Abstract
Staphylococcus aureus can readily form biofilm which enhances the drug-resistance, resulting in life-threatening infections involving different organs. Biofilm formation occurs due to a series of developmental events including bacterial adhesion, aggregation, biofilm maturation, and dispersion, which are controlled by multiple regulatory systems. Rapidly increasing research and development outcomes on natural products targeting S. aureus biofilm formation and/or regulation led to an emergent application of active phytochemicals and combinations. This review aimed at providing an in-depth understanding of biofilm formation and regulation mechanisms for S. aureus, outlining the most important antibiofilm strategies and potential targets of natural products, and summarizing the latest progress in combating S. aureus biofilm with plant-derived natural products. These findings provided further evidence for novel antibiofilm drugs research and clinical therapies.
Keywords: Staphylococcus aureus, Biofilm, Antibiofilm strategies, Targets, Natural products
1. Introduction
Staphylococcus aureus is a drug-resistant pathogen that can cause skin and soft tissue infections, further leading to severe endocarditis, osteomyelitis, pneumonia, and other invasive diseases [1]. In addition to the wide panel of secreted virulence factors, biofilm formation is a significant feature that promotes the development of drug-resistance for S. aureus [2]. It has been reported that biofilm can provide significant survival advantages to microbial communities, resulting in up to a 1500-fold increase in strain resistance [3]. Accordingly, developing new antibiofilm agents against S. aureus is of urgent importance.
Natural products have been extensively explored as important sources of biocompatible antibiofilm agents to avoid the side effects of traditional antibiotics on human health and the environment. For instance, essential oils from plants [4,5], flavonoids [6,7], phenolic acids [8], and terpenoids [9,10] showed biofilm inhibitory and disrupting activity through different mechanisms, including reducing adhesins, destroying biofilm matrix and interrupting bacterial communication. Additionally, an increasing number of new antibiofilm targets of natural products have been studied using molecular biology methods and computer virtual screening [11,12]. The obtained findings provide the basis for screening natural and effective antibiofilm alternatives.
In light of biofilm's complex and rapid adaptability, this review provided an in-depth understanding of biofilm formation and regulation mechanism for S. aureus, outlining the promising antibiofilm strategies and potential targets of natural products. We analyzed the recent advancements, further elucidated the innovative approaches based on plant-derived natural products to combat S. aureus biofilms, and pointed out new directions for future investigations. Combining plant-derived natural products with other antibacterial compounds or techniques to increase the activity was also discussed, adding to the design of novel therapeutics to fight against this clinically significant pathogen.
2. Formation and properties of S. aureus biofilm
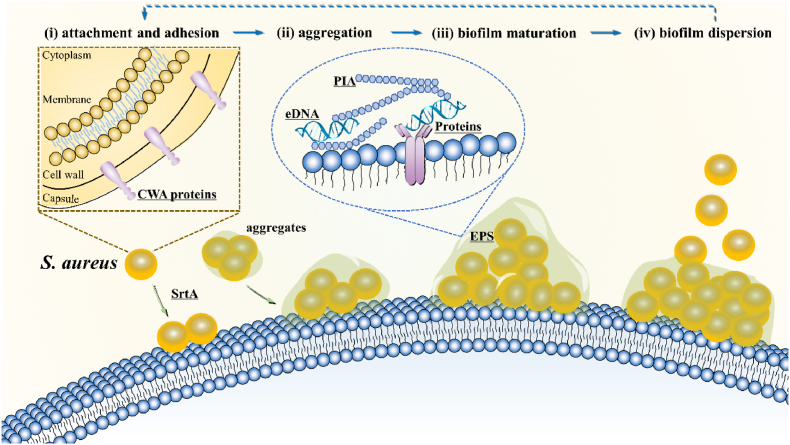
The biofilm development process can be didactically divided into the following four stages: (i) attachment and adhesion; (ii) aggregation with extracellular matrix synthesis and bacterial proliferation; (iii) biofilm structuring and maturation; and (iv) biofilm dispersion with cell detachment [13,14] (Fig. 1).
Fig. 1.
S. aureus biofilm formation process: (i) initial attachment and adhesion, in which single cells or aggregates adhere to surfaces; (ii) aggregation, with cell division and proliferation as well as EPS production; (iii) biofilm structuring and maturation, where microorganisms coexist within polymicrobial interactions; and (iv) biofilm dispersion, with cell detachment from the aggregate biofilm to planktonic state. EPS, extracellular polymeric substances; eDNA, extracellular DNA; PIA, Polysaccharide intercellular adhesin; CWA proteins, cell wall-anchored proteins; SrtA, sortase A. Key molecules of potential antibiofilm targets are underlined.
During the initial adhesion or aggregation stage, S. aureus planktonic cells attach to biotic or abiotic surfaces and are enclosed in complex covering composed of host macromolecules such as proteins [15,16]. During tissue infections, bacteria attach to each other, forming aggregates in viscous mucus (e.g., cystic fibrosis [17]) or on damaged host tissues (e.g., heart valves [18], bones [19], or skin of chronic wounds [20]).
During the maturation and proliferation stage, the attached or aggregated cells multiply and produce an extracellular polymeric substance (EPS), indispensable for constructing three-dimensional biofilm scaffolds [21]. EPS molecules, including polysaccharides, nucleic acids, proteins, and lipiden biofilm cells to provide mechanical stability for the biofilm, thus directly determining the living conditions of the cells [22].
In the biofilm diffusion stage, the biofilm is destabilized with cells and aggregates, dispersing into the environment, attaching to new locations and triggering new biofilm infections. New biofilms locations hinge on the transport opportunity of the detached bacteria, resulting in new infections in nearby tissues, such as implant biofilms causing osteomyelitis [23,24], or farther locations, such as detached biofilms causing endocarditis [25].
Bacterial cells divide and the matrix complexity increase, creating physiological heterogeneity inside the biofilm that is characterized mainly by gradients of nutrients and oxygen [26]. The cells growing within the biofilm are generally categorized into four metabolic states: (i) aerobic (located in the oxygenated and nutrient-rich outer layer); (ii) fermentative (located in the oxygen- and nutrient-poor inner layer); (iii) dormant (located in anoxic layer with slow growth and inactive metabolism); and (iv) dead [[27], [28], [29]]. Dormant cells can result in a drop in intracellular adenosine triphosphate, making bacteria less sensitive to antibiotics [30]. Besides, some other gradients are present in biofilms. For instance, a vertical gradient of viscoelasticity is established during the preliminary stages of S. aureus biofilm development, which also facilitates biofilm dispersal, removing loosely bound bacteria while retaining an entrenched layer in the biofilm structure [31]. Rivera et al. revealed stark lipidomic heterogeneity in S. aureus biofilm, demonstrating that each horizontal layer was molecularly distinct [32]. Future applications of these findings to study spatially localized molecular responses to antibiofilm agents could provide new therapeutic strategies.
3. Key molecules in S. aureus biofilm formation
The degradation of the EPS matrix is of particular relevance for antibiofilm measures. Thus far, various agents have been applied to remove single-species and mixed-population biofilms, primarily by degrading self-produced adhesins, nucleic acids, and polysaccharides [[33], [34], [35], [36]].
3.1. Polysaccharide intercellular adhesin (PIA)
PIA is the majority of EPS components for S. aureus biofilm, which has a vital role in several aspects, including colonization, biofilm formation, immune evasion, and antibiotic resistance [37,38]. PIA production is strongly dependent on the environmental conditions such as anaerobiosis and glucose [37] and controlled by different regulatory systems [39]. Over the years, a variety of regulatory proteins and genes have been found to regulate ica expression, which might underlie the differential expression of PIA in different staphylococcal strains [40]. The PIA-dependent biofilm is predominantly observed in methicillin-sensitive S. aureus strain [37,38]. In this mechanism, PIA makes the polymer process a net positive charge through deacetylation to complete cell attachment and intercellular adhesion, increasing biofilm retention and drug resistance [41]. Biochanin A is a natural isoflavonoid that can prevent biofilm formation by inhibiting the release of PIA and disintegrating the preformed biofilms by dissociated EPS matrix [42]. Furthermore, the natural (+)-nootkatone significantly prevented the S. aureus biofilm formation with an inhibition rate of > 90 % at 50 μg/mL by depressing the icaA expression [43]. Moreover, PIA connects to various proteins with different effects. For example, biofilm-associated protein (Bap) cooperates with PIA to promote cell-to-cell aggregation during the process of biofilm formation [44], and PIA interacts with the accumulation-associated proteins (Aap) to promote the maturation of staphylococcal biofilm [45]. Therefore, ica operon and proteins such as Aap and Bap might be considered potential targets in the PIA-dependent biofilm.
3.2. Extracellular proteins
At the beginning of biofilm formation, adhesion to the surfaces is the main strategy for S. aureus cells to invade host cells [46]. In this process, S. aureus expresses a large number of surface proteins covalently linked to peptidoglycans, also known as cell wall-anchored (CWA) proteins. Microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), the most prevalent CWA proteins, include protein A, fibronectin-binding proteins (FnBPs), clumping factors (ClfA, ClfB), serine-aspartate repeat family proteins (SdrC, SdrD, and SdrE), biofilm-associated protein (Bap), fibrinogen-binding protein (Fib), and S. aureus surface protein (SasG) [47,48]. They facilitate the adhesion of S. aureus cells to EPS and the host cells, thus promoting biofilms accumulation [49]. Natural products, such as luteolin, quercetin, and kaempferol from Euphorbia humifusa have been reported to suppress biofilm formation by down-regulating these surface adhesion proteins encoding genes (e.g., fnbpA, fnbpB) [50].
Transpeptidation mediated by membrane sortase A (SrtA) is essential for anchoring CWA proteins to the cell wall envelope. Knockout of SrtA could inhibit the assembly of surface adhesins in the cell wall envelope and the impact of acute infection [51,52]. Many natural products have been investigated as inhibitors of SrtA. For instance, kaempferol was found to inhibit SrtA activity and down-regulate the expression of CWA proteins-related genes, thus inhibiting the formation of S. aureus biofilm [53]. Likewise, chalcone, quercetin, and myricetin were also found to exert effective inhibitory activity against SrtA [54,55]. Moreover, molecular dynamics simulations and mutagenesis assays of chalcone against SrtA implied that the inhibitory activity lies in the interactions between chalcone and SrtA residues Val168, Ile182, and Arg197 [55]. Therefore, interference with surface proteins anchoring by targeting the corresponding genes or SrtA and modulating the extracellular enzymes by targeting the global regulators are promising antibiofilm strategies to combat S. aureus infections.
3.3. Extracellular DNA (eDNA)
eDNA exerts various biological functions, including adhesion, gene transfer, and DNA damage repair [56,57]. Besides, in Pseudomonas aeruginosa biofilms, eDNA cross-linking with lipoprotein anchored in the membrane can connect the matrix to microbial cells within the biofilm and protect against enzymatic attack from DNase [58]. Moreover, the holin-like protein (CidA) was indicated to positively increase the release of eDNA during biofilm development [59]. Emodin, an anthraquinone derivative obtained from Polygonum cuspidatum and Rheum palmatum, decreased S. aureus biofilm growth by intervening in the release of eDNA and downregulating the expression cidA [60]. Thus, eDNA could be a target for anti-biofilm agent screening.
As recently reported, Z-form eDNA accumulates as biofilm matures, and the compounds driving Z-DNA into B-DNA, such as chloroquine, were found to remarkedly disrupt extant biofilm EPS [61]. Universally, the two-member DNABII family of proteins acts as linchpin proteins to stabilize the structure of eDNA required for the biofilm matrix's stability [62]. Therefore, targeted removal of DNABII proteins could result in rapid biofilm collapse, releasing resident bacteria more susceptible to antibiotics and host immune effectors [61,63].
3.4. Phenolsoluble modulins (PSMs)
PSMs, a class of small peptides with an α-helical structure and resulting surfactant-like characteristics, are produced by most staphylococcal species, especially S. aureus and S. epidermidis [64]. S. aureus secretes four shorter PSMα peptides (∼20 amino acids), two longer PSMβ peptides (∼40 amino acids), and the RNAIII-encoded δ-toxin [65]. PSMs have been documented as determinants of S. aureus virulence [65] and potential peptides to stimulate inflammatory responses [66]. With the given surfactant-like properties, PSMs facilitate biofilm structuring to form channels which makes nutrition available to S. aureus in the deeper layer of the biofilm and promote biofilm detachment to free bacteria [67,68]. In S. aureus, PSMs are soluble and can aggregate into the amyloid fibers to stabilize the biofilm structure [69]. Sinomenine is a natural alkaloid isolated from Sinomenium acutum that significantly reduces S. aureus biofilm dispersal due to cell-cell adhesion, PIA, and PSMs production [70]. Hence, PSMs might represent a potential research target.
4. Promising targets in the regulation of S. Aureus biofilm formation
Biofilm formation includes herd behavior. Multiple regulatory systems strictly control each step, from the initial attachment to the maturation and dispersion of biofilm.
4.1. Quorum ensing(QS) system
QS system is an internal communication system of bacteria, where the expression of relevant genes is initiated when the changes in the signal molecules reach a certain threshold. It involves multiple signal transduction pathways that regulate biofilm formation, virulence, motility, and sporulation [48]. In S. aureus, the research on QS system mainly focused on the accessory regulatory factor (Agr) system and LuxS/autoinducer-2 (AI-2) system [[71], [72], [73]].
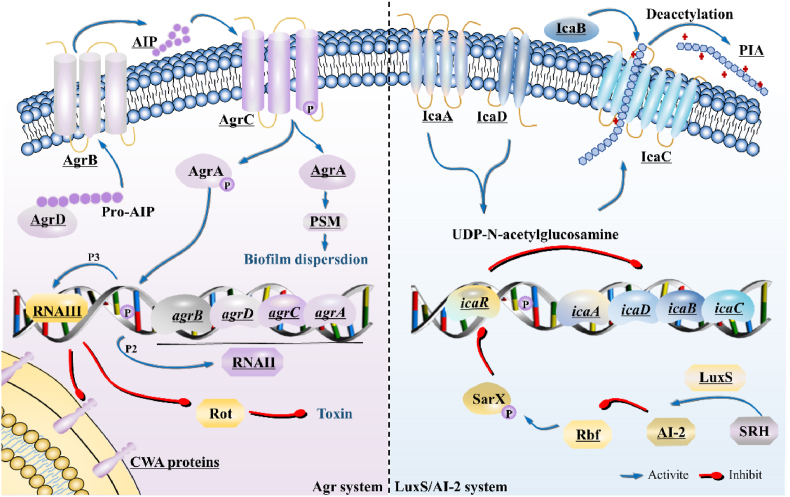
The Agr system in S. aureus (Fig. 2) contains RNAII and RNAIII, activated by the promoters P2 and P3, respectively [74]. The RNAII consists of agrB/D/C/A genes, encoding AgrB/D/C/A proteins required for auto-inducing peptide (AIP) biosynthesis, transport, and regulation of target genes [75]. AgrD, the precursor of AIP, is synthesized and exported to the plasma membrane by a feat of AgrB [76]. The accumulated AIP then binds to and activates the histidine kinase AgrC, leading to autophosphorylation and activating the signal transduction process, and eventually inducing a positive feedback loop on P2 and P3 promoters [77]. In addition, the elevated AIP can promote biofilms to depolymerize by increasing the secretion of extracellular protease [48]. RNAIII, as an important effector, upregulates genes encoding exoproteins, including haemolysins, toxins, and exoproteases, and downregulates genes encoding surface adhesins, such as FnBPs and serine-aspartate repeat family proteins [78,79]. Activation of the Agr system could be considered as an attractive antibiofilm strategy since it does not only inhibit the biofilm formation but also disperse the biofilm. Brazilin, a flavonoid from Caesalpinia sappan, has also been reported to inhibit biofilm formation by manipulating the Agr-related function [80]. However, Agr system can positively increase virulence factors, providing a mechanism for bacteria to rapidly adapt to changing environmental conditions. Hence, precisely manipulating the Agr system to inhibit biofilm formation without increasing virulence poses a great challenge. As practical application of this strategy would turn on the autoinduction of all four Agr subgroups, a promising reagent could be a cocktail of “clean” activators, each of which stimulates one or more agr variants without extensively affecting the rest [77]. The activation of AgrC involves an impediment to an intrasteric inhibitory docking interaction, providing a potential strategy for inhibiting biofilms by activating AgrC [81].
Fig. 2.
Mechanism of QS system for S. aureus biofilm regulation. In the Agr system, RNAII consists of agrB/D/C/A genes, encoding the AgrB/D/C/A proteins required for the biosynthesis, transport, and target gene regulation of AIP. RNAIII, as an essential effector, upregulate genes encoding exoproteins and downregulate genes encoding surface adhesins. In the LuxS/AI-2 system, LuxS catalyzes the synthesis of AI-2, inhibits Rbf and promotes icaR expression to repress IcaA/D/B/C, ultimately reducing the formation of PIA-dependent biofilms. QS, Quorum sensing; Agr, accessory gene regulator protein locus; AIP, auto-inducing peptide; PSM, phenol-soluble modulin; LuxS, S-ribosylhomocysteine lyase; AI-2, interspecies autoinducer. Key molecules of potential antibiofilm targets are underlined.
The S-ribosylhomocysteine lyase (LuxS)/AI-2 system (Fig. 2) regulates the formation of PIA-dependent biofilms by modulating the transcriptional regulation of intercellular adhesin (ica) locus [82]. Specifically, PIA is synthesized via proteins IcaA/D/B/C. IcaA and IcaD synergistically synthesize UDP-N-acetylglucosamine, followed by being exported through IcaC. Afterward, IcaB regulates the partial deacetylation of PIA to increase positive charge, thus improving adhesion [39]. LuxS can catalyze the biosynthesis of AI-2 and suppress the expression of rbf, thus promoting the expression of icaR, reducing the transcription level of icaA, and reducing PIA-dependent biofilm formation [72,83]. Paeoniflorin, a monoterpenoid glycoside extracted from Paeoniaceae plants such as peony, can reduce the luxS/AI-2 system-controlled biofilm formation and virulence [84]. Biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum were found to interfere with the release of AI-2 [11]. Thus, the LuxS/AI-2 QS system can be a competitive target in intercellular communication.
4.2. Two-component signal transduction systems (TCSs)
TCSs comprise two components, i.e., (i) sensory histidine kinase (HK), which senses the external environmental stimuli, and (ii) response regulator (RR), which regulates downstream target gene expression and quickly enhances the adaptive viability of bacteria [85]. Besides the Agr system mentioned above, TCSs in S. aureus, such as the YycFG and SaeRS systems, have emerged as novel targets against biofilms [86,87].
YycFG, also known as VicRK or WalRK, is the only essential two-component regulator contributing to bacterial pathogenicity and biofilm formation in S. aureus [88]. YycF was reported to have the ability to directly regulate the predicted promoter regions of the biofilm formation-related genes (e.g., sarA and icaA), and antisense yycG RNA (ASyycG) strategy successfully inhibited the transcription of sarA and icaA, thus decreasing the biofilm amass [86]. In S. aureus, YycFG also controls the autolysin synthesis related to cell wall metabolism and biofilm formation [89]. Overall, these data imply that inhibition of YycFG could reduce biofilm formation and bacterial pathogenicity, providing a promising target for the management of S. aureus infections. Rhodomyrtone, isolated from Rhodomyrtus tomentosa, has been speculated to interfere with the YycFG system, hence suppressing the expression of MRSA on the secreted proteins such as exoenzymes and antigenic proteins [90].
SaeRS has an important role in the regulation of α-toxin, β-haemolysin, staphylococcal immunoglobulin-binding protein, leucocidin, and toxic shock syndrome toxin-1, etc. [91]. In SaeRS TCS, the HK SaeS controls the expression of several exoproteins, including α-hemolysin and FnBPs [87], which is a potential therapeutic target during S. aureus biofilm formation. Xanthoangelol B, a prenylated chalcone obtained from Angelica keiskei Koidzumi, was found to bind directly to SaeS and inhibit its histidine kinase activity, demonstrating a possibility of a broad-spectrum inhibitor of histidine kinases [92].
4.3. SarA family proteins
The formation of S. aureus biofilm is also controlled by SarA family proteins, mainly including SarA, Rot, and MgrA. SarA can directly upregulate the expression of exoproteins and interconnect with the Agr system, where it can repress the production of extracellular proteases during the biofilm formation [93]. As a transcriptional activator, SarA enhances the transcription of ica operon and the production of PIA precursor to activate biofilm development [93]. Rot, an important regulator of virulence and biofilm-related genes expression in S. aureus, functions as a positive regulator of ClfB, SdrC, and SarS, which increases the surface protein level and decreases the extracellular enzyme level [94]. In S. aureus, RNAIII can regulate the synthesis of Rot via blocking its translation [95]. MgrA acts as a negative regulator against biofilm formation by repressing the production of adhesins [96]. (+)-Nootkatone treatments were found to down-regulate the expression of the sarA gene, thereby decreasing the PIA production and biofilm formation [43].
4.4. Alternative sigma factor σB (SigB)
SigB can promote the initial biofilm formation and repress the biofilm dispersal. Additionally, SigB indirectly regulates the biofilm formation by influencing other regulatory systems, e.g., by repressing RNAIII and SaeRS TCS, and upregulating the sarA expression according to the corresponding environments [27]. A number of natural products have been found to manipulate SigB to inhibit biofilm formation. For example, Ginkgo biloba exocarp extract was reported to downregulate the expression of the MRSA biofilm-associated factors sarA and sigB [97]. Similarly, Scrophularia ningpoensis honey can down-regulate the expression of genes icaA, icaD, sarA, agrA, and sigB in S. aureus [98].
5. Emerging natural products-based therapeutics against S. aureus biofilm
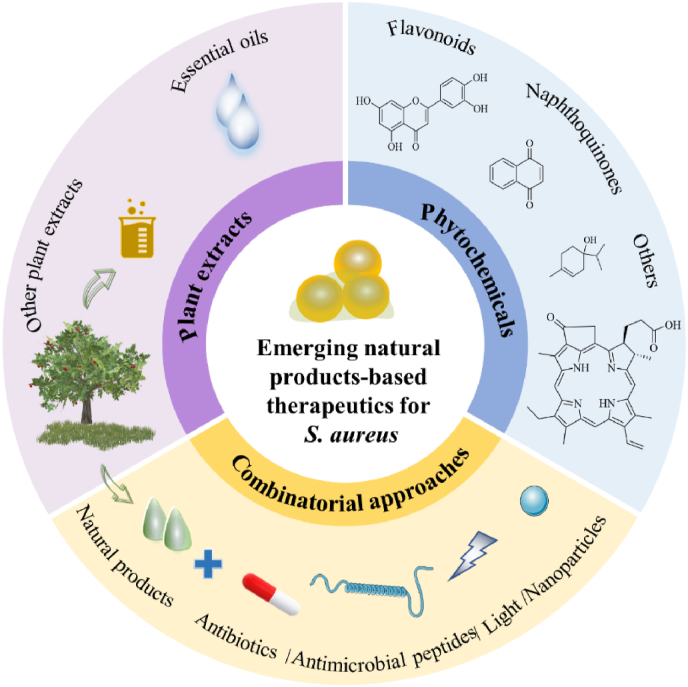
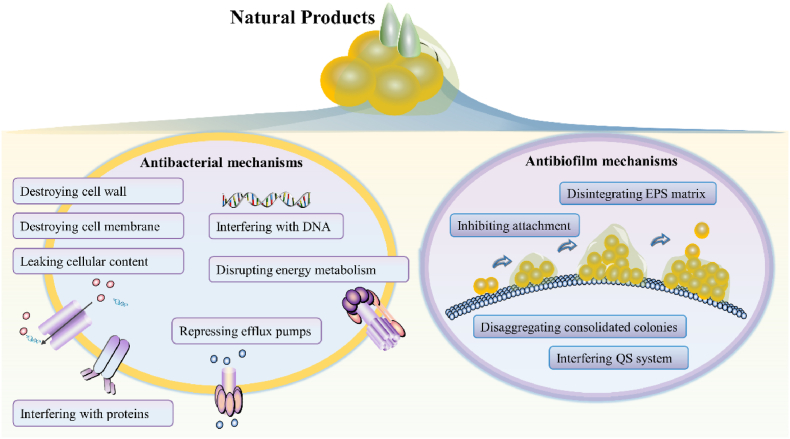
Natural products have represented powerful therapeutics against pathogens since the golden age of antibiotics in the mid-20th century [99]. Since combinatorial approaches have yielded effective drugs, developing new antibiofilm agents around different natural products (Fig. 3), such as plant extracts (Table 1) and phytochemicals (Table 2), is likely to lead to promising new strategies to combat biofilm. The antibiofilm activity of natural compounds primarily lies in certain aspects, including inhibition of bacterial cell attachment and adhesion, suppression of polymer (e.g., ECM) formation, reduction in the generation of pathogenic factors, and interruption of QS system. The current antibacterial and antibiofilm mechanisms of natural products are summarized in Fig. 4. To be note, although natural products have long been considered as nontoxic, on many occasions they have not been as safe as commonly thought [100,101]. Hence, it's definitely needed to study its toxicity before a natural product being nominated as a high-priority candidate in clinical treatments.
Fig. 3.
Novel natural products-based therapeutics for S. aureus.
Table 1.
Recently emerging plant extracts with antibiofilm activity.
| Source | Extract | Strain | MIC (μg/mL) | Biofilm formation inhibition | Mature biofilm disruption | Target | In vivo | Ref. |
|---|---|---|---|---|---|---|---|---|
| Acacia macrostachya | Stem barks, MeOH | S. aureus | >500 | >60 %, 3.9 μg/mL |
NS | QS | N | [118] |
| Allium subhirsutum | Bulbs, H2O | S. aureus | 10000 | 56.21 %, 10 mg/mL |
NS | QS | N | [116] |
| Aphanamixis polystachya | Leaves, DCM, MeOH |
MRSA | 6700 | >40 %, 125 μg/mL |
>20 %, 125 μg/mL |
NS | N | [115] |
| Artocarpus heterophyllus | Heartwood, EtOH | MRSA | 31.2–62.5 | 60 %, 31.2 μg/mL |
NS | NS | N | [134] |
| Azadirachta indica | Leaves, EtOH |
MRSA | 1000 | 78.2–91.8 %, 1∼32 mg/mL |
NS | NS | N | [113] |
| Capsicum annuum | Fruits, MeOH |
S. aureus | 64 | 53.8 %, 64 μg/mL |
NS | NS | Y | [114] |
| Catharanthus roseus | Flowers, EtOH |
MRSA | 1000 | 54.5–99.7 %, 1∼32 mg/mL |
NS | NS | N | [113] |
| Cistus laurifolius, C. monspeliensis, C. parviflorus, C. salviifolius | Leaves, EtOH |
S. aureus | 128∼256 | NS | NS | NS | N | [111] |
| Crithmum maritimum | Leaves, EtOH |
S. aureus | NS | 82 %, 1 mg/mL |
NS | NS | N | [112] |
| Croton blanchetianus | Leaves, H2O |
MRSA | 780 | 100 %, 780 μg/mL |
NS | NS | N | [103] |
| Croton conduplicatus | Leaves, H2O |
MRSA | 512 | 22 %, 512 μg/mL |
27 %, 512 μg/mL |
NS | N | [104] |
| Cuminum cyminum | Seeds, H2O |
MDR S. aureus | 1.25–5 | NS | NS | QS | N | [105] |
| Eruca sativa | Plants, EtOH |
S. aureus | 125 | 67.5 %, 125 μg/mL |
73.45 %, 125 μg/mL |
EPS; SrtA | N | [121] |
| Glycyrrhiza glabra | Plants, EtOH |
S. aureus | NS | NS | 99.7 %, 400 μg/mL |
NS | N | [137] |
| Gmelina arborea | Leaves, H2O |
S. aureus | 90 | 46 %, 1 mg/mL |
NS | NS | N | [149] |
| Illicium verum | Fruits, MeOH |
MDR S. aureus | 4800 | 74 %, 2.4 mg/mL |
NS | AgrA, SarA | N | [119] |
| Melia azedarach | Leaves, DCM, MeOH |
MRSA | 2420 | >50 %, 125 μg/mL |
>60 %, 125 μg/mL |
NS | N | [115] |
| Myrsine umbellata | Leaves, H2O, EtOH |
S. aureus | 1250 | NS | 84.28 %, 1.25 mg/mL |
NS | N | [106] |
| Origanum vulgare | Commercial | S. aureus | 48 | >40 %, 6 μg/mL |
54.05 %, 192 μg/mL |
QS | N | [107] |
| Polyalthia longifolia | Leaves, MeOH, EtOH |
MRSA | 39∼78 | 99.5 %, 39 μg/mL |
NS | NS | N | [135] |
| Viburnum opulus | Fruits/barks, acetone, EtOH | S. aureus | ≥1000 | ∼20.4 %, 100 μg/mL |
NS | SrtA; SpA | N | [120] |
N, no; Y, yes; NS, not specified; DCM, dichloromethane; MeOH, methanol; EtOH, ethanol; MIC, minimum inhibitory concentration; MRSA: methicillin-resistant S. aureus; MDR, multidrug-resistant.
Table 2.
Recently emerging natural compounds with antibiofilm activity.
| Compound | Structure | MIC (μg/mL) | MBIC (μg/mL) | Biofilm formation inhibition | Mature biofilm disruption | Target | In vivo | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1,4-NQ | 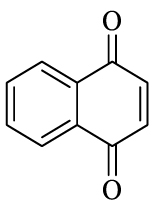 |
≤100 | NS | 55 ± 2 %, 10 μg/mL |
ROS | N | [128] | |
| 3-HBA | 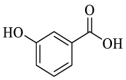 |
400 | NS | 62.69 %, 6.25 μg/mL |
NS | AgrA, SarA | N | [119] |
| Biochanin A | 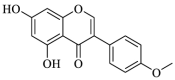 |
64∼256 | 128∼512 | 39.8–58.2 %, 32 μg/mL | 24.8–52.1 %, 128 μg/mL | EPS, icaA, srtA, eno | N | [42] |
| Glabridin | 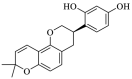 |
6.25 | <50 | NS | NS | NS | N | [137] |
| Kaempferol | 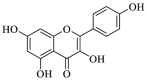 |
512 | 512 | 70∼80 %, 64 μg/mL |
35∼55 %, 128 μg/mL |
fnbpA, fnbpB | N | [50] |
| Luteolin | 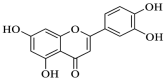 |
32∼64 | 32 | 99.72 %, 64 μg/mL |
88.22 %, 128 μg/mL |
fnbpA, fnbpB, icaA, clfA | N | [50] |
| Menadione | 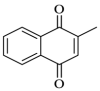 |
64∼256 | NS | >90 %, 64 μg/mL |
>85 %, 1024 μg/mL |
ROS | N | [129] |
| Nerolidol | 1000 | ∼500 | >70 %, 1 mg/mL |
NS | NS | N | [131] | |
| Pyropheophorbide A | 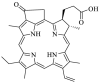 |
NS | NS | NS | NS | SrtA | N | [121] |
| Quercetin | 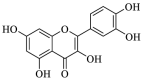 |
256 | 128 | 70∼80 %, 64 μg/mL |
35∼55 %, 128 μg/mL |
fnbpA, fnbpB | N | [50] |
| Shikonin | 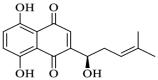 |
15.6 | <15.6 | ∼50 %, 15.6 μg/mL |
NS | icaA, fnbpA, agrA, saeS, sigB | N | [130] |
| Sinomenine | 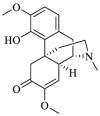 |
>73.2 | NS | NS | 26.29 %, 9.1 μg/mL |
agrA, icaA, PIA, PSM | N | [70] |
| Terpinene-4-ol | 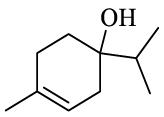 |
48 | NS | >40 %, 12 μg/mL |
70.97 %, 192 μg/mL |
QS | N | [107] |
N, no; NS, not specified; MBIC, minimal biofilm inhibitory concentration.
Fig. 4.
Antibacterial and antibiofilm mechanisms of natural products.
5.1. Plant extracts
5.1.1. Essential oils (EOs)
EOs, volatile compounds extracted from medicinal plants, have shown great therapeutic potential against microbial biofilms [[102], [103], [104], [105]]. The EOs extracted from Croton species (e.g., C. blanchetianus and C. conduplicatus) can inhibit biofilm formation and reduce preformed biofilms of MSSA and MRSA strains [103,104]. Saponins, alkaloids, flavonoids, free steroids, and tannins could be responsible for the eradication of biofilm, since they act by dehydrating the cell wall, disaggregating consolidated colonies, preventing nutrient replacement, and disintegrating the structure of mature biofilm [106]. Sharifi et al. further investigated the antibiofilm mechanism of Cuminum cyminum essential oil (CcEO) against multidrug-resistant (MDR) S. aureus. In the case of QS inhibitory potential, the CcEO at sub-MIC concentrations (0.625–1.25 μL/mL) significantly reduced the hld and ica expression by 3.13- and 2.33-fold, respectively [105]. In addition, Origanum vulgare essential oil at a concentration of 1/8 MIC (6 μg/mL) exerted an anti-attachment effect against S. aureus, while the effect of terpinene-4-ol within this EO was observed at 1/4 MIC [107]. It has been demonstrated that using EOs to inhibit pathogenic bacteria attachment is a strategic way to prevent biofilm formation. Meanwhile, the O. vulgare EO and terpinene-4-ol at the concentrations of MIC∼4 MIC eradicated the mature biofilms by 10.36–54.05 % and 62.28–70.97 %, respectively [107]. Therefore, O. vulgare EO was more effective against S. aureus biofilm formulation, while terpinene-4-ol was superior in eradicating mature biofilms. Additionally, O. vulgare EO presented better anti-QS activity than terpinene-4-ol in a concentration-dependent manner [107]. Overall, the synergistic effects between various compounds in EOs may lead to the interference of the QS system and subsequently affect the ability to develop biofilms and generate virulence factors in bacteria. The superiority of EOs may also originate from the non-polar components with high diffusion coefficients that easily penetrate the biofilm and damage membrane fluidity [108]. Noteworthy, EOs have a low potential to develop microbial resistance due to multiple targets [109] and the inhibition of the function of efflux pumps [110]. Therefore, EOs can be used in combination with antibiotics since they mediate the re-sensitization of the bacteria to some drugs, which helps address the problem of antibiotic resistance.
5.1.2. Other plant extracts
The hydroethanolic extracts of numerous plants have been reported to significantly reduce the S. aureus biofilm formation, which could be explained by the presence of chlorogenic, quercetin, and rutin [[111], [112], [113]]. Likewise, due to rich and diverse phytochemical compositions, the methanolic extract of Capsicum annuum at 64 μg/mL showed an inhibition rate of 53.8 % against S. aureus biofilm, comparable to amoxicillin [114]. Aphanamixis polystachya and Melia azedarach extracts at sub-lethal concentrations were found to inhibit and disrupt the biofilms formed by MRSA, residing in the existence of limonoids, phenolics, and oxygenated triterpenoids [115]. Recently, researchers have been exploring the mechanism of plant extracts against biofilms. For instance, Allium subhirsutum aqueous extract proved to interrupt QS system against a diverse panel of microorganisms. By using gas chromatography-mass spectrometry, the potential bioactive compounds were identified as 5-hydroxymethylfurfural, methyl methanethiolsulfonate, furfural, trisulfide, di-2-propenyl, and diallyl disulfide [116]. In particular, 5-hydroxymethylfurfural at 125 μg/mL have been previously reported to inhibit the S. aureus biofilm formation by 82 %, which might be the contributing component of A. subhirsutum [117]. The possible quenching of QS mechanism was also demonstrated in the crude extract of Acacia macrostachya [118]. In addition, the methanolic extract of Illicium verum showed a biofilm inhibition effect with 3-hydroxybenzoic acid (3-HBA) as the primary active constituent, which strongly interacted with the active site residues of SarA and AgrA of S. aureus [119]. Moreover, the inhibition of SrtA and staphylococcal protein A (SpA) expression, as well as EPS production, are emerging targets of extracts from plants such as Viburnum opulus [120] and Eruca sativa [121].
5.2. Phytochemicals
5.2.1. Flavonoids
Novel natural antibiofilm agents comprise phenolics, terpenoids, alkaloids, polypeptides, lectins, and polyacetylenes [122]. Flavonoids, an important class of phenolics, are well known as antibacterial agents against a wide range of pathogenic microorganism through inhibition of the cell attachment and biofilm formation, alteration of the membrane permeability, inhibition of nucleic acid synthesis, inhibition of energy metabolism, inhibition of the porin on the cell membrane, inhibition of cytoplasmic membrane function, and attenuation of the pathogenicity [123]. Luteolin, quercetin, and kaempferol are common flavonoids found in medical plants, fruits, and vegetables. Quercetin has been found to remarkably reduce the production of elastase, protease and pyocyanin in Pseudomonas aeruginosa, violacein in Chromobacterium violaceum, as well as EPS in Yersinia enterocolitica, inhibiting biofilm formation and disrupting the preformed biofilm [124,125]. Recently, the expression levels of biofilm-related genes in S. aureus were found to be decreased by luteolin, quercetin, and kaempferol at concentrations of 8∼128 μg/mL. In conjugation with luteolin, quercetin, and kaempferol can elicit a more effective antibiofilm effect by synergistically down-regulating fnbpA and fnbpB expression [50]. Another study also revealed that upon luteolin treatment, the expression of key genes involved in virulence (hla) and biofilm formation (icaA, fnbpA, and clfA) was downregulated in the wild-type S. aureus strain [126]. The inefficacy of luteolin with respect to the virulence factor in the agr mutant strain suggested the agr-mediated anti-virulence and antibiofilm potential of flavonoids.
5.2.2. Naphthoquinones
Among various antimicrobial agents, naphthoquinones and their derivatives have received great attention due to their exceptional structural diversity and varied biological properties [127]. 1,4-Naphthoquinone (1,4-NQ) at a concentration of 10 μg/mL was found to inhibit the S. aureus biofilm formation by 55 %, where the microbial motility was reduced. Besides, 1,4-NQ increased the cellular accumulation of reactive oxygen species (ROS), which could impact biofilm formation [128]. In their study, Mone et al. reported that menadione, an analogue of 1,4-NQ, was effective against different MDR strains of S. aureus not only by inhibiting biofilm formation (>90 % at MICs ranging from 64 to 256 μg/mL) but also by removing preformed biofilms (>85 % at 1024 μg/mL), accompanied by enhanced levels of ROS [129]. Likewise, shikonin (SKN), a highly liposoluble 1,4-NQ derivative extracted from the root of Lithospermum erythrorhizon, was found to inhibit the biofilm formation of clinical MRSA strains at concentrations lower than MIC (15.6 μg/mL). Further mechanism studies showed that SKN interferes with the expression of icaA and fnbpA, indicating that SKN could inhibit cell attachment during biofilm formation. In addition, SKN at sub-inhibitory concentrations down-regulated the expression of virulence factor regulatory genes (agrA, saeS, and sigB), subsequently affecting the transcription of exoprotein-coding genes (hla and sea), and further elucidating the relevance of SKN-induced reduction of MRSA virulence factors [130]. Overall, naphthoquinones possessing antibacterial and antibiofilm activities are imperative in the era of drug-resistance developed by bacterial pathogens.
5.2.3. Other natural compounds
Alkaloids, aromatic acids, and sesquiterpenes are natural compounds with antibiofilm activities against S. aureus. For example, the alkaloid sinomenine can significantly upregulate agrA expression and down-regulate icaA level [70]. Aromatic acids such as 3-HBA inhibit S. aureus biofilm activity targeting the Agr and SarA systems [119]. Nerolidol, a sesquiterpene, was found to inhibit S. aureus biofilm by > 70 % at concentrations ranging from 1 to 4 mg/mL [131].
5.3. Combinatorial approaches
5.3.1. Combination of different compounds
Due to limited therapeutic options for S. aureus, identifying effective combinations provides an alternative for infection treatment. For instance, besides having significant biological activity, curcumin-based metal complexes enhance the bioavailability of curcumin. At the concentration of 100 μM, curcumin inhibited S. aureus biofilm formation (55.8 %), whereas oxovanadium complex of curcumin has a significantly stronger effect (82 %), which could reside in the synergistic effect of complex mechanisms, including alkaline phosphatase inhibition and antibacterial mechanism [132].
The antibiofilm effects of combinational terpenes including (−)-trans-caryophyllene, (S)-cis-verbenol, (S)-(−)-limonene, (R)-(+)-limonene, and linalool were also evaluated. As shown in the results, all combinations of terpenes inhibited biofilm formation by > 50 % without affecting bacterial growth. Of note, (−)-trans-caryophyllene and linalool at 500 μg/mL for each acquired the most superior effect with an inhibition percentage of 88 %, resulting from the down-regulation of sdrD, spa, agr, and hld genes related to cell adhesion and QS system, as well as the up-regulation of cap5B and cap5C genes associated with the production of capsular polysaccharides [133]. Taking advantage of different mechanisms, the combinations are expected to achieve enhanced effects, thus overcoming the resistance of S. aureus.
5.3.2. Combination of plant extract and antibiotic
Because pure antibiotic or natural product therapy might be insufficient to combat drug-resistant bacterial infections, combining plant extract and antibiotics is a potential strategy for improving efficacy. Bazmi et al. demonstrated that the combination of artocarpin-rich Artocarpus heterophyllus extract with ampicillin synergistically altered the membrane permeability of MRSA, which led to the release of intracellular materials. Moreover, the tested components revealed that high-level suppression of biofilm formation (62–76 %) at their MICs, which could potentiate the antibacterial effects of each other at their sub-inhibitory concentrations (1/2∼1/16 MIC) in the synergistic cocktails by increasing the penetration of compounds into the matrix of biofilm [134]. Likewise, Polyalthia longifolia leaf extracts showed a synergistic action with penicillin against a clinical MRSA isolate and significantly inhibited the biofilm formation, implying their combinational antibiofilm potential [135]. Sannat et al. explicitly reported that the methanolic extract of Hemidesmus indicus root synergized the antibiofilm activity of amoxicillin and clindamycin against MRSA isolates. Moreover, in the kidney and liver of MRSA-infected mice, the combinations significantly reduced bacterial load, disease activity score, and Gram-positive spots [136]. Therefore, the combination of plant extracts with antibiotics is advocated in the treatment of biofilm-associated infections. In particular, the bioactive compound-rich plant extracts can be used in place of pure compounds in pharmaceutical industries, given the lower cost of production.
5.3.3. Combination of the natural compound and antimicrobial peptide
The ethanol extract of Glycyrrhiza glabra showed potent biofilm eradication activity against S. aureus through glabridin with the minimum biofilm eradication concentration of 50 μg/mL. Furthermore, glabridin combined with the antimicrobial peptide ɛ-poly-l-lysine resulted in broad, more potent biofilm eradication activities. The synergistic effect was speculated to occur due to the enhanced permeability of microbial cell membrane in biofilms by ɛ-poly-l-lysine, allowing more glabridin to transport into the cell, thus generating ROS and causing oxidative damage to cellular structure, lipids, proteins, and DNA [137].
5.3.4. Combination of the natural compound and photodynamic therapy
As a nonantibiotic microbicidal technology, photodynamic therapy relying on blue light has been extensively studied to disinfect various bacteria, including MRSA [138]. Lu et al. reported that blue light and carvacrol synergistically killed various bacteria, including planktonic cells, biofilm cells, and per-sisters. Carvacrol at 0.2 mg/mL and blue light 450 nm at 75 J/cm2 decreased the thickness of in vitro MRSA biofilm from 32.4 μm to 1.7 μm and completely or substantially cured both acute and established MRSA biofilm-associated infections of full thickness murine third-degree burn wounds. Further mechanistic studies showed that carvacrol was photocatalyzed to a series of photoreactive substrates, which underwent photolysis or additional photosensitization reactions and thus generated robust cytotoxic ROS [139]. Recently, Sharma et al. used alginate microfibers as a carrier of curcumin, followed by being irradiated with blue light (450 nm) to assess the efficacy of the combined therapy against MRSA. The curcumin-loaded microfibers exhibited great potential when combined with photodynamic therapy, which resulted in the complete eradication of the MRSA biofilms [140]. These results suggested that the combination of natural products and photodynamic therapy is a potent tool against biofilm-forming bacteria that can reduce their antimicrobial resistance.
5.3.5. Combination based on natural compound and nanoparticle
The rapid development of nanoparticles (NPs) in drug delivery systems has led to many studies exploring the antibacterial and antibiofilm effects of NPs and the potential for resistance [[141], [142], [143], [144], [145]].
Cerium oxide (CeO2) NPs prepared from aqueous leaf extract of Pometia pinnata at 512 μg/mL showed inhibition on S. aureus biofilm by 73 % [146]. However, in the case of Zr/Sn-dual doped CeO2 NPs, only the 10 % Zr/Sn-dual doped sample showed S. aureus biofilm inhibition at 512 μg/mL, whereas the lower concentration of dual doped NPs did not respond to antibiofilm activity [147]. Despite the improvement of transition metal ion dopants on the catalytic and photocatalytic oxidation activity of pure CeO2 NPs, it unexpectedly reduced the antibiofilm activity. Therefore, the oxidative stress and cellular toxicity resulting from the Ce3+ release and ROS generation were proposed as the antibiofilm mechanisms of CeO2 nanoparticles [146], similar to the current antibacterial mechanisms of metallic NPs, such as metal ion release and oxidative stress [148].
Phyto-fabrication of silver nanoparticles (AgNPs) using Gmelina arborea (GA) extract was also studied, where the biofilm inhibition by aqueous leaf extract of GA at 1000 μg/mL was 46 %. GA-AgNPs were more effective than GA-extract, and their antibiofilm activity increased further when loaded on hydrogel as GA-AgNPs-PF127 (59 %), making it a novel distinguishing feature. The acidic-basic affinity between Ag+ and sulfur-containing proteins and phosphorous moieties of DNA can enhance bactericidal capability. Besides, the ability of AgNPs to perturb the bacterial cells could also be due to their atomicity. The smaller the size, the higher the magnitude. GA-AgNPs in size range of 34∼40 nm own high surface area to volume ratio, explaining their stronger inhibitory effect than GA-extract. Furthermore, the synergy between NPs and phytochemicals forms target site-oriented phyto-NPs that disrupt the membrane at the interaction site [149]. Likewise, AgNPs produced from Cedecea and Anthemis species successfully inhibited the biofilms of different pathogens including S. aureus, S. epidermidis, Escherichia coli and Pseudomonas aeruginosa [150,151].
Sonodynamic therapy is a noninvasive and effective therapeutic strategy, which has broadened the way toward dealing with bacteria and biofilms. It uses a combination of ultrasound waves, molecular oxygen, and a sonosensitizer to generate cytotoxic ROS, thereby leading to the death of target microbial cells [152]. Pourhajibagher et al. accessed the combined antibiofilm efficacy of nano-emodin (N-EMO) and sonodynamic therapy against multi-species bacterial biofilms, including S. aureus, Acinetobacter baumannii, and Pseudomonas aeruginosa. As the results demonstrated, there were 71.0 % and 81.5 % reductions in the mixed biofilms following sonodynamic therapy using 0.39 μg/mL and 0.75 μg/mL of N-EMO, respectively. Furthermore, N-EMO-mediated sonodynamic therapy significantly downregulated the expression of agrA by 3.6-fold [153], highlighting the potential of NPs-mediated sonodynamic therapy in antibiofilm and reduction of virulence factors associated with biofilms. Broad spectrum inhibition of bacterial growth and great antibiofilm effect by NPs make them pertinent advocate for curing infectious diseases.
6. Conclusions
Either the biofilm formation inhibition or preformed biofilm disruption could be very effective in the face of bacterial infection. The antibiofilm agents formed by natural products, such as plant extracts and phytochemicals, are promising candidates for treating chronic and persistent biofilm infections. The research on natural products-based antibiofilm agents has been focusing on (i) inhibiting biofilm formation or degrading mature biofilm that targets PIA, eDNA, and proteins; and (ii) interfering with the regulation network of biofilm development that targets the QS system. Natural antibiofilm compounds can inhibit initial bacterial adhesion and/or downregulate the expression of biofilm-related genes, with higher reliability in structure and function than traditional antibacterial agents. On the one hand, an anti-adhesion method can be used as a unique antibiofilm strategy, thus future studies on eradicating biofilms by repressing the adhesion proteins may provide a new dimension for the research and development of antibiofilm agents. On the other hand, different phytochemicals can effectively inhibit the expression of genes associated with biofilm formation or regulation, and further research in this area may prove to be a better way to treat biofilm-associated infections. The current studies revealed that flavonoids can regulate the expression of genes including clfA, fnbpA, fnbpB, icaA, saeS, sigB, srtA, and eno, and degrade EPS matrix, while alkaloids modulate PIA and PSM more, terpenes target QS, as well as aromatic acids interfere with AgrA and SarA (Table 2). In addition, the continuous advances in nanomaterials have also facilitated the development of antibiofilm agents that inhibit the development of resistance by physically destroying bacterial cell membranes and biofilm matrix.
In conclusion, this preview provides valuable information for screening promising targets for antibiofilm agents, while the active compounds/combinations provide attractive candidates for fighting against S. aureus infections. Although some antibiofilm natural products have been identified with preliminarily elucidated mechanisms of action, most of these studies were observational, strongly requiring in-depth exploration of relevant mechanisms. Moreover, the available data on natural products against S. aureus biofilms are generally limited to in vitro studies (Table 1, Table 2), calling for urgently developing in vivo models that can mimic the biofilm in human infections. Future novel and effective antibiofilm therapies will likely include more synergistic actions based on natural products that exhibit antibiofilm properties.
Compliance with ethics requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Xiying Wu: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. Huan Wang: Writing – review & editing, Methodology. Juan Xiong: Writing – review & editing, Methodology. Guo-Xun Yang: Visualization. Jin-Feng Hu: Writing – review & editing, Supervision. Quangang Zhu: Writing – review & editing, Supervision, Conceptualization. Zhongjian Chen: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82003659).
Contributor Information
Jin-Feng Hu, Email: jfhu@tzc.edu.cn, jfhu@fudan.edu.cn.
Quangang Zhu, Email: qgzhu@126.com.
Zhongjian Chen, Email: aajian818@163.com.
Data availability
Data will be made available on request.
References
- 1.Tuon F.F., Suss P.H., Telles J.P., Dantas L.R., Borges N.H., Ribeiro V.S.T. Antimicrobial treatment of Staphylococcus aureus biofilms. Antibiotics. 2023;12(1):87. doi: 10.3390/antibiotics12010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen K., Peng C.T., Chi F., Yu C.D., Yang Q.L., Li Z.J. Antibacterial and antibiofilm activities of chlorogenic acid against Yersinia enterocolitica. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.885092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prosser B.L., Taylor D., Dix B.A., Cleeland R. Method of evaluating effects of antibiotics on bacterial biofilm. Antimicrob Agents Chemother. 1987;31(10):1502–1506. doi: 10.1128/aac.31.10.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X.C., Liu Z.H., Liu Z.J., Meng R.Z., Shi C., Chen X.R., et al. Phenotype and RNA-seq-based transcriptome profiling of Staphylococcus aureus biofilms in response to tea tree oil. Microb Pathog. 2018;123:304–313. doi: 10.1016/j.micpath.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 5.Sharifi A., Mohammadzadeh A., Zahraei Salehi T., Mahmoodi P. Antibacterial, antibiofilm and antiquorum sensing effects of Thymus daenensis and Satureja hortensis essential oils against Staphylococcus aureus isolates. J Appl Microbiol. 2018;124(2):379–388. doi: 10.1111/jam.13639. [DOI] [PubMed] [Google Scholar]
- 6.Silva L.N., Da Hora G.C.A., Soares T.A., Bojer M.S., Ingmer H., Macedo A.J., et al. Myricetin protects Galleria mellonella against Staphylococcus aureus infection and inhibits multiple virulence factors. Sci Rep. 2017;7(1):2823. doi: 10.1038/s41598-017-02712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu D., Xiang H., Dong H.S., Wang D.C., Wang T.D. Isovitexin, a potential candidate inhibitor of Sortase A of Staphylococcus aureus USA300. J Microbiol Biotechnol. 2018;28(9):1426–1432. doi: 10.4014/jmb.1802.02014. [DOI] [PubMed] [Google Scholar]
- 8.Mu D., Luan Y.X., Wang L., Gao Z.Y., Yang P.P., Jing S.S., et al. The combination of salvianolic acid A with latamoxef completely protects mice against lethal pneumonia caused by methicillin-resistant Staphylococcus aureus. Emerg Microb Infect. 2020;9(1):169–179. doi: 10.1080/22221751.2020.1711817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin N., Tan X.J., Jiao Y.M., Liu L., Zhao W.S., Yang S., et al. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep. 2014;4:5467. doi: 10.1038/srep05467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merghni A., Noumi E., Hadded O., Dridi N., Panwar H., Ceylan O., et al. Assessment of the antibiofilm and antiquorum sensing activities of Eucalyptus globulus essential oil and its main component 1,8-cineole against methicillin-resistant Staphylococcus aureus strains. Microb Pathog. 2018;118:74–80. doi: 10.1016/j.micpath.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Yan X., Gu S.S., Cui X.Y., Shi Y.J., Wen S.S., Chen H.Y., et al. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb Pathog. 2019;127:12–20. doi: 10.1016/j.micpath.2018.11.039. [DOI] [PubMed] [Google Scholar]
- 12.Sayed A.M., Alhadrami H.A., El-Hawary S.S., Mohammed R., Hassan H.M., Rateb M.E., et al. Discovery of two brominated oxindole alkaloids as staphylococcal DNA gyrase and pyruvate kinase inhibitors via inverse virtual screening. Microorganisms. 2020;8(2):293. doi: 10.3390/microorganisms8020293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng C.S., Jiang T., Zhang D.W., Wang H.Y., Fang T., Li C.C. Attachment characteristics and kinetics of biofilm formation by Staphylococcus aureus on ready-to-eat cooked beef contact surfaces. J Food Sci. 2023;88(6):2595–2610. doi: 10.1111/1750-3841.16592. [DOI] [PubMed] [Google Scholar]
- 14.Konduri R., Saiabhilash C.R., Shivaji S. Biofilm-forming potential of ocular fluid Staphylococcus aureus and Staphylococcus epidermidis on ex vivo human corneas from attachment to dispersal phase. Microorganisms. 2021;9(6):1124. doi: 10.3390/microorganisms9061124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burel C., Dreyfus R., Purevdorj-Gage L. Physical mechanisms driving the reversible aggregation of Staphylococcus aureus and response to antimicrobials. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-94457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maikranz E., Spengler C., Thewes N., Thewes A., Nolle F., Jung P., et al. Different binding mechanisms of Staphylococcus aureus to hydrophobic and hydrophilic surfaces. Nanoscale. 2020;12(37):19267–19275. doi: 10.1039/d0nr03134h. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Yang J.S., Ji M., Phillips J., Wylam M., Ji Y.D. Identification of cbio gene critical for biofilm formation by MRSA CFSa36 Strain isolated from pediatric patient with cystic fibrosis. Pathogens. 2021;10(11):1363. doi: 10.3390/pathogens10111363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Be'nard L., Litzler P.-Y., Cosette P., Lemeland J.-F., Jouenne T., Junter G.-A. Proteomic analysis of Staphylococcus aureus biofilms grown in vitro on mechanical heart valve leaflets. J Biomed Mater Res. 2009;88(4):1069–1078. doi: 10.1002/jbm.a.31941. [DOI] [PubMed] [Google Scholar]
- 19.Wu S.Z., Liu Y.J., Lei L., Zhang H. An antisense yycF RNA modulates biofilm organization of Methicillin-resistant Staphylococcus aureus and pathogenicity in a rat model of osteomyelitis. Antibiotics. 2021;10(5):603. doi: 10.3390/antibiotics10050603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pouget C., Dunyach-Remy C., Magnan C., Pantel A., Sotto A., Lavigne J.-P. Polymicrobial biofilm organization of Staphylococcus aureus and Pseudomonas aeruginosa in a chronic wound environment. Int J Mol Sci. 2022;23(18) doi: 10.3390/ijms231810761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pei Z.-J., Li C.C., Dai W.N., Lou Z.X., Sun X., Wang H.X., et al. The anti-biofilm activity and mechanism of apigenin-7-O-glucoside against Staphylococcus aureus and Escherichia coli. Infect Drug Resist. 2023;16:2129–2140. doi: 10.2147/IDR.S387157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia Y.Q., Jayathilake P.G., Li B., Zuliani P., Deehan D., Longyear J., et al. Coupled CFD-DEM modeling to predict how EPS affects bacterial biofilm deformation, recovery and detachment under flow conditions. Biotechnol Bioeng. 2022;119(9):2551–2563. doi: 10.1002/bit.28146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu S.P., Jiang B., Jia C., Wu H.M., Shen J., Hu X.M., et al. Investigation of biofilm production and its association with genetic and phenotypic characteristics of OM (osteomyelitis) and non-OM orthopedic Staphylococcus aureus. Ann Clin Microbiol Antimicrob. 2020;19(1):10. doi: 10.1186/s12941-020-00352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monzón M., García-Alvarez F., Laclériga A., Amorena B. Evaluation of four experimental osteomyelitis infection models by using precolonized implants and bacterial suspensions. Acta Orthop Scand. 2002;73(1):11–19. doi: 10.1080/000164702317281341. [DOI] [PubMed] [Google Scholar]
- 25.Herrera A., Vu B.G., Stach C.S., Merriman J.A., Horswill A.R., et al. Staphylococcus aureus β-toxin mutants are defective in biofilm ligase and sphingomyelinase activity, and causation of infective endocarditis and sepsis. Biochemistry. 2016;55(17):2510–2517. doi: 10.1021/acs.biochem.6b00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pabst B., Pitts B., Lauchnor E., Stewart P.S. Gel-entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob Agents Chemother. 2016;60(10):6294–6301. doi: 10.1128/AAC.01336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo H.N., Tong Y.C., Cheng J.H., Abbas Z., Li Z.X., Wang J.Y., et al. Biofilm and small colony variants-an update on Staphylococcus aureus strategies toward drug resistance. Int J Mol Sci. 2022;23(3):1241. doi: 10.3390/ijms23031241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemming H.-C., Wingender J., Szewzyk U., Steinberg P., Rice S.A., Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 29.Ciofu O., Moser C., Jensen P.Ø., Høiby N. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol. 2022;20(10):621–635. doi: 10.1038/s41579-022-00682-4. [DOI] [PubMed] [Google Scholar]
- 30.Conlon B.P., Rowe S.E., Gandt A.B., Nuxoll A.S., Donegan N.P., Zalis E.A., et al. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat Microbiol. 2016;1(5) doi: 10.1038/nmicrobiol.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart J.W., Waigh T.A., Lu J.R., Roberts I.S. Microrheology and spatial heterogeneity of Staphylococcus aureus biofilms modulated by hydrodynamic shear and biofilm-degrading enzymes. Langmuir. 2019;35(9):3553–3561. doi: 10.1021/acs.langmuir.8b04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rivera E.S., Weiss A., Migas L.G., Freiberg J.A., Djambazova K.V., Neumann E.K., et al. Imaging mass spectrometry reveals complex lipid distributions across Staphylococcus aureus biofilm layers. J Mass Spectrom Adv Clin Lab. 2022;26:36–46. doi: 10.1016/j.jmsacl.2022.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pires D.P., Oliveira H., Melo L.D., Sillankorva S., Azeredo J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100(5):2141–2151. doi: 10.1007/s00253-015-7247-0. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Li W., Zhu X.Y., Zhao H.Z., Lu Y.J., Zhang C., et al. Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces. Appl Microbiol Biotechnol. 2019;103(11):4565–4574. doi: 10.1007/s00253-019-09808-w. [DOI] [PubMed] [Google Scholar]
- 35.Yuan G.J., Li P.Y., Xu X.J., Li P.B., Zhong Q.W., He S., et al. Azalomycin F5a eradicates Staphylococcus aureus biofilm by rapidly penetrating and subsequently inducing cell lysis. Int J Mol Sci. 2020;21(3):862. doi: 10.3390/ijms21030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mann E.E., Rice K.C., Boles B.R., Endres J.L., Ranjit D., Chandramohan L., et al. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0005822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cramton S.E., Ulrich M., Götz F., Döring G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69(6):4079–4085. doi: 10.1128/iai.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cramton S.E., Gerke C., Schnell N.F., Nichols W.W., Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67(10):5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen H.T.T., Nguyen T.H., Otto M. The staphylococcal exopolysaccharide PIA-biosynthesis and role in biofilm formation, colonization, and infection. Comput Struct Biotechnol J. 2020;18:3324–3334. doi: 10.1016/j.csbj.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cue D., Lei M.G., Lee C.Y. Genetic regulation of the intercellular adhesion locus in staphylococci. Front Cell Infect Microbiol. 2012;2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vuong C., Kocianova S., Voyich J.M., Yao Y.F., Fischer E.R., DeLeo F.R., et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J Biol Chem. 2004;279(52):54881–54886. doi: 10.1074/jbc.M411374200. [DOI] [PubMed] [Google Scholar]
- 42.Bai X., Shen Y., Zhang T.H., Meng R.Z., Zhang Y., Deng Y.H., et al. Anti-biofilm activity of biochanin A against Staphylococcus aureus. Appl Microbiol Biotechnol. 2023;107:867–879. doi: 10.1007/s00253-022-12350-x. [DOI] [PubMed] [Google Scholar]
- 43.Farha A.K., Yang Q.-Q., Kim G., Zhang D., Mavumengwana V., Habimana O., et al. Inhibition of multidrug-resistant foodborne Staphylococcus aureus biofilms by a natural terpenoid (+)-nootkatone and related molecular mechanism. Food Control. 2020;112 doi: 10.1016/j.foodcont.2020.107154. [DOI] [Google Scholar]
- 44.Cucarella C., Solano C., Valle J., Amorena B., Lasa I., Penadés J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/jb.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaeffer C.R., Woods K.M., Longo G.M., Kiedrowski M.R., Paharik A.E., Büttner H., et al. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect Immun. 2015;83(1):214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drożdż K., Ochońska D., Ścibik Ł., Gołda-Cępa M., Biegun K., Brzychczy-Włoch M. The frequency of occurrence of resistance and genes involved in the process of adhesion and accumulation of biofilm in Staphylococcus aureus strains isolated from tracheostomy tubes. Microorganisms. 2022;10(6):1210. doi: 10.3390/microorganisms10061210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sedarat Z., Taylor-Robinson A.W. Biofilm formation by pathogenic bacteria: applying a Staphylococcus aureus model to appraise potential targets for therapeutic intervention. Pathogens. 2022;11(4):388. doi: 10.3390/pathogens11040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng Q., Tang X.H., Dong W.Y., Sun N., Yuan W.C. A review of biofilm formation of Staphylococcus aureus and its regulation mechanism. Antibiotics. 2022;12(1):12. doi: 10.3390/antibiotics12010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster T.J., Geoghegan J.A., Ganesh V.K., Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12(1):49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X.Y., Ma G.-L., Chen H.-W., Zhao Z.-Y., Zhu Z.-P., Xiong J., et al. Antibacterial and antibiofilm efficacy of the preferred fractions and compounds from Euphorbia humifusa (herba euphorbiae humifusae) against Staphylococcus aureus. J Ethnopharmacol. 2023;306 doi: 10.1016/j.jep.2023.116177. [DOI] [PubMed] [Google Scholar]
- 51.Khodaparast L., Khodaparast L., Shahrooei M., Stijlemans B., Merckx R., Baatsen P., et al. The possible role of Staphylococcus epidermidis LPxTG surface protein SesC in biofilm formation. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss W.J., Lenoy E., Murphy T., Tardio L., Burgio P., Projan S.J., et al. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal models of infection. J Antimicrob Chemother. 2004;53(3):480–486. doi: 10.1093/jac/dkh078. [DOI] [PubMed] [Google Scholar]
- 53.Ming D., Wang D.C., Cao F.J., Xiang H., Mu D., Cao J.J., et al. Kaempferol inhibits the primary attachment phase of biofilm formation in Staphylococcus aureus. Front Microbiol. 2017;8:2263. doi: 10.3389/fmicb.2017.02263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang S.S., Kim J.-G., Lee T.-H., Oh K.-B. Flavonols inhibit sortases and sortase-mediated Staphylococcus aureus clumping to fibrinogen. Biol Pharm Bull. 2006;29(8):1751–1755. doi: 10.1248/bpb.29.1751. [DOI] [PubMed] [Google Scholar]
- 55.Zhang B., Teng Z.H., Li X.H., Lu G.J., Deng X.M., Niu X.D., et al. Chalcone attenuates Staphylococcus aureus virulence by targeting sortase a and alpha-hemolysin. Front Microbiol. 2017;8:1715. doi: 10.3389/fmicb.2017.01715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Svarcova V., Zdenkova K., Sulakova M., Demnerova K., Pazlarova J. Contribution to determination of extracellular DNA origin in the biofilm matrix. J Basic Microbiol. 2021;61(7):652–661. doi: 10.1002/jobm.202100090. [DOI] [PubMed] [Google Scholar]
- 57.Montanaro L., Poggi A., Visai L., Ravaioli S., Campoccia D., Speziale P., et al. Extracellular DNA in biofilms. Int J Artif Organs. 2011;34(9):824–831. doi: 10.5301/ijao.5000051. [DOI] [PubMed] [Google Scholar]
- 58.Flemming H.-C., van Hullebusch E.D., Neu T.R., Nielsen P.H., Seviour T., Stoodley P., et al. The biofilm matrix: multitasking in a shared space. Nat Rev Microbiol. 2022;21:70–86. doi: 10.1038/s41579-022-00791-0. [DOI] [PubMed] [Google Scholar]
- 59.Rice K.C., Mann E.E., Endres J.L., Weiss E.C., Cassat J.E., Smeltzer M.S., et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(19):8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan X., Gu S.S., Shi Y.J., Cui X.Y., Wen S.S., Ge J.W. The effect of emodin on Staphylococcus aureus strains in planktonic form and biofilm formation in vitro. Arch Microbiol. 2017;199(9):1267–1275. doi: 10.1007/s00203-017-1396-8. [DOI] [PubMed] [Google Scholar]
- 61.Buzzo J.R., Devaraj A., Gloag E.S., Jurcisek J.A., Robledo-Avila F., Kesler T., et al. Z-form extracellular DNA is a structural component of the bacterial biofilm matrix. Cell. 2021;184(23):5740–5758. doi: 10.1016/j.cell.2021.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Devaraj A., Buzzo J.R., Mashburn-Warren L., Gloag E.S., Novotny L.A., Stoodley P., et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc Natl Acad Sci USA. 2019;116(50):25068–25077. doi: 10.1073/pnas.1909017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodman S.D., Bakaletz L.O. Bacterial biofilms utilize an underlying extracellular DNA matrix structure that can be targeted for biofilm resolution. Microorganisms. 2022;10(2):466. doi: 10.3390/microorganisms10020466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li S., Huang H., Rao X.C., Chen W., Wang Z.Q., Hu X.M. Phenol-soluble modulins: novel virulence-associated peptides of staphylococci. Future Microbiol. 2014;9(2):203–216. doi: 10.2217/fmb.13.153. [DOI] [PubMed] [Google Scholar]
- 65.Wang R., Braughton K.R., Kretschmer D., Bach T.-H.L., Queck S.Y., Li M., et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13(12):1510–1514. doi: 10.1038/nm1656. [DOI] [PubMed] [Google Scholar]
- 66.Rautenberg M., Joo H.-S., Otto M., Peschel A. Neutrophil responses to staphylococcal pathogens and commensals via the formyl peptide receptor 2 relates to phenol-soluble modulin release and virulence. Faseb J. 2011;25(4):1254–1263. doi: 10.1096/fj.10-175208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Periasamy S., Joo H.-S., Duong A.C., Bach T.-H., Tan V.Y., Chatterjee S.S., et al. How Staphylococcus aureus biofilms develop their characteristic structure. Proc Natl Acad Sci USA. 2012;109(4):1281–1286. doi: 10.1073/pnas.1115006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le K.Y., Dastgheyb S., Ho T.V., Otto M. Molecular determinants of staphylococcal biofilm dispersal and structuring. Front Cell Infect Microbiol. 2014;4:167. doi: 10.3389/fcimb.2014.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwartz K., Syed A.K., Stephenson R.E., Rickard A.H., Boles B.R. Functional amyloids composed of phenol soluble modulins stabilize Staphylococcus aureus biofilms. PLoS Pathog. 2012;8(6) doi: 10.1371/journal.ppat.1002744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yum S.-J., Jeong H.G., Kim S.M. Anti-biofilm effects of sinomenine against Staphylococcus aureus. Food Sci Biotechnol. 2023;32(1):83–90. doi: 10.1007/s10068-022-01174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pei J.J., Huang Y.G., Ren T., Guo Y.D., Dang J., Tao Y.D., et al. The antibacterial activity mode of action of plantaricin YKX against Staphylococcus aureus. Molecules. 2022;27(13):4280. doi: 10.3390/molecules27134280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu D., Zhao L.P., Xue T., Sun B.L. Staphylococcus aureus autoinducer-2 quorum sensing decreases biofilm formation in an icaR-dependent manner. BMC Microbiol. 2012;12:288. doi: 10.1186/1471-2180-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oliveira D., Borges A., Ruiz R.M., Negrín Z.R., Distinto S., Borges F., et al. 2-(2-Methyl-2-nitrovinyl)furan but not furvina interfere with Staphylococcus aureus Agr quorum-sensing system and potentiate the action of fusidic acid against biofilms. Int J Mol Sci. 2021;22(2):613. doi: 10.3390/ijms22020613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reyes D., Andrey D.O., Monod A., Kelley W.L., Zhang G., Cheung A.L. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol. 2011;193(21):6020–6031. doi: 10.1128/jb.05436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gordon C.P., Olson S.D., Lister J.L., Kavanaugh J.S., Horswill A.R. Truncated autoinducing peptides as antagonists of Staphylococcus lugdunensis quorum sensing. J Med Chem. 2016;59(19):8879–8888. doi: 10.1021/acs.jmedchem.6b00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bardelang P., Murray E.J., Blower I., Zandomeneghi S., Goode A., Hussain R., et al. Conformational analysis and interaction of the Staphylococcus aureus transmembrane peptidase AgrB with its AgrD propeptide substrate. Front Chem. 2023;11 doi: 10.3389/fchem.2023.1113885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang B., Muir T.W. Regulation of virulence in Staphylococcus aureus: molecular mechanisms and remaining puzzles. Cell Chem Biol. 2016;23(2):214–224. doi: 10.1016/j.chembiol.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Novick R.P., Ross H.F., Projan S.J., Kornblum J., Kreiswirth B., Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12(10):3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cheung G.Y.C., Wang R., Khan B.A., Sturdevant D.E., Otto M. Role of the accessory gene regulator agr in community-associated Methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79(5):1927–1935. doi: 10.1128/iai.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Peng D., Chen A.L., Shi B., Min X., Zhang T., Dong Z.L., et al. Preliminary study on the effect of brazilin on biofilms of Staphylococcus aureus. Exp Ther Med. 2018;16(3):2108–2118. doi: 10.3892/etm.2018.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie Q., Zhao A.S., Jeffrey P.D., Kim M.K., Bassler B.L., Stone H.A., et al. Identification of a molecular latch that regulates staphylococcal virulence. Cell Chem Biol. 2019;26(4):548–558. doi: 10.1016/j.chembiol.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chopra S., Harjai K., Chhibber S. Antibiotic susceptibility of ica-positive and ica-negative MRSA in different phases of biofilm growth. J Antibiot. 2015;68(1):15–22. doi: 10.1038/ja.2014.96. [DOI] [PubMed] [Google Scholar]
- 83.Ma R.H., Qiu S.W., Jiang Q., Sun H.P., Xue T., Cai G., et al. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int J Med Microbiol. 2017;307:257–267. doi: 10.1016/j.ijmm.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 84.Li J.P., Fan Q.Y., Jin M.Y., Mao C.L., Zhang H., Zhang X.L., et al. Paeoniflorin reduce luxS/AI-2 system-controlled biofilm formation and virulence in Streptococcus suis. Virulence. 2021;12(1):3062–3073. doi: 10.1080/21505594.2021.2010398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu S.Z., Zhang J.Q., Peng Q., Liu Y.J., Lei L., Zhang H. The role of Staphylococcus aureus YycFG in gene regulation, biofilm organization and drug resistance. Antibiotics. 2021;10(12):1555. doi: 10.3390/antibiotics10121555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu S.Z., Liu Y.J., Lei L., Zhang H. Antisense yycG modulates the susceptibility of Staphylococcus aureus to hydrogen peroxide via the sarA. BMC Microbiol. 2021;21(1):160. doi: 10.1186/s12866-021-02218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duan J.J., Li M.L., Hao Z.H., Shen X.F., Liu L., Jin Y., et al. Subinhibitory concentrations of resveratrol reduce alpha-hemolysin production in Staphylococcus aureus isolates by downregulating saeRS. Emerg Microb Infect. 2018;7(1):136. doi: 10.1038/s41426-018-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu S.Z., Liu Y.J., Lei L., Zhang H. Antisense yycG regulation of antibiotic sensitivity of Methicillin-resistant Staphylococcus aureus in chronic osteomyelitis. Surg Infect. 2019;20(6):472–479. doi: 10.1089/sur.2019.016. [DOI] [PubMed] [Google Scholar]
- 89.Dubrac S., Boneca I.G., Poupel O., Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189(22):8257–8269. doi: 10.1128/JB.00645-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Visutthi M., Srimanote P., Voravuthikunchai S.P. Responses in the expression of extracellular proteins in methicillin-resistant Staphylococcus aureus treated with rhodomyrtone. J Microbiol. 2011;49(6):956–964. doi: 10.1007/s12275-011-1115-0. [DOI] [PubMed] [Google Scholar]
- 91.Bleul L., Francois P., Wolz C. Two-component systems of S. aureus: signaling and sensing mechanisms. Genes. 2021;13(1):34. doi: 10.3390/genes13010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mizar P., Arya R., Kim T., Cha S., Ryu K.-S., Yeo W.-S., et al. Total synthesis of xanthoangelol B and its various fragments: toward inhibition of virulence factor production of Staphylococcus aureus. J Med Chem. 2018;61(23):10473–10487. doi: 10.1021/acs.jmedchem.8b01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valle J., Toledo-Arana A., Berasain C., Ghigo J.-M., Amorena B., Penadés J.R., et al. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48(4):1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 94.Saȉd-Salim B., Dunman P.M., McAleese F.M., Macapagal D., Murphy E., et al. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol. 2003;185(2):610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Geisinger E., Adhikari R.P., Jin R., Ross H.F., Novick R.P. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61(4):1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 96.Crosby H.A., Schlievert P.M., Merriman J.A., King J.M., Salgado-Pabón W., Horswill A.R. The Staphylococcus aureus global regulator MgrA modulates clumping and virulence by controlling surface protein expression. PLoS Pathog. 2016;12(5) doi: 10.1371/journal.ppat.1005604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang B., Wei P.-W., Wan S., Yao Y., Song C.-R., Song P.-P., et al. Ginkgo biloba exocarp extracts inhibit S. aureus and MRSA by disrupting biofilms and affecting gene expression. J Ethnopharmacol. 2021;271 doi: 10.1016/j.jep.2021.113895. [DOI] [PubMed] [Google Scholar]
- 98.Lin T.X., Huang L., Cheng N.N., Wang Y.Z., Ning Z., Huang S.K., et al. The in vitro and in vivo antibacterial activities of uniflorous honey from a medicinal plant, Scrophularia ningpoensis Hemsl., and characterization of its chemical profile with UPLC-MS/MS. J Ethnopharmacol. 2022;296 doi: 10.1016/j.jep.2022.115499. [DOI] [PubMed] [Google Scholar]
- 99.Rossiter S.E., Fletcher M.H., Wuest W.M. Natural products as platforms to overcome antibiotic resistance. Chem Rev. 2017;117(19):12415–12474. doi: 10.1021/acs.chemrev.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo X.Q., Mei N. Aloe vera: a review of toxicity and adverse clinical effects. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2016;34(2):77–96. doi: 10.1080/10590501.2016.1166826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kis B., Ifrim F.C., Buda V., Avram S., Pavel I.Z., Antal D., et al. Cannabidiol-from plant to human body: a promising bioactive molecule with multi-target effects in cancer. Int J Mol Sci. 2019;20(23):5905. doi: 10.3390/ijms20235905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kabotso D.E.K., Neglo D., Kwashie P., Agbo I.A., Abaye D.A. GC/MS composition and resistance modulatory inhibitory activities of three extracts of lemongrass: citral modulates the activities of five antibiotics at sub-inhibitory concentrations on Methicillin-resistant Staphylococcus aureus. Chem Biodivers. 2022;19(9) doi: 10.1002/cbdv.202200296. [DOI] [PubMed] [Google Scholar]
- 103.Nunes A.K.A., Araújo Malveira E., Lopes Andrade A., Barbosa da Silva W.M., de Morais S.M., Silva Dos Santos H., et al. Chemical composition determination and evaluation of the antimicrobial activity of essential oil from Croton blanchetianus (Euphorbiaceae) against clinically relevant bacteria. Chem Biodivers. 2023;20(1) doi: 10.1002/cbdv.202200777. [DOI] [PubMed] [Google Scholar]
- 104.de Oliveira G.D., da Rocha W.R.V., Rodrigues J.F.B., da Silva Alves H. Synergistic and antibiofilm effects of the essential oil from Croton conduplicatus (Euphorbiaceae) against Methicillin-resistant Staphylococcus aureus. Pharmaceuticals. 2022;16(1):55. doi: 10.3390/ph16010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sharifi A., Mohammadzadeh A., Salehi T.Z., Mahmoodi P., Nourian A. Cuminum cyminum L. essential oil: a promising antibacterial and antivirulence agent against multidrug-resistant Staphylococcus aureus. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.667833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laskoski L.V., Bandeira D.M., Batista J.M., Costa W.F.D., Baeza L.C., Kuo L.H., et al. Phytochemical prospection and evaluation of antimicrobial, antioxidant and antibiofilm activities of extracts and essential oil from leaves of Myrsine umbellata Mart. (Primulaceae) Braz J Biol. 2022;82 doi: 10.1590/1519-6984.263865. [DOI] [PubMed] [Google Scholar]
- 107.Merghni A., Haddaji N., Bouali N., Alabbosh K.F., Adnan M., Snoussi M., et al. Comparative Study of antibacterial, antibiofilm, antiswarming and antiquorum sensing activities of origanum vulgare essential oil and terpinene-4-ol against pathogenic bacteria. Life. 2022;12(10):1616. doi: 10.3390/life12101616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nazzaro F., Fratianni F., De Martino L., Coppola R., De Feo V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals. 2013;6(12):1451–1474. doi: 10.3390/ph6121451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Panda S.K., Buroni S., Swain S.S., Bonacorsi A., da Fonseca Amorim E.A., Kulshrestha M., et al. Recent advances to combat ESKAPE pathogens with special reference to essential oils. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Álvarez-Martínez F.J., Barrajón-Catalán E., Herranz-López M., Micol V. Antibacterial plant compounds, extracts and essential oils: an updated review on their effects and putative mechanisms of action. Phytomedicine. 2021;90 doi: 10.1016/j.phymed.2021.153626. [DOI] [PubMed] [Google Scholar]
- 111.Nur Onal F., Ozturk I., Aydin Kose F., Der G., Kilinc E., Baykan S. Comparative evaluation of polyphenol contents and biological activities of five Cistus L. species native to Turkey. Chem Biodivers. 2023;20(1) doi: 10.1002/cbdv.202200915. [DOI] [PubMed] [Google Scholar]
- 112.Souid A., Della Croce C.M., Frassinetti S., Gabriele M., Pozzo L., Ciardi M., et al. Nutraceutical potential of leaf hydro-ethanolic extract of the edible halophyte Crithmum maritimum L. Molecules. 2021;26(17):5380. doi: 10.3390/molecules26175380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Neglo D., Adzaho F., Agbo I.A., Arthur R., Sedohia D., Tettey C.O., et al. Antibiofilm activity of Azadirachta indica and Catharanthus roseus and their synergistic effects in combination with antimicrobial agents against fluconazole-resistant Candida albicans strains and MRSA. Evid Based Complement Alternat Med. 2022;2022 doi: 10.1155/2022/9373524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ekom S.E., Tamokou J.-D.-D., Kuete V. Antibacterial and therapeutic potentials of the Capsicum annuum extract against infected wound in a rat model with its mechanisms of antibacterial action. BioMed Res Int. 2021;2021 doi: 10.1155/2021/4303902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang L.L., Ismail M.M., Rocchetti G., Fayek N.M., Lucini L., Saber F.R. The untargeted phytochemical profile of three meliaceae species related to in vitro cytotoxicity and anti-virulence activity against MRSA isolates. Molecules. 2022;27(2):435. doi: 10.3390/molecules27020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Snoussi M., Noumi E., Hajlaoui H., Bouslama L., Hamdi A., Saeed M., et al. Phytochemical profiling of Allium subhirsutum L. aqueous extract with antioxidant, antimicrobial, antibiofilm, and anti-quorum sensing properties: in vitro and in silico studies. Plants. 2022;11(4):495. doi: 10.3390/plants11040495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vijayakumar K., Ramanathan T. Antiquorum sensing and biofilm potential of 5- Hydroxymethylfurfural against Gram positive pathogens. Microb Pathog. 2018;125:48–50. doi: 10.1016/j.micpath.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 118.Barfour A.F., Mensah A.Y., Asante-Kwatia E., Danquah C.A., Anokwah D., Adjei S., et al. Antibacterial, antibiofilm, and efflux pump inhibitory properties of the crude extract and fractions from Acacia macrostachya stem bark. Sci World J. 2021;2021 doi: 10.1155/2021/5381993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ganesh P.S., Veena K., Senthil R., Iswamy K., Ponmalar E.M., Mariappan V., et al. Biofilm-associated agr and sar quorum sensing systems of Staphylococcus aureus are inhibited by 3-hydroxybenzoic acid derived from Illicium verum. ACS Omega. 2022;7(17):14653–14665. doi: 10.1021/acsomega.1c07178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wójcik-Bojek U., Rywaniak J., Bernat P., Podsędek A., Kajszczak D., Sadowska B. An in vitro study of the effect of Viburnum opulus extracts on key processes in the development of staphylococcal Infections. Molecules. 2021;26(6):1758. doi: 10.3390/molecules26061758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Awadelkareem A.M., Al-Shammari E., Elkhalifa A.O., Adnan M., Siddiqui A.J., Mahmood D., et al. Anti-adhesion and antibiofilm activity of Eruca sativa miller extract targeting cell adhesion proteins of food-borne bacteria as a potential mechanism: combined in vitro-in silico approach. Plants. 2022;11(5):610. doi: 10.3390/plants11050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mohamad F., Alzahrani R.R., Alsaadi A., Alrfaei B.M., Yassin A.E.B., Alkhulaifi M.M., et al. An explorative review on advanced approaches to overcome bacterial resistance by curbing bacterial biofilm formation. Infect Drug Resist. 2023;16:19–49. doi: 10.2147/idr.S380883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xie Y.X., Yang W.J., Tang F., Chen X.Q., Ren L.C. Antibacterial activities of flavonoids: structure-activity relationship and mechanism. Curr Med Chem. 2014;22(1):132–149. doi: 10.2174/0929867321666140916113443. [DOI] [PubMed] [Google Scholar]
- 124.Gopu V., Meena C.K., Shetty P.H. Quercetin influences quorum sensing in food borne bacteria: in-vitro and in-silico evidence. PLoS One. 2015;10(8) doi: 10.1371/journal.pone.0134684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ouyang J., Sun F., Feng W., Sun Y., Qiu X., Xiong L., et al. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J Appl Microbiol. 2016;120(4):966–974. doi: 10.1111/jam.13073. [DOI] [PubMed] [Google Scholar]
- 126.Yuan Q., Feng W., Wang Y., Wang Q.M., Mou N., Xiong L.R., et al. Luteolin attenuates the pathogenesis of Staphylococcus aureus by interfering with the agr system. Microb Pathog. 2022;165 doi: 10.1016/j.micpath.2022.105496. [DOI] [PubMed] [Google Scholar]
- 127.Mone N.S., Syed S., Ravichandiran P., Satpute S.K., Kim A.R., Yoo D.J. How structure-function relationships of 1,4-naphthoquinones combat antimicrobial resistance in multidrug-resistant (MDR) pathogens. ChemMedChem. 2023;18(2) doi: 10.1002/cmdc.202200471. [DOI] [PubMed] [Google Scholar]
- 128.Paul P., Chakraborty P., Chatterjee A., Sarker R.K., Dastidar D.G., Kundu T., et al. 1,4-Naphthoquinone accumulates reactive oxygen species in Staphylococcus aureus: a promising approach towards effective management of biofilm threat. Arch Microbiol. 2021;203(3):1183–1193. doi: 10.1007/s00203-020-02117-1. [DOI] [PubMed] [Google Scholar]
- 129.Mone N.S., Kamble E.E., Pardesi K.R., Satpute S.K. Antibacterial and antibiofilm potency of menadione against multidrug-resistant S. aureus. Curr Microbiol. 2022;79(9):282. doi: 10.1007/s00284-022-02975-6. [DOI] [PubMed] [Google Scholar]
- 130.Li Q.-Q., Chae H.-S., Kang O.-H., Kwon D.-Y. Synergistic antibacterial activity with conventional antibiotics and mechanism of action of shikonin against methicillin-resistant Staphylococcus aureus. Int J Mol Sci. 2022;23(14):7551. doi: 10.3390/ijms23147551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.de Moura D.F., Rocha T.A., de Melo Barros D., da Silva M.M., Dos Santos Santana M., Neta B.M., et al. Evaluation of the antioxidant, antibacterial, and antibiofilm activity of the sesquiterpene nerolidol. Arch Microbiol. 2021;203(7):4303–4311. doi: 10.1007/s00203-021-02377-5. [DOI] [PubMed] [Google Scholar]
- 132.Katsipis G., Tsalouxidou V., Halevas E., Geromichalou E., Geromichalos G., Pantazaki A.A. In vitro and in silico evaluation of the inhibitory effect of a curcumin-based oxovanadium (IV) complex on alkaline phosphatase activity and bacterial biofilm formation. Appl Microbiol Biotechnol. 2021;105(1):147–168. doi: 10.1007/s00253-020-11004-0. [DOI] [PubMed] [Google Scholar]
- 133.Salinas C., Florentin G., Rodriguez F., Alvarenga N., Guillen R. Terpenes combinations inhibit biofilm formation in Staphyloccocus aureus by interfering with initial adhesion. Microorganisms. 2022;10(8):1527. doi: 10.3390/microorganisms10081527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bazmi R.R., Panichayupakaranant P. Synergistic interactions between artocarpin-rich extract, lawsone methyl ether and ampicillin on anti-MRSA and their antibiofilm formation. Lett Appl Microbiol. 2022;74(5):777–786. doi: 10.1111/lam.13662. [DOI] [PubMed] [Google Scholar]
- 135.Savu M., Simo M.K., Fopokam G.X., Olaru S.M., Cioanca O., Boyom F.F., et al. New insights into the antimicrobial potential of polyalthia longifolia-antibiofilm activity and synergistic effect in combination with penicillin against Staphylococcus aureus. Microorganisms. 2022;10(10):1943. doi: 10.3390/microorganisms10101943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sannat C., Hirpurkar S.D., Shakya S., Dutta G.K., Roy M., Jolhe D.K., et al. Methanolic extract of Hemidesmus indicus root augments the antibacterial and antibiofilm activity of amoxicillin and clindamycin against methicillin-resistant Staphylococcus aureus of bovine origin. Lett Appl Microbiol. 2022;75(6):1579–1589. doi: 10.1111/lam.13825. [DOI] [PubMed] [Google Scholar]
- 137.Tsukatani T., Kuroda R., Kawaguchi T. Screening biofilm eradication activity of ethanol extracts from foodstuffs: potent biofilm eradication activity of glabridin, a major flavonoid from licorice (Glycyrrhiza glabra), alone and in combination with ɛ-poly-L-lysine. World J Microbiol Biotechnol. 2022;38(2):24. doi: 10.1007/s11274-021-03206-z. [DOI] [PubMed] [Google Scholar]
- 138.Ferrer-Espada R., Wang Y., Goh X.S., Dai T.H. Antimicrobial blue light inactivation of microbial isolates in biofilms. Laser Surg Med. 2020;52(5):472–478. doi: 10.1002/lsm.23159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lu M., Wang S., Wang T., Hu S., Bhayana B., Ishii M., et al. Bacteria-specific phototoxic reactions triggered by blue light and phytochemical carvacrol. Sci Transl Med. 2021;13(575) doi: 10.1126/scitranslmed.aba3571. [DOI] [PubMed] [Google Scholar]
- 140.Sharma K., Pandey S., Sekar H., Alan T., Gundabala V. Microfluidics based generation of curcumin loaded microfibrous mat against Staphylococcus aureus biofilm by photodynamic therapy. ACS Appl Bio Mater. 2023;6(3):1092–1104. doi: 10.1021/acsabm.2c00971. [DOI] [PubMed] [Google Scholar]
- 141.Swolana D., Wojtyczka R.D. Activity of silver nanoparticles against Staphylococcus spp. Int J Mol Sci. 2022;23(8):4298. doi: 10.3390/ijms23084298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marzhoseyni Z., Rashki S., Nazari-Alam A. Evaluation of the inhibitory effects of TiO2 nanoparticle and Ganoderma lucidum extract against biofilm-producing bacteria isolated from clinical samples. Arch Microbiol. 2023;205(2):59. doi: 10.1007/s00203-023-03403-4. [DOI] [PubMed] [Google Scholar]
- 143.Ranpariya B., Salunke G., Karmakar S., Babiya K., Sutar S., Kadoo N., et al. Antimicrobial synergy of silver-platinum nanohybrids with antibiotics. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.610968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tawre M.S., Shiledar A., Satpute S.K., Ahire K., Ghosh S., Pardesi K. Synergistic and antibiofilm potential of Curcuma aromatica derived silver nanoparticles in combination with antibiotics against multidrug-resistant pathogens. Front Chem. 2022;10 doi: 10.3389/fchem.2022.1029056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Abdel-Rafei M.K., Thabet N.M., Abdel Maksoud M.I.A., Abd Elkodous M., Kawamura G., Matsuda A., et al. Influence of Ce3+ substitution on antimicrobial and antibiofilm properties of ZnCexFe2-xO4 nanoparticles (X = 0.0, 0.02, 0.04, 0.06, and 0.08) conjugated with ebselen and its role subsidised with γ-radiation in mitigating human TNBC and colorectal adenocarcinoma proliferation in vitro. Int J Mol Sci. 2021;22(18) doi: 10.3390/ijms221810171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Naidi S.N., Khan F., Harunsani M.H., Tan A.L., Kim Y.-M., Khan M.M. Effect of Zr doping on photoantioxidant and antibiofilm properties of CeO2 NPs fabricated using aqueous leaf extract of Pometia pinnata. Bioproc Biosyst Eng. 2022;45(2):279–295. doi: 10.1007/s00449-021-02656-x. [DOI] [PubMed] [Google Scholar]
- 147.Naidi S.N., Khan F., Tan A.L., Harunsani M.H., Kim Y.-M., Khan M.M. Green synthesis of CeO2 and Zr/Sn-dual doped CeO2 nanoparticles with photoantioxidant and antibiofilm activities. Biomater Sci. 2021;9(14):4854–4869. doi: 10.1039/d1bm00298h. [DOI] [PubMed] [Google Scholar]
- 148.Hulme J. Application of nanomaterials in the prevention, detection, and treatment of methicillin-resistant Staphylococcus aureus (MRSA) Pharmaceutics. 2022;14(4):805. doi: 10.3390/pharmaceutics14040805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chandrasekharan S., Chinnasamy G., Bhatnagar S. Sustainable phyto-fabrication of silver nanoparticles using Gmelina arborea exhibit antimicrobial and biofilm inhibition activity. Sci Rep. 2022;12(1):156. doi: 10.1038/s41598-021-04025-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Singh P., Pandit S., Jers C., Joshi A.S., Garnæs J., Mijakovic I. Silver nanoparticles produced from Cedecea sp. exhibit antibiofilm activity and remarkable stability. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-92006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ajlouni A.-W., Hamdan E.H., Alshalawi R.A.E., Shaik M.R., Khan M., Kuniyil M., et al. Green synthesis of silver nanoparticles using aerial part extract of the Anthemis pseudocotula Boiss. plant and their biological activity. Molecules. 2022;28(1):246. doi: 10.3390/molecules28010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Xu P.-Y., Kumar Kankala R., Wang S.-B., Chen A.-Z. Sonodynamic therapy-based nanoplatforms for combating bacterial infections. Ultrason Sonochem. 2023;100 doi: 10.1016/j.ultsonch.2023.106617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pourhajibagher M., Rahimi-Esboei B., Ahmadi H., Bahador A. The anti-biofilm capability of nano-emodin-mediated sonodynamic therapy on multi-species biofilms produced by burn wound bacterial strains. Photodiagnosis Photodyn Ther. 2021;34 doi: 10.1016/j.pdpdt.2021.102288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.