Abstract
Valuable information on translation initiation is available from biochemical data and recently solved structures. We present a detailed description of current knowledge about the structure, function, and interactions of the individual components involved in bacterial translation initiation. The first section describes the ribosomal features relevant to the initiation process. Subsequent sections describe the structure, function, and interactions of the mRNA, the initiator tRNA, and the initiation factors IF1, IF2, and IF3. Finally, we provide an overview of mechanisms of regulation of the translation initiation event. Translation occurs on ribonucleoprotein complexes called ribosomes. The ribosome is composed of a large subunit and a small subunit that hold the activities of peptidyltransfer and decode the triplet code of the mRNA, respectively. Translation initiation is promoted by IF1, IF2, and IF3, which mediate base pairing of the initiator tRNA anticodon to the mRNA initiation codon located in the ribosomal P-site. The mechanism of translation initiation differs for canonical and leaderless mRNAs, since the latter is dependent on the relative level of the initiation factors. Regulation of translation occurs primarily in the initiation phase. Secondary structures at the mRNA ribosomal binding site (RBS) inhibit translation initiation. The accessibility of the RBS is regulated by temperature and binding of small metabolites, proteins, or antisense RNAs. The future challenge is to obtain atomic-resolution structures of complete initiation complexes in order to understand the mechanism of translation initiation in molecular detail.
INTRODUCTION
Protein biosynthesis occurs on large macromolecular ribonucleoprotein complexes named ribosomes in a process termed translation. The ribosomes are enzymatic complexes that catalyze peptide bond formation and synthesize polypeptides based on the genetic code of the mRNA. Translation is conceptually divided into four phases: initiation, elongation, termination, and ribosome recycling.
The ribosome is composed of a large and a small subunit, which are assembled on the translation initiation region (TIR) of the mRNA during the initiation phase of translation. In the following elongation phase, the mRNA is decoded as it slides through the ribosome and a polypeptide chain is synthesized. Elongation continues until the ribosome encounters a stop codon on the mRNA and the process enters the termination phase of protein synthesis. Newly synthesized protein is released from the ribosome. In the final ribosome recycling phase, the ribosomal subunits dissociate and the mRNA is released. Each phase is regulated by a number of different factors. Reviews of the phases are available (52, 208).
Although the main events of the translation process are universally conserved, major differences in the detailed mechanism of each phase exist. Bacterial translation involves relatively few factors, in contrast to the more complex process in eukaryotes (164). Here we focus on translation initiation in bacteria. Although parallels are drawn to the archaeal and eukaryotic systems where relevant, everything described throughout the rest of this review concerns the bacterial system unless otherwise stated. Archaeal and eukaryotic processes of translation initiation are reviewed elsewhere (7, 44, 177).
BACTERIAL TRANSLATION INITIATION
Ribosomes initiate translation on mRNAs already during transcription. Hence, transcription and translation are tightly coupled cellular processes. Translation initiation is the rate-limiting and most highly regulated phase of the four phases in protein biosynthesis.
The rate at which ribosomes assemble on the mRNA is on the order of seconds, although it is specific for each mRNA. The ribosomes subsequently translate the mRNA at a rate of approximately 12 amino acids per second (89). The ribosome, the aminoacylated and formylated initiator tRNA (fMet-tRNAfMet), mRNA, and the three protein factors, initiation factor IF1, initiation factor IF2, and initiation factor IF3, are involved in the translation initiation phase (Fig. 1).
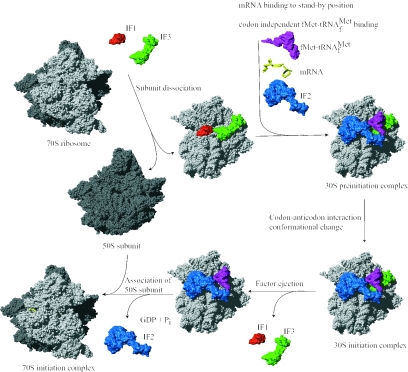
FIG. 1.
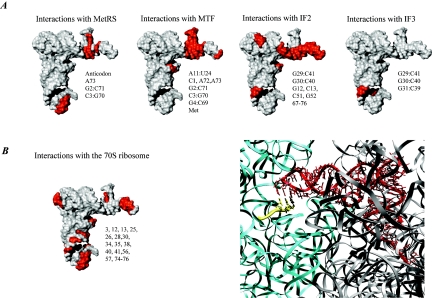
Translation initiation pathway in bacteria. The 30S and 50S ribosomal subunits are shown in light and dark grey, respectively. Translation initiation factors IF1, IF2, and IF3, the mRNA, and the fMet-tRNAfMet are shown in red, blue, green, yellow, and magenta, respectively. The components are placed on the ribosome according to current experimental knowledge. Details of the pathway are given in the text. Structures are derived from PDB entries as follows: 30S ribosomal subunit, 1HR0; 50S ribosomal subunit, 1FFK; IF1, 1HR0; IF2, 1G7T; IF3N, 1TIF; IF3C, 1TIG; mRNA, 1JGQ; fMet-tRNAfMet, 1JGQ. Structural representations in this as well as other figures in this review were made using the program MolMol (93) and Pov-Ray unless otherwise stated.
The bacterial 70S ribosome is composed of a large 50S and a small 30S subunit. It has three tRNA binding sites designated the aminoacyl (A), peptidyl (P), and exit (E) sites. Binding of IF3 to the 30S ribosomal subunit promotes dissociation of the ribosome into subunits and thus couples ribosome recycling and translation initiation (169). Initiation factor IF1 binds specifically to the base of the A-site of the 30S ribosomal subunit and is thought to direct the initiator tRNA to the ribosomal P-site by blocking the A-site (26, 41). IF1 stimulates the activities of IF3 and hence also the dissociation of the ribosomal subunits (63).
Following subunit dissociation, IF2, mRNA, and fMet-tRNAfMet associate with the 30S ribosomal subunit in an unknown and possibly random order. The Shine-Dalgamo (SD) sequence of canonical mRNAs interacts with the anti-SD sequence of the 16S rRNA (258), and the initiation codon is adjusted in the P-site of the ribosome. The initiation factors (especially IF3) seem to be responsible for this adjustment (101). The initiator tRNA is positioned in the P-site of the 30S ribosomal subunit in three steps that are designated codon-independent binding, codon-dependent binding, and fMet-tRNAfMet adjustment (reference 231 and references cited therein). All three steps are probably promoted by IF2, which interacts with fMet-tRNAfMet on the ribosome. Furthermore, IF3 stabilizes the binding of fMet-tRNAfMet to the ribosomal P-site and confers proofreading capability by destabilization of a mismatched codon-anticodon interaction (60).
The 30S preinitiation complex consists of the 30S ribosomal subunit, the three initiation factors, and mRNA in a standby position where fMet-tRNAfMet is bound in a codon-independent manner. This relatively unstable complex undergoes a rate-limiting conformational change that promotes the codon-anticodon interaction and forms the more stable 30S initiation complex (60, 174). Initiation factors IF1 and IF3 are ejected, while IF2 stimulates association of the 50S ribosomal subunit to the complex. Initiator fMet-tRNAfMet is adjusted to the correct position in the P-site, and IF2 is released from the complex. During this process, GTP bound to IF2 is hydrolyzed to GDP and Pi. The newly formed 70S initiation complex holding fMet-tRNAfMet as a substrate for the peptidyltransferase center of the 50S ribosomal subunit is ready to enter the elongation phase of translation.
Reviews of different aspects of bacterial translation initiation can be found in references 12, 61, 63, 208, and 210.
COMPONENTS INVOLVED IN TRANSLATION INITIATION
The translation initiation event is a complex and highly regulated process involving both RNA and protein components. Here we provide a detailed functional and structural description of the individual components.
Ribosome
The ribosome, which is composed of two subunits, is the macromolecular catalyst of protein synthesis. Bacterial ribosomes have a relative sedimentation rate of 70S and a mass of 2.4 MDa. The large subunit has a relative sedimentation rate of 50S and a mass of 1.5 MDa, whereas the small subunit has a relative sedimentation rate of 30S and a mass of 0.8 MDa. Approximately two-thirds of the ribosome consists of RNA and one-third consists of proteins (219). The available structures of ribosomes and ribosomal complexes are from different sources. Escherichia coli residue numbering is used throughout this review.
The first visualizations of ribosomal structures were done by electron microscopy and identified a particle subdivided into two subunits of unequal sizes (82). The first determination of shapes came in the early 1970s (99). Today the resolution of structures of ribosomal particles made by cryoelectron microscopy has increased to 7 Å for the best reconstitutions (49, 247). Atomic resolution structures of ribosomes can, however, be obtained only by X-ray crystallography. High-resolution structures of the intact 70S ribosome (239, 256), the 50S ribosomal subunit (4, 72), and the 30S ribosomal subunit (195, 249) exist. Moreover, the structures of several ribosomal complexes, including different factors, RNAs, and antibiotics, have recently been solved. Only those relevant for translation initiation are discussed here. Reviews describing other complexes may be found elsewhere (5, 180, 219, 253, 259).
Stabilization of the ribosomal structure.
Two-thirds of the ribosome is composed of RNA, the ribosome is thus a large polyanion. Three main types of interactions stabilize the tertiary structure of the rRNA: (i) Mg2+ bridges, (ii) RNA-RNA interactions, and (iii) RNA-protein interactions. The magnesium ions form neutralizing bridges between two or more phosphate groups from secondary-structure elements remote in sequence. RNA-RNA interactions of different types exist: (i) base pairing between nucleotides associated with secondary-structure elements remote in sequence, and (ii) A-minor motifs. The A-minor motif is an interaction between an adenine that inserts its minor groove face into the minor groove of a base pair in a helix. This is most often a GC pair. The adenine forms hydrogen bonds with one or both of the backbone 2′ hydroxyl groups of the RNA duplex (146). Different types of helix-helix packing interactions occur, involving the insertion of a ridge of phosphates into the minor groove of another helix or using an unpaired purine base to mediate the perpendicular packing of one helix against the minor groove of another (249).
RNA-protein interactions occur mainly via the sugar-phosphate backbone of the RNA. Thus, the ribosomal proteins recognize the unique shape of the rRNA rather than the bases, and the interactions are therefore sequence unspecific (146). Many of the ribosomal proteins have nonglobular extensions that are highly conserved in sequence. These tails penetrate into the ribosome and fill the gaps between RNA helices. In isolation, these protein tails, which contain approximately 26% arginine and lysine residues, look like random coils that probably only assume the conformation they have on the ribosome when bound (4, 219).
Prior to peptide bond formation, an aminoacyl-tRNA is bound in the ribosomal A-site, a peptidyl-tRNA is bound in the P-site, and a deacylated tRNA, which is ready for ejection from the ribosome, is bound to the E-site. Translation moves the tRNA from the A-site through the P- and E-sites before they exit the ribosome again, with the exception of the initiator tRNA, which binds directly to the P-site. The small ribosomal subunit contains the decoding center, where the triplet codons of the mRNA are base-paired with the anticodons of the cognate tRNA, and hence determines the sequence of amino acids to be incorporated in the synthesized protein. The large subunit contains the peptidyltransferase center and is thus the catalytic unit.
Small ribosomal subunit
The small ribosomal subunit is composed of 21 proteins and an RNA of approximately 1,500 nucleotides sedimenting at 16S. Two individual research groups determined the structure of the Thermus thermophilus subunit at 3- and 3.3-Å resolution (195, 249). The shape of the subunit is determined largely by the RNA component, which forms four secondary-structure domains (Fig. 2) (249). Traditionally, the subunit has been divided into an upper third, called the head, connected by the neck to the body with a shoulder and platform. A protrusion in the lower part of the body is called the spur (or toe). The side of the 30S subunit facing the 50S subunit is called the front, whereas the solvent-exposed side is called the back. A complete description of the domains and the location of proteins and their interactions with RNA can be found in reference 18.
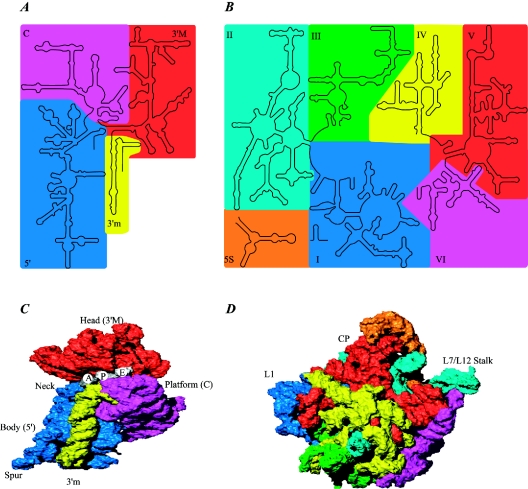
FIG. 2.
Structures of the ribosomal subunits. (A) Overview of the 16S rRNA secondary structure. The domains are shown in colors according to the secondary structure: blue, 5′ domain (bulk of body); magenta, central domain (platform); red, 3′ major domain (head); yellow, 3′ minor domain (helices 44 and 45 located at the subunit interface). (B) Overview of the 23S and 5S rRNA secondary structures. The RNA domains are shown in colors according to the secondary structure of the 23S rRNA: blue, domain I; cyan, domain II; green, domain III; yellow, domain IV; red, domain V; magenta, domain VI. The 5S rRNA is shown in orange. (C) Three-dimensional structure of the 30S ribosomal subunit from T. thermophilus at 3-Å resolution (PDB entry 1J5E). RNA secondary-structure domains are colored as in panel A. Note that the secondary-structure domains of the RNA correspond well to the tertiary domains. Proteins are omitted for clarity. The tRNA binding sites A, P, and E are indicated. (D) Three-dimensional structure of the 50S ribosomal subunit from H. marismortui at 2.4-Å resolution (PDB entry 1FFK). Colors are the same as in panel B. Note that the secondary-structure domains of the RNA do not correspond to the tertiary domains, unlike for the 30S subunit. Proteins are omitted for clarity. The L1 stalk, the central protuberance (CP), and the L7-L12 stalk are indicated.
The small subunit binds mRNA and the anticodon loop and stem of tRNAs. Translational fidelity is controlled on the subunit by monitoring the base pairing between the codon and anticodon in the process known as decoding (56). The decoding center located at the upper part of the body and lower part of the head of the subunit is constructed entirely of RNA and contains, among other elements, the upper part of helix 44 and the 3′ and 5′ ends of the 16S rRNA (195). An interaction that is important for translation initiation occurs at the 3′ end of the 16S rRNA (also known as the anti-SD [ASD] sequence) that base-pairs with the SD sequence of the mRNA.
E. coli has 41 different tRNA species with different anticodons. The ribosome must select the tRNA with an anticodon complementary to the codon of the mRNA. This is termed the cognate tRNA. The error rate of tRNA selection in the decoding process is 10−3 to 10−4. Pre-steady-state kinetics showed that in addition to having lower dissociation rates from the ribosome, cognate tRNAs have higher rates of elongation factor EF1A GTPase activation and accommodation (movement of the aminoacyl end of tRNA into the A-site of the 50S ribosomal subunit) than do near-cognate tRNAs. Based on this result, it was proposed that binding of cognate tRNA induces a conformational change of the ribosome (162). Crystal structures of the 30S subunit complexed with mRNA and cognate tRNA in the A-site reveal an induced fit mechanism. Bases A1492 and A1493 of the 16S rRNA flip out of helix 44 and interact with the correctly base-paired codon-anticodon helix in an A-minor motif type interaction. A1492 and A1493 interact with the minor groove of a correctly paired codon-anticodon but not with incorrectly paired codons and anticodons. Binding of cognate tRNA also causes a flip of G530 of the 16S rRNA from syn to anti conformation (156). Bases A1492 and A1493 interact with the first and second base pairs of the codon-anticodon helix, respectively. G530 interacts with the second position of the anticodon as well as the third position of the codon. The result is a strictly monitored codon-anticodon interaction in the first two positions, whereas the ribosome is able to tolerate noncanonical base pairs at the third position (156). During decoding, the flipping of the 30S subunit bases A1492, A1493, and G530 translates to other parts of the subunit and leaves it in a closed conformation in which the shoulder and head domains are rotated toward the subunit center, compared to the more open structure when the A-site is unoccupied. The transition to the more closed state is unfavorable for near-cognate tRNAs (158). However, X-ray crystal structures of the intact 70S E. coli ribosome reveal that the closing of the head domain of the 30S subunit is connected to formation of the intact 70S ribosome and not to decoding. These structures do, however, indicate a movement of the small-subunit body connected with decoding (239). Thus, ribosomes play an active role in tRNA selection by direct recognition of the codon-anticodon base pairing. The extent to which conformational changes occur needs further investigation. An extensive review of decoding is available (157).
Large ribosomal subunit.
The large ribosomal subunit is composed of 34 proteins and two RNAs sedimenting at 5S and 23S, containing about 120 and 2,900 nucleotides, respectively. Six secondary-structure domains are defined by the 23S rRNA (149), whereas the 5S rRNA is regarded as the seventh domain of the subunit (219). A direct relationship between secondary structure elements and morphological domains is not present in the large subunit, which presents a more compact structure than the small subunit (Fig. 2). The 50S subunit consists of a rounded base with three protuberances called the L1 protuberance, the central protuberance, and the L7/L12 stalk (Fig. 2) (248). A tunnel starts at the peptidyltransferase center (PTC), where the formation of peptide bonds occurs. The nascent polypeptide is thought to exit at the base of the cytoplasmic side of the subunit through the approximately 100-Å-long tunnel, which has an average diameter of 15 Å (145).
During the peptidyl transfer reaction, the α-amino group of A-site tRNA attacks the carbonyl group of the P-site peptidyl group, which is linked to the tRNA via an ester bond. The reaction proceeds via a tetrahedral intermediate to form a peptidyl bond. The reaction occurs in the PTC, where the amino acid of the A-site tRNA has been properly positioned relative to the nascent peptide chain bound to the P-site tRNA. Peptide bond formation is then catalyzed. The PTC was identified by soaking crystals of Haloarcula marismortui 50S subunits with a transition state analogue, the so-called Yarus inhibitor. Surprisingly, the subunit is completely devoid of protein within 18 Å of the PTC, and the ribosome is thus a ribozyme (145). It is beyond the scope of this review to go into detail about the mechanism of the peptidyl transferase reaction. Reviews can be found elsewhere (55, 219, 248).
After the peptidyltransferase reaction has occurred, a deacylated tRNA is left in the P-site and the A-site tRNA is covalently bound to a peptide chain extended by one residue. For elongation to proceed, the P-site tRNA has to move into the E-site ready for ejection from the ribosome and the A-site peptidyl-tRNA has to move to the P-site. The E-site is specific for deacylated tRNAs (196). The movement of tRNAs must be accompanied by a precise movement of the mRNA to preserve the reading frame. A review of the translocation process can be found in reference 150.
In the following sections, the binding of mRNA, tRNA, and the translation initiation factors to the ribosome is described, along with the properties of each individual component.
mRNA
mRNA interacts specifically with tRNA as well as the 30S ribosomal subunit during translation initiation. The mRNA covered by the ribosome in the translation initiation phase is called the ribosomal binding site (RBS) and extends over about 30 nucleotides (218). Bacterial mRNAs are normally polycistronic and possess multiple signals for initiation and termination of protein synthesis.
TIRs on mRNAs are not only characterized by the presence of a putative initiation codon. Additional elements are necessary to promote correct initiation and avoid initiation from, for instance, AUG codons encoding internal methionines of a protein. Upstream from the initiation codon is the 5′ untranslated region (5′ UTR). This region contains the SD sequence, which can undergo base-pairing to the 3′ of the 16S rRNA of the 30S ribosomal subunit (207). A direct consequence of the SD interaction is the adjustment of the initiation codon to the ribosomal P-site, where it interacts with fMet-tRNAfMet. E. coli mRNAs typically have the SD sequence GGAGG located 7 ± 2 nucleotides upstream from the initiation codon, which can be AUG, GUG or UUG (123). The exceptional AUU initiation codon has been observed in infC (encoding IF3) and pcnB [encoding E. coli poly(A) polymerase] (10). Initiation codons in E. coli occur at a frequency of 90, 8, and 1% for AUG, GUG, and UUG, respectively (198).
Ribosomal protein S1 interacts with a pyrimidine-rich region 5′ to the SD region on mRNAs. This pyrimidine-rich region acts as a ribosome recognition site (15). A direct interaction has been confirmed by cryoelectron microscopy (EM) studies of S1 on the 30S ribosomal subunit with a bound mRNA (200).
A region downstream from the initiation codon of several mRNAs was found to show complementarity to bases 1469 to 1483 within helix 44 of the 16S rRNA. This region was named the downstream box (DB), and there appeared to be a correlation between the degree of complementarity to the 16S rRNA and the translational efficiency of the mRNA. A mechanism similar to the SD-ASD interaction was proposed (212). The presence of a DB-anti-DB (ADB) interaction has been a matter of debate, and a recent review concludes that there is no biochemical or genetic evidence in support of the proposed role of the DB-ADB interaction in ribosomal recruitment of mRNA (133, 134). This is supported by the crystal structure of the T. thermophilus 30S ribosomal subunit, which reveals that the shoulder of the subunit is located between the putative DB of the mRNA and the proposed anti-DB of the 16S rRNA (249).
Bacterial mRNAs are either canonical or leaderless, although the latter is rare, with no more than ∼40 identified cases in bacteria (133). Canonical mRNAs contain the 5′ UTR elements described above, whereas leaderless mRNAs start at, or a few nucleotides 5′ upstream of, the initiation codon. A clear mechanism for binding of canonical mRNAs and the order in which mRNA and fMet-tRNAfMet enter the 30S ribosomal subunit has not been established (Fig. 3). Initiation factors do not affect the SD-ASD interaction or the association between the 30S ribosomal subunit and canonical mRNA (61). However, site-directed cross-linking experiments have shown that mRNA is partially relocated on the 30S ribosomal subunit from a “standby site” to a site closer to the P-site in a process influenced by IF1, IF2, and especially IF3 (101).
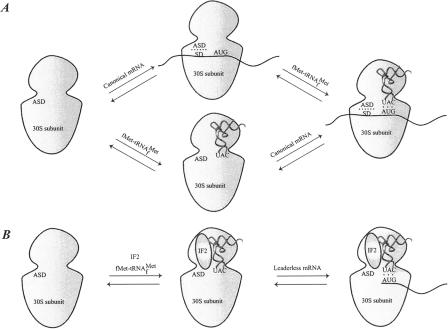
FIG. 3.
Binding of mRNA to the 30S ribosomal subunit. (A) Binding of a canonical mRNA to the 30S ribosomal subunit. Two alternative pathways are shown where either the mRNA or the fMet-tRNAfMet binds first, followed by the other component. The mRNA is bound via the SD-ASD interaction as well as the codon-anticodon interaction. (B) Binding of a leaderless mRNA to the 30S ribosomal subunit. The mRNA is bound to the ribosome mainly via the codon-anticodon interaction. IF2 stimulates the binding of leaderless mRNAs, presumably by recruitment of fMet-tRNAfMet to the subunit.
Binding of leaderless mRNAs to the ribosome involves a mechanism that is somewhat different from binding of canonical mRNAs. The binding is dependent on the presence of the initiator tRNA, whereas canonical mRNAs bind independently of the initiator tRNA, as observed for the archaea Sulfolobus solfataricus (9). Studies with E. coli revealed that the ratio of IF2 to IF3 plays an important role in translation initiation of leaderless mRNAs. It was suggested that leaderless mRNA is recognized by a 30S-IF2-fMet-tRNAfMet complex equivalent to that formed during translation initiation in eukaryotes (Fig. 3) (57, 58, 227). This was based on the finding that an increase in the concentration of IF2 enhances the efficiency of leaderless mRNA translation, possibly by recruitment of fMet-tRNAfMet to 30S ribosomal subunits, thus enabling codon-anticodon interaction. Recently, a cell-free translation system was used to show that leaderless mRNAs preferentially interact with 70S ribosomes and are able to proceed from the initiation to the elongation phase even in the absence of initiation factors (233).
Biochemical experiments and immuno- as well as cryo-EM studies establish a model in which the mRNA wraps around the neck of the 30S ribosomal subunit, with its 5′ end on the platform side and its 3′ end near the shoulder (50, 206). Structural data for the interaction between mRNA and the ribosome are now available from X-ray crystallographic studies (156, 258). The new data confirm the general features of the previous models. Interaction between the ASD and SD sequences is located at a large cleft between the head and the back of the platform on the 30S ribosomal subunit (Fig. 4). The mRNA wraps around the 30S ribosomal subunit while it passes through the up- and downstream tunnels (38). A latch-like closure between the head and body on activation of the subunit forms the tunnels (195). Early studies indicated that binding of mRNA to the ribosome through the SD interaction melts the mRNA secondary structure in the TIR of the mRNA (161). The mRNA is probably unwound by mRNA helicases before entering the downstream tunnel, since an RNA helix is too large to pass.
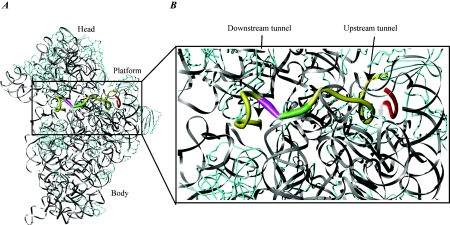
FIG. 4.
mRNA bound to the 30S ribosomal subunit. (A) A 36-nucleotide mRNA is bound to the 30S ribosomal subunit. rRNA is shown in grey, mRNA is shown in yellow, and protein is shown in cyan. The ASD sequence of the 16S rRNA is shown in red to indicate the SD-ASD interaction. The P-site initiation codon is shown in green, and the A-site codon is shown in magenta. Note the kink in the mRNA between the two codons. (B) Close-up of the region indicated in panel A. The upstream and downstream tunnels are marked by arrows. Colors are the same as in panel A. The structure is derived from PDB entry 1JGQ, prepared using the program Ribbons (25), and rendered in Pov-Ray.
Initiator tRNA
The first amino acid of a polypeptide chain is always a methionine that enters the ribosome bound to the initiator tRNAfMet. Methionine incorporated internally in the polypeptide is bound to elongator tRNAMet and carried to the ribosome by elongation factor EF1A. The role of the initiator tRNA is to ensure correct initiation of translation at the TIR of the mRNA. In bacteria as well as chloroplasts and mitochondria, the methionine bound to the tRNAfMet is N- formylated, which selectively excludes the fMet-tRNAfMet from the elongation phase of translation (95, 178, 179). As mentioned in the previous section, alternative initiation codons related to AUG by a single base change are found in some genes. These codons are all decoded by the initiator fMet-tRNAfMet and translated as formylmethionine.
Two genes in the E. coli genome encode tRNAfMet (83). The major fraction of cellular initiator tRNA (tRNAf1Met) is encoded by the metZ gene. Three identical copies of the gene occur in tandem repeats within the operon known as the metZ operon (90). A relatively small fraction of tRNAfMet (tRNAf2Met) is encoded by the metY gene, located at the beginning of the nusA/infB operon (83). The presence of adenosine instead of 7-methylguanosine at position 46 is the only difference between tRNAf2Met and tRNAf1Met (84).
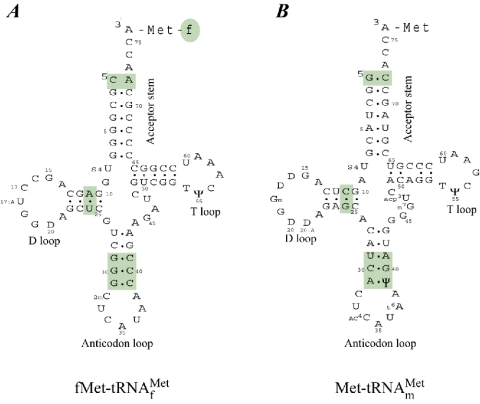
Initiator tRNAs bind directly to the P-site of the small subunit of the ribosome, whereas elongator tRNAs enter the ribosome at the A-site and subsequently translocate to the P-site. Binding of tRNAs to the P-site and the A-site is controlled by the initiation and elongation factors, respectively. Therefore, the initiator tRNAs have structural features that are recognized by initiation factors and discriminated against by elongation factors. The initiator tRNA determinants are located in the anticodon stem, the acceptor stem, and the dihydrouridine (D)-stem (Fig. 5). The significant features include (i) absence of a Watson-Crick base pair between positions 1 and 72 in the acceptor stem, (ii) three conserved consecutive GC base pairs in the anticodon stem, and (iii) the presence of a purine-11-pyrimidine-24 in contrast to the pyrimidine-11-purine-24 base pair found in other tRNAs (235). The GC pairs make the anticodon loop less flexible compared to the anticodon loop in elongator tRNAs and are important for targeting the initiator tRNA to the ribosomal P-site (199, 201).
FIG. 5.
Initiator and elongator methionine-accepting tRNAs. Cloverleaf representation of methionine-accepting tRNAs: (A) initiator tRNA and (B) elongator tRNA. The regions important for initiator tRNA identity are highlighted. Details are given in the text.
Methionine-isoaccepting initiator and elongator tRNAs are both aminoacylated by methionyl tRNA synthetase. Two identical subunits form the dimeric native enzyme, which binds two tRNAMet molecules in an anticooperative manner (125, 178). MetRS interacts with part of the acceptor stem and the anticodon loop of the tRNA (Fig. 6). The major determinant for MetRS in tRNAMet binding is the anticodon. Aminoacylation with methionine is not possible if this triplet is mutated, whereas other tRNAs provided with a CAU anticodon can be methionylated by MetRS. Recognition of the anticodon by MetRS occurs through the helical C-terminal region of the synthetase (125).
FIG. 6.
Interactions of the initiator tRNA. (A) Surface representations of the initiator tRNA. The regions that interact with the indicated component of the translational machinery are highlighted in red, and the nucleotide positions on the tRNA are indicated next to the structure. The structure of the initiator tRNA is derived from PDB entry 2FMT. (B) Detailed view of the interaction between the initiator tRNA and the ribosome. On the left is a surface representation showing the important sites of interaction in red. On the right is the initiator tRNA on the 70S ribosome. The tRNA is shown in red, the mRNA is shown in yellow, and the 16S and 23S rRNA are shown in cyan and grey, respectively. The codon-anticodon interaction is shown (the structure is derived from PDB entries 1GIX and 1GIY, prepared using the program Ribbons [25], and rendered in Pov-Ray).
Aminoacylated initiator tRNA is formylated by methionyl-tRNA transformylase (MTF). The enzyme catalyzes the transfer of a formyl group from N10-formyltetrahydrofolate to the α-amino group of the methionine of Met-tRNAfMet. The most important determinant for formylation of the initiator tRNA is the absence of a 1:72 base pair. This provides the tRNA with a 5-nucleotide single-stranded 3′ end of the acceptor arm just long enough to reach the active site of MTF (197). Both the initiator tRNA and MTF undergo structural changes in an induced-fit mechanism upon binding (122, 181). The regions on Met-tRNAfMet that interact with MTF are shown in Fig. 6.
Peptidyl-tRNA hydrolase (PTH) is a 21-kDa monomeric enzyme that recycles all N-blocked aminoacyl-tRNA molecules accumulating from abortive translation. Initiator fMet-RNAfMet is not a substrate for PTH. The presence of a 5′-terminal phosphate at the end of a fully base-paired acceptor stem is crucial for hydrolase activity (197). Therefore, the absence of a base pair between nucleotides 1 and 72 protects fMet-RNAfMet against hydrolytic cleavage by PTH. The amino acid attached to tRNAfMet is also important since fMet-tRNAfMet is completely resistant to hydrolysis by PTH whereas fGln-tRNAfMet is not (228).
In most cases, the formyl group and the methionine residue are removed posttranslationally or even as the nascent polypeptide chain emerges from the ribosomal exit tunnel (94). Formylation of Met-tRNAfMet is important for protein synthesis in E. coli. Mutant initiator tRNAs defective in formylation are extremely poor in initiation of protein synthesis, and a strain of E. coli carrying disruptions in the fmt gene encoding MTF has severe growth defects (69, 126, 236). Formylation is, however, not necessary for translation initiation in all bacteria, as exemplified by Pseudosomonas aeruginosa (144).
Formylation favors selection of the fMet-tRNAfMet by IF2 (223), and blocks binding to the elongation factor EF1A and thus the function as elongator tRNA (71, 147, 202). The nature of the amino acid attached to the tRNA is less important for IF2 binding than is formylation. Hence, IF2 is able to bind to fVal-tRNA and fGln-tRNA but not to the unformylated tRNAs (251). However, moderate overexpression of IF2 leads to translation initiation without formylation of Met-tRNAfMet, and IF2 stimulates the binding of unformylated Met-tRNAfMet to 30S ribosomal subunits in vitro (68, 250).
IF2 protects the ester bond in fMet-tRNAfMet against spontaneous hydrolysis but does not protect the unformylated Met-tRNAfMet (167). Discrimination by IF2 against the unformylated Met-tRNAfMet has also been demonstrated by footprinting experiments performed with Met-tRNAfMet and fMet-tRNAfMet in the presence of IF2. The experiments indicate binding of IF2 to the acceptor stem, position 12 to 13 in the D-stem, two sites in the anticodon stem, and parts of the T-loop and the minor groove of the fMet-tRNAfMet T-stem (Fig. 6) (166, 243). Based on a similar cleavage pattern of RNase VI in the anticodon stem of Met-tRNAfMet bound to MTF, it was proposed that the interaction between fMet-tRNAfMet and IF2 induces a conformational change in the anticodon stem (122). However, site-directed mutagenesis and nuclear magnetic resonance (NMR) spectroscopy studies reveal that essentially all thermodynamic determinants governing the stability and specificity of the interaction are located within the fMet-3′ ACCAC part of fMet-tRNAfMet (66).
Original proposals suggested that IF2 carries fMet-tRNAfMet to the ribosome by analogy to EF1A for aminoacyl-tRNAs and eIF2 for Met-tRNAiMet (reviewed in reference 94). A specific binary complex can be formed between fMet-tRNAfMet and IF2 in vitro, but the interaction is weak, with a Kd of 1.8 μM, and the complex dissociates readily in the presence of magnesium ions (113, 118, 119, 167, 251). Most evidence suggests that IF2 performs its interactions with fMet-tRNAfMet on the 30S ribosomal subunit (251).
IF3 appears to inspect the anticodon stem of the P-site bound tRNA (73). However, this inspection may occur through indirect and not direct interactions, as discussed in the section on IF3 (see below).
Binding of fMet-tRNAfMet to the P-site of the 30S ribosomal subunit is highly influenced by the conformation of the anticodon stem. Site-directed mutagenesis has shown that the three consecutive GC base pairs in the anticodon stem influence the unique conformation of fMet-tRNAfMet as well as P-site binding (202). The structure of the 70S ribosome in complex with P-site-bound fMet-tRNAfMet maps the interactions with 16S rRNA primarily to the anticodon stem and loop, whereas the 23S rRNA interacts with the CCA acceptor arm and part of the D-stem of fMet-tRNAfMet. The T-loop interacts with ribosomal protein L23 (256) (Fig. 6). A conformational change of the rRNA on P-site tRNA binding similar to that observed for A-site tRNA binding has not been found. However, C1400 of the 16S rRNA appears to stabilize the wobble base pair of the codon-anticodon interaction by stacking on base 34 of the tRNA (256). Thus, the ribosome itself does not seem to perform as stringent a proofreading of the codon-anticodon interaction in the P-site as in the A-site. As described in the following sections, the initiation factors play a key role in the selection and adjustment of the initiator tRNA in the P-site.
Translation Initiation Factors
Initiation factor IF1.
Initiation factor IF1 is the smallest of the three bacterial translation initiation factors with a molecular mass of 8.2 kDa in E. coli. IF1 is encoded by the infA gene. Two promoters control the transcription of the E. coli gene as monocistronic mRNAs, both ending at the same transcriptional terminator. infA transcription is not physically linked to any other genes as are infB and infC (the genes encoding IF2 and IF3) (40). Proteins with IF1-like structure and function are present in all three phylogenetic domains. The archaeal and eukaryotic homologues are referred to as aIF1A and eIF1A, respectively (98, 209). One model suggests that the bacterial initiation factors evolved from an ancestral IF1-type protein through consecutive duplication and fusion events (35).
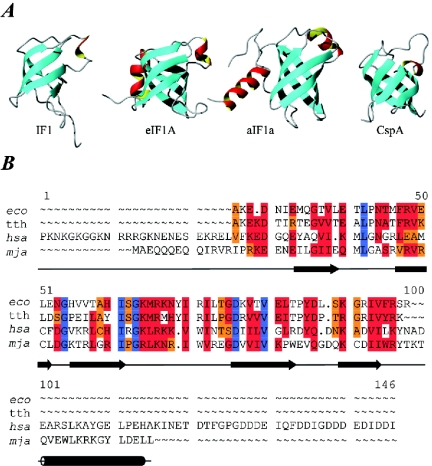
The structure of IF1 in solution has been determined by NMR spectroscopy (204). IF1 belongs to the family of oligonucleotide binding (OB) fold proteins. It consists of a five-strand beta barrel with the loop between strands 3 and 4, capping one end of the barrel (Fig. 7). Structures of the archaeal and eukaryotic IF1 homologues (aIF1A and eIF1A) have also been determined (6, 111). These structures are highly similar with respect to the OB fold (Fig. 7). The C terminus, however, contains α-helical structures that are important for the archaeal and eukaryotic scanning mechanism and interactions with the small ribosomal subunit.
FIG. 7.
Initiation factor IF1 and structural homologues. (A) Structures of IF1 and homologues: IF1 from E. coli (PDB entry 1AH9); human eIF1A, residues 40 to 125 (PDB entry 1D7Q); aIF1A from Methanococcus jannaschii (PDB entry 1JT8); and cold shock protein A (CspA) from E. coli (PDB entry 1MJC). eIF1A and aIF1A have an additional helix located at the C terminus. (B) Sequence alignment of selected IF1 sequences. Abbreviations: eco, E. coli; tth, T. thermophilus; hsa, H. sapiens; mja, M. jannaschii. Positions with 100% identity are shown in blue. If more than 50% are identical or highly similar, the residues are highlighted in red; if more than 50% of the residues are weakly similar, they are highlighted in orange.
The OB fold proteins include RNA binding proteins such as ribosomal protein S1, the cold shock proteins CspA and CspB (20), domain II of eIF5A (91), eIF2α (151), and aspartyl-tRNA synthetase (187). It has been shown that the cellular defects resulting from a double deletion in the genes encoding cold shock proteins CspB and CspC in Bacillus subtilis can be complemented by heterologous expression of E. coli IF1 (244). This confirms the structural resemblance and functionality between IF1 and the cold shock proteins.
The binding of IF1 to the 30S ribosomal subunit has been extensively mapped. IF1 binds in a cleft between the 530 loop and helix 44 of 16S rRNA and ribosomal protein S12 (Fig. 8). Cleavage of 16S rRNA with cloacin DF13 between A1493 and A1494, two positions located in the 30S ribosomal A-site, specifically disrupts the function of IF1 (2a). Binding of IF1 to the 30S ribosomal subunit protects A1492 and A1493 from modification by dimethyl sulfate and protects G530 from attack by kethoxal (132). These positions are protected by A-site-bound tRNA, strongly supporting the notion that IF1 is located at an overlapping binding site. Mutational analysis demonstrated that the C1408-G1494 base pair and the three adenosines A1408, A1492, and A1493 are required for optimal IF1 binding, and it appeared that the internal loop of helix 44 is more important for IF1 binding than the identity of the nucleotide present at a certain position in the interacting part of helix 44 (41).
FIG. 8.
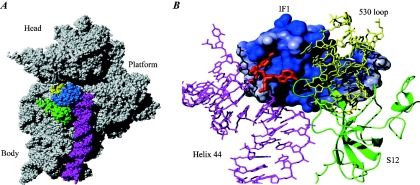
IF1 bound to the 30S ribosomal subunit. (A) Structure of IF1 on the 30S ribosomal subunit. IF1 is shown in blue, helix 44 is shown in magenta, the 530 loop is shown in yellow, and protein S12 is shown in green. The structure is derived from PDB entry 1HR0. (B) Close-up of the interaction of IF1 with the 30S subunit. IF1 is shown in a surface representation colored according to the electrostatic potential (positive charges, blue; negative charges, red). Helix 44 and the 530 loop of 16S rRNA are shown in magenta and yellow, respectively. Protein S12 is shown in a green ribbon representation. Bases A1492 and A1493 of the 16S rRNA are indicated in red. Note that they have flipped out of helix 44 and are buried in a pocket in IF1 and a pocket between IF1 and S12, respectively.
The structure of the 30S ribosomal subunit from T. thermophilus in complex with IF1 agrees well with most biochemical and mutagenesis data. A1492 and A1493 are buried in the part of the IF1 surface responsible for 16S rRNA interaction. In addition, C519 and G530 of the 530 loop and the amino acids V40 and W42 of ribosomal protein S12 are important for the interaction. A number of amino acids crucial for IF1 function were identified by site-directed mutagenesis (37, 65, 216). Moreover, specific signal changes in NMR spectroscopy experiments on titration of IF1 with 30S ribosomal subunits have been used to identify positions on IF1 involved in the interaction (204) (Fig. 8). As IF1 binds to the 30S ribosomal subunit, it inserts the loop containing residues 17 to 25 into helix 44 and thereby flips out the bases A1492 and A1493 (Fig. 8). This induces a conformational change over a long distance in the subunit that may represent a transition state in the equilibrium between subunit association and dissociation (180). Biochemical data support the conformational change in 16S rRNA. IF1 binding alters the reactivity of sites in 16S rRNA protected by tRNA, 50S ribosomal subunits, or aminoglycoside antibiotics (41).
Several functions have been attributed to bacterial IF1. It enhances the dissociation and association rate for 70S ribosomes (46, 59), primarily through the stimulating effect on the activity of IF2 and IF3 (174). Interaction between IF2 and the 30S ribosomal subunit is favored when IF1 is bound, and the release of IF2 is indirectly promoted when IF1 is ejected (27, 136, 222). IF1 cooperates with IF2 to ensure that only the initiator tRNA binds to the P-site and that it interacts with the initiation codon of the mRNA (24, 73, 127, 251). IF1 occlude tRNAs from the A-site until the 70S initiation complex has formed. Ejection of IF1 consequently opens the A-site for incoming aminoacyl-tRNAs. In vivo studies have shown that IF1-depleted cells have low growth rates and short polysomes (39). These data demonstrate that IF1 is essential for cell viability and suggest that one or more of its functions are crucial. However, no clear function has been assigned to the initiation factor yet (37).
Initiation factor IF2.
IF2 is the largest of the initiation factors. It is encoded by the infB gene. The infB gene is part of the polycistronic nusA operon containing metY (minor form of the initiator tRNA), ylxC (protein of unknown function), nusA (a transcriptional termination factor), infB (translation initiation factor IF2), rbfA (ribosome binding factor A), truB (tRNA pseudouridine 5S synthase), rpsO (ribosomal protein S15), and pnp (polynucleotide phosphorylase). All these genes are transcribed from metY toward pnp on a part of the DNA that contains several transcriptional promoters (172, 173, 189, 193). Transcription of the nusA/infB operon occurs primarily from a promoter separated by three genes upstream from infB. This promoter is autogenously controlled by the translation product of nusA (143, 171). The organization of nusA and infB in bacteria reveals that they are simultaneously present in operons. The rather conserved distribution of the genes within the operon leads to the proposal that this organization may be important for regulation (241).
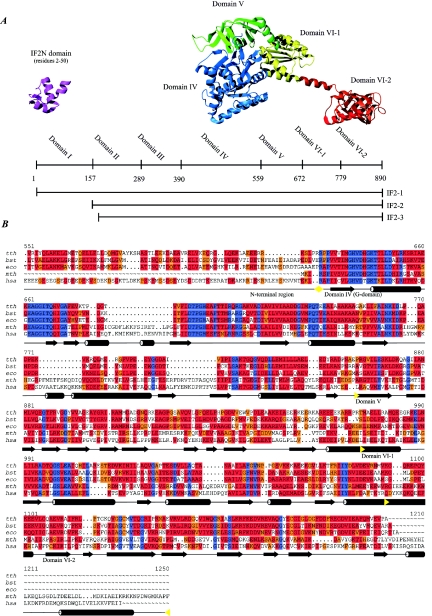
Three isoforms of the initiation factor, named IF2-1 (97.3 kDa), IF2-2 (79.7 kDa), and IF2-3 (78.8 kDa), exist in E. coli and other members of the family Enterobacteriaceae (107, 154). Bacillus subtilis is the only organism that does not belong to the Enterobacteriaceae where more than one isoform of IF2 has been experimentally demonstrated (81). IF2-1, IF2-2, and IF2-3 are translated from three independent but in-frame translational start sites of the infB mRNA. This feature has been referred to as tandem translation. Hence, IF2-2 and IF2-3 differ from IF2-1 only by the absence of the first 157 and 164 amino acid residues, respectively (Fig. 9) (139). The presence of both the large and smaller isoforms is required for optimal growth of E. coli. The cellular content of IF2-2 and IF2-3 is close to the level of IF2-1 at optimal growth conditions (79, 191), but the ratio of IF2-2 and IF2-3 to IF2-1 increases as a response to cold shock (53). An open single-stranded structure is present in the intracistronic TIR of the infB mRNA. We suggest that this structure is required for the translation of IF2-2 and IF2-3 (107).
FIG. 9.
IF2 and structural homologues. (A) Schematic representation of the E. coli IF2 primary structure. The domain boundaries and the lengths of the three IF2 isoforms are indicated. Ribbon diagrams of the structures of the IF2N domain from E. coli (PDB entry 1ND9) and the IF2 homologue aIF5B from M. thermoautotrophicum (PDB entry 1G7T) are shown. The domains are indicated in different colors, and the E. coli domain nomenclature is used. (B) Sequence alignment of selected bacterial IF2 and archaeal and eukaryotic homologues. Only a small part of the N-terminal nonconserved region is shown. Abbreviations: tth; T. thermophilus, bst, B. stearothermophilus; eco, E. coli; mth, M. thermoautotrophicum; hsa, H. sapiens. Secondary-structure elements defined from the structure of aIF5B from M. thermoautotrophicum are indicated by cylinders for helical segments and arrows for segments in β-strand conformation. The domain boundaries are indicated by yellow arrows. Color codes are as in Fig. 7.
IF2 can be divided into domains based on interspecies homology. The domain nomenclature differs somewhat among species. Throughout this review, the nomenclature for E. coli suggested by Mortensen et al. is used (Fig. 9) (140). Domain VI was subsequently divided into two subdomains, designated VI-1 and VI-2.
The factor can be divided into a conserved C-terminal region consisting of domains IV to VI and a less highly conserved N-terminal region corresponding to domains I to III (209, 217). Homologues of IF2 have been found in archaeabacteria and eukaryotes, where the factor is referred to as aIF5B and eIF5B, respectively (98). Remarkable interspecies homology in the C-terminal region is present among the homologues (209). The homologues have functions similar to those of bacterial IF2, including GTPase activity, promotion of ribosomal subunit association, and probably interaction with the initiator tRNA (30, 165).
No direct tertiary-structure information is available for domains IV to VI of E. coli IF2, but the structure of the homologous protein aIF5B from the archaeon Methanobacterium thermoautotrophicum has been solved (Fig. 9) (185). Amino acid sequence homology predicts a similar structure for domains IV to VI of bacterial IF2, although there are two additional helical segments at the C-terminus of the archaeal factor (Fig. 9).
The conserved C-terminal region of the protein begins with the domain responsible for GTP binding: the G-domain (E. coli domain IV). Overall homology among bacterial, eukaryotic, and archaeabacterial IF2, eIF5B, and aIF5B is highest in the G-domain area of the factor (Fig. 9). Significant sequence homology to other proteins is found only for this domain. The GTP binding motif is shared with at least four other proteins involved in translation, namely, EF1A, EF2, RF3, and SelB (184, 238). Following the G-domain is an EF1A-like β-barrel (domain V) and a novel αβα sandwich (domain VI-I) connected via a long α-helix to the C-terminal domain (domain VI-2). The structure of the C-terminal domain of Bacillus stearothermophilus IF2 has been solved by NMR spectroscopy methods (128). It is similar to domain VI-2 from M. thermoautotrophicum with the exception of the two C-terminal helices found in the M. thermoautotrophicum factor. These helical segments are not present in any of the bacterial sequences (Fig. 9).
In contrast to the C-terminal region, the N-terminal region of IF2 is highly variable in both primary structure and length. The region can be divided into three separate domains in E. coli IF2 based on sequence and biochemical data (Fig. 9) (140). Domain I contains a small subdomain of approximately 50 residues, found in all bacterial and some plastid IF2s. We solved the structure of the domain by NMR spectroscopy methods (105). The subdomain is now designated IF2N in the protein families database (PFAM). IF2N has homology to the stem contact fold domains of the methionyl- and glutaminyl-tRNA synthetases and the B5 domain of the phenylalanine-tRNA synthetase. However, no specific function has been assigned to the IF2N domain (105). NMR spectroscopy of the full-length E. coli IF2-1 revealed that the IF2N domain is connected to domain IV by a highly flexible linker region (104). Domain I is extremely soluble and has been applied as a solubility-enhancing fusion partner for the expression of proteins prone to aggregate in the E. coli cytoplasm (211).
Macromolecular interactions of the domains in the N-terminal region of IF2 has been demonstrated only in E. coli, where a fragment consisting of the combined domains I and II, but not a fragment of isolated domain I, binds to the 30S ribosomal subunit (136, 137). Furthermore, we have identified domains I and II of E. coli IF2 as an interaction partner for the infB mRNA (107).
B. stearothermophilus and T. thermophilus IF2 possess a single domain in the region N-terminal to the G-domain, whereas myxobacterial IF2, which has the longest N-terminal regions characterized in bacteria, is composed of several domains with a highly unusual amino acid composition. The latter is a general feature of the N-terminal regions of IF2 (16, 64, 225, 230). Regions N-terminal to the G-domain of eukaryotic IF2 homologues are generally long, up to ∼700 amino acids, whereas the regions of the archaeal IF2 homologues are generally short. Conclusively, the functional importance of the N-terminal region of IF2 and its homologues remains unresolved.
The role of the GTPase activity of IF2 has been a matter of debate for decades. Available structures of aIF5B in the nucleotide free form, the inactive form with GDP bound, and the active form with GTP bound reveal that binding of Mg2+-GTP induces movements in domains V and VI-2 over a distance of more than 90 Å (185, 186).
Elongation factor EF1A associates approximately 100-fold more strongly with GDP than with GTP, whereas IF2 associates only 10-fold more strongly with GDP (163, 175). A guanine nucleotide exchange factor may therefore not be required for IF2 as in the case of EF1A, where nucleotide exchange involves the factor EF1B. Hydrolysis of GTP by IF2 depends on the presence of ribosomes, and the factor has no intrinsic GTPase activity by itself. However, B. stearothermophilus IF2 can hydrolyze GTP in the absence of ribosomes when 20% ethanol is included in the reaction mixture (64). GTP hydrolysis in translation initiation has been suggested to be important for the release of IF2 from the 70S initiation complex (108, 114), and for the adjustment of initiator tRNA in the ribosomal P-site (103). The crucial importance of GTP hydrolysis in translation initiation and its direct relation to cell viability have been confirmed by several studies of G-domain mutants of IF2 (98a, 100, 106, 115).
The G-domains of IF2 and EF1A are thought to have overlapping binding sites. Fluorescence stopped-flow experiments showed that binding of EF1A to ribosomes probed with IF2 was independent of the bound nucleotide. It was thus concluded that neither the ejection of IF2 from the ribosome nor its recycling requires GTP hydrolysis (102, 231). A recent study of initiation complex formation by using stopped-flow experiments with light scattering gave contradictory results. The conclusions from these experiments are that the GTP-bound form of IF2 accelerates association of the ribosomal subunits and that GTP hydrolysis accelerates ejection of IF2 from the 70S ribosome (2). Further studies are needed to achieve a detailed understanding of the role of GTP hydrolysis in the translation initiation event.
Early cross-linking experiments show that parts of IF1 and IF2 are in close proximity on the ribosome (13). The interaction between the two factors maps to a region between domains III and V of E. coli IF2 (136). Interactions between the eukaryotic homologues eIF5B and eIF1A have been mapped by using the yeast two-hybrid system, coimmunoprecipitation, in vitro binding assays, and NMR spectroscopy (31, 120, 159). The C-terminal unstructured region of eIF1A, which is not present in bacterial IF1, interacts with the C terminus of eIF5B. Domain V of bacterial IF2 was suggested to interact with IF1 on the ribosome (120, 136).
As mentioned above, the interactions between fMet-tRNAfMet and IF2 have been studied extensively. Formation of the binary complex is strongly dependent on the formylation of Met-tRNAfMet (223) but independent of GTP (175). The C-terminal domain VI-2 of IF2 has been suggested to contain the entire binding site for fMet-tRNAfMet (215). The interaction of the domain with fMet-tRNAfMet has been studied using mutagenesis as well as Raman and NMR spectroscopy (66, 96, 130, 225). Functionally essential residues of the B. stearothermophilus domain are C668, K699, R700, Y701, K702, E713, C714, and G715 (Fig. 10). Cross-linking experiments indicate interactions between E. coli residues N611-R645 (belonging to domain V) and the T-stem of fMet-tRNAfMet as well as residues W215-R237 (domain II) and the anticodon stem of fMet-tRNAfMet (243, 257). Finally, fMet-tRNAfMet protects a position in domain IV and weakly in domain V of B. stearothermophilus IF2 against digestion by trypsin (205). A stable interaction between the archaeal and eukaryotic IF2 homologues and Met-tRNAiMet has not been observed in vitro, but overexpression of the gene encoding tRNAiMet partially suppresses the severe slow-growth phenotype of yeast strains lacking the IF2 homologue (30).
FIG. 10.
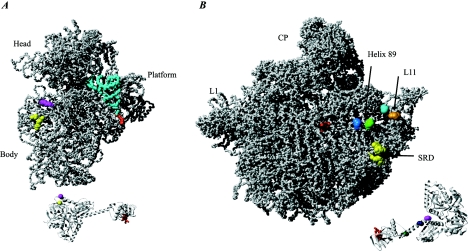
IF2 Interactions with fMet-tRNAfMet and the ribosome. (A) The 30S ribosomal subunit from T. thermophilus (PDB entry 1J5E) in complex with a P-site fMet-tRNAfMet (derived and docked based on PDB entry 1GIX). The fMet-ACC region of the tRNA that interacts with the C-terminal region of IF2 is shown in red. The corresponding region is red on the aIF5B structure shown at the bottom part (PDB entry 1G7T). Two residues in domain V of IF2 (V451 and S520 of B. stearothermophilus IF2) are shown in yellow and magenta, respectively. A nuclease at position V451 cleaves positions 38 to 40 and 498, and a nuclease at position S520 cleaves positions 538 to 540 of the E. coli 16S rRNA in a 70S ribosomal complex. The corresponding positions are indicated in yellow and magenta on the ribosomal subunit. Note that these cleavages are absent in a 30S-IF2 initiation complex. (B) The 50S ribosomal subunit from H. marismortui (PDB entry 1JJ2) is shown along with aIF5B. Red indicates the position of the fMet ACC in the decoding center of the ribosomal subunit and the corresponding region on a IF5B that interacts with the fMET ACC. Pale blue and light green on the subunit indicate two positions in helix 89 of the 23S rRNA (U2474 and A2482 in E. coli numbering) that are cleaved by nucleases attached at the residue located at the interface between domains VI-1 and VI-2 of IF2 (shown in dark green on the aIF5b structure) (E644 of the B. stearothermophilus IF2). A nuclease attached to a residue in domain VI-1 of IF2 (dark blue) (E632 of the B. stearothermophilus IF2) cleaves both the C1076 (cyan) and G1068 (cyan) positions in the L11 region. The nuclease (shown in purple) on aIF5B (position Y625 of the VI-1 domain) cleaves weakly at C1076 (cyan) and U2474 (light blue). Data were derived from reference 121. Another study showed that IF2 protects residues in the sarcin-ricin domain (SRD) (G2655, A2665, and G2661) against chemical modification (100). These residues are indicated in yellow on the subunit. No attempt was made to dock IF2 on the ribosomal subunit, since it is likely that conformational changes occur in the subunit as a result of IF2 binding.
IF2 is the only one of the three initiation factors that displays a relatively high and specific affinity for both ribosomal subunits. However, structures of IF2 in complex with the ribosome or the fMet-tRNAfMet remain absent. Models in which IF2 and IF1 in concert mimic the anticodon loop, anticodon stem, D-loop, and D-stem of A-site-bound tRNA have been suggested (17, 138). However, these models were proposed prior to the elucidation of the structure of the IF2 homologue from M. thermoautotrophicum (185). The new structural knowledge and recent biochemical data reveal that the macromolecular mimicry model for IF2 and IF1 is inaccurate.
The GTPases involved in translation probably occupy partly overlapping binding sites on the ribosome. A site located on the 50S ribosomal subunit, termed the factor binding site, is composed of the α-sarcin loop, the L11-binding region, and the L7-L12 stalk. EF1A, EF2, and IF2 all interact with the ribosome at the factor binding site (23, 131, 182). An additional constraint to place IF2 on the ribosome is the interaction with fMet-tRNAfMet, which places domain VI-2 of IF2 in proximity of the fMet-CCA end of the P-site-bound initiator tRNA.
IF2 has been cross-linked to S13, L7-L12, IF1, and IF3 (13), as well as S1, S2, S11, S12, and S19 on the ribosome (14). Chemical probing experiments with 23S rRNA indicated that IF2 protects A2476 and A2478 in helix 89 of domain V as well as G2655, A2660, G2661, and A2665 of the sarcin-ricin domain positioned in domain VI (Fig. 10) (102). These footprints were generated regardless of the presence of GTP, IF1, mRNA, and fMet-tRNAfMet. Unfortunately, the results are unclear with respect to the 30S ribosomal subunit, since IF2 affects the reactivity of residues spread all over the subunit. This is consistent with an observed rearrangement of the subunit induced by IF2 (242). Recently, a model of IF2 binding to the ribosome was presented, based on cleavage of the rRNA by chemical nucleases tethered to cysteine residues introduced at specific sites of IF2 (121). No cleavage of the 16S rRNA was observed when IF2 was bound to 30S ribosomal subunits or to the complete 30S initiation complex. However, cleavage of the 16S rRNA was observed when IF2 was bound to the 70S initiation complex (Fig. 10). These data indicate that domain V of IF2 is localized toward the 30S subunit in the 70S initiation complex. As described above, cross-linking data of the 30S complex and footprinting data on the binary fMet-tRNAfMet-IF2 complex place domain V of IF2 in proximity to the elbow of the P-site-bound fMet-tRNAfMet. The distance between the 16S rRNA and the elbow of the fMet-tRNAfMet appears to be too far for domain V of IF2 to establish contact with both simultaneously. Conclusively, IF2 changes localization during the transition from the 30S to the 70S initiation complex (121). The cleavage experiments were performed in the presence of excess GTP. Large domain movements take place in IF2 during GTP hydrolysis (185), and the cleavage patterns in the rRNA might be dependent on whether IF2 is in the GTP- or GDP-bound form. To fully understand the function of IF2 during translation initiation, detailed atomic resolution structures of both the 30S and 70S initiation complexes as well as a better understanding of the timing and not least the consequences of GTP hydrolysis are needed.
Besides the function as a translation factor, IF2 has the properties of a chaperone. It promotes functional folding of proteins and forms stable complexes with unfolded proteins (22). Furthermore, the expression of IF2 is upregulated during the cold shock response (3), and the factor is important for the translation of leaderless transcripts (57). Finally, we have introduced the use of IF2 sequence data for the classification of organisms of closely related organisms (75-77, 152, 209, 226).
Initiation factor IF3.
E. coli IF3 is a 20.4-kDa protein composed of 180 amino acids encoded by the essential infC gene (160, 190). The infC-rpmI-rplT operon contains the genes encoding IF3 and the two ribosomal proteins L35 and L20 (28, 110). These genes are transcribed from four promoters and terminated by two transcriptional terminators (110, 245). At the translational level, the expression from the operon is regulated by two different control circuits, discussed further in the last section of this review. Whereas IF1 and IF2 are universally present and important for the function of all living cells, IF3 is limited to a number of bacterial species and has been found in some plastids (112, 254, 255). The human mitochondrial IF3mt has short extensions in the N and C termini surrounding a region homologous to bacterial IF3. It promotes initiation complex formation on mitochondrial ribosomes (92).
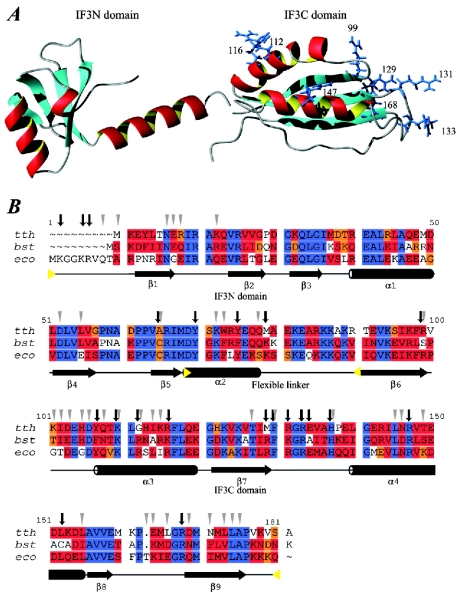
IF3 is composed of two structural domains of approximately equal size (Fig. 11) (48, 97). The two domains, called the IF3N and IF3C, are separated by a ∼ 45-Å lysine-rich flexible linker (80, 135). The IF3N domain consists of a globular α/β-fold, with helix α1 packed against a mixed five-strand β-sheet (Fig. 11). This fold is followed by helix α2, which connects IF3N to IF3C. The length of α2 was found to be different in the structures derived from NMR spectroscopy (51) and X-ray diffraction (11) experiments and has been the subject of debate (reviewed in reference 12). The linker is essential for IF3 function, but variation of its length and composition does not considerably change the activity (43).
FIG. 11.
IF3 structure and alignment. (A) Structures of the IF3N domain from B. stearothermophilus (PDB entry 1TIF) and the IF3C domain from E. coli (PDB entry 2IFE). The side chains of the arginine residues in the IF3C domain are shown and labeled with the residue number. Mutations in the arginine residues that affect binding to the 30S ribosomal subunit are residue numbers 99, 112, 116, 147, and possibly 168. These roughly define the surface that binds to the 30S ribosomal subunit. Mutations of arginine residues reducing IF3 activity involved in mRNA-related functions define a surface comprising residues 129, 131, and 133. (B) Sequence alignment of selected sequences of IF3. Abbreviations, color codes, and secondary-structure nomenclature are as in Fig. 7. Secondary-structure elements are as defined in reference 203. Black vertical arrows indicate residues that have been identified as interacting with the 30S ribosomal subunit by mutagenesis and/or chemical modification. Grey triangles indicate residues whose intensity was most strongly affected by titration with 30S ribosomal subunits in NMR spectroscopy studies (reference 203 and references cited therein). Yellow triangles indicate approximate domain boundaries.
The structure of IF3C has been solved by NMR spectroscopy and X-ray diffraction (11, 51). It consists of a two-layer α/β sandwich fold composed of a four-strand mixed β-sheet packed against two parallel α-helices (α3 and α4), leading to a βαβαββ topology (Fig. 11). The structure is similar to U1A (RNA binding protein involved in RNA splicing) (51) and YppH (a protein involved in cell division) (88).
IF3 perform several different functions. (i) It prevents association of the ribosomal subunits by binding to the 30S subunit, thereby blocking binding of the 50S subunit (59, 188). (ii) It monitors the codon-anticodon interaction by promoting the dissociation of fMet-tRNAfMet from initiation complexes formed at the 5′ initiation codon of leaderless mRNAs (227). Likewise, initiation complexes with an incorrectly bound aminoacyl-tRNA (noninitiator tRNA) (73, 74) and complexes with triplets other than AUG, GUG, and UUG in the P-site are dissociated by IF3 (70, 127, 224). (iii) It stimulates the rapid formation of codon-anticodon interaction at the ribosomal P-site (60, 250). (iv) It is involved in the adjustment of the mRNA from the standby site to the decoding P-site of the 30S ribosomal subunit (101). Finally, a role for IF3 in recycling of subunits has been proposed. It was observed to enhance the dissociation of deacylated tRNAs from posttermination complexes and to dissociate 70S ribosomes into subunits (78, 87).
All functions of the native IF3 can be accomplished by the isolated IF3C domain in vitro, while the IF3N domain probably serves the purpose of modulating the thermodynamic stability of the IF3-30S complexes (169). Site-directed mutagenesis of the eight arginine residues in the IF3C domain has been used to map the active sites (168). The arginines at positions 99, 112, 116, 147, and 168 are important for the binding to the 30S ribosomal subunit (Fig. 11). The ability of IF3 to dissociate the ribosome into subunits was affected mainly by mutations of R112 and R147 (and less extensively by mutations of R99 and R116). The stimulation of the pseudoinitiation complex dissociation (with a noninitiator tRNA bound) was affected by mutations of R99 and R112 (and less extensively of the arginine residues at positions 116, 129, 133, and 147). Dissociation of noncanonical initiation complexes (initiation codons other than AUG, GUG, and UUG) was not affected in any of the mutants. Stimulation of translation was affected by mutations of R116 and R129 (and less extensively of the arginine residues at positions 99, 112, and 131), whereas inhibition of noncanonical mRNA translation was affected by mutations of R99, R112, and R168 (and less extensively of the arginine residues at positions 116, 129, and 131). Finally, the repositioning of the mRNA from the standby site to the P-decoding site was weakly affected by mutations of the arginine residues at positions 129, 131, 133, 147, and 168. The data indicate that IF3C contains at least two active surfaces, one embedded in the 30S subunit and the other facing the mRNA (Fig. 11) (168).
Both IF3 domains are RNA binding and interact independently with the 30S ribosomal subunit. IF3C interacts with the highest affinity through a large surface of symmetrically distributed residues in loops and α-helices, whereas IF3N interacts mainly via a small number of asymmetrically distributed residues (203). Results of mapping of IF3 residues implicated in binding to the 30S ribosomal subunit by NMR spectroscopy, site-directed mutagenesis, and other chemical methods are in excellent agreement (203) (Fig. 11).
The localization of IF3 on the 30S ribosomal subunit has been studied by various methods, with conflicting results. Immuno-EM located the factor at the cleft of the 30S ribosomal subunit (220). The ribosomal proteins S7, S11, S12, S13, S18, S19, and S21 have been cross-linked to IF3 but are spread over a wide area of the 30S subunit (13, 33, 34, 117). Helices 26 (central domain) and 45 (3′ minor domain) of the 16S rRNA have been cross-linked to IF3 (47). Chemical probing revealed IF3 contacts to helices 23 and 24 in the central domain of the 16S rRNA (132, 141), and NMR spectroscopy indicated that IF3 interacts with the 3′ end of the 16S rRNA (246). Cryo-EM located the IF3C domain to the interface side of the small ribosomal subunit (124). However, X-ray diffraction of 30S ribosomal subunit crystals soaked with IF3C places the domain at the solvent side of the platform (170).
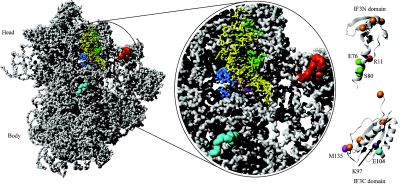
A model based on hydroxyl radical footprinting and directed probing from Fe(II)-derivatized IF3 has been presented (42). This model is in agreement with the cryo-EM data, and the results are summarized in Fig. 12. It was suggested that the observations in the crystallographic studies represent binding to a secondary site in the crystal-soaking experiments as a result of blockage of the primary binding site by crystal contacts. IF3C is located in the same area as helix 69 of the 23S rRNA in the 70S ribosome, which explains why IF3 blocks subunit association.
FIG. 12.
Interaction of IF3 with the 30S ribosomal subunit. The interactions between IF3 and the 30S ribosomal subunit identified by hydroxyl radical footprinting and directed probing are shown (data from reference 42). (A) 30S ribosomal subunit from T. thermophilus with P-site-bound tRNA (derived from PDB entries 1J5E and 1GIX). The tRNA is shown in yellow. (B) Close-up of the ribosomal subunit. Sites in the 16S rRNA and on the tRNA that are cleaved by nucleases attached to IF3 are indicated. The IF3N and IF3C domains (PDB entries 1TIF and 2IFE) are shown in a ribbon representation, with the modified cysteines indicated by spheres. Orange spheres on the IF3 domains indicate residues where an attached nuclease does not cleave the 16S rRNA or tRNA. The magenta spheres in the IF3C domain indicate residues K97 and MI35 from where nucleases cleave in the 790 loop of the 16S rRNA (magenta) seen below the acceptor arm of the tRNA. Nucleases at these positions also cleave the side of the tRNA marked in blue (residues 3 to 5 and 12 to 24). The cyan sphere in the IF3C domain indicates E104 from which nucleases cleave in the 790 loop (magenta) and at residues 1482 to 1487, indicated in cyan on the 30S subunit. Green spheres in the linker region on the IF3N domain indicate residues (E76 and S80) on which nucleases cleave on the other side of the tRNA, marked in green (residues 26 to 29 and 35 to 37), the 790 loop region (magenta), and the 690 loop region (red) of the 16S rRNA. A nuclease attached to position R11 in the IF3N domain (red) cleaves at positions in the 690 loop (red). No attempt was made to dock IF3 on the ribosomal subunit, since conformational changes most probably take place in the subunit as a result of IF3 binding.
The two domains of IF3 were shown to be on opposite sides of the fMet-tRNAfMet (42). IF3 has been thought to interact with the anticodon stem and loop of fMet-tRNAfMet (73). However, IF3 is unable to reach the three conserved discriminator GC base pairs in the anticodon stem of fMet-tRNAfMet in the current model. Hence, discrimination against elongator tRNAs promoted by IF3 is probably indirect (42).
REGULATION OF TRANSLATION INITIATION
Bacteria must be able to adjust to environmental changes in temperature, the availability of nutrients and water, presence of toxic molecules, etc. A prerequisite for induction of an appropriate stress response is precise monitoring of internal and external parameters. Although transcriptional regulation is the primary mechanism in stress responses, regulation of translation is faster and consequently important. Post-transcriptional regulation occurs at different stages including mRNA stability and translation initiation. Here we focus on responses involving the translation initiation phase.
Regulation occurs by a variety of events that control the formation of elongation-competent translation initiation complexes. The only variable component in translation initiation is the mRNA. The sequence and structure of the mRNA determine its interaction with the translational machinery and hence the efficiency and frequency of translation. A highly expressed mRNA contains some or all of the following elements at the RBS: (i) a cognate initiation codon for interaction with the fMet-tRNAfMet (183, 237); (ii) an SD sequence complementary to the ASD sequence of the 16S rRNA (207); (iii) a pyrimidine tract for interaction with ribosomal protein S1 (15, 232, 260); and (iv) base-specific enhancer elements upstream (155) or downstream (213, 214) of the initiation codon. The interdependence and relative importance of these mRNA elements are poorly understood (234). Translational regulation can involve cis-acting elements of the mRNA that form secondary or tertiary structures which sequester the ribosomal binding site. trans-Acting elements include protein, antisense RNA, and other factors that control the alternative structures of the RBS and thus affect the efficiency of initiation complex formation. A recent review describes the translational repression mechanisms (194).
Initial binding of the mRNA RBS to the ribosome occurs primarily through interactions with the ribosomal protein S1 and the ASD sequence of the 16S rRNA. Both interactions require local single-stranded mRNA (reference 194 and references cited therein). Secondary structures in the RBS can lower the translational efficiency of an mRNA. Figure 13 summarizes some mechanisms for translational repression and activation caused by changes in the secondary structure of the mRNA. The thermodynamic stability of secondary structures in the RBS plays a crucial role, but the kinetics of the mRNA folding is also an important factor (reference 194 and references cited therein). This was demonstrated for the phage MS2 maturation protein. The mRNA contains a leader sequence forming a cloverleaf structure, which inhibits translation initiation. The effects of mRNA renaturation time on translation initiation were studied, and it was observed that the formation of the cloverleaf structure is slower than the formation of initiation complexes on the mRNA (176).
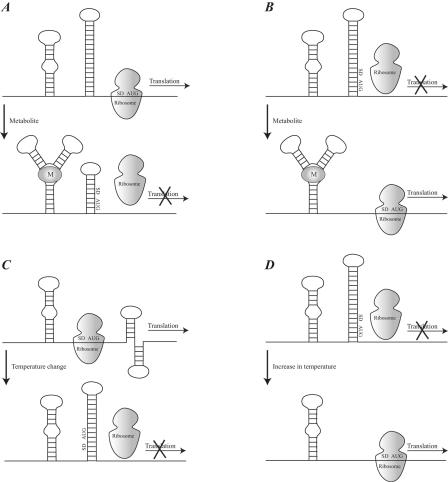
FIG. 13.
Examples of translational regulation mechanisms. (A) Repression of translation by binding of a metabolite that stabilizes an alternative mRNA secondary structure and leaves the SD sequence and initiation codon (AUG) in a base-paired region. (B) Activation of translation by binding of a metabolite that stabilizes an alternative mRNA secondary structure and leaves the SD sequence and initiation codon (AUG) in an unpaired region, thus providing ribosomal access. (C) Repression of translation by the formation of an alternative mRNA secondary structure as a result of a change in temperature. (D) Activation of translation by an increase in temperature, causing a local melting of the mRNA secondary structure covering the SD and AUG region.
The interactions involved in forming the secondary and tertiary structures of mRNA are sensitive to temperature. Structural changes of the mRNA as a consequence of temperature changes may regulate the translational activity of mRNAs. This is especially seen for the mRNAs involved in the expression of heat shock genes. The E. coli heat shock factor σ32 is the best-characterized example (reference 194 and references cited therein). The structure of the RBS of this mRNA is extremely sensitive to changes in temperature near 42°C.
Regulation is also mediated by trans-acting factors that stabilize or destabilize mRNA structures. A typical example is the autoregulation of the S10 operon mediated by the ribosomal protein L4. L4 is encoded by the S10 operon and stabilizes a hairpin in the mRNA, which represses translation (45). A protein is usually responsible for the regulation, but recent examples show that other molecules can control the expression of an mRNA. For example, thiamine (also known as vitamin B1) controls the expression of the genes involved in thiamine biosynthesis via the thi box of the mRNA (129). Similar models have been proposed for cobalamin and vitamin B12, indicating that regulation can be conferred not exclusively by proteins but also by small molecules (142, 221). The mRNA elements that directly monitor environmental conditions have been termed riboswitches and are involved in several metabolic pathways (for example, biosynthesis of vitamins and metabolism of methionine, lysine, and purines) (240). Intermolecular RNA interactions also play an important regulatory role. For example, the translation initiation site of the mRNA of outer membrane protein F (OmpF) in E. coli can be blocked by a natural antisense RNA that is transcribed from the micF gene in response to changes in osmolarity (142, 221).
Protein synthesis needs to be tightly coupled to the nutritional conditions met by the cell. The cellular content of GTP is an indicator of the overall nutritional conditions. GTP levels have been proposed to be directly coupled to the activity of IF2, which is active only in the GTP-bound form (12). mRNA-mediated detection of environmental conditions has been reviewed previously (32, 153, 240).
Regulation by competition is also a common method of regulation. For example, the threonyl-tRNA synthetase represses its own expression by binding to the RBS of the mRNA (214). The homodimeric tRNA synthetase recognizes two domains in the mRNA that structurally mimic the anticodon arm of tRNAThr. Thus, if excess threonyl-tRNA synthetase is present it binds to its own mRNA and represses expression, whereas if excess tRNAThr is present the synthetase binds to the tRNAThr instead of the mRNA and translation will be promoted. The mechanism of the repression caused by proteins that bind to their own mRNA is in some cases more complicated. Ribosomal protein S15 selectively binds to its own mRNA when it is in a pseudoknot conformation; an inactive ternary complex of S15, the mRNA, and the 30S ribosomal subunit is then formed. This prevents the formation of active translation initiation complexes (8).
Bacterial translation initiation can occur at multiple sites on a polycistronic mRNA. Translation of the individual cistrons is often coupled by base pairing between the TIR of a downstream cistron and part of the preceding coding sequence. The base pairing is disrupted when the ribosome advances toward the TIR of the downstream cistron that, as a consequence, becomes activated (109). Translational coupling also occurs by a reinitiation mechanism in which a ribosome translating a cistron reinitiates at the next cistron when a stop codon is reached (54, 192). Overlapping between the end of the first cistron and the beginning of the second cistron is often involved in reinitiation. When the terminating ribosome loosens, it may slip forward or backward to locate a reinitiation site in a process with potential modulating roles for the termination factors (1, 36, 85).
Other features including the initiation and even termination codons are involved in translational control. Expression from the infC-rpmI-rplT operon, which contains the genes encoding IF3, L35, and L20, is regulated at the translational level by two different control circuits. The initiation codon on the infC mRNA is the unusual AUU codon. As discussed above, IF3 discriminates against non-cognate initiation codons and hence represses the expression of its own gene (19). The second control circuit in the infC-rpmI-rplT operon concerns L20. L20 directly represses the translation of rpmI and indirectly represses that of its own gene via translational coupling (109). The L20-mediated repression requires a base-pairing interaction between nucleotides within infC and nucleotides in the TIR of rpmI (28, 29). The result is a pseudoknot recognized by L20. An irregular stem located upstream from rpmI is also recognized by L20, and it was suggested that L20 binds to its mRNA and rRNA partners in a similar manner (67). Recently it was suggested that the stop codon could influence the initiation process (86). An inefficient termination codon was suggested to cause ribosomal pausing and queuing along the upstream mRNA region, thus blocking the translation initiation of short genes.
The examples mentioned above are concerned mostly with regulation of translation from specific mRNAs. However, more nonspecific control mechanisms also exist. These primarily involve regulation of a whole class of genes in response to, for example, environmental changes. A temperature downshift causes a transient inhibition of protein synthesis, which results in a growth lag called the acclimation phase, where protein synthesis is arrested. However, a certain class of cold shock proteins is synthesized during this phase. The most extensively induced genes have an unusually long 5′ UTR in common (229). This 5′ UTR contains a conserved cold-box sequence that stabilizes the mRNA at low temperature. Moreover, the 5′ UTR has a remarkably high affinity for ribosomes at low temperature (252). After the acclimation phase, ribosomes become cold adapted, presumably by the binding of RbfA, a 30S ribosomal subunit-associated protein, and CsdA, a member of the DEAD box family of helicases. Subsequently, synthesis of non-cold-shock proteins is induced (229). This induction is caused in part by the cold shock proteins of the CspA family, which are able to destabilize mRNA secondary structures at low temperature (reference 148 and references cited therein).
The relative levels of the initiation factors may also play a role in regulation. Expression from leaderless mRNAs is suppressed by IF3 and stimulated by IF2 (58). This led to the proposal that the relative IF3 deficiency present when an increase in growth rate alters the ribosome/IF3 ratio could favor an increased translational rate of leaderless mRNAs. Moreover, the stoichiometry of the three translation initiation factors relative to ribosomes is more than doubled during the acclimation period of the cold shock response. These factors selectively stimulate translation of cold shock mRNAs at low temperature (62).
CONCLUDING REMARKS
During the last few years, a vast amount of structural and functional knowledge about the components involved in initiation of protein synthesis have accumulated. The interactions between some of the components have been mapped at atomic resolution, whereas other interactions have been mapped at low resolution. Binding of tRNA, mRNA, and initiation factors to the small ribosomal subunit as well as subunit association are associated with conformational changes of the ribosome. To fully understand the mechanism of translation initiation and its regulation, atomic-resolution structures of functional states of the initiation complexes, along with supporting biochemical data, are necessary. This task is the next challenge in the elucidation of the exact mechanism of translation initiation.
Acknowledgments
This work was funded by grants from the Familien Hede Nielsens Fund (grant 9901722), The Carlsberg Foundation (grants 0987/40 and 1649/40), and the Danish Natural Science Research Council (grants 21-03-0592 and 21-03-0465) to H. U. Sperling-Petersen and K. K. Mortensen.
REFERENCES
- 1.Adhin, M. R., and J. van Duin. 1990. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 213:811-818. [DOI] [PubMed] [Google Scholar]
- 2.Antoun, A., M. Y. Pavlov, K. Andersson, T. Tenson, and M. Ehrenberg. 2003. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 22:5593-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Baan, R. A., J. J. Duijfjes, E. van Leerdam, P. H. van Knippenberg, and L. Bosch. 1976. Specific in situ cleavage of 16S ribosomal RNA of Escherichia coli interferes with the function of initiation factor IF-1. Proc. Natl. Acad. Sci. USA 73:702-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 5.Bashan, A., R. Zarivach, F. Schluenzen, I. Agmon, J. Harms, T. Auerbach, D. Baram, R. Berisio, H. Bartels, H. A. Hansen, P. Fucini, D. Wilson, M. Peretz, M. Kessler, and A. Yonath. 2003. Ribosomal crystallography: peptide bond formation and its inhibition. Biopolymers 70:19-41. [DOI] [PubMed] [Google Scholar]
- 6.Battiste, J. L., T. V. Pestova, C. U. Hellen, and G. Wagner. 2000. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell 5:109-119. [DOI] [PubMed] [Google Scholar]
- 7.Bell, S. D., and S. P. Jackson. 1998. Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features. Trends Microbiol. 6:222-228. [DOI] [PubMed] [Google Scholar]
- 8.Benard, L., C. Philippe, B. Ehresmann, C. Ehresmann, and C. Portier. 1996. Pseudoknot and translational control in the expression of the S15 ribosomal protein. Biochimie 78:568-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benelli, D., E. Maone, and P. Londei. 2003. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol. Microbiol. 50:635-643. [DOI] [PubMed] [Google Scholar]
- 10.Binns, N., and M. Masters. 2002. Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol. Microbiol. 44:1287-1298. [DOI] [PubMed] [Google Scholar]
- 11.Biou, V., F. Shu, and V. Ramakrishnan. 1995. X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix. EMBO J. 14:4056-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boelens, R., and C. O. Gualerzi. 2002. Structure and function of bacterial initiation factors. Curr. Protein Pept. Sci. 3:107-119. [DOI] [PubMed] [Google Scholar]
- 13.Boileau, G., P. Butler, J. W. Hershey, and R. R. Traut. 1983. Direct cross-links between initiation factors 1, 2, and 3 and ribosomal proteins promoted by 2-iminothiolane. Biochemistry 22:3162-3170. [DOI] [PubMed] [Google Scholar]
- 14.Bollen, A., R. L. Heimark, A. Cozzone, R. R. Traut, and J. W. Hershey. 1975. Cross-linking of initiation factor IF-2 to Escherichia coli 30 S ribosomal proteins with dimethylsuberimidate. J. Biol. Chem. 250:4310-4314. [PubMed] [Google Scholar]
- 15.Boni, I. V., D. M. Isaeva, M. L. Musychenko, and N. V. Tzareva. 1991. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bremaud, L., S. Laalami, B. Derijard, and Y. Cenatiempo. 1997. Translation initiation factor IF2 of the myxobacterium Stigmatella aurantiaca: presence of a single species with an unusual N-terminal sequence. J. Bacteriol. 179:2348-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brock, S., K. Szkaradkiewicz, and M. Sprinzl. 1998. Initiation factors of protein biosynthesis in bacteria and their structural relationship to elongation and termination factors. Mol. Microbiol. 29:409-417. [DOI] [PubMed] [Google Scholar]
- 18.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, B. T. Wimberly, and V. Ramakrishnan. 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316:725-768. [DOI] [PubMed] [Google Scholar]
- 19.Butler, J. S., M. Springer, J. Dondon, M. Graffe, and M. Grunberg-Manago. 1986. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J. Mol. Biol. 192:767-780. [DOI] [PubMed] [Google Scholar]
- 20.Bycroft, M., T. J. Hubbard, M. Proctor, S. M. Freund, and A. G. Murzin. 1997. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell 88:235-242. [DOI] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Caldas, T., S. Laalami, and G. Richarme. 2000. Chaperone properties of bacterial elongation factor EF-G and initiation factor IF2. J. Biol. Chem. 275:855-860. [DOI] [PubMed] [Google Scholar]
- 23.Cameron, D. M., J. Thompson, P. E. March, and A. E. Dahlberg. 2002. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the Escherichia coli ribosome. J. Mol. Biol. 319:27-35. [DOI] [PubMed] [Google Scholar]
- 24.Canonaco, M. A., R. A. Calogero, and C. O. Gualerzi. 1986. Mechanism of translational initiation in prokaryotes. Evidence for a direct effect of IF2 on the activity of the 30 S ribosomal subunit. FEBS Lett. 207:198-204. [DOI] [PubMed] [Google Scholar]
- 25.Carson, M. 1997. Ribbons. Methods Enzymol. 277:493-505. [PubMed] [Google Scholar]
- 26.Carter, A. P., W. M. Clemons, Jr., D. E. Brodersen, R. J. Morgan-Warren, T. Hartsch, B. T. Wimberly, and V. Ramakrishnan. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291:498-501. [DOI] [PubMed] [Google Scholar]
- 27.Celano, B., R. T. Pawlik, and C. O. Gualerzi. 1988. Interaction of Escherichia coli translation-initiation factor IF-1 with ribosomes. Eur. J. Biochem. 178:351-355. [DOI] [PubMed] [Google Scholar]
- 28.Chiaruttini, C., M. Milet, and M. Springer. 1996. A long-range RNA-RNA interaction forms a pseudoknot required for translational control of the IF3-L35-L20 ribosomal protein operon in Escherichia coli. EMBO J. 15:4402-4413. [PMC free article] [PubMed] [Google Scholar]
- 29.Chiaruttini, C., M. Milet, and M. Springer. 1997. Translational coupling by modulation of feedback repression in the IF3 operon of Escherichia coli. Proc. Natl. Acad. Sci. USA 94:9208-9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi, S. K., J. H. Lee, W. L. Zoll, W. C. Merrick, and T. E. Dever. 1998. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280:1757-1760. [DOI] [PubMed] [Google Scholar]
- 31.Choi, S. K., D. S. Olsen, A. Roll-Mecak, A. Martung, K. L. Remo, S. K. Burley, A. G. Hinnebusch, and T. E. Dever. 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 20:7183-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooperman, B. S., J. Dondon, J. Finelli, M. Grunberg-Manago, and A. M. Michelson. 1977. Photosensitized cross-linking of IF-3 to Escherichia coli 30 S subunits. FEBS Lett. 76:59-63. [DOI] [PubMed] [Google Scholar]
- 34.Cooperman, B. S., A. Expert-Bezancon, L. Kahan, J. Dondon, and M. Grunberg-Manago. 1981. IF-3 crosslinking to Escherichia coli ribosomal 30 S subunits by three different light-dependent procedures: identification of 30 S proteins crosslinked to IF-3-utilization of a new two-stage crosslinking reagent, p-nitrobenzylmaleimide. Arch. Biochem. Biophys. 208:554-562. [DOI] [PubMed] [Google Scholar]
- 35.Cousineau, B., F. Leclerc, and R. Cedergren. 1997. On the origin of protein synthesis factors: a gene duplication/fusion model. J. Mol. Evol. 45:661-670. [DOI] [PubMed] [Google Scholar]
- 36.Crawford, D. J., K. Ito, Y. Nakamura, and W. P. Tate. 1999. Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3. EMBO J. 18:727-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croitoru, V., M. Bucheli-Witschel, P. Hagg, F. Abdulkarim, and L. A. Isaksson. 2004. Generation and characterization of functional mutants in the translation initiation factor IF1 of Escherichia coli. Eur. J. Biochem. 271:534-544. [DOI] [PubMed] [Google Scholar]
- 38.Culver, G. M. 2001. Meanderings of the mRNA through the ribosome. Structure (Cambridge) 9:751-758. [DOI] [PubMed] [Google Scholar]
- 39.Cummings, H. S., and J. W. Hershey. 1994. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cummings, H. S., J. F. Sands, P. C. Foreman, J. Fraser, and J. W. Hershey. 1991. Structure and expression of the infA operon encoding translational initiation factor IF1. Transcriptional control by growth rate. J. Biol. Chem. 266:16491-16498. [PubMed] [Google Scholar]
- 41.Dahlquist, K. D., and J. D. Puglisi. 2000. Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J. Mol. Biol. 299:1-15. [DOI] [PubMed] [Google Scholar]
- 42.Dallas, A., and H. F. Noller. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8:855-864. [DOI] [PubMed] [Google Scholar]
- 43.de Cock, E., M. Springer, and F. Dardel. 1999. The interdomain linker of Escherichia coli initiation factor IF3: a possible trigger of translation initiation specificity. Mol. Microbiol. 32:193-202. [DOI] [PubMed] [Google Scholar]
- 44.Dennis, P. P. 1997. Ancient ciphers: translation in Archaea. Cell 89:1007-1010. [DOI] [PubMed] [Google Scholar]
- 45.de Smit, M. H., and J. van Duin. 1990. Control of prokaryotic translational initiation by mRNA secondary structure. Prog. Nucleic Acid Res. Mol. Biol. 38:1-35. [DOI] [PubMed] [Google Scholar]
- 46.Dottavio-Martin, D., D. P. Suttle, and J. M. Ravel. 1979. The effects of initiation factors IF-1 and IF-3 on the dissociation of Escherichia coli 70 S ribosomes. FEBS Lett. 97:105-110. [DOI] [PubMed] [Google Scholar]
- 47.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J. P. Ebel, and B. Ehresmann. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fortier, P. L., J. M. Schmitter, C. Garcia, and F. Dardel. 1994. The N-terminal half of initiation factor IF3 is folded as a stable independent domain. Biochimie 76:376-383. [DOI] [PubMed] [Google Scholar]
- 49.Frank, J. 2003. Electron microscopy of functional ribosome complexes. Biopolymers 68:223-233. [DOI] [PubMed] [Google Scholar]
- 50.Frank, J., J. Zhu, P. Penczek, Y. Li, S. Srivastava, A. Verschoor, M. Radermacher, R. Grassucci, R. K. Lata, and R. K. Agrawal. 1995. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature 376:441-444. [DOI] [PubMed] [Google Scholar]
- 51.Garcia, C., P. L. Fortier, S. Blanquet, J. Y. Lallemand, and F. Dardel. 1995. Solution structure of the ribosome-binding domain of E. coli translation initiation factor IF3. Homology with the U1A protein of the eukaryotic spliceosome. J. Mol. Biol. 254:247-259. [DOI] [PubMed] [Google Scholar]
- 52.Garret, R. A., S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller. 2000. The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 53.Giuliodori, A. M., A. Brandi, C. O. Gualerzi, and C. L. Pon. 2004. Preferential translation of cold-shock mRNAs during cold adaptation. RNA 10:265-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Govantes, F., E. Andujar, and E. Santero. 1998. Mechanism of translational coupling in the nifLA operon of Klebsiella pneumoniae. EMBO J. 17:2368-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green, R., and J. R. Lorsch. 2002. The path to perdition is paved with protons. Cell 110:665-668. [DOI] [PubMed] [Google Scholar]
- 56.Green, R., and H. F. Noller. 1997. Ribosomes and translation. Annu. Rev. Biochem. 66:679-716. [DOI] [PubMed] [Google Scholar]
- 57.Grill, S., C. O. Gualerzi, P. Londei, and U. Blasi. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 19:4101-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grill, S., I. Moll, D. Hasenohrl, C. O. Gualerzi, and U. Blasi. 2001. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett. 495:167-171. [DOI] [PubMed] [Google Scholar]
- 59.Grunberg-Manago, M., P. Dessen, D. Pantaloni, T. Godefroy-Colburn, A. D. Wolfe, and J. Dondon. 1975. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol. 94:461-478. [DOI] [PubMed] [Google Scholar]
- 60.Gualerzi, C., G. Risuleo, and C. L. Pon. 1977. Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry 16:1684-1689. [DOI] [PubMed] [Google Scholar]
- 61.Gualerzi, C. O., L. Brandi, E. Caserta, A. L. Teana, R. Spurio, J. Tomsic, and C. L. Pon. 2000. Translation initiation in bacteria, p. 477-494. In R. A. Garret, S. R. Douthwaite, A. Liljas, A. T. Matheson, P. B. Moore, and H. F. Noller (ed.), The ribosome: structure, function, antibiotics, and cellular interactions. ASM Press, Washington, D.C.
- 62.Gualerzi, C. O., A. M. Giuliodori, and C. L. Pon. 2003. Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331:527-539. [DOI] [PubMed] [Google Scholar]
- 63.Gualerzi, C. O., and C. L. Pon. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 29:5881-5889. [DOI] [PubMed] [Google Scholar]
- 64.Gualerzi, C. O., M. Severini, R. Spurio, A. La Teana, and C. L. Pon. 1991. Molecular dissection of translation initiation factor IF2. Evidence for two structural and functional domains. J. Biol. Chem. 266:16356-16362. [PubMed] [Google Scholar]
- 65.Gualerzi, C. O., R. Spurio, A. La Teana, R. Calogero, B. Celano, and C. L. Pon. 1989. Site-directed mutagenesis of Escherichia coli translation initiation factor IF1. Identification of the amino acid involved in its ribosomal binding and recycling. Protein Eng. 3:133-138. [DOI] [PubMed] [Google Scholar]
- 66.Guenneugues, M., E. Caserta, L. Brandi, R. Spurio, S. Meunier, C. L. Pon, R. Boelens, and C. O. Gualerzi. 2000. Mapping the fMet-tRNA(f)(Met) binding site of initiation factor IF2. EMBO J. 19:5233-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guillier, M., F. Allemand, S. Raibaud, F. Dardel, M. Springer, and C. Chiaruttini. 2002. Translational feedback regulation of the gene for L35 in Escherichia coli requires binding of ribosomal protein L20 to two sites in its leader mRNA: a possible case of ribosomal RNA-messenger RNA molecular mimicry. RNA 8:878-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guillon, J. M., S. Heiss, J. Soutourina, Y. Mechulam, S. Laalami, M. Grunberg-Manago, and S. Blanquet. 1996. Interplay of methionine tRNAs with translation elongation factor Tu and translation initiation factor 2 in Escherichia coli. J. Biol. Chem. 271:22321-22325. [DOI] [PubMed] [Google Scholar]
- 69.Guillon, J. M., Y. Mechulam, J. M. Schmitter, S. Blanquet, and G. Fayat. 1992. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol. 174:4294-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haggerty, T. J., and S. T. Lovett. 1997. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J. Bacteriol. 179:6705-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hansen, P. K., F. Wikman, B. F. Clark, J. W. Hershey, and H. Uffe Petersen. 1986. Interaction between initiator Met-tRNAfMet and elongation factor EF-Tu from E. coli. Biochimie 68:697-703. [DOI] [PubMed] [Google Scholar]
- 72.Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679-688. [DOI] [PubMed] [Google Scholar]
- 73.Hartz, D., J. Binkley, T. Hollingsworth, and L. Gold. 1990. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes Dev. 4:1790-1800. [DOI] [PubMed] [Google Scholar]
- 74.Hartz, D., D. S. McPheeters, and L. Gold. 1989. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev. 3:1899-1912. [DOI] [PubMed] [Google Scholar]
- 75.Hedegaard, J., M. Hauge, J. Fage-Larsen, K. K. Mortensen, M. Kilian, H. U. Sperling-Petersen, and K. Poulsen. 2000. Investigation of the translation-initiation factor IF2 gene, infB, as a tool to study the population structure of Streptococcus agalactiae. Microbiology 146:1661-1670. [DOI] [PubMed] [Google Scholar]
- 76.Hedegaard, J., H. Okkels, B. Bruun, M. Kilian, K. K. Mortensen, and N. Norskov-Lauritsen. 2001. Phylogeny of the genus Haemophilus as determined by comparison of partial infB sequences. Microbiology 147:2599-2609. [DOI] [PubMed] [Google Scholar]
- 77.Hedegaard, J., S. A. Steffensen, N. Norskov-Lauritsen, K. K. Mortensen, and H. U. Sperling-Petersen. 1999. Identification of Enterobacteriaceae by partial sequencing of the gene encoding translation initiation factor 2. Int. J. Syst. Bacteriol. 49:1531-1538. [DOI] [PubMed] [Google Scholar]
- 78.Hirokawa, G., M. C. Kiel, A. Muto, M. Selmer, V. S. Raj, A. Liljas, K. Igarashi, H. Kaji, and A. Kaji. 2002. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 21:2272-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Howe, J. G., and J. W. Hershey. 1983. Initiation factor and ribosome levels are coordinately controlled in Escherichia coli growing at different rates. J. Biol. Chem. 258:1954-1959. [PubMed] [Google Scholar]
- 80.Hua, Y., and D. P. Raleigh. 1998. On the global architecture of initiation factor IF3: a comparative study of the linker regions from the Escherichia coli protein and the Bacillus stearothermophilus protein. J. Mol. Biol. 278:871-878. [DOI] [PubMed] [Google Scholar]
- 81.Hubert, M., N. R. Nyengaard, K. Shazand, K. K. Mortensen, S. F. Lassen, M. Grunberg-Manago, and H. U. Sperling-Petersen. 1992. Tandem translation of Bacillus subtilis initiation factor IF2 in E. coli. Over-expression of infBB.su in E. coli and purification of alpha- and beta-forms of IF2B.su. FEBS Lett. 312:132-138. [DOI] [PubMed] [Google Scholar]
- 82.Huxley, H. E., and G. Zubay. 1960. Electron microscope observations on the structure of microsomal particles from Escherichia coli. J. Mol. Biol. 2:10-18. [Google Scholar]
- 83.Ikemura, T., and H. Ozeki. 1977. Gross map location of Escherichia coli transfer RNA genes. J. Mol. Biol. 117:419-446. [DOI] [PubMed] [Google Scholar]
- 84.Ishii, S., K. Kuroki, and F. Imamoto. 1984. tRNAMetf2 gene in the leader region of the nusA operon in Escherichia coli. Proc. Natl. Acad. Sci. USA 81:409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Janosi, L., S. Mottagui-Tabar, L. A. Isaksson, Y. Sekine, E. Ohtsubo, S. Zhang, S. Goon, S. Nelken, M. Shuda, and A. Kaji. 1998. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 17:1141-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jin, H., A. Bjornsson, and L. A. Isaksson. 2002. Cis control of gene expression in E. coli by ribosome queuing at an inefficient translational stop signal. EMBO J. 21:4357-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karimi, R., M. Y. Pavlov, R. H. Buckingham, and M. Ehrenberg. 1999. Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell 3:601-609. [DOI] [PubMed] [Google Scholar]
- 88.Katoh, E., T. Hatta, H. Shindo, Y. Ishii, H. Yamada, T. Mizuno, and T. Yamazaki. 2000. High precision NMR structure of YhhP, a novel Escherichia coli protein implicated in cell division. J. Mol. Biol. 304:219-229. [DOI] [PubMed] [Google Scholar]
- 89.Kennell, D., and H. Riezman. 1977. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol. 114:1-21. [DOI] [PubMed] [Google Scholar]
- 90.Kenri, T., F. Imamoto, and Y. Kano. 1994. Three tandemly repeated structural genes encoding tRNA(f1Met) in the metZ operon of Escherichia coli K-12. Gene 138:261-262. [DOI] [PubMed] [Google Scholar]
- 91.Kim, K. K., L. W. Hung, H. Yokota, R. Kim, and S. H. Kim. 1998. Crystal structures of eukaryotic translation initiation factor 5A from Methanococcus jannaschii at 1.8 A resolution. Proc. Natl. Acad. Sci. USA 95:10419-10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koc, E. C., and L. L. Spremulli. 2002. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J. Biol. Chem. 277:35541-35549. [DOI] [PubMed] [Google Scholar]
- 93.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:29-32, 51-55. [DOI] [PubMed] [Google Scholar]
- 94.Kozak, M. 1983. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 47:1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 96.Krafft, C., A. Diehl, S. Laettig, J. Behlke, U. Heinemann, C. L. Pon, C. O. Gualerzi, and H. Welfle. 2000. Interaction of fMet-tRNA(fMet) with the C-terminal domain of translational initiation factor IF2 from Bacillus stearothermophilus. FEBS Lett. 471:128-132. [DOI] [PubMed] [Google Scholar]
- 97.Kycia, J. H., V. Biou, F. Shu, S. E. Gerchman, V. Graziano, and V. Ramakrishnan. 1995. Prokaryotic translation initiation factor IF3 is an elongated protein consisting of two crystallizable domains. Biochemistry 34:6183-6187. [DOI] [PubMed] [Google Scholar]
- 98.Kyrpides, N. C., and C. R. Woese. 1998. Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. USA 95:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98a.Laalami, S., A. V. Timofeev, H. Putzer, J. Leautey, and M. Grunberg-Manago. 1994. In vivo study of engineered G-domain mutants of Escherichia coli translation initiation factor IF2. Mol. Microbiol. 11:293-302. [DOI] [PubMed] [Google Scholar]
- 99.Lake, J. A. 1976. Ribosome structure determined by electron microscopy of Escherichia coli small subunits, large subunits and monomeric ribosomes. J. Mol. Biol. 105:131-139. [DOI] [PubMed] [Google Scholar]
- 100.Larigauderie, G., S. Laalami, N. R. Nyengaard, M. Grunberg-Manago, Y. Cenatiempo, K. K. Mortensen, and H. U. Sperling-Petersen. 2000. Mutation of Thr445 and Ile500 of initiation factor 2 G-domain affects Escherichia coli growth rate at low temperature. Biochimie 82:1091-1098. [DOI] [PubMed] [Google Scholar]
- 101.La Teana, A., C. O. Gualerzi, and R. Brimacombe. 1995. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA 1:772-782. [PMC free article] [PubMed] [Google Scholar]
- 102.La Teana, A., C. O. Gualerzi, and A. E. Dahlberg. 2001. Initiation factor IF 2 binds to the alpha-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.La Teana, A., C. L. Pon, and C. O. Gualerzi. 1996. Late events in translation initiation. Adjustment of fMet-tRNA in the ribosomal P-site. J. Mol. Biol. 256:667-675. [DOI] [PubMed] [Google Scholar]
- 104.Laursen, B. S., A. C. Kjærgaard, K. K. Mortensen, D. W. Hoffman, and H. U. Sperling-Petersen. 2004. The N-terminal domain (IF2N) of bacterial translation initiation factor IF2 is connected to the conserved C-terminal domains by a flexible linker. Protein Sci. 13:230-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Laursen, B. S., K. K. Mortensen, H. U. Sperling-Petersen, and D. W. Hoffman. 2003. A conserved structural motif at the N terminus of bacterial translation initiation factor IF2. J. Biol. Chem. 278:16320-16328. [DOI] [PubMed] [Google Scholar]
- 106.Laursen, B. S., I. Siwanowicz, G. Larigauderie, J. Hedegaard, K. Ito, Y. Nakamura, K. K. Mortensen, and H. U. Sperling-Petersen. 2002. Characterization of mutations in the GTP-binding domain of IF2 resulting in cold-sensitive growth of Escherichia coli. J. Mol. Biol. 326:543-551. [DOI] [PubMed] [Google Scholar]
- 107.Laursen, B. S., S. A. Steffensen, J. Hedegaard, J. M. Moreno, K. K. Mortensen, and H. U. Sperling-Petersen. 2002. Structural requirements of the mRNA for intracistronic translation initiation of the enterobacterial infB gene. Genes Cells 7:901-910. [DOI] [PubMed] [Google Scholar]
- 108.Lelong, J. C., M. Grunberg-Manago, J. Dondon, D. Gros, and F. Gros. 1970. Interaction between guanosine derivatives and factors involved in the initiation of protein synthesis. Nature 226:505-510. [DOI] [PubMed] [Google Scholar]
- 109.Lesage, P., C. Chiaruttini, M. Graffe, J. Dondon, M. Milet, and M. Springer. 1992. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J. Mol. Biol. 228:366-386. [DOI] [PubMed] [Google Scholar]
- 110.Lesage, P., H. N. Truong, M. Graffe, J. Dondon, and M. Springer. 1990. Translated translational operator in Escherichia coli. Auto-regulation in the infC-rpmI-rplT operon. J. Mol. Biol. 213:465-475. [DOI] [PubMed] [Google Scholar]
- 111.Li, W., and D. W. Hoffman. 2001. Structure and dynamics of translation initiation factor aIF-1A from the archaeon Methanococcus jannaschii determined by NMR spectroscopy. Protein Sci. 10:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin, Q., N. J. Yu, and L. L. Spremulli. 1996. Expression and functional analysis of Euglena gracilis chloroplast initiation factor 3. Plant Mol. Biol. 32:937-945. [DOI] [PubMed] [Google Scholar]
- 113.Lockwood, A. H., P. R. Chakraborty, and U. Maitra. 1971. A complex between initiation factor IF2, guanosine triphosphate, and fMet-tRNA: an intermediate in initiation complex formation. Proc. Natl. Acad. Sci. USA 68:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lockwood, A. H., P. Sarkar, and U. Maitra. 1972. Release of polypeptide chain initiation factor IF-2 during initiation complex formation. Proc. Natl. Acad. Sci. USA 69:3602-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luchin, S., H. Putzer, J. W. Hershey, Y. Cenatiempo, M. Grunberg-Manago, and S. Laalami. 1999. In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J. Biol. Chem. 274:6074-6079. [DOI] [PubMed] [Google Scholar]
- 116.Reference deleted.
- 117.MacKeen, L. A., L. Kahan, A. J. Wahba, and I. Schwartz. 1980. Photochemical cross-linking of initiation factor-3 to Escherichia coli 30 S ribosomal subunits. J. Biol. Chem. 255:10526-10531. [PubMed] [Google Scholar]
- 118.Majumdar, A., K. K. Bose, and N. K. Gupta. 1976. Specific binding of Escherichia coli chain initiation factor 2 to fMet-tRNAfMet. J. Biol. Chem. 251:137-140. [PubMed] [Google Scholar]
- 119.Mangroo, D., X. Q. Wu, and U. L. RajBhandary. 1995. Escherichia coli initiator tRNA: structure-function relationships and interactions with the translational machinery. Biochem. Cell Biol. 73:1023-1031. [DOI] [PubMed] [Google Scholar]
- 120.Marintchev, A., V. G. Kolupaeva, T. V. Pestova, and G. Wagner. 2003. Mapping the binding interface between human eukaryotic initiation factors IA and 5B: a new interaction between old partners. Proc. Natl. Acad. Sci. USA 100:1535-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marzi, S., W. Knight, L. Brandi, E. Caserta, N. Soboleva, W. E. Hill, C. O. Gualerzi, and J. S. Lodmell. 2003. Ribosomal localization of translation initiation factor IF2. RNA 9:958-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mayer, C., C. Kohrer, E. Kenny, C. Prusko, and U. L. RajBhandary. 2003. Anticodon sequence mutants of Escherichia coli initiator tRNA: effects of overproduction of aminoacyl-tRNA synthetases, methionyl-tRNA formyltransferase, and initiation factor 2 on activity in initiation. Biochemistry 42:4787-4799. [DOI] [PubMed] [Google Scholar]
- 123.McCarthy, J. E., and R. Brimacombe. 1994. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 10:402-407. [DOI] [PubMed] [Google Scholar]
- 124.McCutcheon, J. P., R. K. Agrawal, S. M. Philips, R. A. Grassucci, S. E. Gerchman, W. M. Clemons, Jr., V. Ramakrishnan, and J. Frank. 1999. Location of translational initiation factor IF3 on the small ribosomal subunit. Proc. Natl. Acad. Sci. USA 96:4301-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mechulam, Y., E. Schmitt, L. Maveyraud, C. Zelwer, O. Nureki, S. Yokoyama, M. Konno, and S. Blanquet. 1999. Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J. Mol. Biol. 294:1287-1297. [DOI] [PubMed] [Google Scholar]
- 126.Meinnel, T., J. M. Guillon, Y. Mechulam, and S. Blanquet. 1993. The Escherichia coli fmt gene, encoding methionyl-tRNA(fMet) formyltransferase, escapes metabolic control. J. Bacteriol. 175:993-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Meinnel, T., C. Sacerdot, M. Graffe, S. Blanquet, and M. Springer. 1999. Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol. 290:825-837. [DOI] [PubMed] [Google Scholar]
- 128.Meunier, S., R. Spurio, M. Czisch, R. Wechselberger, M. Guenneugues, C. O. Gualerzi, and R. Boelens. 2000. Structure of the fMet-tRNA(fMet)-binding domain of B. stearothermophilus initiation factor IF2. EMBO J. 19:1918-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miranda-Rios, J., M. Navarro, and M. Soberon. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. USA 98:9736-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Misselwitz, R., K. Welfle, C. Krafft, H. Welfle, L. Brandi, E. Caserta, and C. O. Gualerzi. 1999. The fMet-tRNA binding domain of translational initiation factor IF2: role and environment of its two Cys residues. FEBS Lett. 459:332-336. [DOI] [PubMed] [Google Scholar]
- 131.Moazed, D., J. M. Robertson, and H. F. Noller. 1988. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature 334:362-364. [DOI] [PubMed] [Google Scholar]
- 132.Moazed, D., R. R. Samaha, C. Gualerzi, and H. F. Noller. 1995. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 248:207-210. [DOI] [PubMed] [Google Scholar]
- 133.Moll, I., S. Grill, C. O. Gualerzi, and U. Blasi. 2002. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 43:239-246. [DOI] [PubMed] [Google Scholar]
- 134.Moll, I., M. Huber, S. Grill, P. Sairafi, F. Mueller, R. Brimacombe, P. Londei, and U. Blasi. 2001. Evidence against an interaction between the mRNA downstream box and 16S rRNA in translation initiation. J. Bacteriol. 183:3499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moreau, M., E. de Cock, P. L. Fortier, C. Garcia, C. Albaret, S. Blanquet, J. Y. Lallemand, and F. Dardel. 1997. Heteronuclear NMR studies of E. coli translation initiation factor IF3. Evidence that the inter-domain region is disordered in solution. J. Mol. Biol. 266:15-22. [DOI] [PubMed] [Google Scholar]
- 136.Moreno, J. M., L. Drskjotersen, J. E. Kristensen, K. K. Mortensen, and H. U. Sperling-Petersen. 1999. Characterization of the domains of E. coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Lett. 455:130-134. [DOI] [PubMed] [Google Scholar]
- 137.Moreno, J. M., J. Kildsgaard, I. Siwanowicz, K. K. Mortensen, and H. U. Sperling-Petersen. 1998. Binding of Escherichia coli initiation factor IF2 to 30S ribosomal subunits: a functional role for the N-terminus of the factor. Biochem. Biophys. Res. Commun. 252:465-471. [DOI] [PubMed] [Google Scholar]
- 138.Moreno, J. M., H. P. Sørensen, K. K. Mortensen, and H. U. Sperling-Petersen. 2000. Macromolecular mimicry in translation initiation: a model for the initiation factor IF2 on the ribosome. IUBMB Life 50:347-354. [DOI] [PubMed] [Google Scholar]
- 139.Mortensen, K. K., E. Hajnsdorf, P. Regnier, and H. U. Sperling-Petersen. 1995. Improved recombinant tandem expression of translation initiation factor IF2 in RNASE E deficient E. coli cells. Biochem. Biophys. Res. Commun. 214:1254-1259. [DOI] [PubMed] [Google Scholar]
- 140.Mortensen, K. K., J. Kildsgaard, J. M. Moreno, S. A. Steffensen, J. Egebjerg, and H. U. Sperling-Petersen. 1998. A six-domain structural model for Escherichia coli translation initiation factor IF2. Characterisation of twelve surface epitopes. Biochem. Mol. Biol. Int. 46:1027-1041. [DOI] [PubMed] [Google Scholar]
- 141.Muralikrishna, P., and E. Wickstrom. 1989. Escherichia coli initiation factor 3 protein binding to 30S ribosomal subunits alters the accessibility of nucleotides within the conserved central region of 16S rRNA. Biochemistry 28:7505-7510. [DOI] [PubMed] [Google Scholar]
- 142.Nahvi, A., N. Sudarsan, M. S. Ebert, X. Zou, K. L. Brown, and R. R. Breaker. 2002. Genetic control by a metabolite binding mRNA. Chem. Biol. 9:1043. [DOI] [PubMed] [Google Scholar]
- 143.Nakamura, Y., J. Plumbridge, J. Dondon, and M. Grunberg-Manago. 1985. Evidence for autoregulation of the nusA-infB operon of Escherichia coli. Gene 36:189-193. [DOI] [PubMed] [Google Scholar]
- 144.Newton, D. T., C. Creuzenet, and D. Mangroo. 1999. Formylation is not essential for initiation of protein synthesis in all eubacteria. J. Biol. Chem. 274:2214-22146. [DOI] [PubMed] [Google Scholar]
- 145.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 146.Nissen, P., J. A. Ippolito, N. Ban, P. B. Moore, and T. A. Steitz. 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. USA 98:4899-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nissen, P., M. Kjeldgaard, S. Thirup, G. Polekhina, L. Reshetnikova, B. F. Clark, and J. Nyborg. 1995. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464-1472. [DOI] [PubMed] [Google Scholar]
- 148.Nogueira, T., and M. Springer. 2000. Post-transcriptional control by global regulators of gene expression in bacteria. Curr. Opin. Microbiol 3:154-158. [DOI] [PubMed] [Google Scholar]
- 149.Noller, H. F., J. Kop, V. Wheaton, J. Brosius, R. R. Gutell, A. M. Kopylov, F. Dohme, W. Herr, D. A. Stahl, R. Gupta, and C. R. Waese. 1981. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 9:6167-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Noller, H. F., M. M. Yusupov, G. Z. Yusupova, A. Baucom, and J. H. Cate. 2002. Translocation of tRNA during protein synthesis. FEBS Lett. 514:11-16. [DOI] [PubMed] [Google Scholar]
- 151.Nonato, M. C., J. Widom, and J. Clardy. 2002. Crystal structure of the N-terminal segment of human eukaryotic translation initiation factor 2alpha. J. Biol. Chem. 277:17057-17061. [DOI] [PubMed] [Google Scholar]
- 152.Norskov-Lauritsen, N., D. Sandvang, J. Hedegaard, V. Fussing, K. K. Mortensen, H. U. Sperling-Petersen, and H. C. Schonheyder. 2001. Clonal origin of aminoglycoside-resistant Citrobacter freundii isolates in a Danish county. J. Med. Microbiol. 50:636-641. [DOI] [PubMed] [Google Scholar]
- 153.Nudler, E., and A. S. Mironov. 2004. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29:11-17. [DOI] [PubMed] [Google Scholar]
- 154.Nyengaard, N. R., K. K. Mortensen, S. F. Lassen, J. W. Hershey, and H. U. Sperling-Petersen. 1991. Tandem translation of E. coli initiation factor IF2 beta: purification and characterization in vitro of two active forms. Biochem. Biophys. Res. Commun. 181:1572-1579. [DOI] [PubMed] [Google Scholar]
- 155.O'Connor, M., T. Asai, C. L. Squires, and A. E. Dahlberg. 1999. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl. Acad. Sci. USA 96:8973-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ogle, J. M., D. E. Brodersen, W. M. Clemons, Jr., M. J. Tarry, A. P. Carter, and V. Ramakrishnan. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897-902. [DOI] [PubMed] [Google Scholar]
- 157.Ogle, J. M., A. P. Carter, and V. Ramakrishnan. 2003. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 28:259-266. [DOI] [PubMed] [Google Scholar]
- 158.Ogle, J. M., F. V. Murphy, M. J. Tarry, and V. Ramakrishnan. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111:721-732. [DOI] [PubMed] [Google Scholar]
- 159.Olsen, D. S., E. M. Savner, A. Mathew, F. Zhang, T. Krishnamoorthy, L. Phan, and A. G. Hinnebusch. 2003. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 22:193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Olsson, C. L., M. Graffe, M. Springer, and J. W. Hershey. 1996. Physiological effects of translation initiation factor IF3 and ribosomal protein L20 limitation in Escherichia coli. Mol. Gen. Genet. 250:705-714. [DOI] [PubMed] [Google Scholar]
- 161.Olsthoorn, R. C., S. Zoog, and J. van Duin. 1995. Coevolution of RNA helix stability and Shine-Dalgarno complementarity in a translational start region. Mol. Microbiol. 15:333-339. [DOI] [PubMed] [Google Scholar]
- 162.Pape, T., W. Wintermeyer, and M. Rodnina. 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18:3800-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parmeggiani, A., G. W. Swart, K. K. Mortensen, M. Jensen, B. F. Clark, L. Dente, and R. Cortese. 1987. Properties of a genetically engineered G domain of elongation factor Tu. Proc. Natl. Acad. Sci. USA 84:3141-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pestova, T. V., and C. U. Hellen. 2000. The structure and function of initiation factors in eukaryotic protein synthesis. Cell Mol. Life Sci 57:651-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pestova, T. V., I. B. Lomakin, J. H. Lee, S. K. Choi, T. E. Dever, and C. U. Hellen. 2000. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403:332-335. [DOI] [PubMed] [Google Scholar]
- 166.Petersen, H. U., T. A. Kruse, H. Worm-Leonhard, G. E. Siboska, B. F. Clark, A. S. Boutorin, P. Remy, J. P. Ebel, J. Dondon, and M. Grunberg-Manago. 1981. Study of the interaction of Escherichia coli initiation factor IF2 with formylmethionyl-tRNAMetf by partial digestion with cobra venom ribonuclease. FEBS Lett. 128:161-165. [DOI] [PubMed] [Google Scholar]
- 167.Petersen, H. U., T. Roll, M. Grunberg-Manago, and B. F. Clark. 1979. Specific interaction of initiation factor IF2 of E. coli with formylmethionyl-tRNAfMet Biochem. Biophys. Res Commun. 91:1068-1074. [DOI] [PubMed] [Google Scholar]
- 168.Petrelli, D., C. Garofalo, M. Lammi, R. Spurio, C. L. Pon, C. O. Gualerzi, and A. La Teana. 2003. Mapping the active sites of bacterial translation initiation factor IF3. J. Mol. Biol. 331:541-556. [DOI] [PubMed] [Google Scholar]
- 169.Petrelli, D., A. LaTeana, C. Garofalo, R. Spurio, C. L. Pon, and C. O. Gualerzi. 2001. Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J. 20:4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Pioletti, M., F. Schlunzen, J. Harms, R. Zarivach, M. Gluhmann, H. Avila, A. Bashan, H. Bartels, T. Auerbach, C. Jacobi, T. Hartsch, A. Yonath, and F. Franceschi. 2001. Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J. 20:1829-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Plumbridge, J. A., J. Dondon, Y. Nakamura, and M. Grunberg-Manago. 1985. Effect of NusA protein on expression of the nusA, infB operon in E. coli. Nucleic Acids Res. 13:3371-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Plumbridge, J. A., J. G. Howe, M. Springer, D. Touati-Schwartz, J. W. Hershey, and M. Grunberg-Manago. 1982. Cloning and mapping of a gene for translational initiation factor IF2 in Escherichia coli. Proc. Natl. Acad. Sci. USA 79:5033-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Plumbridge, J. A., and M. Springer. 1983. Organization of the Escherichia coli chromosome around the genes for translation initiation factor IF2 (infB) and a transcription termination factor (nusA). J. Mol. Biol. 167:227-243. [DOI] [PubMed] [Google Scholar]
- 174.Pon, C. L., and C. O. Gualerzi. 1984. Mechanism of protein biosynthesis in prokaryotic cells. Effect of initiation factor IF1 on the initial rate of 30 S initiation complex formation. FEBS Lett. 175:203-207. [DOI] [PubMed] [Google Scholar]
- 175.Pon, C. L., M. Paci, R. T. Pawlik, and C. O. Gualerzi. 1985. Structure-function relationship in Escherichia coli initiation factors. Biochemical and biophysical characterization of the interaction between IF-2 and guanosine nucleotides. J. Biol. Chem. 260:8918-8924. [PubMed] [Google Scholar]
- 176.Poot, R. A., N. V. Tsareva, I. V. Boni, and J. van Duin. 1997. RNA folding kinetics regulates translation of phage MS2 maturation gene. Proc. Natl. Acad. Sci. USA 94:10110-10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Preiss, T., and M. W. Hentze. 2003. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays 25:1201-1211. [DOI] [PubMed] [Google Scholar]
- 178.RajBhandary, U. L. 1994. Initiator transfer RNAs. J. Bacteriol. 176:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rajbhandary, U. L., and C. Ming Chow. 1995. Initiator tRNAs and initiation of protein synthesis, p. 511-528. In D. Söll and U. L. Rajbhandary (ed.), tRNA: structure, biosynthesis, and function. American Soceity for Microbiology, Washington, D.C.
- 180.Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell 108:557-572. [DOI] [PubMed] [Google Scholar]
- 181.Ramesh, V., C. Mayer, M. R. Dyson, S. Gite, and U. L. RajBhandary. 1999. Induced fit of a peptide loop of methionyl-tRNA formyltransferase triggered by the initiator tRNA substrate. Proc. Natl. Acad. Sci. USA 96:875-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Richman, N., and J. W. Bodley. 1972. Ribosomes cannot interact simultaneously with elongation factors EF Tu and EF G. Proc. Natl. Acad. Sci. USA 69:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ringquist, S., S. Shinedling, D. Barrick, L. Green, J. Binkley, G. D. Stormo, and L. Gold. 1992. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6:1219-1229. [DOI] [PubMed] [Google Scholar]
- 184.Rodnina, M. V., H. Stark, A. Savelsbergh, H. J. Wieden, D. Mohr, N. B. Matassova, F. Peske, T. Daviter, C. O. Gualerzi, and W. Wintermeyer. 2000. GTPases mechanisms and functions of translation factors on the ribosome. Biol. Chem. 381:377-387. [DOI] [PubMed] [Google Scholar]
- 185.Roll-Mecak, A., C. Cao, T. E. Dever, and S. K. Burley. 2000. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103:781-792. [DOI] [PubMed] [Google Scholar]
- 186.Roll-Mecak, A., B. S. Shin, T. E. Dever, and S. K. Burley. 2001. Engaging the ribosome: universal IFs of translation. Trends Biochem. Sci. 26:705-709. [DOI] [PubMed] [Google Scholar]
- 187.Ruff, M., S. Krishnaswamy, M. Boeglin, A. Poterszman, A. Mitschler, A. Podjarny, B. Rees, J. C. Thierry, and D. Moras. 1991. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science 252:1682-1689. [DOI] [PubMed] [Google Scholar]
- 188.Sacerdot, C., C. Chiaruttini, K. Engst, M. Graffe, M. Milet, N. Mathy, J. Dondon, and M. Springer. 1996. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 21:331-346. [DOI] [PubMed] [Google Scholar]
- 189.Sacerdot, C., P. Dessen, J. W. Hershey, J. A. Plumbridge, and M. Grunberg-Manago. 1984. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc. Natl. Acad. Sci. USA 81:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sacerdot, C., G. Fayat, P. Dessen, M. Springer, J. A. Plumbridge, M. Grunberg-Manago, and S. Blanquet. 1982. Sequence of a 1:26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J. 1:311-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sacerdot, C., G. Vachon, S. Laalami, F. Morel-Deville, Y. Cenatiempo, and M. Grunberg-Manago. 1992. Both forms of translational initiation factor IF2 (alpha and beta) are required for maximal growth of Escherichia coli. Evidence for two translational initiation codons for IF2 beta. J. Mol. Biol. 225:67-80. [DOI] [PubMed] [Google Scholar]
- 192.Saito, K., L. C. Mattheakis, and M. Nomura. 1994. Post-transcriptional regulation of the str operon in Escherichia coli. Ribosomal protein S7 inhibits coupled translation of S7 but not its independent translation. J. Mol. Biol. 235:111-124. [DOI] [PubMed] [Google Scholar]
- 193.Sands, J. F., P. Regnier, H. S. Cummings, M. Grunberg-Manago, and J. W. Hershey. 1988. The existence of two genes between infB and rpsO in the Escherichia coli genome: DNA sequencing and S1 nuclease mapping. Nucleic Acids Res. 16:10803-10816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schlax, P. J., and D. J. Worhunsky. 2003. Translational repression mechanisms in prokaryotes. Mol. Microbiol. 48:1157-1169. [DOI] [PubMed] [Google Scholar]
- 195.Schluenzen, F., A. Tocilj, R. Zarivach, J. Harms, M. Gluehmann, D. Janell, A. Bashan, H. Bartels, I. Agmon, F. Franceschi, and A. Yonath. 2000. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell 102:615-623. [DOI] [PubMed] [Google Scholar]
- 196.Schmeing, T. M., P. B. Moore, and T. A. Steitz. 2003. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schmitt, E., M. Panvert, S. Blanquet, and Y. Mechulam. 1998. Crystal structure of methionyl-tRNAfMet transformylase complexed with the initiator formyl-methionyl-tRNAfMet. EMBO J. 17:6819-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Schneider, T. D., G. D. Stormo, L. Gold; and A. Ehrenfeucht. 1986. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188:415-431. [DOI] [PubMed] [Google Scholar]
- 199.Schweisguth, D. C., and P. B. Moore. 1997. On the conformation of the anticodon loops of initiator and elongator methionine tRNAs. J. Mol. Biol. 267:505-519. [DOI] [PubMed] [Google Scholar]
- 200.Sengupta, J., R. K. Agrawal, and J. Frank. 2001. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. USA 98:11991-11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Seong, B. L., and U. L. RajBhandary. 1987. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc. Natl. Acad. Sci. USA 84:334-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Seong, B. L., and U. L. RajBhandary. 1987. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc. Natl. Acad. Sci. USA 84:8859-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sette, M., R. Spurio, P. van Tilborg, C. O. Gualerzi, and R. Boelens. 1999. Identification of the ribosome binding sites of translation initiation factor IF3 by multidimensional heteronuclear NMR spectroscopy. RNA 5:82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sette, M., P. van Tilborg, R. Spurio, R. Kaptein, M. Paci, C. O. Gualerzi, and R. Boelens. 1997. The structure of the translational initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Severini, M., T. Choli, A. La Teana, and C. O. Gualerzi. 1992. Proteolysis of Bacillus stearothermophilus IF2 and specific protection by fMet-tRNA. FEBS Lett. 297:226-228. [DOI] [PubMed] [Google Scholar]
- 206.Shatsky, I. N., A. V. Bakin, A. A. Bogdanov, and V. D. Vasiliev. 1991. How does the mRNA pass through the ribosome? Biochimie. 73:937-945. [DOI] [PubMed] [Google Scholar]
- 207.Shine, J., and L. Dalgarno. 1974. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 71:1342-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sonenberg, N., J. W. Hershey, and M. B. Mathews. 2000. Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 209.Sørensen, H. P., J. Hedegaard, H. U. Sperling-Petersen, and K. K. Mortensen. 2001. Remarkable conservation of translation initiation factors: IF1/eIF1A and IF2/eIF5B are universally distributed phylogenetic markers. IUBMB Life 51:321-327. [DOI] [PubMed] [Google Scholar]
- 210.Sørensen, H. P., B. S. Laursen, K. K. Mortensen, and H. U. Sperling-Petersen. 2002. Bacterial translation initiation—mechanism and regulation. Recent Res. Dev. Biophys. Biochem. 2:243-270. [Google Scholar]
- 211.Sørensen, H. P., H. Sperling-Petersen, and K. K. Mortensen. 2003. A favorable solubility partner for the recombinant expression of streptavidin. Protein Exp. Purif. 32:252-259. [DOI] [PubMed] [Google Scholar]
- 212.Sprengart, M. L., H. P. Fatscher, and E. Fuchs. 1990. The initiation of translation in E. coli: apparent base pairing between the 16s rRNA and downstream sequences of the mRNA. Nucleic Acids Res. 18:1719-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sprengart, M. L., E. Fuchs, and A. G. Porter. 1996. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 15:665-674. [PMC free article] [PubMed] [Google Scholar]
- 214.Springer, M., C. Portier, and M. Grunberg-Manago. 1998. RNA mimicry in the translational apparatus, p. 377-413. In R. W. Simons and M. Grunberg-Manago (ed.), RNA structure and function. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 215.Spurio, R., L. Brandi, E. Caserta, C. L. Pon, C. O. Gualerzi, R. Misselwitz, C. Krafft, K. Welfle, and H. Welfle. 2000. The C-terminal subdomain (IF2 C-2) contains the entire fMet-tRNA binding site of initiation factor IF2. J. Biol. Chem. 275:2447-2454. [DOI] [PubMed] [Google Scholar]
- 216.Spurio, R., M. Paci, R. T. Pawlik, A. La Teana, B. V. DiGiacco, C. L. Pon, and C. O. Gualerzi. 1991. Site-directed mutagenesis and NMR spectroscopic approaches to the elucidation of the structure-function relationships in translation initiation factors IF1 and IF3. Biochimie 73:1001-1006. [DOI] [PubMed] [Google Scholar]
- 217.Steffensen, S. A., A. B. Poulsen, K. K. Mortensen, and H. U. Sperling-Petersen. 1997. E. coli translation initiation factor IF2—an extremely conserved protein. Comparative sequence analysis of the infB gene in clinical isolates of E. coli. FEBS Lett. 419:281-284. [DOI] [PubMed] [Google Scholar]
- 218.Steitz, J. A. 1969. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature 224:957-964. [DOI] [PubMed] [Google Scholar]
- 219.Steitz, T. A., and P. B. Moore. 2003. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem. Sci. 28:411-418. [DOI] [PubMed] [Google Scholar]
- 220.Stoffler, G., and M. Stoffler-Meilicke. 1984. Immunoelectron microscopy of ribosomes. Annu. Rev. Biophys. Bioeng. 13:303-330. [DOI] [PubMed] [Google Scholar]
- 221.Stormo, G. D., and Y. Ji. 2001. Do mRNAs act as direct sensors of small molecules to control their expression? Proc. Natl. Acad. Sci. USA 98:9465-9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Stringer, E. A., P. Sarkar, and U. Maitra. 1977. Function of initiation factor 1 in the binding and release of initiation factor 2 from ribosomal initiation complexes in Escherichia coli. J. Biol. Chem. 252:1739-1744. [PubMed] [Google Scholar]
- 223.Sundari, R. M., E. A. Stringer, L. H. Schulman, and U. Maitra. 1976. Interaction of bacterial initiation factor 2 with initiator tRNA. J. Biol. Chem. 251:3338-3345. [PubMed] [Google Scholar]
- 224.Sussman, J. K., E. L. Simons, and R. W. Simons. 1996. Escherichia coli translation initiation factor 3 discriminates the initiation codon in vivo. Mol. Microbiol. 21:347-360. [DOI] [PubMed] [Google Scholar]
- 225.Szkaradkiewicz, K., T. Zuleeg, S. Limmer, and M. Sprinzl. 2000. Interaction of fMet-tRNAfMet and fMet-AMP with the C-terminal domain of Thermus thermophilus translation initiation factor 2. Eur. J. Biochem. 267:4290-4299. [DOI] [PubMed] [Google Scholar]
- 226.Takahashi, S., S. Detrick, A. A. Whiting, A. J. Blaschke-Bonkowksy, Y. Aoyagi, E. E. Adderson, and J. F. Bohnsack. 2002. Correlation of phylogenetic lineages of group B Streptococci, identified by analysis of restriction-digestion patterns of genomic DNA, with infB alleles and mobile genetic elements. J. Infect. Dis. 186:1034-1038. [DOI] [PubMed] [Google Scholar]
- 227.Tedin, K., I. Moll, S. Grill, A. Resch, A. Graschopf, C. O. Gualerzi, and U. Blasi. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31:67-77. [DOI] [PubMed] [Google Scholar]
- 228.Thanedar, S., N. V. Kumar, and U. Varshney. 2000. The fate of the initiator tRNAs is sensitive to the critical balance between interacting proteins. J. Biol. Chem. 275:20361-20367. [DOI] [PubMed] [Google Scholar]
- 229.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. Bioessays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 230.Tiennault-Desbordes, E., Y. Cenatiempo, and S. Laalami. 2001. Initiation factor 2 of Myxococcus xanthus, a large version of prokaryotic translation initiation factor 2. J. Bacteriol. 183:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tomsic, J., L. A. Vitali, T. Daviter, A. Savelsbergh, R. Spurio, P. Striebeck, W. Wintermeyer, M. V. Rodnina, and C. O. Gualerzi. 2000. Late events of translation initiation in bacteria: a kinetic analysis. EMBO J. 19:2127-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tzareva, N. V., V. I. Makhno, and I. V. Boni. 1994. Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337:189-194. [DOI] [PubMed] [Google Scholar]
- 233.Udagawa, T., Y. Shimizu, and T. Ueda. 2004. Evidence for the translation initiation of leaderless mRNAs by the intact 70 S ribosome without its dissociation into subunits in eubacteria. J. Biol. Chem. 279:8539-8546. [DOI] [PubMed] [Google Scholar]
- 234.Van Etten, W. J., and G. R. Janssen. 1998. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol. Microbiol. 27:987-1001. [DOI] [PubMed] [Google Scholar]
- 235.Varshney, U., C. P. Lee, and U. L. RajBhandary. 1993. From elongator tRNA to initiator tRNA. Proc. Natl. Acad. Sci. USA 90:2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Varshney, U., and U. L. RajBhandary. 1992. Role of methionine and formylation of initiator tRNA in initiation of protein synthesis in Escherichia coli. J. Bacteriol. 174:7819-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]
- 238.Vetter, I. R., and A. Wittinghofer. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294:1299-1304. [DOI] [PubMed] [Google Scholar]
- 239.Vila-Sanjurjo, A., W. K. Ridgeway, V. Seymaner, W. Zhang, S. Santoso, K. Yu, and J. H. Cate. 2003. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl. Acad. Sci. USA 100:8682-8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2004. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 20:44-50. [DOI] [PubMed] [Google Scholar]
- 241.Vornlocher, H. P., R. Kreutzer, and M. Sprinzl. 1997. Organization of the Thermus thermophilus nusA/infB operon and overexpression of the infB gene in Escherichia coli. Biochimie 79:195-203. [DOI] [PubMed] [Google Scholar]
- 242.Wakao, H., P. Romby, J. P. Ebel, M. Grunberg-Manago, C. Ehresmann, and B. Ehresmann. 1991. Topography of the Escherichia coli ribosomal 30S subunit-initiation factor 2 complex. Biochimie 73:991-1000. [DOI] [PubMed] [Google Scholar]
- 243.Wakao, H., P. Romby, E. Westhof, S. Laalami, M. Grunberg-Manago, J. P. Ebel, C. Ehresmann, and B. Ehresmann. 1989. The solution structure of the Escherichia coli initiator tRNA and its interactions with initiation factor 2 and the ribosomal 30 S subunit. J. Biol. Chem. 264:20363-20371. [PubMed] [Google Scholar]
- 244.Weber, M. H., C. L. Beckering, and M. A. Marahiel. 2001. Complementation of cold shock proteins by translation initiation factor IF1 in vivo. J. Bacteriol. 183:7381-7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wertheimer, S. J., R. A. Klotsky, and I. Schwartz. 1988. Transcriptional patterns for the thrS-infC-rplT operon of Escherichia coli. Gene 63:309-320. [DOI] [PubMed] [Google Scholar]
- 246.Wickstrom, E., H. A. Heus, C. A. Haasnoot, and P. H. van Knippenberg. 1986. Circular dichroism and 500-MHz proton magnetic resonance studies of the interaction of Escherichia coli translational initiation factor 3 protein with the 16S ribosomal RNA 3′ cloacin fragment. Biochemistry 25:2770-2777. [DOI] [PubMed] [Google Scholar]
- 247.Wilson, D. N., G. Blaha, S. R. Connell, P. V. Ivanov, H. Jenke, U. Stelzl, Y. Teraoka, and K. H. Nierhaus. 2002. Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Pept. Sci. 3:1-53. [DOI] [PubMed] [Google Scholar]
- 248.Wilson, D. N., and K. H. Nierhaus. 2003. The ribosome through the looking glass. Angew. Chem. Int. Ed. Engl. 42:3464-3486. [DOI] [PubMed] [Google Scholar]
- 249.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 250.Wintermeyer, W., and C. Gualerzi. 1983. Effect of Escherichia coli initiation factors on the kinetics of N-Acphe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry 22:690-694. [DOI] [PubMed] [Google Scholar]
- 251.Wu, X. Q., and U. L. RajBhandary. 1997. Effect of the amino acid attached to Escherichia coli initiator tRNA on its affinity for the initiation factor IF2 and on the IF2 dependence of its binding to the ribosome. J. Biol. Chem. 272:1891-1895. [DOI] [PubMed] [Google Scholar]
- 252.Xia, B., H. Ke, W. Jiang, and M. Inouye. 2002. The Cold Box stem-loop proximal to the 5′-end of the Escherichia coli cspA gene stabilizes its mRNA at low temperature. J. Biol. Chem. 277:6005-6011. [DOI] [PubMed] [Google Scholar]
- 253.Yonath, A. 2002. The search and its outcome: high-resolution structures of ribosomal particles from mesophilic, thermophilic, and halophilic bacteria at various functional states. Annu. Rev. Biophys. Biomol. Struct. 31:257-273. [DOI] [PubMed] [Google Scholar]
- 254.Yu, N. J., and L. L. Spremulli. 1998. Regulation of the activity of chloroplast translational initiation factor 3 by NH2- and COOH-terminal extensions. J. Biol. Chem. 273:3871-3877. [DOI] [PubMed] [Google Scholar]
- 255.Yu, N. J., and L. L. Spremulli. 1997. Structural and mechanistic studies on chloroplast translational initiation factor 3 from Euglena gracilis. Biochemistry 36:14827-14835. [DOI] [PubMed] [Google Scholar]
- 256.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]
- 257.Yusupova, G., J. Reinbolt, H. Wakao, S. Laalami, M. Grunberg-Manago, P. Romby, B. Ehresmann, and C. Ehresmann. 1996. Topography of the Escherichia coli initiation factor 2/fMet-tRNA(f)(Met) complex as studied by cross-linking. Biochemistry 35:2978-2984. [DOI] [PubMed] [Google Scholar]
- 258.Yusupova, G. Z., M. M. Yusupov, J. H. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]
- 259.Zarivach, R., A. Bashan, F. Schluenzen, J. Harms, M. Pioletti, F. Franceschi, and A. Yonath. 2002. Initiation and inhibition of protein biosynthesis studies at high resolution. Curr. Protein Pept. Sci. 3:55-65. [DOI] [PubMed] [Google Scholar]
- 260.Zhang, J., and M. P. Deutscher. 1992. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc. Natl. Acad. Sci. USA 89:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]