Abstract
Certain monoclonal antibodies and polyclonal antisera directed to pMGA, the major protein of Mycoplasma gallisepticum, were tested for the ability to influence the surface phenotype of the cell population which resulted from their inclusion in growth medium. The polyclonal antiserum and one monoclonal antibody (MAb 66) resulted in an alteration of surface phenotype; specifically, populations of cells grown either on plates or in broth cultures which contained these reagents ceased the expression of pMGA and instead expressed an antigenically unrelated new polypeptide (p82). Upon the removal of antibody, the progeny of these cells regained pMGA expression and produced antigenically sectored colonies. The basis of this switch between pMGA+ and pMGA− states was shown to be transcriptional. The p82 polypeptide, the expression of which resulted from growth of cells in antibodies, was another member of the pMGA gene family and was located just downstream from the pMGA gene normally expressed by the M. gallisepticum cells used. Collectively the results of this work suggest that this organism has evolved an unusual means of altering the antigenic composition of its surface in response to antibodies or to other environmental cues.
Mycoplasmas are the simplest bacterial pathogens, yet they typically cause chronic diseases, which suggests that they may avoid their host’s immune response (10, 17). Antigenic variation has been observed in a number of Mycoplasma species (20) and studied in detail in Mycoplasma hyorhinis (15, 16, 21). Investigations in this laboratory have shown that the gene for a major surface protein of Mycoplasma gallisepticum S6 (pMGA1.1) is a member of a multigene family (12, 13). The pMGA gene family in strain S6 contains 33 members comprising a total of 7.7% of the 1,030-kb genome (1). Only one pMGA gene is expressed at any one time within strain S6, and unique pMGA molecules are expressed in different strains (9). The gene expressed by the cells used in the present work (strain S6) is termed pMGA1.1. Previous studies described three monoclonal antibodies (MAbs), 66, 71, and 86, which were directed to distinct epitopes on pMGA1.1, and showed that the pMGA molecules expressed in two other M. gallisepticum strains, R and F, contained epitopes bound by MAbs 86 and 71 but not by MAb 66 (9, 11). Given the existence of a sizeable repertoire of pMGA genes and the possibility that they are differentially expressed in different strains of this organism, it seemed possible that variation of pMGA gene expression occurs within a single strain as part of an immune evasion strategy. It therefore seemed germane to investigate if antibodies directed to pMGA would affect the growth of M. gallisepticum cells and the expression of pMGA in vitro.
MATERIALS AND METHODS
Antibodies.
The construction and serologic specificities of the three MAbs, 66, 71, and 86, used in this study were described in a previous publication from this laboratory (11). A rabbit antiserum directed to pMGA1.1 was elicited with a sample of antigen purified by immunoaffinity chromatography using MAb 86 attached to a Sepharose-4B matrix to select pMGA from a detergent lysate of M. gallisepticum S6 cells. Elution was with 0.2 M glycine, pH 2.7 (HCl). A 100-μg sample of electrophoretically pure pMGA1.1, dialyzed against phosphate-buffered saline (PBS) and emulsified in Freund’s complete adjuvant, was injected intramuscularly into a New Zealand White rabbit. Two further injections of the same antigen in Freund’s incomplete adjuvant were subsequently given at 2 weekly intervals. Preliminary experiments established that this reagent bound not only the pMGA1.1 antigen but also the pMGA antigens of two other M. gallisepticum strains, R and F (9). To take this cross-reactivity into account, this reagent is referred to as rabbit anti-pMGA.
Growth of M. gallisepticum cells.
The cells used throughout this study were the S6 strain of M. gallisepticum and have been used in previous studies from this laboratory (1, 9, 11–13). The composition of the culture medium used has also been described previously (8, 19). Growth inhibition in liquid culture medium was assessed by inoculation of dilute aliquots of M. gallisepticum cells into broth medium containing either 5 μg of MAb 66 ml−1, 5 μg of MAb 86 ml−1, or an equivalent volume of PBS. Cultures were incubated until the pH of the culture medium fell to 6.7 as judged by a color change of the phenol red indicator dye.
For experiments in which the effects of antibodies on colony growth were investigated, appropriate dilutions from a stock of M. gallisepticum S6 cells were dispersed to a single-cell suspension by passage through a 0.45-μm-pore-size filter (Schleicher & Schuell). Aliquots of these cells were inoculated in triplicate onto agar plates containing MAb 66 (5 μg ml−1), MAb 86 (5 μg ml−1), or no MAb. In some experiments, agar plates were first inoculated with dispersed M. gallisepticum cells and then a disc of filter paper (10 mm in diameter), impregnated with 25 μl of either MAb 86, MAb 71, or MAb 66 (1 mg ml−1 in all cases), was placed onto the plates. Colonies were then allowed to grow as usual.
Blotting techniques.
Nitrocellulose impressions of agar plates containing colonies were made by placing a nitrocellulose disc membrane (Amersham) directly onto the agar surface for 1 min. The membranes were removed, allowed to air dry, and incubated for 1 h at room temperature in PBS containing 5% bovine serum albumin (BSA). They were then washed three times for 5 min each in PBS containing 0.05% Tween 20 (PBS-T) and 0.1% BSA. The membranes were then incubated for 1 h at room temperature in a 1:750 dilution of rabbit anti-pMGA serum in PBS-T containing 1% BSA. The membranes were washed three times as before and incubated at room temperature for 1 h in a 1:1,000 dilution of swine anti-rabbit immunoglobulin (Ig)-horseradish peroxidase (HRPO) conjugate (Dako) in PBS-T–1% BSA. The membranes were washed three times, and binding of the conjugate was visualized by incubation in a solution containing 3,3′-diaminobenzidine (10 mg ml−1; Sigma) in 0.02 M Tris–137 mM NaCl, pH 7.6 (HCl)–0.03% (vol/vol) H2O2. Color development was stopped by extensive washing in distilled water. In some experiments, HRPO conjugates of rabbit anti-mouse Ig (Dako) and of purified MAb 86 were used to develop membrane impressions of M. gallisepticum colonies. Both of these reagents were used at 1:1,000 in the same manner as for the swine anti-rabbit Ig-HRPO conjugate.
SDS-PAGE and Western blotting.
Cells were pelleted in a microcentrifuge (12,000 rpm for 5 min), the supernatant was discarded, and the cell pellet was washed by resuspension in PBS. This washing step was performed twice more, and the cell pellet was finally resuspended in sodium dodecyl sulfate (SDS) lysis buffer. Mycoplasma cell proteins were separated in SDS-polyacrylamide gel electrophoresis (PAGE) (using 10% acrylamide gels) and either stained with Coomassie brilliant blue or transferred onto polyvinyl difluoride (PVDF) membranes (Millipore Waters). Membranes were incubated overnight at 4°C in PBS containing 5% BSA and washed three times in PBS-T–0.1% BSA for 10 min before immunostaining essentially as described above for nitrocellulose membranes.
Isolation of mycoplasma RNA and Northern blotting.
Isolation and Northern blotting of mycoplasma RNA was carried out essentially as described previously (9). M. gallisepticum and M. pullorum cells were grown to late log phase and harvested by centrifugation at 10,000 × g for 30 min at 4°C. M. gallisepticum cells were grown in broth medium either containing MAb 66 (5 μg ml−1) or without antibody. The harvested cells were resuspended in PBS and immediately processed for RNA extraction (8, 14). The quality and concentration of the isolated RNA were determined by gel electrophoresis and absorbance at 260 nm. RNA samples from M. gallisepticum and M. pullorum cells were subjected to gel electrophoresis under denaturing conditions and transferred to Hybond-N (Amersham). The construction of synthetic RNA copies of the pMGA1.1, -1.2, -1.4, and -’1.8’ genes and of Tuf has previously been described along with the conditions used to conduct filter hybridization using the DNA probes of this study (9).
Southern blotting using p82 and p45 probes.
Isolation of M. gallisepticum genomic DNA, restriction digestion, gel electrophoresis, and filter hybridization have previously been described (8, 12). Probes for p82 and p45 (Fig. 1) were both oligonucleotides which were 5′ labeled with 32P as previously described (12). In both cases, filter hybridization reactions were conducted in Church buffer (5) at 50°C.
FIG. 1.
Nucleotide sequences used for the oligonucleotide probes, which were based on amino acid sequences.
Protein sequencing.
Polypeptides to be sequenced were purified by SDS-PAGE and transferred to PVDF membranes which were stained with Coomassie brilliant blue for detection, and excised strips were subjected to automated Edman degradation as described in previous publications from this laboratory (9, 11).
Nucleotide sequence accession number.
GenBank accession no. L28423 has been assigned to the nucleotide sequence which predicts the pMGA1.9 amino acid sequence (Fig. 8). The same accession number applies to the sequence data in Fig. 9.
FIG. 8.
Predicted amino acid sequences of the pMGA1.9 polypeptide and its homology to pMGA1.1. The predicted amino acid sequence of pMGA1.9 is aligned to the known sequence of pMGA1.1 to maximize homology. Residues identical with counterparts in pMGA1.1 are marked with asterisks and conservative substitutions are indicated with dots. The predicted amino termini of the p82 and p45 polypeptides are indicated by arrows.
FIG. 9.
Sequence alignment between 5′ noncoding regions of pMGA genes. The nucleotide sequences compared extend from the stop codons of preceding coding sequences and extend to the translational start codons of the pMGA genes compared. The −35 and −10 regions of putative promoters are boxed, and the GAA repeat segments of each region occur 5′ to the −35 regions.
RESULTS
An antibody to pMGA affects the metabolism of M. gallisepticum cells.
The effects of two pMGA-specific MAbs (66 and 86) on the metabolism of M. gallisepticum cells in liquid culture were first examined. Three parallel M. gallisepticum cultures were established, using equal numbers of cells to initiate growth; one culture contained growth medium alone, one contained MAb 86 (5 μg ml−1) as an additive, and the third contained MAb 66 (5 μg ml−1). The relative metabolism rates in each culture were determined by the time taken to produce a color change of the phenol red indicator dye. The relevant times were 68 h for the control culture, 68 h for the MAb 86 culture, and 120 h for the MAb 66 culture. The metabolism rates of cultures of Mycoplasma synoviae were not affected by the inclusion of MAb 66 within the growth medium, ruling out a nonspecific growth inhibition. Experiments using enzyme-linked immunosorbent assay techniques in which purified pMGA was used as a capture antigen on plastic microtiter plates revealed that both antibodies were present at saturating levels at least at the outset of the experiment. Clearly MAb 66 but not MAb 86 caused some reduction in the growth or metabolic activity of cells compared to the control culture lacking MAb. Direct measurements of cell growth using viable counting techniques were not possible due to the ability of both MAb 66 and MAb 86 to agglutinate M. gallisepticum cells.
MAb 66 causes a cessation of pMGA expression in growing colonies.
To determine whether MAb 66 inclusion in growth medium had caused the selection of rare pMGA− variants within the pMGA+ starting population, the following experiment was conducted. Aliquots of an M. gallisepticum culture were dispensed onto agar plates onto which were placed filter discs impregnated with various additives that diffused into the surrounding agar. One disc contained PBS and others contained MAb 86, MAb 66, a third pMGA-specific antibody, MAb 71 (11), rabbit antiserum to pMGA, or preimmune rabbit serum. After colonies became apparent, nitrocellulose colony blots were taken from plates and then immunostained with the polyclonal rabbit antiserum directed to affinity-purified pMGA1.1. The results of this experiment are shown in Fig. 2. The fields selected for photography in Fig. 2 were either those closest to the discs or those which depicted relevant changes in pMGA1.1 expression. As is evident from Fig. 2, colonies grown close to discs impregnated with MAb 66 or rabbit anti-pMGA expressed little or no pMGA whereas colonies more distant from the central discs and all colonies from control plates were intensely immunostained. Colonies with sectored staining patterns were quite common in an intermediate annular zone around MAb 66- and rabbit anti-pMGA-containing discs, and one such colony from the MAb 66 plate is depicted at high magnification in Fig. 2. Despite the ability of MAbs 86 and 71 to bind pMGA1.1 (11), little or no alteration in pMGA1.1 staining pattern within their colonies was apparent. To further characterize the effect of pMGA antibodies on the growth of M. gallisepticum colonies, equal numbers of dispersed single cells were seeded onto agar plates containing 5 μg of MAb 66 ml−1, 5 μg of MAb 86 ml−1, or PBS. The numbers of colonies formed on triplicate plates containing MAb 66 (75 ± 7) or MAb 86 (58 ± 3) were similar to those without antibody (85 ± 9). Thus, neither pMGA-specific MAb prevented the growth of M. gallisepticum cells which initially expressed pMGA, as judged at least by the ability of single cells to form colonies. The diameters of the colonies obtained from plates containing either MAb were 60 to 80% of those of control colonies at day 3, but no diameter differences were apparent at day 6. To rule out the possibility that MAb 66, when included directly in the agar medium, had denatured, nitrocellulose colony blots were taken from representative plates and then immunostained with the polyclonal rabbit antiserum directed to affinity-purified pMGA1.1. Colonies obtained from plates without additive or those containing MAb 86 exhibited uniform staining (not shown). In contrast, nearly all colonies grown on plates containing MAb 66 exhibited little or no reactivity to rabbit anti-pMGA. Faint, vestigial staining was apparent in the centers of some colonies, and infrequent variant colonies (<0.1%) exhibited normal staining. Thus, MAb 66 at least had retained its ability to alter the pMGA expression pattern of colonies even if it did not significantly diminish the number of colonies which grew in its presence.
FIG. 2.
Nitrocellulose blots of colonies grown on agar culture plates around discs containing antibodies. Discs containing the reagents shown were placed at the centers of plates seeded with equal numbers of M. gallisepticum cells. After 6 days of growth, membrane lifts of each plate were immunostained with rabbit anti-pMGA (RαpMGA) as the primary detection reagent. In all cases, filter discs were located to the left of the colonies shown. The plate to the right of the MAb 66 panel depicts a single colony from this plate at a 10-fold-higher magnification.
The MAb 66 effect is associated with a cessation of transcription.
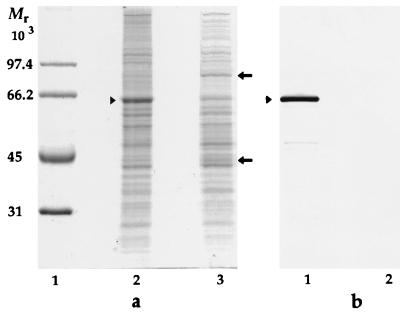
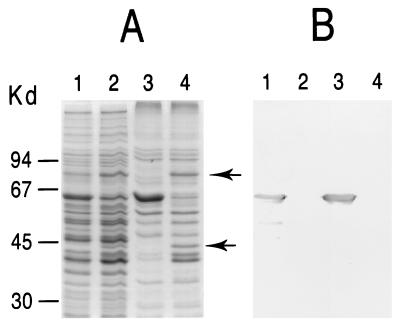
To obtain a cell population lacking pMGA1.1, M. gallisepticum cells were grown in broth medium containing MAb 66 and compared to a population of cells grown without a medium additive. Extracts of both types of cells were subjected to SDS-PAGE and Western immunoblotting with rabbit anti-pMGA as the primary detection reagent. As is apparent in Fig. 3, cells grown in the presence of MAb 66 contained greatly reduced amounts of a 67-kDa species (pMGA), as revealed by both Coomassie blue staining after SDS-PAGE (Fig. 3a) and by Western blots immunostained with rabbit anti-pMGA (Fig. 3b). Two protein species of 45 and 82 kDa, both antigenically unrelated to pMGA, were apparent in cells grown in the presence of MAb 66 (Fig. 2a, lane 3) but not from cells cultured without it. In contrast, the 67-kDa pMGA species was prominent in cell populations grown in medium either with MAb 86 (results not shown) or without any added MAb but was absent from cells cultured in MAb 66 medium (Fig. 3). The nature of the 45- and 82-kDa polypeptides expressed in pMGA− cells is considered later.
FIG. 3.
SDS-PAGE and Western blot analysis of M. gallisepticum cultured in the presence of a pMGA-specific MAb. (a) Coomassie blue-stained molecular weight standards (lane 1) and cells grown in broth medium alone (lane 2) or grown in broth containing MAb 66 (lane 3). (b) Rabbit anti-pMGA serum was used to immunostain a Western transfer of cellular proteins from mycoplasma cells grown in broth alone (lane 1) or in broth containing MAb 66 (lane 2). Arrowheads indicate the location of the pMGA protein. Arrows indicate the positions of 45- and 82-kDa bands referred to in the text. Molecular weight protein standards (Bio-Rad) are phosphorylase b (97,400), BSA (66,200), ovalbumin (45,000), and carbonic anhydrase (31,000).
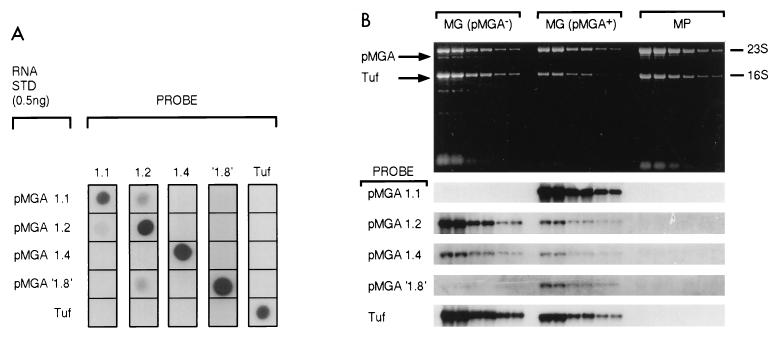
The failure of the cell population grown in pMGA-specific antibodies to express pMGA could, in principle, be due to any of a number of factors, including enhanced intracellular degradation induced by antibody-mediated cross-linking or surface shedding of pMGA promoted by antibody attachment or by a translational or transcriptional block. To more closely examine the latter possibility, cells grown in broth medium containing MAb 66 were compared by Northern blotting to normal cells (Fig. 4). Equivalent amounts of RNAs from both types of cells were compared for the ability to bind labeled probes for a number of related pMGA gene transcripts. In Fig. 4A, the hybridization specificities of five probes, four of them for specific pMGA mRNA molecules and the fifth for the Tuf transcript of M. gallisepticum, were verified. The construction of the synthetic, sense-strand RNA standards used for Fig. 4A has previously been described (9). Clearly the probes used in all cases efficiently detected their RNA complements and exhibited acceptably low levels of cross-hybridization with other synthetic RNA molecules. The upper panel of Fig. 4B depicts the patterns produced by electrophoresis of duplicate dilutions of total RNA samples from pMGA+ and pMGA− M. gallisepticum cells and from M. pullorum cells. Autoradiograms of membrane replicas of this pattern hybridized with the labeled probes indicated are shown in the lower panel of Fig. 4B. In contrast to control M. gallisepticum cells which expressed abundant pMGA1.1 mRNA, cells grown in the presence of MAb 66 expressed little or none. As anticipated, the negative control RNA sample from M. pullorum cells bound no pMGA gene probe. Quantitative analysis of the data of Fig. 4 (using a phosphorimager) revealed that pMGA+ cells contained at least 100-fold more pMGA1.1 mRNA than their pMGA− counterparts. Thus, growth of cells in the presence of MAb 66 profoundly diminished the abundance of pMGA1.1 mRNA. The abundance of one of the minor transcripts also present normally in M. gallisepticum cells, pMGA′1.8′ (9), was also substantially diminished in abundance in pMGA− cells. Another transcript, pMGA1.4, was slightly diminished, but the pMGA1.2 transcript was present at a slightly elevated level.
FIG. 4.
Growth of M. gallisepticum cells in MAb 66-containing medium results in a selective cessation of transcription of the pMGA1.1 gene. (A) Synthetic sense-strand RNA samples (0.5 ng) derived from the four pMGA genes indicated and from the elongation factor Tuf gene were blotted (horizontally) onto nylon membrane strips. Each strip was hybridized with the indicated labeled probes (vertically). Details of synthetic RNA molecules, probes, and hybridization conditions are from reference 8. STD, standard. (B) Twofold serial dilutions of purified RNA from M. gallisepticum cells grown in medium to which MAb 66 had been added (5 μg ml−1) [MG (pMGA−1)] or in the medium without additive [MG (pMGA+)] or from Mycoplasma pullorum cells (MP) were subjected to denaturing electrophoresis, and replicate gels were either photographed to detect ethidium bromide patterns (upper panel) or subjected to Northern blot analysis using the labeled probes indicated alongside strips on the lower panel. Hybridization conditions used for individual probes were identical to those in panel A. Binding of probes was detected and then quantified with a phosphorimager.
Cessation of pMGA transcription by growth of cells in MAb 66-containing medium is reversible.
To determine whether the inhibition of pMGA expression by MAb 66 was reversible, M. gallisepticum cells were grown in broth medium containing MAb 66 (5 μg ml−1) and harvested. To remove clumps or aggregates, the suspension was passed through a 0.45-μm-pore-size filter before inoculation onto agar plates without antibody. Nitrocellulose blots were made of the resultant colonies and then immunostained. The colonies obtained exhibited a sectored appearance when immunostained with rabbit anti-pMGA1.1 (Fig. 5). Each colony originally consisted of pMGA1.1− cells, but as growth progressed, events occurred whereby single cells within growing colonies reacquired the pMGA1.1+ phenotype, which was retained by their progeny. Up to 20 independent separate sectors per colony were observed, and almost all colonies were sectored.
FIG. 5.
Immunostaining of nitrocellulose blots of M. gallisepticum colonies derived from pMGA− cells. The expression of pMGA in colonies derived from cells originally grown in broth containing MAb 66 was detected by immunostaining nitrocellulose blots of colonies with rabbit anti-pMGA serum. Sectorial reacquisition of pMGA expression is apparent.
The gene expressed within M. gallisepticum S6 cells, pMGA1.1, is one of a large family (1, 12, 13). To investigate whether the pMGA gene reexpressed within the sectors of Fig. 5 was the same as that expressed before MAb 66 treatment, four sectored colonies were each inoculated into antibody-free broth medium and the resultant cells were analyzed by SDS-PAGE and Western blotting. In every case, reacquisition of a 67-kDa pMGA species was demonstrable by Coomassie blue staining as well as by Western blotting using MAb 66, MAb 86, and rabbit anti-pMGA1.1 as detection reagents. Previous studies have shown that MAb 86 and the rabbit polyclonal antiserum bind pMGA proteins from strains R and F as well as S6 but MAb 66 detects a pMGA epitope which is strain specific and which may be specific for the product of the pMGA1.1 gene (11, 13). Samples of the 67-kDa species from the four colonies were subjected to protein sequencing. In all cases, the amino-terminal sequence obtained was (C)TTPTPSPAPNPNPPSN---. This sequence is identical to that of the pMGA1.1 protein but is distinct from that encoded by any other characterized pMGA gene (9, 11). Thus, the effect on pMGA1.1 expression caused by MAb 66 appears to be reversible.
The genes for p82 and p45 are closely linked to one another and to pMGA1.1.
Figure 3 demonstrates that cells grown in MAb 66 medium are deficient in a prominent constituent of approximately 67 kDa conspicuously present in control cells; conversely, two polypeptides of molecular masses 82 and 45 kDa (p82 and p45, respectively) were present in the former culture but absent or greatly reduced in the latter. When Triton X-114 lysates of these samples were used to obtain detergent-rich phases for SDS-PAGE (3), these polypeptide differences between normal cells and cells grown in MAb 66 medium were much more pronounced (Fig. 6). Thus, the p82 and p45 polypeptides, like pMGA1.1, were likely to be membrane proteins by virtue of their selective affinity for Triton X-114. When the same samples as in Fig. 6A were subjected to immunoblot analysis using a polyclonal rabbit anti-pMGA serum as the detection reagent, the 67-kDa pMGA1.1 structure intensely immunostained but neither of the new polypeptides in the former culture was detected, indicating a lack of antigenic cross-reactivity with pMGA1.1. To study the structures of p45 and p82, both polypeptides were purified by SDS-PAGE from pMGA− cells followed by electrophoretic transfer onto a PVDF membrane. These membrane strips were used to obtain amino-terminal sequences of both p45 and p82 polypeptides, which are shown in Table 1. Clearly the two sequences were not closely related either to one another or to the amino terminus of pMGA1.1. Synthetic oligonucleotides based on the p82 and p45 amino-terminal sequences in Table 1 were constructed, labeled, and used as probes to clone a 13-kb ClaI fragment of M. gallisepticum DNA which bound both p82 and p45 oligonucleotides. Subsequent experiments detected the presence of the pMGA1.1 gene on the same ClaI fragment. A restriction map of this fragment depicting the subfragments which bound the p82 and p45 probes and the locations of the three pMGA genes, pMGA1.1, pMGA1.7, and pMGA1.9, relevant to the remainder of this work is shown in Fig. 7. The nucleotide sequence of the p82-p45 segment of the ClaI fragment was determined, and the complete predicted amino acid sequence of the p82 polypeptide is presented in Fig. 8, which depicts a sequence comparison of the predicted polypeptide sequences of pMGA1.1 and p82. The sequence encoding the p82 polypeptide is hereafter termed pMGA1.9. The locations of the amino-terminal sequences of the p82 and p45 polypeptides are indicated in Fig. 8. Their predicted amino-terminal sequences concur perfectly in both cases with the directly determined sequences in Table 1. The number of amino acids predicted for the mature form of the pMGA1.9 gene product (p82) is 678, and its predicted molecular mass (approximately 74,000 Da) is in fair agreement with its size as estimated by SDS-PAGE. The predicted chain size of the p45 polypeptide is 367 residues, and its expected molecular mass of approximately 40,000 Da agrees well with its SDS-PAGE mobility. Comparison between the nucleotide sequences 5′ to the pMGA1.1, pMGA1.7, and pMGA1.9 genes revealed, in common with all other known pMGA genes, the presence of a tract of GAA repeats followed by related −35 and −10 promoter motifs in all three genes (Fig. 9).
FIG. 6.
The p82 and p45 polypeptides are partitioned into Triton X-114. M. gallisepticum cells were grown either in broth medium containing no antibody additive (lanes 1 and 3) or in medium containing 5 μg of MAb 66 ml−1 (lanes 2 and 4). Recovered cells were boiled in SDS-containing buffer and subjected to SDS-PAGE (lanes 1 and 2). Alternatively, cells were lysed with Triton X-114 and subjected to partition into detergent-rich phases before electrophoresis (lanes 3 and 4). (A) Coomassie blue-stained gel; (B) identical gel transferred to a PVDF membrane and then immunostained with rabbit anti-pMGA followed by HRPO-conjugated anti-rabbit Ig. Arrowheads indicate the p45 and p82 species referred to in the text.
TABLE 1.
Amino-terminal sequences of pMGA1.1, p82, and p45
| Protein | Sequence |
|---|---|
| pMGA1.1a | CTTPTPSPAPNPN... |
| p82 | -TSATIPTLNPTP... |
| p45 | -ETQEVSPTPAAEVQ.. |
From reference 11.
FIG. 7.
Restriction map of a ClaI fragment containing pMGA1.1, pMGA1.7, p82, and p45 sequences. Single and double digests of the cloned 13-kb ClaI fragment containing the pMGA1.1 coding sequence were conducted with the restriction enzymes ClaI (C), EcoRI (E), HindIII (H), and PstI (P). Digests were subjected to agarose gel electrophoresis for fragment size measurements, and the resultant gels were subjected to Southern blotting. The p82 and p45 N-terminal oligonucleotide probes used in Southern blot studies were based on the amino acid sequences in Table 1 and are listed in Fig. 1. Informative fragments which bound the p82 and p45 probes are indicated. The approximate lengths and locations of the three pMGA gene coding sequences referred to in the text are shown. The shaded bar toward the 3′ end of the pMGA1.7 gene depicts an unusual DNA insertion which has not yet been definitively sequenced.
The comparative sequence montages of Fig. 8 and 9 leave no doubt that the pMGA1.9 gene is a member of the same family as pMGA1.1. In the first place, the signal sequence appending the mature p82 amino terminus is identical to its counterpart in pMGA1.1 but for one amino acid. The mature sequences also exhibit clear homology; the p82 and pMGA1.1 polypeptides are 42% identical.
DISCUSSION
This study initially investigated the effects of antibodies directed to a major surface protein, pMGA, upon growing M. gallisepticum cells. The results in Fig. 2 demonstrated that the presence of certain pMGA-specific antibodies during the growth of M. gallisepticum cells resulted in the growth of colonies which lacked pMGA. No overall reduction in colony numbers occurred when pMGA-specific antibodies were included as mycoplasma growth medium additives. Instead, certain antibodies, MAb 66 in particular, acted by promoting the growth of pMGA− colonies from pMGA+ progenitor cells. Either MAb 66 attachment to pMGA transmitted a message which affected pMGA expression or cells of the pMGA+ phenotype were selected against by the presence of MAb 66 and overgrowth by pMGA1.1− cells occurred. Studies using M. hominis revealed that growth inhibition by a particular surface-binding MAb was due to its ability to agglutinate cells (7), and it is possible that MAb 66 and rabbit anti-pMGA1.1 act in a similar fashion; i.e., their attachment to M. gallisepticum cells imposes a growth inhibition which is alleviated by the loss of pMGA1.1 expression. Neither of these explanations is entirely acceptable without additional provisos (see below).
Transcription of the pMGA1.1 gene was shown to have ceased in cell populations grown in MAb 66-containing medium (Fig. 4). Two other pMGA genes which in normal cells were transcribed at low levels, pMGA1.4 and pMGA′1.8′, were also diminished in abundance, whereas the pMGA1.2 transcript was slightly increased. These observations suggested either that pMGA1.1− cells carried pleiotropic mutations of some transcriptional control element which affected the expression of more than one pMGA gene or alternatively that mutations which alter transcriptional activity are common events within the pMGA gene family. Alternative explanations for the MAb 66 effect such as translational or posttranslational inhibition of pMGA expression were made redundant by the experiment of Fig. 4.
Cultures in which the pMGA1.1 gene had been transcriptionally silenced by growth in MAb 66 medium were shown to revert to its expression when grown on medium lacking MAb 66 (Fig. 5). Most colonies exhibited sectorial staining patterns when either rabbit anti-pMGA or MAb 66 was used as the immunodetection reagent. The known selectivity for the pMGA1.1 polypeptide of the MAb 66 reagent in particular suggested that the pMGA1.1 gene rather than another family member was reexpressed in growing colonies established mainly or exclusively by pMGA1.1− progenitor cells. The distinctive sectorial reexpression patterns of most of these colonies suggested that related or identical reversion events occurred frequently and independently during colony growth. The amino-terminal sequence analyses of the 67-kDa proteins purified from four revertant colonies leave little doubt that the pMGA gene reexpressed by cells recovering from growth in MAb 66 medium was indeed pMGA1.1.
The work herein reveals a further important characteristic of the MAb 66 effect of which account must be taken by any explanatory model. The cessation of pMGA1.1 transcription caused by MAb 66 in M. gallisepticum cells was accompanied by the expression of another member of the pMGA gene family, pMGA1.9. Initially the products of this gene were detected in lysates of cells grown in MAb 66-containing medium (Fig. 3 and 6). Studies on the p82 and p45 polypeptides revealed that they were probably plasma membrane proteins, as judged by their selective partition into the detergent Triton X-114 (Fig. 6). Determination of the p82 and p45 amino-terminal sequences facilitated the construction of probes which enabled the molecular cloning of the gene that encoded both polypeptides. The p45 polypeptide is likely to be a proteolytic fragment of p82. The gene which encodes p82 was shown to be a member of the pMGA gene family by virtue of its sequence homology to pMGA1.1 (Fig. 8). It was designated pMGA1.9 in this work and shown to occur on the same restriction fragment as the pMGA1.1 gene. Specifically it was shown to be separated from pMGA1.1 by a third gene, pMGA1.7, and all three genes were in the same transcriptional orientation. The preliminary sequence of the pMGA1.7 region suggests that the coding sequence may be interrupted by the insertion of a DNA segment which bears no homology to any pMGA sequence (results not shown). The approximate location of this unusual DNA segment is shown in Fig. 7. Analysis of the 5′ noncoding region of the pMGA1.9 gene revealed that it contained acceptable −10 and −35 promoter motifs in addition to a series of trinucleotide (GAA) repeats, 5′ to the promoter, which seem to be a property of most or all pMGA genes (1).
Collectively these observations are compatible with the existence of two alternate heritable states (pMGA1.1+ and pMGA1.1−) with respect to pMGA gene transcription in M. gallisepticum. It is difficult to avoid invoking a change in DNA structure between these states to account for the pMGA1.1 transcription differences and for the apparent heritability of the pMGA+ phenotype which the sectorial reacquisition patterns of Fig. 5 evidence. Some relatively high frequency alteration to the pMGA1.1 promoter site rather than the coding sequence would account for the rapid extinction of the pMGA1.1+ phenotype in cultures containing MAb 66, as well as for the pMGA1.1−-to-pMGA1.1+ reversion events which occur when antibody selection is removed. There is no conceptual difficulty with such a switching mechanism. In M. hyorhinis, for example, four members of the vlp gene family are variably expressed (15, 16, 20). Transcription of each gene appears to be independently controlled by variation in the number of residues in a polyadenosine tract between the −10 and −35 regions of the promoter. In Salmonella species, two flagellin genes are expressed as strict alternatives, and switching between phases is mediated by the site-specific inversion of a 995-bp DNA element (18). In one orientation the H1 flagellin variant is expressed, and in the other orientation the H2 variant is expressed. The hsd 1 locus of Mycoplasma pulmonis (20) is another example of an invertible element which in one orientation facilitates the expression of restriction/modification functions but in the other does not. Site-specific inversions of DNA elements also account for the rapid phase variation of M. pulmonis V-1 surface antigens (2).
The unique feature of the pMGA+-to-pMGA− transition in M. gallisepticum is that MAb 66 seems to specifically influence the switching process in the sense that the presence of MAb 66 in agar medium causes the pMGA+-to-pMGA− switch in colony phenotype without altering the overall number of colonies. In particular, at the single-cell level the (pMGA+) progenitors of (pMGA−) colonies are not killed or inactivated by MAb 66. Two alternative explanations for the switch might be considered. First, ligation of the MAb 66 epitope may activate cytoplasmic events, the end result of which is a targeted change to the pMGA1.1 gene promoter. This instructive model is difficult to reconcile with the nonuniform distribution of the pMGA1.1 antigen within colonies derived either from pMGA1.1+ or pMGA1.1− founder cells (Fig. 2 or 5, respectively). The singularly unattractive feature of this model is its requirement for surface-bound antibody to instruct specific changes in DNA sequence. No such mechanism is known in any other organism, including closely related mycoplasmas.
There is a more orthodox, alternative explanation which accounts for the role of pMGA-specific antibodies in the pMGA1.1-to-pMGA1.9 switch and which avoids the need to directly link an environmental stimulus with specific changes in DNA sequence. According to this model, surface ligation of pMGA by MAb 66 may potently inhibit the growth of pMGA1.1+ cells but without directly transmitting any specific instruction to the cytoplasm. Affected cells would not die but would not replicate either. A change in pMGA gene expression would, given time, occur and release the cells from growth inhibition. The frequency of these events would need to be sufficiently high to enable pMGA1.1+ progenitor cells of pMGA1.1− colonies to change phenotype within the time frame of the experiment. Such a model accounts well for the phenotypes of colonies grown on MAb 66-containing agar but requires supplementary explanations for three salient experimental facts: first, for the fact that specifically pMGA1.9 rather than some other pMGA gene(s) is expressed concomitant with the cessation of pMGA1.1 expression; second, for the unusually high frequency of the pMGA−-to-pMGA+ reversion events; and third, for the fact that expression of pMGA1.1, rather than a different pMGA gene, is reacquired when MAb 66 is removed. Studies on the high-frequency oscillation between pMGA+ and pMGA− states are now in progress to determine if DNA sequence changes 5′ to the promoter sites of relevant pMGA genes are important as a primary cause for the switches in pMGA gene expression documented herein. The reason for the apparently selective expression of either the pMGA1.1 or the pMGA1.9 gene from the pMGA family is as yet unclear. Perhaps the S6 strain of M. gallisepticum used in this work has an obligate survival requirement subserved by pMGA1.1 or pMGA1.9 but by no other pMGA family member. This explanation would account for the switching between the two apparently discrete states documented in this work. Studies are now in progress to distinguish the two explanations by determining the nature of the DNA differences between the pMGA1.1− and pMGA1.1+ states.
Irrespective of whether pMGA-specific antibodies act in-structively or selectively to operate the switch in pMGA phenotype documented herein, the advantage to the pathogen of this switch may be considerable. The ability to alter surface antigenicity within the same time frame as the elicitation of a host immune response is a common hallmark of many successful pathogens, but the ability of M. gallisepticum to accomplish this without the loss or permanent alteration of a gene which might be essential in a subsequent infection confers an additional teleological advantage to this bacterium in the colonization of its host.
ACKNOWLEDGMENTS
We thank C. J. Morrow for help with photomicrography.
This work was supported by project grants from the NH and MRC (to I.D.W.), the Australian Research Council (I.D.W., G.F.B., and K.G.W.), and Bioproperties Australia Pty. Ltd. (G.F.B. and K.G.W.).
REFERENCES
- 1.Bassegio N B, Glew M D, Markham P F, Whithear K G, Browning G F. Size and genomic location of the pMGA multigene family of Mycoplasma gallisepticum. Microbiology. 1995;142:1429–1435. doi: 10.1099/13500872-142-6-1429. [DOI] [PubMed] [Google Scholar]
- 2.Bhugra B, Voelker L L, Zou N, Yu H, Dybwig K. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 3.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 4.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dybvig K, Yu H. Regulation of a restriction and modification system via DNA inversion in Mycoplasma pulmonis. Mol Microbiol. 1994;12:547–560. doi: 10.1111/j.1365-2958.1994.tb01041.x. [DOI] [PubMed] [Google Scholar]
- 7.Feldmann R C, Henrich B, Kolb B V, Hadding U. Decreased metabolism and viability of Mycoplasma hominis induced by monoclonal antibody-mediated agglutination. Infect Immun. 1992;60:166–174. doi: 10.1128/iai.60.1.166-174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey M L, Hanson R P, Anderson D P. A medium for the isolation of avian mycoplasmas. Am J Vet Res. 1968;29:2163–2171. [PubMed] [Google Scholar]
- 9.Glew M D, Markham P F, Browning G B, Walker I D. Expression studies on four members of the pMGA multigene family in Mycoplasma gallisepticum strain S6. Microbiology. 1995;141:3005–3014. doi: 10.1099/13500872-141-11-3005. [DOI] [PubMed] [Google Scholar]
- 10.Krause D C, Taylor-Robinson D. Mycoplasmas which infect humans. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 417–444. [Google Scholar]
- 11.Markham P F, Glew M, Brandon M R, Walker I D, Whithear K G. Characterization of a major haemagglutinin protein from Mycoplasma gallisepticum. Infect Immun. 1992;60:3885–3891. doi: 10.1128/iai.60.9.3885-3891.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markham P F, Glew M D, Whithear K G, Walker I D. Molecular cloning of a member of the gene family that encodes pMGA, a hemagglutinin of Mycoplasma gallisepticum. Infect Immun. 1993;61:903–909. doi: 10.1128/iai.61.3.903-909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markham P F, Glew M D, Sykes J E, Bowden T R, Pollocks T D, Browning G F, Whithear K G, Walker I D. The organisation of the multigene family which encodes the major cell surface protein, pMGA, of M. gallisepticum. FEBS Lett. 1994;352:347–352. doi: 10.1016/0014-5793(94)00991-0. [DOI] [PubMed] [Google Scholar]
- 14.Markham P F. Ph.D. thesis. Parkville, Victoria, Australia: The University of Melbourne; 1995. [Google Scholar]
- 15.Rosengarten R, Wise K S. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science. 1990;247:315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- 16.Rosengarten R, Wise K S. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J Bacteriol. 1991;173:4782–4793. doi: 10.1128/jb.173.15.4782-4793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simecka J W, Davis J K, Davidson M K, Ross S E, Städtlander C T K-H, Cassell G H. Mycoplasma diseases of animals. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 391–415. [Google Scholar]
- 18.van De Putte P, Goosen N. DNA inversions in phages and bacteria. Trends Genet. 1992;8:457–462. doi: 10.1016/0168-9525(92)90331-w. [DOI] [PubMed] [Google Scholar]
- 19.Whithear K G, Bowtell D D, Ghiocas E, Hughes K L. Evaluation and use of a micro broth dilution procedure for testing sensitivity of fermentative avian mycoplasmas to antibiotics. Avian Dis. 1983;27:937–949. [PubMed] [Google Scholar]
- 20.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 473–489. [Google Scholar]
- 21.Yogev D, Rosengarten R, Watson M R, Wise K S. Molecular basis of mycoplasma surface antigenic variation: a novel set of divergent genes undergo spontaneous mutation of periodic coding regions and 5′ regulatory sequences. EMBO J. 1991;10:4069–4080. doi: 10.1002/j.1460-2075.1991.tb04983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]