.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrikopoulou A., Zografos E., Apostolidou K., Kyriazoglou A., Papatheodoridi A.M., Kaparelou M., Koutsoukos K., Liontos M., Dimopoulos M.A., Zagouri F. Germline and somatic variants in ovarian carcinoma: a next-generation sequencing (NGS) analysis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.1030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eoh K.J., Kim H.M., Lee J.Y., Kim S., Kim S.W., Kim Y.T., Nam E.J. Mutation landscape of germline and somatic BRCA1/2 in patients with high-grade serous ovarian cancer. BMC Cancer. 2020;20:204. doi: 10.1186/s12885-020-6693-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stordal B., Timms K., Farrelly A., Gallagher D., Busschots S., Renaud M., Thery J., Williams D., Potter J., Tran T., et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 2013;7:567–579. doi: 10.1016/j.molonc.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen M., Yang L., Lei T., Xiao L., Li L., Zhang P., Feng W., Ye F., Bu H. BRCA1/2 mutation spectrum in Chinese early-onset breast cancer. Transl. Cancer Res. 2019;8:483–490. doi: 10.21037/tcr.2019.03.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler T., Maravent S., Boisselle J., Valdes J., Fellner C. A review of 2014 cancer drug approvals, with a look at 2015 and beyond. P T. 2015;40:191–205. [PMC free article] [PubMed] [Google Scholar]
- 37.Deeks E.D. Olaparib. First global approval. Drugs. 2015;75:231–240. doi: 10.1007/s40265-015-0345-6. [DOI] [PubMed] [Google Scholar]
- 38.Swisher E.M., Lin K.K., Oza A.M., Scott C.L., Giordano H., Sun J., Konecny G.E., Coleman R.L., Tinker A.V., O'Malley D.M., et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 39.Mirza M.R., Monk B.J., Herrstedt J., Oza A.M., Mahner S., Redondo A., Fabbro M., Ledermann J.A., Lorusso D., Vergote I., et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 40.Kim G., Ison G., McKee A.E., Zhang H., Tang S., Gwise T., Sridhara R., Lee E., Tzou A., Philip R., et al. FDA approval summary: olaparib monotherapy in patients with deleterious germline BRCA-mutated advanced ovarian cancer treated with three or more lines of chemotherapy. Clin. Cancer Res. 2015;21:4257–4261. doi: 10.1158/1078-0432.CCR-15-0887. [DOI] [PubMed] [Google Scholar]
- 41.Balasubramaniam S., Beaver J.A., Horton S., Fernandes L.L., Tang S., Horne H.N., Liu J., Liu C., Schrieber S.J., Yu J., et al. FDA approval summary: Rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin. Cancer Res. 2017;23:7165–7170. doi: 10.1158/1078-0432.CCR-17-1337. [DOI] [PubMed] [Google Scholar]
- 42.Scott L.J. Niraparib. First global approval. Drugs. 2017;77:1029–1034. doi: 10.1007/s40265-017-0752-y. [DOI] [PubMed] [Google Scholar]
- 43.Pennington K.P., Walsh T., Harrell M.I., Lee M.K., Pennil C.C., Rendi M.H., Thornton A., Norquist B.M., Casadei S., Nord A.S., et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin. Cancer Res. 2014;20:764–775. doi: 10.1158/1078-0432.CCR-13-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schettini F., Giudici F., Bernocchi O., Sirico M., Corona S.P., Giuliano M., Locci M., Paris I., Scambia G., De Placido S., et al. Poly (ADP-ribose) polymerase inhibitors in solid tumours: systematic review and meta-analysis. Eur. J. Cancer. 2021;149:134–152. doi: 10.1016/j.ejca.2021.02.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.