Abstract
T-cell responses to dengue viruses may be important in both protective immunity and pathogenesis. This study of 48 Vietnamese adults with secondary dengue virus infections defined the breadth and magnitude of peripheral T-cell responses to 260 overlapping peptide antigens derived from a dengue virus serotype 2 (DV2) isolate. Forty-seven different peptides evoked significant gamma interferon enzyme-linked immunospot (ELISPOT) assay responses in 39 patients; of these, 34 peptides contained potentially novel T-cell epitopes. NS3 and particularly NS3200-324 were important T-cell targets. The breadth and magnitude of ELISPOT responses to DV2 peptides were independent of the infecting dengue virus serotype, suggesting that cross-reactive T cells dominate the acute response during secondary infection. Acute ELISPOT responses were weakly correlated with the extent of hemoconcentration in individual patients but not with the nadir of thrombocytopenia or overall clinical disease grade. NS3556-564 and Env414-422 were identified as novel HLA-A*24 and B*07-restricted CD8+ T-cell epitopes, respectively. Acute T-cell responses to natural variants of Env414-422 and NS3556-564 were largely cross-reactive and peaked during disease convalescence. The results highlight the importance of NS3 and cross-reactive T cells during acute secondary infection but suggest that the overall breadth and magnitude of the T-cell response is not significantly related to clinical disease grade.
Dengue viruses are positive-strand RNA viruses belonging to the family Flaviviridae. Many tropical and subtropical regions have become areas where dengue viruses are endemic; such endemicity is an important cause of morbidity and mortality (17, 40). Primary infection with any of the four serotypes of dengue virus (DV1 to DV4) typically results in either an asymptomatic illness or dengue fever (DF), a self-limiting acute viral infection characterized by high fever and myalgia. Epidemiological studies suggest that individuals who are infected a second time with a different dengue virus serotype are at significantly greater risk of developing dengue hemorrhagic fever (DHF), a serious illness characterized by plasma leakage, thrombocytopenia, and occasionally hypovolemic shock (5, 12, 43, 45). While children carry most of the symptomatic disease burden in countries where dengue viruses are endemic, young adults are also significantly affected.
Antibody-dependent enhancement (ADE) of secondary viral infection is a prominent explanation for the association between severe disease and preexisting dengue virus immune responses (19). It is hypothesized that ADE promotes higher systemic viral loads, a theory supported by studies of cultured cells (20, 21) and of experimentally infected primates (19). Correspondingly, high dengue viral loads (31, 47) and antigenemia (32) have been associated with DHF in children. However, the mechanisms through which relatively higher dengue viral loads might mediate the vascular leakage that is so prominent in patients with severe disease remains unclear. Other parameters, such as the age, nutritional status, and genetic background of the host, may also be important (1, 10, 30, 34, 35, 44, 46). Excessive immune activation during secondary infection has also been hypothesized to act in concert with these other risk factors and to promote the development of severe vascular leakage. In particular, the rapid mobilization of serotype cross-reactive memory T cells that release vasodilatory inflammatory molecules has been suggested to explain some aspects of the clinical syndrome (28). Accordingly, relatively higher frequencies of activated CD8+ T cells in peripheral blood have been detected in patients with DHF than in those with DF (14, 41, 51). Other plasma markers of cellular immune activation, including tumor necrosis factor (23), gamma interferon (IFN-γ) (26), interleukin-6 (IL-6) (22), soluble IL-2 receptor (15, 26), and tumor necrosis factor receptor II (15) levels, are also elevated in DHF compared to DF. Collectively, these studies suggest a role for the immune response in disease pathogenesis. How T cells contribute to this process is incompletely defined, however, since only a few studies have directly examined T-cell responses during acute disease (14, 41). A more comprehensive understanding of the relationship between T-cell response and disease severity in acute dengue illness would be obtained by measuring ex vivo responses to a spectrum of dengue virus antigens in a patient population with a mixed HLA background. To this end, this study measured the magnitude and specificity of T-cell responses to 260 peptide antigens in 48 Vietnamese adults experiencing a secondary dengue virus infection. Results from these experiments were used as a foundation from which to define two novel T-cell epitopes and to assess the relationship between cellular immune response and clinical and virological parameters.
MATERIALS AND METHODS
Study design and patients.
Venous blood samples were obtained from adult patients enrolled in a prospective study of dengue virus infection at The Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam. Patients were recruited if there was a clinical suspicion of dengue virus infection. World Health Organization classification criteria (49) were applied to each case after a review of the study notes. However, the strict classification scheme could not be applied to two cases because of conflicting clinical presentations. For example, two subjects had clinical manifestations of plasma leakage (e.g., pleural effusions) and mucosal bleeding, but platelet counts were between 100,000 and 120,000. We classified these cases as DHF II. Several cases also demonstrated hemoconcentration of <20% during hospitalization but nevertheless had clinical signs of plasma leakage, e.g., pleural effusions or ascites. These cases were classified as DHF I or II, depending on whether or not hemorrhagic manifestations were present.
Peripheral blood samples were collected from patients between 8 and 11 a.m. on the first morning after admission (study day 1), on study day 3, and again on study day 5 unless the patient had been discharged earlier. Convalescent samples were obtained at 2 weeks and 1 month postadmission. Platelet counts and hematocrit values were recorded regularly during hospitalization. The extent of hemoconcentration during symptomatic illness was determined by comparing the maximum hematocrit recorded during hospitalization with the value recorded at convalescence (study day 14 or 30). These data were available for 24 patients. Written informed consent was obtained from the patient. The study protocol was approved by the Scientific and Ethical Committee at The Hospital for Tropical Diseases and the Oxford Tropical Research Ethical Committee.
Dengue virus PCR.
Dengue virus RNA in acute plasma samples was isolated with RNAgents (Promega, Madison, Wis.). RNA was reverse transcribed, and two rounds of PCR were performed using primers and methods described previously (29). In samples containing virus, the PCR yielded DNA products the unique sizes of which were diagnostic for each dengue virus serotype.
Serology, cell isolation, and HLA typing.
Dengue virus infection was confirmed via serological testing of acute- and early-convalescent-phase plasma samples collected at least 3 days apart with a commercial capture-immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) (Panbio, Brisbane, Australia). The ELISA was performed and the results were interpreted according to the manufacturer's instructions. This ELISA assay has been validated as both sensitive and specific for primary and secondary dengue virus infections (48). In this study, the term “secondary infection” is used to describe the nature of the serological response and does not imply that this was necessarily the second dengue virus infection experienced by the patient. Peripheral blood mononuclear cells (PBMC) were isolated from whole blood by Ficoll-Hypaque (Lymphoprep; Axis-Shield, Oslo, Norway) density gradient centrifugation and used directly in enzyme-linked immunospot (ELISPOT) assays or cryopreserved for future use. Molecular HLA typing was performed on most study subjects with amplification refractory mutation system-PCR with sequence-specific primers, as described previously (4). With samples from some subjects, a commercial typing application, the Dynal RELI SSO HLA typing kit (Dynal Biotech, Wirral, United Kingdom) was used.
ELISPOT and peptides.
In this study, we were unable to collect enough blood from patients to study T-cell responses to peptide antigens spanning the entire genome. Consequently, we focused our investigations on structural antigens (capsid, preM/M, and Env) and those nonstructural viral antigens previously nominated as being T-cell immunogens (NS3 and NS4a). A total of 260 peptides (220 15mers and 40 20mers, all overlapping by 10 amino acids [aa]), spanning the capsid, preM, M, Env, NS3, and NS4a sequences from a dengue virus serotype 2 isolate (strain 16681) were synthesized by standard, solid-phase 9-fluorenylmethoxy carbonyl chemistry. Purity ranged from 30 to 90% as determined by high-performance liquid chromatography (2). PBMC samples were obtained between study days 5 to 14 from 48 patients experiencing a secondary dengue virus infection. PBMC samples were tested in IFN-γ ELISPOT assays against the 260 peptides arranged into a matrix of 32 peptide pools, with 16 peptides in each pool (each at 1 μM) and each peptide present in two different pools. This approach allows rapid identification of the peptide within a pool that is responsible for evoking a response in the IFN-γ ELISPOT assay, since this peptide should drive a response in two different pools. In every case, peptides identified as being antigenic in this manner were retested as individual peptides. Thus, each positive response was confirmed twice, by means of the internal control afforded by the matrix and then individually by a confirmatory assay. The ELISPOT assay was performed essentially according to the manufacturer's instructions (Mabtech AB, Stockholm, Sweden). Briefly, 96-well polyvinylidene difluoride-backed plates (MAIPS45; Millipore), precoated with 15 μg of anti-IFN-γ monoclonal antibody 1-D1K (Mabtech)/ml, were blocked with R10 (RPMI 1640 containing 10% heat-inactivated fetal calf serum, 2 mM glutamine, 100 μg of streptomycin/ml, and 100 U of penicillin) for 2 h. A total of 1 × 105 to 3 × 105 PBMC were added in 100 μl of R10 per well, and peptide pools or individual peptides were subsequently added to a final concentration of 16 μM (peptide pools) or 1 μM (individual peptides). After an overnight incubation at 37°C and 5% CO2 in air, plates were washed with phosphate-buffered saline (PBS) containing 0.05% Tween-20 (Sigma). Next, 100 μl of PBS containing 1 μg of biotinylated anti-IFN-γ monoclonal antibody 7-B6-1-biotin (Mabtech)/ml was added. After 2 h, plates were washed, and streptavidin-alkaline phosphatase conjugate (Mabtech) was added at a dilution of 1:1,000 in PBS. The number of spot-forming units (SFU) in each well was counted with the aid of a dissecting microscope, and the background (no antigen stimulation) was subtracted. Wells containing PBMC stimulated with phytohemagglutinin (Sigma, Poole, United Kingdom) served as a positive assay control. A positive response to a pool of peptides was defined as having >50 SFU per million PBMC after subtraction of background. This cutoff was used on the basis that 50 SFU per million PBMC was 2 standard deviations (SD) above the mean of unstimulated PBMC from early-convalescent-phase dengue virus infection cases.
Cell depletions.
T-cell depletions from PBMC were performed using anti-CD8 or anti-CD4 antibody-coated immunomagnetic beads (Dynal, Oslo, Norway) according to the manufacturer's instructions. Flow cytometry analysis of the depleted cells indicated this approach routinely achieved >95% depletion of the target cells.
Cytotoxicity assay.
A T-cell line from BC503 was generated by pulsing 2 × 106 PBMC with 100 μM of Env414-422 for 1 h. Cells were cultured in R10 supplemented with 25 ng of IL-7/ml for 3 days, and then 100 U of IL-2/ml was added every 3 to 4 days thereafter. After 14 days, cells were harvested and used as effectors in a 51Cr-labeled release assay. Targets cells in the chromium release assay consisted of a 51Cr-labeled B-cell line (BCL) that was HLA matched with effectors only at the B*07 locus. The BCL was pulsed for 1 h with the Env414-422 peptide, washed, and aliquoted in microtiter plates (5,000 cells/well), and then effector cells were added at a range of effector-to-target ratios. Unpulsed cells were used as negative controls. The 51Cr release was calculated from the following equation: ([experimental release−spontaneous release]/[maximum release−spontaneous release]) × 100%. Nonspecific killing of unpulsed target cells was subtracted from that of pulsed target cells.
Short-term stimulation of PBMC with peptide-pulsed B-cell lines.
A rapid, flow cytometry-based method was employed to define the HLA restriction of candidate T-cell epitopes as described previously (11). Firstly, Epstein-Barr virus-transformed B-cell lines (BCL) that matched or mismatched the effector cells to be studied (patients' PBMC) at given HLA alleles were selected. These BCLs were pulsed with test or control peptides (10 μM) for 1 h and then washed three times. A total of 5 × 105 peptide-pulsed and washed BCL were then added to 2 × 106 PBMC (effectors) in a 0.3-ml volume and cultured at 37°C with 5% CO2 in air for 6 h. During the last 4 h, brefeldin A (10 μg/ml) was added. After 6 h, cells were washed, surface stained for CD3 and CD8, then fixed (FACSlyse; Becton Dickinson, San Diego, Calif.), and made permeable with Facsperm buffer (Becton Dickinson). Cells were then washed and stained for intracellular IFN-γ and the early activation marker CD69. After being stained and fixed, CD3+ CD8+ cells were gated on a FACSCalibur flow cytometer (Becton Dickinson), and the percentage of cells that was CD69+ IFN-γ+ was determined for each BCL-peptide combination. PBMC stimulated with phorbol myristate acetate-ionomycin were used as positive controls, and PBMC stimulated with medium alone were used as negative controls.
Statistics.
Spearman correlation analysis was used to measure associations between nonnormally distributed variables. Analysis of variance was used for comparisons across multiple groups. A P value of <0.05 was regarded as significant. The SPSS software package SPSS, version 10 (SPSS, Inc., Chicago, Ill.), was used for all analyses.
RESULTS
Characteristics of study population.
This study was conducted at The Hospital for Tropical Diseases, Ho Chi Minh City, Vietnam. The Hospital for Tropical Diseases serves the local community and acts as a tertiary referral hospital for patients in southern Vietnam with infectious diseases. Between June 2002 and April 2003, we recruited 48 patients suspected of having a dengue virus infection from the adult intensive care unit into a prospective study of cellular immune responses. The mean age was 19 years (SD, 3 years; range, 15 to 27 years) and the mean length of illness before admission was 5 days (SD, 1 day; range, 1 to 10 days). Serology of paired plasma samples indicated that all patients were experiencing a secondary dengue virus infection. During hospital admission, the average maximum hematocrit attained was 49% (SD, 4.7%; range, 41 to 60%) and the mean nadir of the platelet count was 47,700 × 106/ml (SD, 33,900 × 106; range, 13,900 × 106 to 115,000 × 106).
IFN-γ ELISPOT responses against a panel of peptides spanning the capsid, preM, M, Env, NS3, and NS4a viral antigens.
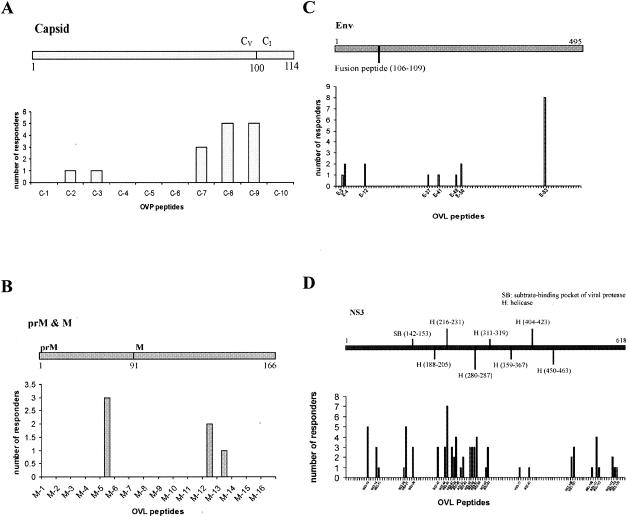
To measure the breadth and magnitude of T-cell responses to multiple dengue virus antigens, overlapping peptides spanning the capsid, preM, M, Env, NS3, and NS4a viral antigens from a DV2 isolate were employed in IFN-γ ELISPOT assays with PBMC samples from dengue virus-immune Vietnamese adults. In 39 patients, we identified one or more individual peptides that reproducibly evoked an IFN-γ response in the ELISPOT assay that exceeded 50 SFU/million PBMC. In total, T cells from 39 patients recognized 47 different peptides. The magnitude of responses to individual peptides ranged from 50 to 500 SFU/million PBMC. The breadth of the response in individual patients ranged from 1 to 10 peptides. A summary of the individual peptides that reproducibly evoked responses in IFN-γ ELISPOT assays is provided in Table 1. In several cases, two adjacent and overlapping peptides evoked a response in the same patient, suggesting the peptides contained a shared epitope or alternatively suggesting the presence of overlapping epitopes. For several of the peptide antigens, we used IFN-γ ELISPOT assays with PBMC depleted of either CD4+ or CD8+ T cells to identify the responding T-cell subset. These experiments identified several peptides (capsid62-81, preM41-60, Env411-425, NS345-59, NS3285-299, and NS3550-564) that contain novel CD4+ or CD8+ T-cell determinants (Table 1). Thirteen (23%) of the 47 peptides recognized by PBMC contain sequences that have previously been characterized as dengue virus- or flavivirus-specific CD4+ or CD8+ T-cell epitopes (Table 1).
TABLE 1.
Summary of antigenic peptides recognized by T cells from patients with secondary dengue virus infection
| Sequencea | Location | nb | ELISPOT frequencyc | Subsetd | Minimum epitope, HLA restriction (reference)e |
|---|---|---|---|---|---|
| PFNMLKRERNRVSTVQQLTK | Capsid12-31 | 1 | 100 | ||
| RVSTVQQLTKRFSLGMLQGR | Capsid22-41 | 1 | 70 | ||
| TAGILKRWGTIKKSKAINVL | Capsid62-81 | 3 | 97 (60-130) | CD4 | |
| IKKSKAINVLRGFRKEIGRM | Capsid72-91 | 5 | 58 (30-120) | CD4 | Capsid83-92 (GFRKEIGRML)/DPw4 (9) |
| RGFRKEIGRMLNILNRRRRS | Capsid82-101 | 5 | 58 (50-80) | CD4 | Capsid83-92 (GFRKEIGRML)/DPw4 (9) |
| LGELCEDTITYKCPLLRQNE | preM41-60 | 3 | 202 (68-500) | CD4 | |
| MSSEGAWKHVQRIETWILRH | M20-39 | 2 | 95 (60-130) | ||
| QRIETWILRHPGFTMMAAI | M30-49 | 1 | 120 | ||
| FVEGVSGGSWVDIVL | Env11-25 | 1 | 500 | ||
| SGGSWVDIVLEHGSC | Env16-30 | 2 | 230 (60-400) | ||
| LRKYCIEAKLTNTTT | Env56-70 | 2 | 115 (80-150) | ||
| TLVTFKNPHAKKQDV | Env136-150 | 1 | 66 | ||
| VTMECSPRTGLDFNE | Env181-195 | 1 | 120 | ||
| MENKAWLVHRQWFLD | Env201-215 | 1 | 80 | ||
| KKQDVVVLGSQEGAM | Env246-260 | 2 | 96 (75-117) | ||
| RMAILGDTAWDFGSLf | Env411-425 | 7 | 162 (60-300) | CD8 | |
| TFHTMWHVTRGAVLM | NS345-59 | 5 | 151 (55-390) | CD4 | |
| IEPSWADVKKDLISY | NS365-79 | 3 | 151 (100-194) | CD8 | NS371-79 (SVKKDLISY)/B*62 (52) |
| ADVKKDLISYGGGWK | NS370-84 | 1 | 97 | NS371-79 (SVKKDLISY)/B*62 (52) | |
| AVSLDFSPGTSGSPI | NS3125-139 | 1 | 125 | ||
| FSPGTSGSPIIDKKG | NS3130-144 | 4 | 148 (53-230) | CD8 | NS3133-142 (GTSGSPIIK)/A*11 (41) |
| KVVGLYGNGVVTRSG | NS3145-159 | 3 | 140 (100-200) | CD4 | NS3146-154 (VIGLYGNGV)/HLA-DR15 (25) |
| TKRYLPAIVREAIKR | NS3200-214 | 3 | 63 (49-85) | CD4 | NS3202-211 (RKYLPAIVRE)/HLA-DR15 (50) |
| GLRTLIAPTRVVAA | NS3215-229 | 3 | 92 (75-100) | ||
| ILAPTRVVAAEMEEA | NS3220-234 | 7 | 150 (40-350) | CD8 | NS3221-232 (LAPTRVVAAEME)/B*07 (3) |
| EMEEALRGLPIRYQT | NS3230-244 | 3 | 213 (120-260) | CD8 | NS3235-243 (AMKGLPIRY)/B*62 (52) |
| LRGLPIRYQTPAIRA | NS3235-249 | 2 | 57 (56-58) | ||
| IRYQTPAIRAEHTGR | NS3240-254 | 3 | 76 (47-100) | ||
| EHTGREIVDLMCHAT | NS3250-264 | 1 | 200 | CD4 | NS3255-264 (EIVDLMCHAT)/HLA-DPw2 (13) |
| EIVDLMCHATFTMRL | NS3255-269 | 1 | 100 | CD4 | NS3255-264 (EIVDLMCHAT)/HLA-DPw2 (13) |
| LSPVRVPNYNLIIMD | NS3270-284 | 2 | 145 (90-200) | ||
| VPNYNLIIMDEAHFT | NS3275-289 | 2 | 230 (210-250) | ||
| LIIMDEAHFTDPASI | NS3280-294 | 2 | 198 (85-310) | ||
| EAHFTDPASIAARGY | NS3285-299 | 3 | 147 (60-200) | CD8 | |
| EMGEAAGIFMTATPP | NS3305-319 | 1 | 70 | ||
| AGIFMTATPPGSRDP | NS3310-324 | 3 | 105 (80-153) | ||
| KKVIQLSRKTFDSEY | NS3380-394 | 1 | 118 | ||
| NDWDFVVTTDISEMG | NS3400-414 | 1 | 100 | ||
| LDNINTPEGIIPSMF | NS3495-509 | 2 | 155 (80-230) | CD8 | NS3500-508 (TPEGIIPTL)/B*35 (33) |
| TPEGIIPSMFEPERE | NS3500-514 | 2 | 155 (70-240) | CD8 | NS3500-508 (TPEGIIPTL)/B*35 (33) |
| GDLPVWLAYRVAAEG | NS3540-554 | 1 | 60 | ||
| VAAEGINYADRRWCFf | NS3550-564 | 4 | 222 (100-310) | CD8 | |
| INYADRRWCFDGVKN | NS3555-569 | 1 | 310 | ||
| EGERKKLKPRWLDAIY | NS3585-599 | 2 | 68 (48-97) | ||
| KLKPRWLDARIYSDP | NS3590-604 | 1 | 85 | ||
| WLDARIYSDPLALKE | NS3595-609 | 1 | 250 | ||
| LATVTGGIFLFLMSGRGIGK | NS4a61-80 | 1 | 58 |
Sequences are derived from DV2 isolate 16681.
n, number of patients who responded to each peptide.
Shown are the mean (range) numbers of SFU/million PBMC evoked by each peptide.
The T-cell subset responding to peptide stimulation was determined by bead depletion of CD4+ and CD8+ T cells.
The peptide sequence and HLA restriction of published epitopes that are present in the larger, overlapping 15- or 20mer peptide are shown.
Peptides shown in boldface were studied further.
Antigen specificity of IFN-γ ELISPOT responses and their relationship to recognized structural motifs.
The spectrum of peptides recognized by T cells ex vivo was examined in the context of known biological features in each of the respective viral antigens. Several antigenic peptides were identified in the capsid protein; these were primarily located near to the conserved cleavage point of the C-terminal hydrophobic signal sequence (Table 1 and Fig. 1A). Antigenic peptides were also detected in preM/M (Table 1 and Fig. 1B), Env (Table 1 and Fig. 1C), and NS4a (Table 1 and data not shown). These antigenic peptides were not associated with any recognized conserved structural motifs. NS3, a 618-aa-long serine protease and helicase, contained the highest number (n = 30) of antigenic peptides (Table 1 and Fig. 1D). A total of 14 (47%) out of 30 of these antigenic peptides were clustered within a 124-aa-long stretch of NS3 (NS3200-324) (Fig. 1D). Alignment of consensus amino acid sequences from all four dengue virus serotypes indicated that this region of NS3 is more conserved (78%) than NS3 as a whole (68%). NS3 was the most frequently recognized viral antigen, with 56% of all tested patients responding to at least one antigenic peptide in NS3 compared to 27% for Env, 23% for capsid, 10% for preM/M, and 2% for NS4a.
FIG. 1.
T-cell responses by viral antigen. Shown is the frequency with which individual antigenic peptides in the (A) capsid, (B) preM/M, (C) Env, and (D) NS3 viral antigens were recognized by PBMC collected on study days 5 to 14 from 48 patients with secondary dengue. Shown within each viral protein are recognized structural features and (in parentheses) their amino acid locations with respect to the start of the protein. The relative position of the overlapping peptide (OVL) in each antigen that evoked a response is indicated on the x axis. (A) CV, capsid virion; CI, intracellularly anchored capsid.
Relationship between the serotype of the infecting virus and the breadth and magnitude of responses to overlapping DV2 peptides in IFN-γ ELISPOT assays.
A dengue virus serotype-specific PCR performed on acute plasma samples collected from all 48 patients identified the serotype of the infecting virus in 33 individuals (68%). Among these 32 patients, DV1 was detected in 9 (28%), DV2 was detected in 15 (47%), DV3 was detected in 1 (3%), and DV4 was detected in 7 (22%). One patient had a mixed infection comprising DV1 and DV4 (3%). We sought to determine whether a relationship existed between the breadth and magnitude of the T-cell response to individual peptides during early convalescence (study day 5) and the serotype of the infecting virus. Study day 5 was selected because it allowed us to assess the breadth and magnitude of the T-cell response as close as possible to when the patient was ill but the viremia had nonetheless been resolved. It also allowed us to measure responses in a large number of patients, since not all patients returned for follow-up visits at study day 14. We did not include the patient with a mixed DV1-DV4 infection in this analysis. In cases where patients made responses to adjacent peptides, we included responses to both in the analysis. Among the 47 patients analyzed, the breadth of the response was not significantly associated with the serotype of the infecting virus. Thus, patients infected with DV2 (n = 15) did not respond to significantly more peptides (median, 3; range, 0 to 10) than patients infected with either DV1 (n = 9, median, 1.5; range, 1 to 4), DV3 (n = 1, median, 0), DV4 (n = 7; median, 2; range, 0 to 4) or patients in whom a dengue virus could not be detected (n = 15; median, 3; range, 0 to 5). Like the breadth of the response, the mean sum of ELISPOT frequencies to individual DV2 peptides in patients infected with DV2 did not significantly exceed (93 ± 42 SFU/million PBMC [values are means ± SD) those in patients infected with either DV1 (185 ± 135 SFU) or DV4 (112 ± 60 SFU) or in patients in whom a dengue virus could not be detected (108 ± 78 SFU).
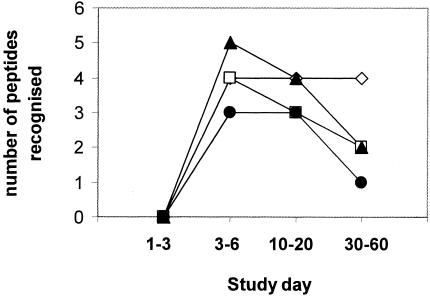
One possible explanation for the absence of significantly stronger responses to DV2 peptides in patients infected with a serotype 2 virus is that the DV2-specific response had not sufficiently matured at the time of analysis (study day 5). To explore this hypothesis further, ELISPOT responses to pools of overlapping DV2 peptides were measured in serial PBMC samples collected from five prospectively recruited patients infected with DV2. Responses detected against pooled peptides were verified using individual peptides. In the four patients who had responses to one or more peptide antigens, the breadth of the response actually contracted rather than expanded as time since infection elapsed (Fig. 2). The narrowing of the breadth of responses coincided with a reduction in the magnitude of responses to each antigenic peptide (data not shown). This suggests that a significant DV2-specific response does not evolve with increased time since secondary infection.
FIG. 2.
Breadth and magnitude of the ELISPOT response to DV2 peptides in patients infected with a DV2 virus at different times postpresentation. The number of individual peptide antigens that evoked significant responses (>50 SFU/million PBMC) in PBMC samples collected from four prospectively recruited patients infected with DV2 was determined at different times postinfection. Shown are the number of peptides recognized in each patient at different times postdiagnosis.
Relationship between clinical parameters and the breadth and magnitude of responses to overlapping DV2 peptides in IFN-γ ELISPOT assays.
Clinical disease grade had no apparent relationship to the breadth or magnitude of the T-cell response. With respect to the breadth of responses, samples from patients with DHF grade I (n = 11) recognized 2.5 peptides on average (range, 0 to 5) in ELISPOT assays, which was virtually identical to samples from patients with grade II (n = 16; 2.3 peptides; range, 0 to 7) or grade III (n = 20; 2.4 peptides; range, 0 to 5). Only one patient with DHF IV was recruited to the study, although interestingly, PBMC from this individual recognized 10 different peptides. Like the breadth of the response, there were no significant differences between the mean sum of ELISPOT responses recorded for patients in each of the disease grades (grade I, 138 ± 148; grade II, 143 ± 71; grade III, 103 ± 74; grade IV, 110).
The magnitude of responses to individual peptides in each patient was also compared to the minimum platelet count and maximum relative increase in hemoconcentration recorded during hospitalization. The extent of acute hemoconcentration was determined by comparing the maximum hematocrit recorded during hospitalization to the hematocrit recorded at follow-up in each patient; these data were available for 24 patients, all of whom had detectable ELISPOT responses during the acute phase of illness. Correlation analysis revealed a significant relationship (r = 0.41; P = 0.04) between the magnitude of IFN-γ ELISPOT responses measured on study day 5 and the extent of hemoconcentration in individual patients. Conversely, the nadir of thrombocytopenia did not significantly correlate with the sum of responses to peptides in individual patients (r = 0.13; P = 0.56).
The minimum determinant and HLA restriction of T cells responding to Env411-425.
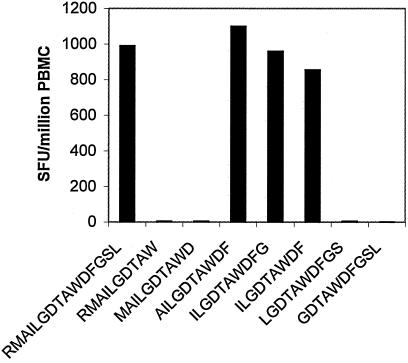
The peptide RMAILGDTAWDFGSL(Env411-425) was among the most frequently recognized by patient T cells in ELISPOT assays. Seven of 39 patients recognized Env411-425 with a mean response of 153 ± 76 SFU per 106 PBMC. HLA typing revealed that HLA-B*07 was the only HLA antigen shared by all seven responders. In one patient (BC503), depletion of CD4+ or CD8+ T cells from PBMC prior to the ELISPOT assay revealed that only the CD8+ T-cell-enriched fraction responded to Env414-422 peptide stimulation (data not shown). Truncated synthetic peptides and ELISPOT assays indicated ILGDTAWDF (Env414-422) was the smallest peptide recognized by CD8+ T cells from patient BC503 (Fig. 3). The HLA restriction of Env414-422 in patient BC503 was confirmed via a cytotoxic T lymphocyte killing assay. A short term T-cell line generated from Env414-422 peptide-stimulated PBMC from BC503 was lytic to a peptide-pulsed BCL that shared only B*07 in common with the effector cells (25% specific lysis at an effector:target ratio of 10:1 for Env414-422 peptide pulsed targets versus −2.5% for targets alone). Env411-425 did not elicit ELISPOT responses in B*07-positive, flavivirus-naïve individuals (United Kingdom residents).
FIG. 3.
Recognition by CD8+ T cells of truncated peptide variants of RMAILGDTAWDFGSL (Env411-425). Shown are the frequencies of IFN-γ SFU elicited by truncated Env411-425 peptides in IFN-γ ELISPOT assays against PBMC from BC503 after enrichment for CD8+ T cells.
Kinetics and cross-reactivity of responses to Env414-422.
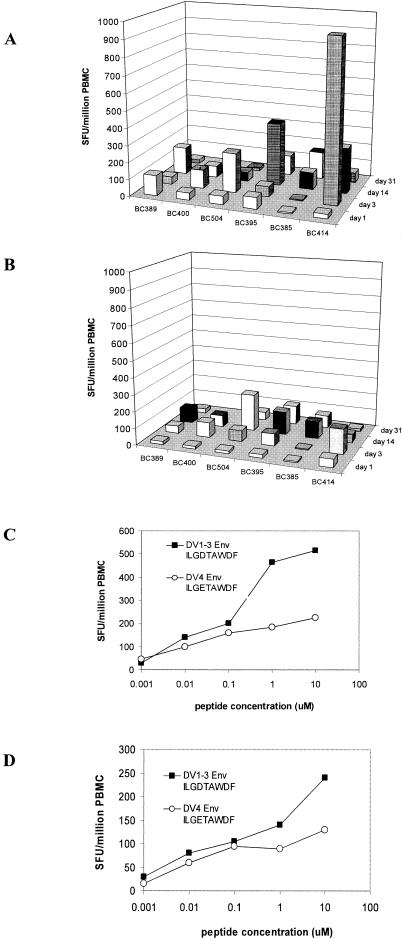
We sought to define the kinetics of T-cell responses to the B*07-restricted Env414-422 epitope in prospectively recruited dengue patients. First, the extent of sequence variation present at Env414-422 across all four dengue virus serotypes was examined. The sequence of Env414-422 in DV1 to DV3 was invariant (ILGDTAWDF), while DV4 Env414-422 differed by only 1 aa (ILGETAWDF). The dynamic of T-cell responses to these variant peptides were measured in six prospectively identified HLA-B*07-positive patients with serologically confirmed secondary dengue virus infections. All six patients responded to the HLA-B*07-restricted epitopes Env414-422 (Fig. 4A and B) in ELISPOT assays, with the peak response measured on study day 3 or 14. ELISPOT responses to each of the epitope variants were generally similar. In samples from patient BC414, however, we observed much stronger responses (∼6-fold) to the DV1 to DV3 Env414-422 (ILGDTAWDF) epitope variant than to DV4 Env414-422 (ILGETAWDF). Stronger responses to DV1 to DV3 Env414-422 were also observed in patient BC504 (3.5 fold at days 3 and 14) and patient BC395 (2-fold at day 14) but were less pronounced. Measurement of T-cell responses to these variant peptides at lower peptide concentrations, akin to those likely to be found in vivo, revealed much more homogenous responses, however. For example, at peptide concentrations of <1 μm, PBMC from patients BC414 and BC504 recognized Env414-422 peptide variants equally well; similar results were observed for PBMC from the other study subjects (Fig. 4C and D).
FIG. 4.
Kinetics and cross-reactivity of responses to Env414-422. The data depict the number of IFN-γ SFU detected in PBMC enriched for CD8+ T cells from six HLA-B*07-positive patients with secondary dengue virus infections. PBMC were incubated with peptides corresponding to DV1-3Env414-422 (A) and DV4Env414-422 (B). Patients BC385, BC389, BC400, BC395, and BC504 were DHF grade II. Patient BC414 was DHF grade III. Patients BC389, BC395, and BC385 were infected with DV2. Patient BC504 was infected with DV4. Virus was not detected in patient BC400 or BC414. Peptide titration experiments suggested that T cells from patients BC414 (C) and BC504(D) preferentially recognized the DV1 to DV3 Env414-422 variant at high peptide concentrations but were cross-reactive for the variant peptides DV1 to DV3 Env414-422 and DV4 Env414-422 at peptide concentrations of <1 μM.
The minimum determinant and HLA restriction of T cells responding to NS3550-564.
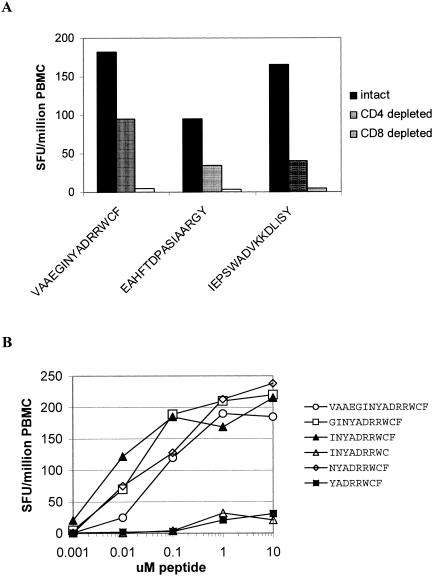
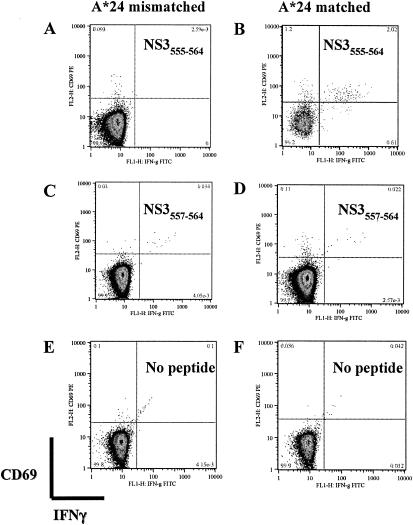
In IFN-γ ELISPOT assays, subject BC313 (HLA-A*24, HLA-A*26, HLA-B38, HLA-B15, HLA-Cw7, and HLA-Cw3) responded to three nonoverlapping NS3 peptides: VAAEGINYADRRWCF (NS3550-564), EAHFTDPASIAARGY (NS3285-299), and IEPSWADVKKDLISY (NS365-79). Depletion of CD4+ or CD8+ T cells from PBMC prior to ELISPOT assay indicated the responding cells were CD8+ T cells (Fig. 5A). The peptide spanning NS365-79 contained a previously defined HLA-B*15-restricted epitope, NS371-79 (52); since patient BC313 was also HLA-B*15-positive, these findings were not investigated further. Two lines of evidence suggested the NS3550-564 peptide contained an HLA-A*24 epitope. First, a peptide containing this sequence had previously been shown to evoke T-cell responses in HLA-A*24-positive Vietnamese dengue patients (35). Second, the sequence NYADRRWCF (NS3556-564) conformed to the consensus A*24 sequence, which predicts peptides with a tyrosine or phenylalanine at the second position and an uncharged amino acid at the C-terminal anchor position. The minimum determinant in NS3550-564 that elicited a response from patient BC313, who was HLA-A*24 positive, was identified by using truncated peptides spanning the predicted epitope in ELISPOT assays. INYADRRWCF (NS3555-564) was the minimum peptide that evoked the strongest response at the lowest peptide concentration in samples from BC313 (Fig. 5B). We used intracellular IFN-γ staining to confirm the HLA restriction of DV2NS3556-564. PBMC from BC313 was cocultured with brefeldin A and peptide-pulsed BCLs matched or mismatched to effectors at the A*24 locus. After 6 h, the frequency of IFN-γ-staining CD8+ T cells was determined for each peptide and BCL combination. The results confirmed that functional CD8+ T-cell responses to DV2 NS3555-564 were dependent on peptide presentation through the A*24 locus (Fig. 6). Presentation of DV2 NS3555-564 by targets matched to effectors at A*26, B38, B15, or Cw7 did not elicit functional responses (Fig. 6 and data not shown). NS3555-564 did not elicit ELISPOT responses in A*24-positive, flavivirus-naïve individuals (United Kingdom residents).
FIG. 5.
NS3555-564 is a CD8+ T-cell epitope. (A) In IFN-γ ELISPOT assays, PBMC (study day 3) from subject BC313 responded to the DV2 NS3 peptide VAAEGINYADRRWCF (NS3550-564) in a CD8-dependent fashion. (B) The truncated peptide INYADRRWCF (NS3550-564) was the minimum peptide that evoked the strongest ELISPOT response at the lowest peptide concentration.
FIG. 6.
NS3555-564 is recognized only in the context of HLA-A*24. PBMC (study day 5) from subject BC313 (A*24, A*26, B38, B15, Cw7, and Cw3) were cocultured with peptide-pulsed BCLs that were HLA matched (A*24 only) or mismatched to BC313 as described in Materials and Methods. After coculture, the frequency of CD3+ CD8+ T cells that expressed the early activation marker CD69+ and IFN-γ (upper right) was determined in combinations of PBMC and peptide-pulsed BCL targets. (A) Presentation of DV2 NS3555-564 by targets matched to effectors at A*26 and B38, B15, or Cw7 (data not shown) did not elicit functional responses. (B) Activated (CD69+), IFN-γ-producing CD8+ T cells were only observed when DV2NS3555-564 was presented to PBMC in an HLA-A*24-matched BCL. Functional CD8 T-cell responses were not observed when the truncated peptide DV2NS3557-564 (YADRRWCF) (C and D) or no peptide (E and F) was presented by HLA-A*24-matched or mismatched BCLs.
Kinetics and cross-reactivity of responses to NS3556-564.
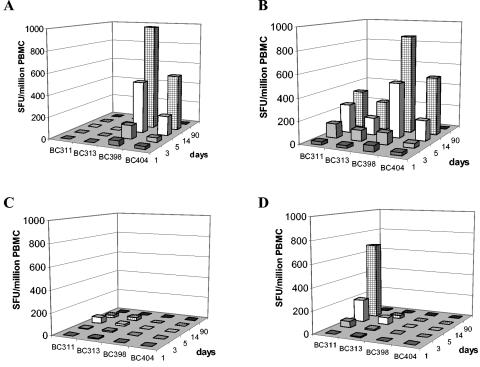
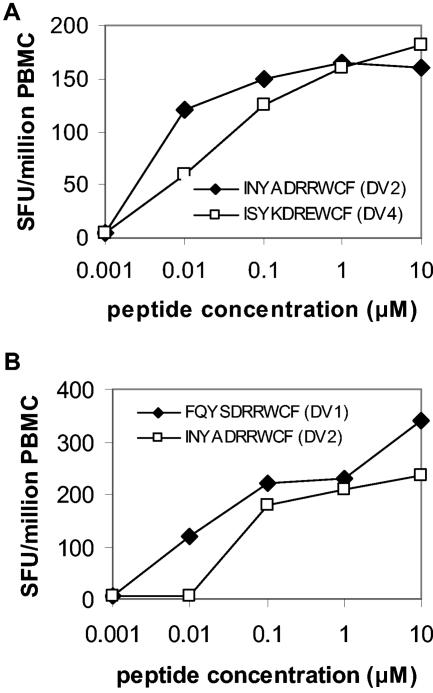
T-cell responses to peptides corresponding to all known natural variants of the A*24-restricted NS3556-564 epitope were measured in 16 prospectively recruited, HLA-A*24-positive dengue patients during early convalescence (i.e., at hospital discharge). T-cell responses to any one of the NS3556-564 variants were detected in 4 (25%) of 16 subjects, suggesting that this was not a dominant epitope. In all patients, the peak response to each of the epitope variants was recorded on study day 14 (Fig. 7). The spectrum of responses detected in each of the four patients varied according to the peptide antigen (Fig. 7). Thus, some patients made monotypic responses (patient BC313), while others had responses against two peptide variants (patient s BC311, BC398, and BC404). In both patients where information on the infecting viral serotype was available (BC311 [DV4] and BC398 [DV2]), there was an ELISPOT response to the NS3556-564 epitope matching the serotype of virus mediating the current infection, but also a response to a second natural NS3556-564 epitope sequence (Fig. 8). BC311 and BC398 also responded to the second NS3556-564 epitope sequence at low-peptide concentrations that better reflected in vivo conditions (Fig. 8).
FIG. 7.
Kinetics of T-cell responses to NS3555-564. Shown are the kinetics and magnitude of IFN-γ ELISPOT responses in samples from four patients with secondary dengue virus infection to each of the recognized peptide variants of NS3555-564. PBMC from each patient were stimulated with DV1NS3555-564 (FQYSDRRWCF) (A), DV2NS3555-564 (INYADRRWCF) (B), DV3NS3555-564 (IKYTDRKWCF) (C), and DV4NS3555-564 (ISYKDREWCF) (D). Subject BC311 was infected with DV4, and subject BC398 was infected with DV2.
FIG. 8.
T-cell responses to NS3555-564 variants during acute secondary dengue virus infection. (A) Peptide titration curves in IFN-γ ELISPOT assays for PBMC collected in the acute phase of infection (study day 5) from patient BC311 (infected with DV4) and stimulated with DV4NS3555-564 (ISYKDREWCF) or DV2NS3555-564 (INYADRRWCF). (B) Peptide titration curves in IFN-γ ELISPOT assays for PBMC collected in the acute phase of infection (study day 5) from patient BC398 (infected with DV2) and stimulated with DV2NS3555-564 (INYADRRWCF) and DV1NS3555-564 (FQYSDRRWCF).
DISCUSSION
The breadth and specificity of T-cell responses to dengue viral antigens in populations living in areas where dengue is endemic remain relatively poorly characterized. This serological and virological study of Vietnamese adults with secondary dengue virus infection measured the breadth and frequency of T-cell responses to 260 overlapping peptides from a DV2 virus. The large numbers of subjects and peptides included represents a strength of this study. Neither the breadth nor magnitudes of the recorded peptide-specific T-cell responses were significantly associated with clinical disease grade or the serotype of dengue virus mediating the current infection. A second key finding was the identification of 34 different peptide sequences that potentially contained many novel T-cell epitopes. These assays facilitated the identification of a novel HLA-B*07-restricted epitope in Env and an HLA-A*24-restricted epitope in NS3.
Most studies of dengue virus-specific T cells have occurred in the context of T-cell clones generated from live attenuated dengue virus vaccinees, or less frequently, from dengue patients (6, 9, 13, 24, 27, 37). This study represents the first attempt to define the relative antigenicity of peptides from multiple dengue viral antigens in a large number of patients from a hyperendemic country. NS3 was recognized by more than half of all Vietnamese adult patients and also contained the largest number of antigenic peptides. These results are consistent with previous studies that have emphasized the importance of NS3 as a T-cell target (37). Almost half the antigenic peptides in NS3 were clustered within an amino acid region (NS3200-324) that represents just 20% of the whole protein. NS3200-324 is highly conserved (78%) across all four serotypes of dengue virus and contains motifs and charged residues that are essential for helicase activity and viral replication (38, 39). Given the conserved nature of NS3 (68%), and particularly NS3200-324, it seems likely that many of the NS3-derived antigenic peptides identified in this study were targets of reactivated, dengue virus cross-reactive T cells that were primed by a previous dengue virus infection. That Vietnamese patients infected with either DV1 or DV4 mounted ELISPOT responses to DV2 peptides that were equivalent in breadth and strength to patients infected with a DV2 virus supports the contention that serotype cross-reactive, rather than serotype-specific, T cells dominate the acute response during secondary infections. In patients infected with DV2, there was no evidence that the T-cell response to DV2 peptides increased in breadth with time since secondary infection. This suggests the early, cross-reactive T-cell response remains dominant in late convalescence when the memory T-cell pool has been established. This is supported by a previous study that demonstrated the dominance of cross-reactive T cells among the dengue virus-specific memory T-cell population in Thai children 12 months after secondary dengue virus infection (36).
An important question is whether T-cell responses to dengue virus antigens are associated with clinical disease. In this study, no significant differences were observed in either the breadth or magnitude of the total T-cell response between patients with different clinical disease grades. We did, however, observe that the sum of peptide-specific responses measured in ELISPOT assays correlated, albeit weakly, with the extent of hemoconcentration recorded in each patient. Other studies have found significant associations between the magnitude of epitope-specific T-cell responses and clinical disease grade. For example, relatively higher frequencies of activated, cross-reactive CD8+ T cells (41, 51) and a range of direct and indirect markers of CD4+ and CD8+ T-cell activation have been associated with increasing disease severity during secondary infection (8, 14-16, 22, 23, 26). An important difference between this study and that of others is that we performed a comprehensive analysis of the functional T-cell response in adult patients to 260 peptides spanning five viral antigens as opposed to an analysis of responses in children to a single CD8+ T-cell epitope (41, 51). Other confounding influences, such as ethnicity, HLA background, and viral factors, may also explain differences between this study and those that have been reported previously.
Two novel CD8+ T-cell epitopes were identified in this study. The rationale for studying the NS3550-564 peptide was threefold. First, previous studies had suggested this peptide might contain an HLA-A*24 epitope (35). Second, A*24 is one of the commonest alleles present in the Vietnamese population (∼33%) and hence an A*24 epitope might be widely recognized. Third, A*24 has been associated with susceptibility to DHF in Vietnamese children (35), suggesting immune responses restricted through this allele might be poorly protective and/or pathogenic. Responses to the A*24-restricted NS3556-564 epitope were detected in a minority of HLA-A*24-positive patients, suggesting this is not a dominant epitope. Consistent with other studies on unrelated epitopes (41, 51), responses to the CD8+ T-cell epitopes defined in this study were generally strongest after the patient had been discharged from hospital, suggesting that antigen may continue to persist, possibly in the form of immune complexes, well after the resolution of viremia. The establishment of a library of defined T-cell epitopes from dengue viruses will help to define the role of T cells in immunity and disease pathogenesis. They should also prove useful in measuring the immunogenicity of live dengue virus vaccine candidates (7, 18, 42).
Two Vietnamese adults whose samples recognized the A*24-restricted NS3556-564 epitope also had samples that recognized an NS3556-564 epitope different from that present in the infecting dengue virus serotype. Mongkolsapaya et al. have recently suggested that infection history and what they termed “original antigenic sin” by dengue virus-specific memory T cells may explain the hierarchy of responses to an A*11-restricted dengue virus T-cell epitope following secondary infection (41). Such skewed T-cell responses could hypothetically limit or delay viral elimination, leading to higher viral loads, increased immune activation, and immunopathology. In the context of A*24-restricted NS3556-564-specific T cells, further studies of T-cell affinity and functional phenotype will be required to determine whether this represents original antigenic sin in the T-cell response.
Future studies of CD8+ T-cell responses to dengue virus antigens are likely to employ HLA class I tetramers that can be used to evaluate the functional phenotype of serotype-specific and cross-reactive CD8+ T cells. Integrating these cellular data with viremia parameters and carefully collected clinical phenotypes should reveal novel insights into disease pathogenesis and immunity.
Acknowledgments
The generous support of the Wellcome Trust is acknowledged. S.R.-J. and T. D. are funded by Medical Research Council, UK.
Special thanks to the patients, nurses, and clinicians who made this study possible. We acknowledge Gavin Screaton and Juthathip Mongkolsapaya for sharing the overlapping peptides and Kati DiGleria for synthesis of peptides.
REFERENCES
- 1.Bethell, D. B., J. Gamble, P. L. Pham, M. D. Nguyen, T. H. Tran, T. H. Ha, T. N. Tran, T. H. Dong, I. B. Gartside, N. J. White, and N. P. Day. 2001. Noninvasive measurement of microvascular leakage in patients with dengue hemorrhagic fever. Clin. Infect. Dis. 32:243-253. [DOI] [PubMed] [Google Scholar]
- 2.Blok, J., S. M. McWilliam, H. C. Butler, A. J. Gibbs, G. Weiller, B. L. Herring, A. C. Hemsley, J. G. Aaskov, S. Yoksan, and N. Bhamarapravati. 1992. Comparison of a dengue-2 virus and its candidate vaccine derivative: sequence relationships with the flaviviruses and other viruses. Virology 187:573-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinton, M. A., I. Kurane, A. Mathew, L. Zeng, P. Y. Shi, A. Rothman, and F. A. Ennis. 1998. Immune mediated and inherited defences against flaviviruses. Clin. Diagn. Virol. 10:129-139. [DOI] [PubMed] [Google Scholar]
- 4.Bunce, M., C. M. O'Neill, M. C. Barnardo, P. Krausa, M. J. Browning, P. J. Morris, and K. I. Welsh. 1995. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 and DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens 46:355-367. [DOI] [PubMed] [Google Scholar]
- 5.Burke, D. S., A. Nisalak, D. E. Johnson, and R. M. Scott. 1988. A prospective study of dengue infections in Bangkok. Am. J. Trop. Med. Hyg. 38:172-180. [DOI] [PubMed] [Google Scholar]
- 6.Dharakul, T., I. Kurane, N. Bhamarapravati, S. Yoksan, D. W. Vaughn, C. H. Hoke, and F. A. Ennis. 1994. Dengue virus-specific memory T cell responses in human volunteers receiving a live attenuated dengue virus type 2 candidate vaccine. J. Infect. Dis. 170:27-33. [DOI] [PubMed] [Google Scholar]
- 7.Edelman, R., S. S. Wasserman, S. A. Bodison, R. J. Putnak, K. H. Eckels, D. Tang, N. Kanesa-Thasan, D. W. Vaughn, B. L. Innis, and W. Sun. 2003. Phase I trial of 16 formulations of a tetravalent live-attenuated dengue vaccine. Am. J. Trop. Med. Hyg. 69:48-60. [DOI] [PubMed] [Google Scholar]
- 8.Gagnon, S. J., M. Mori, I. Kurane, S. Green, D. W. Vaughn, S. Kalayanarooj, S. Suntayakorn, F. A. Ennis, and A. L. Rothman. 2002. Cytokine gene expression and protein production in peripheral blood mononuclear cells of children with acute dengue virus infections. J. Med. Virol. 67:41-46. [DOI] [PubMed] [Google Scholar]
- 9.Gagnon, S. J., W. Zeng, I. Kurane, and F. A. Ennis. 1996. Identification of two epitopes on the dengue 4 virus capsid protein recognized by a serotype-specific and a panel of serotype-cross-reactive human CD4+ cytotoxic T-lymphocyte clones. J. Virol. 70:141-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamble, J., D. Bethell, N. P. Day, P. P. Loc, N. H. Phu, I. B. Gartside, J. F. Farrar, and N. J. White. 2000. Age-related changes in microvascular permeability: a significant factor in the susceptibility of children to shock? Clin. Sci. (London) 98:211-216. [PubMed] [Google Scholar]
- 11.Goulder, P. J., M. M. Addo, M. A. Altfeld, E. S. Rosenberg, Y. Tang, U. Govender, N. Mngqundaniso, K. Annamalai, T. U. Vogel, M. Hammond, M. Bunce, H. M. Coovadia, and B. D. Walker. 2001. Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by ELISPOT and intracellular cytokine staining assays. J. Virol. 75:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham, R. R., M. Juffrie, R. Tan, C. G. Hayes, I. Laksono, C. Ma'roef, Erlin, Sutaryo, K. R. Porter, and S. B. Halstead. 1999. A prospective seroepidemiologic study on dengue in children four to nine years of age in Yogyakarta, Indonesia I. Studies in 1995-1996. Am. J. Trop. Med. Hyg. 61:412-419. [DOI] [PubMed] [Google Scholar]
- 13.Green, S., I. Kurane, R. Edelman, C. O. Tacket, K. H. Eckels, D. W. Vaughn, C. H. Hoke, Jr., and F. A. Ennis. 1993. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J. Virol. 67:5962-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Green, S., S. Pichyangkul, D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, A. Nisalak, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J. Infect. Dis. 180:1429-1435. [DOI] [PubMed] [Google Scholar]
- 15.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, R. Lew, B. L. Innis, I. Kurane, A. L. Rothman, and F. A. Ennis. 1999. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J. Infect. Dis. 179:755-762. [DOI] [PubMed] [Google Scholar]
- 16.Green, S., D. W. Vaughn, S. Kalayanarooj, S. Nimmannitya, S. Suntayakorn, A. Nisalak, A. L. Rothman, and F. A. Ennis. 1999. Elevated plasma interleukin-10 levels in acute dengue correlate with disease severity. J. Med. Virol. 59:329-334. [PubMed] [Google Scholar]
- 17.Guzman, M. G., and G. Kouri. 2002. Dengue: an update. Lancet Infect. Dis. 2:33-42. [DOI] [PubMed] [Google Scholar]
- 18.Gwinn, W., W. Sun, B. L. Innis, J. Caudill, and A. D. King. 2003. Serotype-specific T(H)1 responses in recipients of two doses of candidate live-attenuated dengue virus vaccines. Am. J. Trop. Med. Hyg. 69:39-47. [DOI] [PubMed] [Google Scholar]
- 19.Halstead, S. B. 1979. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J. Infect. Dis. 140:527-533. [DOI] [PubMed] [Google Scholar]
- 20.Halstead, S. B., and E. J. O'Rourke. 1977. Antibody-enhanced dengue virus infection in primate leukocytes. Nature 265:739-741. [DOI] [PubMed] [Google Scholar]
- 21.Halstead, S. B., and E. J. O'Rourke. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 146:201-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juffrie, M., G. M. Meer, C. E. Hack, K. Haasnoot, Sutaryo, A. J. Veerman, and L. G. Thijs. 2001. Inflammatory mediators in dengue virus infection in children: interleukin-6 and its relation to C-reactive protein and secretory phospholipase A2. Am. J. Trop. Med. Hyg. 65:70-75. [DOI] [PubMed] [Google Scholar]
- 23.Kittigul, L., W. Temprom, D. Sujirarat, and C. Kittigul. 2000. Determination of tumor necrosis factor-alpha levels in dengue virus infected patients by sensitive biotin-streptavidin enzyme-linked immunosorbent assay. J. Virol. Methods 90:51-57. [DOI] [PubMed] [Google Scholar]
- 24.Kurane, I., M. A. Brinton, A. L. Samson, and F. A. Ennis. 1991. Dengue virus-specific, human CD4+ CD8− cytotoxic T-cell clones: multiple patterns of virus cross-reactivity recognized by NS3-specific T-cell clones. J. Virol. 65:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurane, I., B. L. Innis, C. H. Hoke, Jr., K. H. Eckels, A. Meager, J. Janus, and F. A. Ennis. 1995. T cell activation in vivo by dengue virus infection. J. Clin. Lab. Immunol. 46:35-40. [PubMed] [Google Scholar]
- 26.Kurane, I., B. L. Innis, S. Nimmannitya, A. Nisalak, A. Meager, J. Janus, and F. A. Ennis. 1991. Activation of T lymphocytes in dengue virus infections. High levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-gamma in sera of children with dengue. J. Clin. Investig. 88:1473-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurane, I., Y. Okamoto, L. C. Dai, L. L. Zeng, M. A. Brinton, and F. A. Ennis. 1995. Flavivirus-cross-reactive, HLA-DR15-restricted epitope on NS3 recognized by human CD4+ CD8− cytotoxic T lymphocyte clones. J. Gen. Virol. 76:2243-2249. [DOI] [PubMed] [Google Scholar]
- 28.Kurane, I., A. L. Rothman, P. G. Livingston, S. Green, S. J. Gagnon, J. Janus, B. L. Innis, S. Nimmannitya, A. Nisalak, and F. A. Ennis. 1994. Immunopathologic mechanisms of dengue hemorrhagic fever and dengue shock syndrome. Arch. Virol. Suppl. 9:59-64. [DOI] [PubMed] [Google Scholar]
- 29.Lanciotti, R. S., C. H. Calisher, D. J. Gubler, G. J. Chang, and A. V. Vorndam. 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J. Clin. Microbiol. 30:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. Villalobos, C. de, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Libraty, D. H., T. P. Endy, H. S. Houng, S. Green, S. Kalayanarooj, S. Suntayakorn, W. Chansiriwongs, D. W. Vaughn, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J. Infect. Dis. 185:1213-1221. [DOI] [PubMed] [Google Scholar]
- 32.Libraty, D. H., P. R. Young, D. Pickering, T. P. Endy, S. Kalayanarooj, S. Green, D. W. Vaughn, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J. Infect. Dis. 186:1165-1168. [DOI] [PubMed] [Google Scholar]
- 33.Livingston, P. G., I. Kurane, L. C. Dai, Y. Okamoto, C. J. Lai, R. Men, S. Karaki, M. Takiguchi, and F. A. Ennis. 1995. Dengue virus-specific, HLA-B35-restricted, human CD8+ cytotoxic T lymphocyte (CTL) clones. Recognition of NS3 amino acids 500 to 508 by CTL clones of two different serotype specificities. J. Immunol. 154:1287-1295. [PubMed] [Google Scholar]
- 34.Loke, H., D. Bethell, C. X. T. Phuong, N. Day, N. White, J. Farrar, and A. Hill. 2002. Susceptibility to dengue hemorrhagic fever in Vietnam: evidence of an association with variation in the vitamin D receptor and Fcγ receptor IIa genes. Am. J. Trop. Med. Hyg. 67:102-106. [DOI] [PubMed] [Google Scholar]
- 35.Loke, H., D. B. Bethell, C. X. Phuong, M. Dung, J. Schneider, N. J. White, N. P. Day, J. Farrar, and A. V. Hill. 2001. Strong HLA class I-restricted T cell responses in dengue hemorrhagic fever: a double-edged sword? J. Infect. Dis. 184:1369-1373. [DOI] [PubMed] [Google Scholar]
- 36.Mathew, A., I. Kurane, S. Green, H. A. Stephens, D. W. Vaughn, S. Kalayanarooj, S. Suntayakorn, D. Chandanayingyong, F. A. Ennis, and A. L. Rothman. 1998. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J. Virol. 72:3999-4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathew, A., I. Kurane, A. L. Rothman, L. L. Zeng, M. A. Brinton, and F. A. Ennis. 1996. Dominant recognition by human CD8+ cytotoxic T lymphocytes of dengue virus nonstructural proteins NS3 and NS1.2a. J. Clin. Investig. 98:1684-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matusan, A. E., P. G. Kelley, M. J. Pryor, J. C. Whisstock, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the dengue virus type 2 NS3 proteinase and the production of growth-restricted virus. J. Gen. Virol. 82:1647-1656. [DOI] [PubMed] [Google Scholar]
- 39.Matusan, A. E., M. J. Pryor, A. D. Davidson, and P. J. Wright. 2001. Mutagenesis of the Dengue virus type 2 NS3 protein within and outside helicase motifs: effects on enzyme activity and virus replication. J. Virol. 75:9633-9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mongkolsapaya, J., W. Dejnirattisai, X. N. Xu, S. Vasanawathana, N. Tangthawornchaikul, A. Chairunsri, S. Sawasdivorn, T. Duangchinda, T. Dong, S. Rowland-Jones, P. T. Yenchitsomanus, A. McMichael, P. Malasit, and G. Screaton. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921-927. [DOI] [PubMed] [Google Scholar]
- 42.Sabchareon, A., J. Lang, P. Chanthavanich, S. Yoksan, R. Forrat, P. Attanath, C. Sirivichayakul, K. Pengsaa, C. Pojjaroen-Anant, L. Chambonneau, J. F. Saluzzo, and N. Bhamarapravati. 2004. Safety and immunogenicity of a three dose regimen of two tetravalent live-attenuated dengue vaccines in five- to twelve-year-old Thai children. Pediatr. Infect. Dis. J. 23:99-109. [DOI] [PubMed] [Google Scholar]
- 43.Sangkawibha, N., S. Rojanasuphot, S. Ahandrik, S. Viriyapongse, S. Jatanasen, V. Salitul, B. Phanthumachinda, and S. B. Halstead. 1984. Risk factors in dengue shock syndrome: a prospective epidemiologic study in Rayong, Thailand. I. The 1980 outbreak. Am. J. Epidemiol. 120:653-669. [DOI] [PubMed] [Google Scholar]
- 44.Stephens, H. A., R. Klaythong, M. Sirikong, D. W. Vaughn, S. Green, S. Kalayanarooj, T. P. Endy, D. H. Libraty, A. Nisalak, B. L. Innis, A. L. Rothman, F. A. Ennis, and D. Chandanayingyong. 2002. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens 60:309-318. [DOI] [PubMed] [Google Scholar]
- 45.Thein, S., M. M. Aung, T. N. Shwe, M. Aye, A. Zaw, K. Aye, K. M. Aye, and J. Aaskov. 1997. Risk factors in dengue shock syndrome. Am. J. Trop. Med. Hyg. 56:566-572. [DOI] [PubMed] [Google Scholar]
- 46.Thisyakorn, U., and S. Nimmannitya. 1993. Nutritional status of children with dengue hemorrhagic fever. Clin. Infect. Dis. 16:295-297. [DOI] [PubMed] [Google Scholar]
- 47.Vaughn, D. W., S. Green, S. Kalayanarooj, B. L. Innis, S. Nimmannitya, S. Suntayakorn, T. P. Endy, B. Raengsakulrach, A. L. Rothman, F. A. Ennis, and A. Nisalak. 2000. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J. Infect. Dis. 181:2-9. [DOI] [PubMed] [Google Scholar]
- 48.Vaughn, D. W., A. Nisalak, T. Solomon, S. Kalayanarooj, M. D. Nguyen, R. Kneen, A. Cuzzubbo, and P. L. Devine. 1999. Rapid serologic diagnosis of dengue virus infection using a commercial capture ELISA that distinguishes primary and secondary infections. Am. J. Trop. Med. Hyg. 60:693-698. [DOI] [PubMed] [Google Scholar]
- 49.World Health Organization,. 1997. Dengue haemorrhagic fever: diagnosis, treatment, prevention and control. World Health Organization, Geneva, Switzerland.
- 50.Zeng, L., I. Kurane, Y. Okamoto, F. A. Ennis, and M. A. Brinton. 1996. Identification of amino acids involved in recognition by dengue virus NS3-specific, HLA-DR15-restricted cytotoxic CD4+ T-cell clones. J. Virol. 70:3108-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zivna, I., S. Green, D. W. Vaughn, S. Kalayanarooj, H. A. Stephens, D. Chandanayingyong, A. Nisalak, F. A. Ennis, and A. L. Rothman. 2002. T cell responses to an HLA-B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J. Immunol. 168:5959-5965. [DOI] [PubMed] [Google Scholar]
- 52.Zivny, J., M. DeFronzo, W. Jarry, J. Jameson, J. Cruz, F. A. Ennis, and A. L. Rothman. 1999. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J. Immunol. 163:2754-2760. [PubMed] [Google Scholar]