Abstract
A broad variety of herpes simplex virus type 1 clones was selected under a single round of high-dose selection with brivudin. Mutations in the thymidine kinase (TK) genes consisted of 42% frameshift mutations within homopolymer repeats of G's and C's and single nucleotide substitutions (58%) that produced stop codons (Q261 and R281) or a new codon at the site of the substitution (A168T, R51W, G59W, G206R, R220H, Y239S, and T287 M). The A168T change, associated with an altered TK phenotype, proved to be the most commonly selected substitution. For the different mutants, a correlation between phenotype, genotype, and in vivo neurovirulence was observed.
Herpesviruses encode their own thymidine kinases (TK) which differ considerably from the human cellular TK. In particular, the herpes simplex virus type 1 (HSV-1) TK is a multisubstrate enzyme which is able to phosphorylate a broad spectrum of pyrimidine and purine nucleoside substrate analogues, including the antiviral drugs acyclovir (ACV), penciclovir (PCV), ganciclovir (GCV), and brivudin (BVDU) (39). The HSV-1 TK phosphorylates ACV, PCV, and GCV to their monophosphate forms, which are subsequently converted to the diphosphorylated and triphosphorylated derivatives by cellular kinases. The antivirally active triphosphate metabolites then inhibit viral DNA synthesis. In contrast, BVDU and its closely related analogue BVaraU are phosphorylated to both their 5′ monophosphate and 5′ diphosphate metabolites by HSV-1 TK and its associated thymidilate kinase activity and are converted to the 5′ triphosphate form by cellular kinases (25).
Since the introduction of ACV, now 20 years ago, for the treatment of herpesvirus infections (2), many studies have characterized drug-resistant herpesvirus mutants isolated either in vitro or in vivo. Different mechanisms exist by which HSV can acquire resistance to ACV. Three of these mechanisms involve the viral TK: (i) viral TK protein with altered substrate specificity (TKaltered), (ii) complete deficiency in viral TK (TKnegative [TK−]), and (iii) decreased activity or production of small amounts of viral TK (TKpartial) (7, 12, 26, 27, 28). Alternatively, alterations at the level of the viral DNA polymerase gene have also been observed (13, 26, 29, 41). Although viruses of all four different phenotypes have been isolated from patients, the predominant drug-resistant phenotype recovered in vivo (similar to the in vitro situation) exhibited TK deficiency (TKpartial, albeit with rather low activity, or TKnegative), as shown by several investigators (9, 26, 28, 40).
In contrast to usually self-limiting infections in healthy individuals, in immunocompromised individuals, HSV infection can be severe and persistent. In these cases, prolonged antiviral therapy is required for the management of the infection, resulting in the emergence of drug-resistant mutants in approximately 4 to 7% of immunocompromised patients (3, 6, 8).
The TK gene is not essential for virus replication in cell culture, although in vivo, it is involved in HSV virulence, pathogenicity, and reactivation from latency (10, 20, 31). Nevertheless, about 95% of clinical HSV isolates resistant to ACV contain mutations in the viral TK and not in the viral DNA polymerase (8, 41). Mutations in the TK gene that are associated with ACV resistance are due mostly to the addition or deletion of nucleotides in long homopolymer runs of G's and C's resulting in frameshift mutations and, consequently, a truncated enzyme (12, 30, 42, 43). In fact, two studies have demonstrated that about 50% of the clinical ACV-resistant (ACVr) HSV strains contain such types of mutations and the other half harbor single nucleotide substitutions in conserved and/or nonconserved regions of the TK gene (26, 35). Mutations identified in PCVr mutant herpesviruses isolated in vitro were generally not found within homopolymeric G and C nucleotide stretches (42). However, in a subsequent study, Suzutani and colleagues (44) reported that mutations in the TK genes from ACVr HSV-1 mutants consisted of 50% single nucleotide substitutions and 50% frameshift mutations while the corresponding figures for the PCVr mutants were 4 and 96%, respectively.
BVDU has been recently approved in Europe for the treatment of HSV-1 and varicella-zoster virus infections. In contrast to ACV and PCV, much less information is known concerning resistance to BVDU in cell culture as well as in patients. Only a few studies have described the isolation of BVDUr strains of HSV-1 in cell culture (17, 23, 33). In this study, we characterized the TK genotype and phenotype of a large panel of BVDUr HSV-1 strains isolated after a single round of selection.
To select the different BVDUr clones, confluent Vero cell cultures prepared in six-well microplates were infected with a one-fifth serial dilution of the HSV-1 laboratory strain KOS for 2 h at 37°C. The inoculum was then removed, and overlay medium containing 50 μg of BVDU/ml was added. After 3 to 4 days of incubation, overlay medium containing neutral red was added and the cultures were further incubated overnight. Individual plaques were picked up, and virus from single plaques was grown in Vero cells. Virus stocks were prepared, titrated, and used in the antiviral assays.
A total of 36 clones were isolated and screened in a first step for sensitivity to ACV, BVDU, and foscarnet (PFA). The drug susceptibilities of the different virus clones were determined by virus cytopathic effect reduction assays in human embryonic lung (HEL) fibroblasts as previously described (1). The 50% inhibitory concentration (IC50) was defined as the drug concentration required to reduce the virus cytopathic effect by 50%. All selected clones were highly resistant to BVDU; however, they differed in their sensitivity to ACV. Twenty-eight clones proved to be resistant to both ACV and BVDU (ACVr/BVDUr) and remained sensitive to PFA, while eight clones were resistant to only BVDU but not to ACV or PFA (ACVs/BVDUr). Among the ACVr/BVDUr clones, significant differences in the degree of resistance to ACV and BVDU were noticed. Three clones had an increase in the 50% effective concentration (EC50) for BVDU of 25 to 50 times that of the wild-type strain, while for the remaining number of clones, the drug resistance level varied 500- to 4,000-fold.
To determine the TK gene sequence of the different HSV-1 clones, DNA from HSV-infected cells was first extracted with a QIAamp Bloodkit (QIAGEN) according to the manufacturer's instructions. The entire HSV-1 TK gene coding sequence was PCR amplified from DNA extracted from infected cells with the forward primer 1 (5′-TGGCGTGAAACTCCCGCACCTC-3′) and the reverse primer 2 (5′-TCTGTCTTTTTATTGCCGTCATAGC-3′). All PCRs were performed under the following conditions: one denaturation step for 2 min at 95°C; 45 cycles of melting at 95°C for 1 min, annealing at 61°C for 30 s, and extension at 72°C for 2 min; and a final elongation step for 10 min at 72°C. The PCR products were purified with a DNA extraction kit (High Pure PCR product purification kit; Roche), and then the TK gene amplicons were directly sequenced using a cycle sequencing kit (DYEnamic dye terminator kit; Amersham Biosciences) on a capillary DNA sequencing system (MegaBACE 500; Amersham Biosciences). Together with forward primer 1 and reverse primer 2, a set of six primers spanning the entire coding region of the TK gene was utilized for DNA sequencing of both strains. The sequencing results were computer assembled and compared with the TK sequence from the KOS reference strain (VectorNTI; Informax).
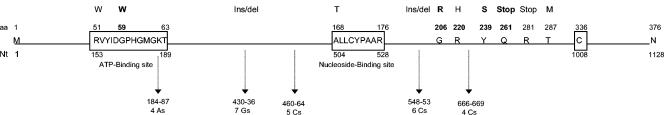
The HSV TK gene coding region is 1,128 nucleotides (nt) in length and encodes a protein of 376 amino acids (aa) (Fig. 1). It exhibits an ATP-binding site (aa 51 through 63) and a nucleoside-binding site (aa 168 through 176) (21, 32). All 36 clones contained distinct mutations in the coding region of the TK gene. Eight clones had an insertion or a deletion of one G in a series of seven G's (nt 430 through 436), located between the putative ATP- and nucleoside-binding sites of the TK. Seven clones had an insertion or a deletion of one C in a series of six C's (nt 548 through 553), located downstream of the nucleoside-binding site of the enzyme.
FIG. 1.
Mutations identified in the TK gene of HSV-1 mutants resistant to BVDU (not to scale). ATP-binding site, nucleoside-binding site, and cysteine-336 are indicated by open boxes. The codon (aa) and nucleotide (Nt) numbers of the wild-type HSV-1 sequence are indicated. Mutational homopolymer runs and the nucleotides involved in drug resistance are indicated by vertical arrows. Amino acid changes and the relative positions of stop codons (Stop), insertions (Ins), and deletions (del) are indicated above the diagram of the protein and the gene. Novel mutations described in the present study (G59W, G206R, Y239S, and G261stop) are indicated in bold letters.
All the other 21 clones (58%) contained different single-base substitutions all within the TK gene, which resulted in a stop codon or a new codon at the site of the substitution. Nucleotide substitutions in the TK appeared to occur as frequently as the homopolymer insertion and deletion mechanism in BVDUr clones. Point mutations were observed at two positions within the ATP-binding site: R51W (two clones) and G59W (one clone). Four different substitutions (G206R [one clone], R220H [three clones], Y239S [three clones], and T287 M [one clone]) located downstream of the nucleoside-binding site were also detected. Two other nucleotide substitutions lead to a stop codon, resulting in the premature termination of the peptide chain downstream of aa 261 (one clone) and 281 (one clone). Interestingly, all eight ACVs/BVDUr virus clones showed the A168T mutation within the nucleoside-binding site.
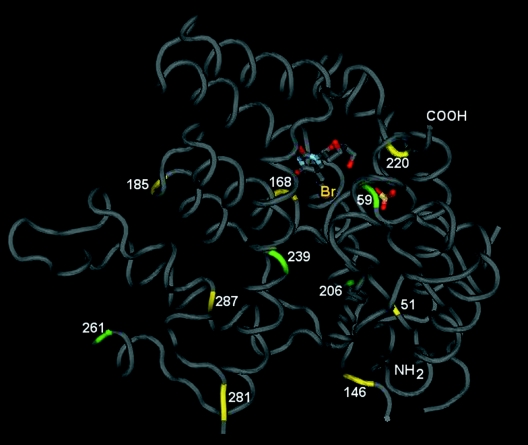
Figure 2 shows the model of the HSV-1 TK protein with different mutations found in the BVDUr clones, including known mutations and novel mutations reported here. The R51W mutation has been previously described both in vitro (42) and in clinical samples (26, 35). A G59A mutation, but not a G59W mutation, has been reported in an in vitro-selected ACVr mutant in combination with a frameshift 146 change (42). Changes at positions 220, 281, and 287, but not at positions 206, 239, and 261, have been previously reported (26, 42, 44). The Arg220, Tyr239, and Thr287 appeared to be conserved residues among herpesviral TKs (21).
FIG. 2.
Model of the HSV-1 TK protein with indication of the known (yellow) and new (green) changes, leading to altered drug sensitivity. The model was constructed using the Molecular Modeling Database (5), based on the crystal structure of TK from HSV-1 complexed with BVDU (4).
Representative clones from the entire panel of HSV mutants described here were then tested for susceptibility to a broad spectrum of antiherpesvirus compounds, including TK-dependent antiviral agents (i.e., BVDU, BVaraU, ACV, GCV, and PCV) and TK-independent antiviral agents (i.e., PFA, the 2-phosphonylmethoxyethyl derivatives of adenine adefovir [PMEA] and 2,6-diaminopurine [PMEDAP], and the 3-hydroxy-2-phosphonylmethoxypropyl derivatives of cytosine cidofovir [HPMPC] and adenine [HPMPA]). All BVDUr clones remained fully susceptible to TK-independent drugs, i.e., the pyrophosphate analogue PFA and the acyclic nucleoside phosphonates PMEA, PMEDAP, HPMPC, and HPMPA suggesting a lack of mutations in the viral DNA polymerase gene (Table 1). The Cl-1 and Cl-5 clones, bearing the A168T mutation, were resistant to the pyrimidine nucleoside derivatives BVDU and its closely related analogue BVaraU but remained sensitive to the acyclic purine nucleoside derivatives ACV, GCV, and PCV. These results are consistent with a TK-altered (TKa) phenotype. According to the three-dimensional model of the A168T mutant described by Kussmann-Gerber and colleagues (32), this amino acid change does indeed not affect the binding of substrates with a substitution on C5 of the pyrimidine ring equal to, or smaller than, a methyl group in size. Thus, no difference for thymidine binding to the wild-type and mutant TK enzymes are expected, whereas the lack of sufficient space between the 168Thr and pyrimidine ring does not allow a correct positioning of heterocycles with a bulkier substituent at C5 such as 2-bromovinyl.
TABLE 1.
Drug susceptibility profilea of selected BVDUr clones and comparative neurovirulence in NMRI mice
| Clone or wild type | Mutation |
n-Fold increase in IC50b
|
Neurovirulenced log (PFU/LD50) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BVDU | BVaraU | ACV | GCV | PCV | PFA | PMEA | PMEDAP | HPMPC | HPMPA | |||
| CI-1 | Ala 168 to Thr | ≥2,420 | >3,450 | 0.9 | 1.0 | 0.92 | 1.3 | 1.5 | 1.7 | 0.7 | 1.3 | 0.05 |
| CI-5 | Ala 168 to Thr | ≥2,260 | ≥3,450 | 1.0 | 0.9 | 0.73 | 1.2 | 1.0 | 1.1 | 0.8 | 0.9 | 0.03 |
| CI-24 | Arg 51 to Trp | 565 | >3,450 | 1,000 | 3,400 | 500 | 0.9 | 1.1 | 1.2 | 1.2 | 1.0 | 2.92 |
| CI-28 | Arg 51 to Trp | 500 | >3,450 | 984 | 10,670 | 450 | 1.1 | 1.5 | 1.8 | 1.4 | 1.6 | 2.92 |
| CI-3 | Gly 59 to Trp | ≥2,550 | >3,450 | 605 | 1,400 | 73 | 0.6 | 0.7 | 0.6 | 2.0 | 0.9 | 2.54 |
| CI-27 | Gly 206 to Arg | ≥3,230 | >3,450 | ≥979 | 20,670 | >769 | 1.2 | 1.5 | 2.2 | 1.1 | 1.6 | 3.30 |
| CI-2 | Arg 220 to His | >3,230 | >3,450 | 147 | 57 | 5.4 | 0.7 | 1.2 | 1.0 | 0.7 | 1.1 | 0.94 |
| CI-22 | Tyr 239 to Ser | 113 | 931 | 247 | 180 | 20 | 1.0 | 1.5 | 1.5 | 0.8 | 1.1 | NDe |
| CI-23 | Tyr 239 to Ser | 23 | 1,400 | 126 | 63 | 24 | 1.1 | 1.2 | 1.4 | 1.0 | 1.9 | 1.40 |
| CI-29 | Gln 261 to stop | ≥3,230 | >3,450 | 468 | 667 | ≥769 | 1.5 | 1.5 | 2.5 | 1.8 | 2.5 | 3.68 |
| CI-25 | Arg 281 to stop | ≥1,770 | >3,450 | 316 | 333 | ≥438 | 1.1 | 1.2 | 1.3 | 0.9 | 1.0 | 4.22 |
| CI-34 | Thr 287 to Met | >3,230 | >3,450 | ≥1,050 | ≥33,330 | >769 | 1.0 | 0.8 | 1.5 | 1.3 | 2.3 | 2.07 |
| CI-4 | 146 frameshift | ≥2,020 | >3,450 | 789 | 4,600 | >769 | 0.5 | 1.1 | 0.9 | 1.1 | 1.7 | 3.43 |
| CI-15 | 185 frameshift | ≥3,230 | >3,450 | ≥947 | 6,270 | ≥627 | 1.1 | 1.5 | 1.0 | 1.3 | 0.7 | 2.5 |
| CI-30 | 185 frameshift | >3,230 | ND | >1,050 | 14,000 | ≥769 | 1.3 | 1.5 | 1.6 | 0.9 | 1.1 | ND |
| Wild type | 0.0062c | 0.0058 | 0.019 | 0.00015 | 0.026 | 19.6 | 7.4 | 1.91 | 0.114 | 0.035 | 0.05 | |
Drug susceptibility profiles of BVDUr HSV-1 clones were determined in HEL cells.
The increase in IC50s was calculated from at least two independent experiments.
Mean IC50s were from seven independent experiments.
Neurovirulence of the different mutants was evaluated by intracerebral inoculation of the viruses into mice. The log (PFU/LD50) was determined as a reciprocal parameter of neurovirulence.
ND, not determined.
The other clones were highly resistant to the different TK-dependent drugs tested, i.e., ACV, BVDU, BVaraU, GCV, and PCV, although differences among the IC50s were observed. Clones with the 146 and 185 frameshift mutations and clones with single amino acid changes in the viral TK at residue 206 or 287 showed the highest increase in EC50 for all TK-dependent drugs (Table 1). Clones with a single nucleotide substitution leading to a stop codon at position 261 or 281 showed a high level of resistance to BVDU, BVaraU, and PCV but an intermediate increase of their IC50s for ACV and GCV (Table 1). Further investigations are necessary to elucidate why stop codon mutations exhibit intermediate resistance to ACV and GCV.
Remarkably, the clones with the R51W and G59W mutations, both located in the ATP-binding site, showed high-level resistance to ACV and BVaraU, but they differed in their degree of resistance to GCV and PCV (Table 1). The R51W mutants possessed higher levels of resistance to GCV and PCV and lower levels of resistance to BVDU than the G59W mutant. Although 51Arg and 59Gly are 2 aa positions located in the ATP-binding site that confer resistance when mutated, our results suggested that the molecular mechanisms are different in each case.
Clones with the Y239S mutation showed the lowest levels of resistance to all TK-dependent drugs. The R220H mutant also demonstrated low levels of resistance to ACV, GCV, and PCV but proved to be fully resistant to BVDU and BVaraU. It has been somewhat surprising that the R220H mutant showed varying resistance profiles against several purine nucleoside analogues (e.g., fivefold resistance to PCV but 147-fold resistance to ACV) and that the Y239S mutants showed varying resistance profiles toward the several pyrimidine nucleoside analogues (i.e., lower sensitivity to BVDU than BVaraU). Obviously, the sugar or acyclic aliphatic moiety of the antiherpetic drugs may slightly differ in their positioning in the TK active site and therefore may interact differently with the mutant 220His and 239Ser amino acids located downstream the nucleoside-binding site. The analysis of the structure of the HSV-1 TK performed by Evans and colleagues (21) indicated that several nonsequential residue contacts and long-range effects appear to influence the well-known breath of the range of nucleoside substrates.
To determine whether lower concentrations of BVDU resulted in selection of mutants with a similar resistance and sensitivity phenotype, viral clones from the HSV-1/KOS strain were selected in the presence of 5 and 10 μg/ml of the compound. As a result, 2 out of 11 clones (18%) and 2 out of 13 clones (15%) selected under pressure of 5 and 10 μg of BVDU/ml, respectively, showed the ACVs/BVDUr phenotype compared to 8 out of 37 clones (22%) isolated in the presence of high-dose BVDU (50 μg/ml). The remaining virus clones displayed the ACVr/BVDUr phenotype. When a plaque-purified stock of the KOS strain was used in similar studies, all clones proved to be resistant to BVDU, 1 out of 14 clones (7%) being ACVs/BVDUr. The selection of BVDUr clones after a single round of selection with 50 μg of BVDU/ml also occurred when a different HSV-1 strain (F) was used, 1 out of 15 clones (7%) being ACVs/BVDUr. Thus, irrespective of the drug concentration, the nature, and the purity of the virus strain used during the selection process, a comparable fraction of the virus mutants was of the mutant ACVs/BVDUr phenotype.
To demonstrate that the TK-resistant phenotypes observed in the mutant viruses could be ascribed to mutations in the TK gene, clones with a TK deficiency were tested in OstTK-negative, HSV-1 TK-positive cells, which stably express the HSV-1 TK product, for susceptibility to ACV, PCV, GCV, and PFA. BVDU and BVaraU could not be tested in this system due to their potent cytostatic and cytototic activities against OstTK-negative, HSV-1TK-positive cells, as has been previously shown by Degrève and colleagues (18). All BVDUr clones showed IC50s for ACV, PCV and GCV in the OstTK−/HSV-1TK+ cells that were comparable with those obtained for the wild-type KOS strain, while the mutant strains exhibited a TK-resistant phenotype in the corresponding nontransfected OstTK-negative cells (Table 2). As expected, no differences in the IC50s were observed for PFA between the cell lines. These results are consistent with rescue of the TK-negative phenotype of the different mutants and point to a lack of mutation(s) in the viral DNA polymerase gene. It should be noted that HEL cells appeared to be more sensitive to determine differences in the drug susceptibility profiles of drug-resistant mutants than the osteosarcoma TK-negative cells. Thus, only the mutants R220H and Y239S, which showed the lowest levels of resistance to TK-dependent drugs in HEL cells, also showed decreased levels of resistance in the osteosarcoma TK-negative cells compared to the other mutants. The level of resistance varied between the HEL cells and the osteosarcoma TK-negative cells used in this study. Thus, while the R220H mutants showed 147-, 57-, and 5.4-fold resistance to, respectively, ACV, GCV, and PCV in HEL cells, the corresponding values of increase in severalfold resistance were ≥266, 1,364, and 68 in the osteosarcoma TK-negative cells. Similarly, the increase in severalfold resistance for the Y239S mutants was higher in the osteosarcoma TK-negative cells than in HEL cells. Therefore, differences in levels of resistance to the various TK-dependent drugs observed with the other BVDUr mutants in HEL cells could not be detected in the osteosarcoma TK-negative cells. These results point to the awareness of comparing levels of resistance among different cell lines, confirming previous reports showing differences in drug susceptibility profiles for HSV among different cell lines (34, 37). A comparative study of drug phosphorylation and drug susceptibilities in different cell lines infected with wild-type virus and the different mutants should help to understand the differences in levels of resistance among various cell lines.
TABLE 2.
Antiviral susceptibilities of BVDUr clones in Ost TK− cells versus Ost TK−/HSV-1TK+ cells
| Clone or wild type | Mutation | IC50 (μg/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Ost TK− cells
|
Ost TK−/HSV-1TK+ cells
|
||||||||
| ACV | GCV | PCV | PFA | ACV | GCV | PCV | PFA | ||
| CI-24 | R51W | ≥12.5 | 20 | 20 | 11.3 | 0.035 | 0.0013 | 0.035 | 20 |
| CI-28 | R51W | >20 | >20 | >20 | 14.3 | 0.029 | 0.0015 | 0.032 | 20 |
| CI-3 | G59W | ≥15 | ≥20 | ≥15 | 16 | 0.026 | 0.0026 | 0.042 | 25.1 |
| CI-27 | G206R | ≥15 | ≥20 | ≥20 | 22.6 | 0.045 | 0.0014 | 0.07 | 18 |
| CI-2 | R220H | ≥18.6 | 14.7 | 8.2 | 27.3 | 0.034 | 0.002 | 0.037 | 19 |
| CI-22 | Y239S | 5.7 | 0.95 | 7 | 19.6 | 0.043 | 0.004 | 0.07 | 25.1 |
| CI-23 | Y239S | 4.7 | 0.7 | 12 | 22.9 | 0.051 | 0.009 | 0.07 | 25.1 |
| CI-29 | Q261 stop | ≥20 | ≥15 | >20 | 12.1 | 0.036 | 0.0051 | 0.096 | 22.6 |
| CI-25 | R281 stop | 20.0 | >20 | >20 | 20.8 | 0.036 | 0.0033 | 0.06 | 25.1 |
| CI-34 | T287M | ≥20 | ≥15 | >20 | 16.3 | 0.02 | 0.002 | 0.032 | 20 |
| CI-4 | 146 frameshift | >20 | 16 | >20 | 15 | 0.033 | 0.0032 | 0.032 | 15 |
| CI-15 | 185 frameshift | >20 | >20 | >20 | 15 | 0.02 | 0.005 | 0.02 | 20 |
| CI-30 | 185 frameshift | 5 | ≥15 | ≥20 | 11.3 | 0.02 | 0.0032 | 0.032 | 20 |
| Wild type (KOS) | 0.07 | 0.0094 | 0.12 | 15 | 0.04 | 0.0034 | 0.065 | 26 | |
The results are mean values of two independent experiments.
Neurovirulence of the different BVDUr HSV-1 mutants was evaluated by intracerebral inoculation of the virus strains into mice. Different virus clones were titrated in HEL cell cultures by plaque formation, and the virus titer was expressed in PFU/ml. In parallel, adult NMRI mice were inoculated intracerebrally with 10-fold dilutions of each virus. Four mice were used per virus dilution. Mortality was recorded over a period of 20 days, and the 50% lethal doses (LD50s) for the different isolates were calculated by Reed and Muench (40a). The PFU/LD50 ratios were monitored as a reciprocal parameter of neurovirulence (Table 1). The virus clones with the ACVs/BVDUr phenotype, bearing the A168T mutation in their TK, showed high neurovirulence similar to that of the parental neurovirulent KOS strain. In contrast, virus clones with frameshift mutations in the coding sequence of the TK (i.e., frameshift mutations at residues 146 and 185) or stop codons (i.e., at positions 261 and 281) showed a marked reduction in pathogenicity (log PFU/LD50 of ≥2.5). Also, virus clones with mutations in the ATP-binding site (R51W and G59W) were significantly reduced in neurovirulence. Virus clones with substitutions located downstream of the nucleotide-binding site showed different degrees of neuropathogenicity. Virus clones harboring the G206R or the T287 M mutations were markedly reduced in neuropathogenicity, and these mutations were associated with a high level of resistance to all TK-dependent drugs (Table 1). Interestingly, virus clones with the R220H or Y239S mutations, which had pronounced neuropathogenicity (log ratio PFU/LD50 of 0.94 to 1.4) also showed the lowest levels of resistance to ACV, GCV, and PCV (Table 1). Thus, for the different BVDUr virus clones, a close correlation between the degree of in vitro resistance to antiviral drugs, TK genotype, and in vivo neurovirulence in mice was found.
HSV pathogenesis has been studied mostly in mouse models via intracerebral inoculation, which leads to encephalitis and death (neurovirulence), and via inoculation at a peripheral site, where the virus replicates and, following axonal transport, it reaches the trigeminal ganglia (secondary site of replication) and establishes a latent infection (11). Following intracerebral inoculation, which can be considered the assay most sensitive to drug resistance, mutants described as TKnegative and TKpartial have been shown to be significantly less virulent than wild-type viruses (14, 15, 22, 45). However, Pelosi and colleagues (37) have reported a TK mutant with a large deletion which was only slightly impaired in neurovirulence and one TKpartial mutant which was fully neurovirulent following intracerebral inoculation. The degrees of attenuation of TKaltered mutants and some polymerase mutants may vary substantially (16, 22, 24, 37, 38).
In the present study, we assessed the degrees of pathogenicity of the different HSV-1 clones resistant to BVDU in terms of neurovirulence. Not surprinsingly, only the A168T TK mutated virus strains retained full neurovirulence, while the others did not. The A168T virus mutants were the only virus strains that had retained pronounced TK catalytic activity in the presence of the natural substrate dThd, but not when BVDU was used as substrate (data not shown), confirming previous reports associating this mutation with a TKaltered phenotype (32, 46). Darby and collegues (15) described the isolation of an HSV-1 ACVr mutant which induced a TK of altered substrate specificity and showed that the virus retained pathogenicity for mice with an only slight attenuation of neurovirulence. Also, a BVDUr strain which induced normal levels of TK and retained virulence for mice has been described previously (23). Studies are in progress to determine the ability of the different TK mutants to replicate in the peripheral nervous system during the acute phase of infection and to spread to secondary peripheral sites.
The A168T substitution was as frequent as a frameshift mutation caused by nucleotide insertions or deletions at one of the homopolymer repeat domains, G7 and C6. We showed that the appearance of the ACVs/BVDUr TK profile was not strictly linked to the high-drug-dose selection procedure or to the virus strain used. It appears that in all circumstances, the A168T mutation found in the ACVs/BVDUr HSV-1 TK phenotype may be among those mutations that emerge at a relatively high frequency during the mutant virus selection process, in contrast to the emergence of ACVr mutant viruses with a TKaltered phenotype (9, 26, 36). It is interesting to note that an A168T mutant virus has been isolated in a patient who had not received any prior drug therapy (19, 46). However, it is obvious that such mutant virus infections, if occurring in BVDU-treated HSV-1-infected patients, can still be efficiently treated by purine nucleoside derivatives such as ACV, GCV, and PCV.
The fact that no changes in sensitivity to TK-independent drugs were observed for the different BVDUr clones, together with rescue of their TK-phenotype for ACV, PCV, and GCV in a cell line constitutively expressing the product of the HSV-1 TK gene, point to a lack of mutations in the viral DNA polymerase genes of these mutants.
In conclusion, single-round selection with BVDU allowed the isolation of different drug-resistant HSV-1 phenotypes, i.e., ACVs/BVDUr and ACVr/BVDUr. Mutations associated with BVDU resistance occurred in the homopolymer stretches of G's and C's of the TK gene. Novel nucleotide substitutions producing stop codons or a new codon at the site of substitution were also identified. Previously described mutations in TK were also found, namely, the A168T mutation, which is associated with a TKaltered phenotype, the most common substitution observed. Different alterations in the viral TK gene could be correlated with different degrees of resistance to various TK-dependent drugs and in vivo neurovirulence.
Acknowledgments
This work was supported by Fortis Insurances AB, Brussels, Belgium; the Fund for Scientific Research Flanders (FWO—Vlaanderen); the Charcot Foundation; and the Belgian Geconcerteerde Onderzoeksacties (GOA).
We thank C. Callebaut for her fine editorial help and L. van Berckelaer and W. Zeegers for technical assistance.
REFERENCES
- 1.Andrei, G., R. Snoeck, E. De Clercq., R. Esnouf, P. Fiten, and G. Opdenakker. 2000. Resistance of herpes simplex virus type 1 against different phosphonylmethoxyalkyl derivatives of purines and pyrimidines due to specific mutations in the viral DNA polymerase gene. J. Gen. Virol. 81:639-648. [DOI] [PubMed] [Google Scholar]
- 2.Bacon, T. H., M. J. Levin, J. J. Leary, R. T. Sarisky, and D. Sutton. 2003. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin. Microbiol. Rev. 16:114-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakrabarti, S., D. Pillay, D. Ratcliffe, P. A. Cane, K. E. Collinghan, and D. W. Milligan. 2000. Resistance to antiviral drugs in herpes simplex virus infections among allogeneic stem cell transplant recipients: risk factors and prognostic significance. J. Infect. Dis. 181:2055-2058. [DOI] [PubMed] [Google Scholar]
- 4.Champness, J. N., M. S. Bennett, F. Wien, R. Visse, W. C. Summers, P. Herdewijn, E. de Clerq, T. Ostrowski, R. L. Jarvest, and M. R. Sanderson. 1998. Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands. Proteins 32:350-361. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, A. Marchler-Bauer, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, B. S. Rao, A. R. Panchenko, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yamashita, J. J. Yin, and S. H. Bryant. 2003. MMDB: Entrez's 3D-structure database. Nucleic Acids Res. 31:474-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y., C. Scieux, V. Garrait, G. Socié, V. Rocha, J. M. Molina, D. Thouvenot, F. Morfin, I. Hocqueloux, L. Garderet, H. Esperou, F. Selimi, A. Dervergie, G. Leleu, M. Aymard, F. Morinet, E. Gluckman, and P. Ribaud. 2000. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clin. Infect. Dis. 31:927-935. [DOI] [PubMed] [Google Scholar]
- 7.Chibo, D., J. Druce, J. Sasadeusz, and C. Birch. 2004. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antivir. Res. 61:83-91. [DOI] [PubMed] [Google Scholar]
- 8.Christophers, J., J. Clayton, J. Craske, R. Ward, P. Collins, M. Trowbridge, and G. Darby. 1998. Survey of resistance of herpes simplex virus to acyclovir in northwest England. Antimicrob. Agents Chemother. 42:868-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coen, D. M., P. A. Schaffer, P. A. Furman, P. M. Keller, and M. H. St. Clair. 1982. Biochemical and genetic analysis of acyclovir-resistant mutants of herpes simplex virus type 1. Am. J. Med. 73:351-360. [DOI] [PubMed] [Google Scholar]
- 10.Coen, D. M., M. Kosz-Vnenchak, J. G. Jacobson, D. A. Leib, C. L. Bogard, P. A. Schaffer, K. L. Tyler, and D. M. Knipe. 1989. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. USA 86:4736-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coen, D. M. 1994. Acyclovir-resistant, pathogenic herpesviruses. Trends Microbiol. 2:481-485. [DOI] [PubMed] [Google Scholar]
- 12.Collins, P., and G. Darby. 1991. Laboratory studies of herpes simplex virus strains resistant to acyclovir. Rev. Med. Virol. 1:19-28. [Google Scholar]
- 13.Collins, P., B. A. Larder, N. M. Oliver, S. Kemp, I. W., Smith, and G. Darby. 1989. Characterization of a DNA polymerase mutant of herpes simplex virus from a severely immunocompromised patient receiving acyclovir. J. Gen. Virol. 70:375-382. [DOI] [PubMed] [Google Scholar]
- 14.Chrisp, C. E., J. C. Sunstrum, D. R. Averill, Jr., M. Levine, and J. C. Glorioso. 1989. Characterization of encephalitis in adult mice induced by intracerebral inoculation of herpes simplex virus type 1 (KOS) and comparison with mutants showing decreased virulence. Lab. Investig. 60:822-830. [PubMed] [Google Scholar]
- 15.Darby, G., H. J. Field, and S. A. Salisbury. 1981. Altered substrate specificity of herpes simplex virus thymidine kinase confers acyclovir-resistance. Nature 289:81-83. [DOI] [PubMed] [Google Scholar]
- 16.Darby, G., M. J. Churcher, and B. A. Larder. 1984. Cooperative effects between two acyclovir resistance loci in herpes simplex virus. J. Virol. 50:838-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darby, G., B. A. Larder, and M. M. Inglis. 1986. Evidence that the ‘active centre’ of the herpes simplex virus thymidine kinase involves an interaction between three distinct regions of the polypeptide. J. Gen. Virol. 67:753-758. [DOI] [PubMed] [Google Scholar]
- 18.Degrève, B., E. De Clercq, and J. Balzarini. 1999. Bystander effect of purine nucleoside analogues in HSV-1 tk gene therapy is superior to that of pyrimidine nucleoside analogues. Gene Ther. 6:162-170. [DOI] [PubMed] [Google Scholar]
- 19.Docherty, J. J., A. T. Dobson, J. J. Trimble, and B. A. Jennings. 1991. Herpes simplex virus type 1 that exhibits herpes simplex virus type 2 sensitivity to (E)-5-(bromovinyl)-2′-deoxyuridine. Intervirology 32:308-315. [DOI] [PubMed] [Google Scholar]
- 20.Efstathiou, S., S. Kemp, G. Darby, and A. C. Minson. 1989. The role of herpes simplex virus type 1 thymidine kinase in pathogenesis. J. Gen. Virol. 70:869-879. [DOI] [PubMed] [Google Scholar]
- 21.Evans, J. S., K. P. Lock, B. A. Levine, J. N. Champness, M. R. Sanderson, W. C. Summers, P. J. McLeish, and A. Buchan. 1998. Herpesviral thymidine kinases: laxity and resistance by design. J. Gen. Virol. 79:2083-2092. [DOI] [PubMed] [Google Scholar]
- 22.Field, H. J., and G. Darby. 1980. Pathogenicity in mice of strains of herpes simplex virus which are resistant to acyclovir in vitro and in vivo. Antimicrob. Agents Chemother. 17:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Field, H. J., and J. Neden. 1982. Isolation of bromovinyldeoxyuridine-resistant strains of herpes simplex virus and successful chemotherapy of mice infected with one such strain by using acyclovir. Antiviral Res. 2:243-254. [DOI] [PubMed] [Google Scholar]
- 24.Field, H. J., and D. M. Coen. 1986. Pathogenicity of herpes simplex virus mutants containing drug resistance mutations in the viral DNA polymerase gene. J. Virol. 60:286-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fyfe, J. A. 1982. Differential phosphorylation of (E)-5-(2-bromovinyl)-2′-deoxyuridine monophosphate by thymidylate kinases from herpes simplex viruses types 1 and 2 and varicella zoster virus. Mol. Pharmacol. 21:432-437. [PubMed] [Google Scholar]
- 26.Gaudreau, A., E. Hill, H. H. Balfour, Jr., A. Erice, and G. Boivin. 1998. Phenotypic and genotyic characterization of acyclovir-resistant herpes simplex viruses from immunocompromised patients. J. Infect. Dis. 178:297-303. [DOI] [PubMed] [Google Scholar]
- 27.Harris, W., P. Collins, F. J. Fenton, W. Snowden, M. Sowa, and G. Darby. 2003. Phenotypic and genotypic characterization of clinical isolates of herpes simplex virus resistant to acyclovir. J. Gen. Virol. 84:1393-1401. [DOI] [PubMed] [Google Scholar]
- 28.Hill, E. L., G. A. Hunter, and M. N. Ellis. 1991. In vitro and in vivo characterization of herpes simplex virus clinical isolates recovered from patients infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 35:2322-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang, C. B. C., K. L. Ruffner, and D. M. Coen. 1992. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J. Virol. 66:1774-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang, C. B. C., and H. J. H. Chen. 1995. An altered spectrum of herpes simplex virus mutations mediated by an antimutator DNA polymerase. Gene 152:191-193. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson, J. G., K. L., Ruffner, M. Kosz-Vnenchak, C. B. C. Hwang, K. K. Wobe, D. M. Knipe, and D. M. Coen. 1993. Herpes simplex virus thymidine kinase and specific stages of latency in murine trigeminal ganglia. J. Virol. 67:6903-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kussmann-Gerber, S., O. Kuonen, G. Folkers, B. D. Pilger, and L. Scapozza. 1998. Drug resistance of herpes simplex virus type 1—structural considerations at the molecular level of the thymidine kinase. Eur. J. Biochem. 255:472-481. [DOI] [PubMed] [Google Scholar]
- 33.Larder, B. A., Y. C. Cheng, and G. Darby. 1983. Characterization of abnormal thymidine kinases induced by drug-resistant strains of herpes simplex virus type 1. J. Gen. Virol. 64:523-532. [DOI] [PubMed] [Google Scholar]
- 34.Leary, J. J., R. Wittrock, R. T. Sarisky, A. Weinberg, and M. J. Levin. 2002. Susceptibilities of herpes simplex viruses to penciclovir and acyclovir in eight cell lines. Antimicrob. Agents Chemother. 46:762-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morfin, F., G. Souillet, K. Bilger, T. Ooka, M. Aymard, and D. Thouvenot. 2000. Genetic characterization of thymidine kinase from acyclovir-resistant and -susceptible herpes simplex virus type 1 isolated from bone marrow transplant recipients. J. Infect. Dis. 182:290-293. [DOI] [PubMed] [Google Scholar]
- 36.Nugier, F., J. N. Colin, M. Aymard, and M. Langlois. 1992. Occurrence and characterization of acyclovir-resistant herpes simplex virus isolates: report on a two-year sensitivity screening survey. J. Med. Virol. 36:1-12. [DOI] [PubMed] [Google Scholar]
- 37.Pelosi, E., G. B. Mulamba, and D. M. Coen. 1998. Penciclovir and pathogenesis phenotypes of drug-resistant herpes simplex virus mutants. Antivir. Res. 37:17-28. [DOI] [PubMed] [Google Scholar]
- 38.Pelosi, E., F. Rozenberg, D. M. Coen, and K. L. Tyler. 1998. A herpes simplex virus DNA polymerase mutation that specifically attenuates neurovirulence in mice. Virology 252:364-372. [DOI] [PubMed] [Google Scholar]
- 39.Pilger, B. D., R. Perozzo, F. Alber, C. Wurth, G. Folkers, L. Scapozza. 1999. Substrate diversity of herpes simplex virus thymidine kinase. Impact of the kinematics of the enzyme. J. Biol. Chem. 274:31967-31973. [DOI] [PubMed] [Google Scholar]
- 40.Pottage, J. C., Jr., and H. A. Kessler. 1995. Herpes simplex virus resistance to acyclovir: clinical relevance. Infect. Agents Dis. 4:115-124. [PubMed] [Google Scholar]
- 40a.Reed, J. H., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 41.Sacks, S. L., R. J. Wanklin, D. E. Reece, K. A. Hicks, K. L. Tyler, and D. M. Coen. 1989. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann. Intern. Med. 111:893-899. [DOI] [PubMed] [Google Scholar]
- 42.Sarisky, R. T., M. R. Quail, P. E. Clark, T. T. Nguyen, W. S. Halsey, R. J. Wittrock, J. O'Leary Bartus, M. M. Van Horn, G. M. Sathe, S. Van Horn, M. D. Kelly, T. H. Bacon, J. J. Leary. 2001. Characterization of herpes simplex viruses selected in culture for resistance to penciclovir or acyclovir. J. Virol. 75:1761-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasadeusz, J. J., F. Tufaro, S. Safrin, K. Schubert, M. M. Hubinette, P. K. Cheung, and S. L. Sacks. 1997. Homopolymer mutational hot spots mediate herpes simplex virus resistance to acyclovir. J. Virol. 71:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzutani, T., K. Ishioka, E. De Clercq, K. Ishibashi, H. Kaneko, T. Kira, K.-I. Hashimoto, M. Ogasawara, K. Ohtani, N. Wakamiya, and M. Saijo. 2004. Differential mutation patterns in thymidine kinase and DNA polymerase genes of herpes simplex virus type 1 clones passaged in the presence of acyclovir and penciclovir. Antimicrob. Agents Chemother. 47:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenser, R. B. 1983. Intracerebral inoculation of newborn and adult mice with thymidine kinase-deficient mutants of herpes simplex virus type 1. J. Infect. Dis. 147:956. [DOI] [PubMed] [Google Scholar]
- 46.Wilber, B. A., and J. J. Docherty. 1994. Analysis of the thymidine kinase of herpes simplex virus type 1 isolate that exhibits resistance to (E)-5-(2-bromovinyl)-2′-deoxyuridine. J. Gen. Virol. 75:1743-1747. [DOI] [PubMed] [Google Scholar]