Abstract
Legionella pneumophila is able to survive inside phagocytic cells by an internalization route that bypasses fusion of the nascent phagosome with the endocytic pathway to allow formation of a replicative phagosome. The dot/icm genes, a major virulence system of L. pneumophila, encode a type IVB secretion system that is required for intracellular growth. One Dot protein, DotL, has sequence similarity to type IV secretion system coupling proteins (T4CPs). In other systems, coupling proteins are not required for viability of the organism. Here we report the first example of a strain, L. pneumophila Lp02, in which a putative T4CP is essential for viability of the organism on bacteriological media. This result is particularly surprising since the majority of the dot/icm genes in Lp02 are dispensable for growth outside of a host cell, a condition that does not require a functional Dot/Icm secretion complex. We were able to isolate suppressors of the ΔdotL lethality and found that many contained mutations in other components of the Dot/Icm secretion system. A systematic analysis of dot/icm deletion mutants revealed that the majority of them (20 of 26) suppressed the lethality phenotype, indicating a partially assembled secretion system may be the source of ΔdotL toxicity in the wild-type strain. These results are consistent with a model in which the DotL protein plays a role in regulating the activity of the L. pneumophila type IV secretion apparatus.
The gram-negative bacterium Legionella pneumophila is the causative agent of a potentially fatal form of pneumonia called Legionnaires' disease. L. pneumophila is found in freshwater environments, where it parasitizes many different species of protozoa (17). Humans become infected with L. pneumophila by inhaling aerosols generated from contaminated water sources. Upon entry into the human lung, L. pneumophila is internalized into bactericidal, alveolar macrophages. In contrast to phagosomes bearing most bacterial species, the compartment harboring L. pneumophila does not traffic into the lysosomal network and is not significantly acidified in the first few hours after uptake (26, 27). Instead, the phagosome interacts with early secretory vesicles at endoplasmic reticulum exit sites (29) and then undergoes a series of maturation events in which it sequentially associates with small vesicles, mitochondria, and eventually becomes surrounded by the rough endoplasmic reticulum (25, 60). Formation of this specialized compartment, called a “replicative phagosome,” allows the microorganism to grow intracellularly (25, 28). Later in the infective cycle, a majority of the replicative phagosomes fuse with acidified compartments containing late endocytic markers, and this is believed to play an important role in the replicative cycle of this pathogen prior to exit from its host cell (59).
The key to L. pneumophila's virulence is its ability to form a replicative phagosome, since mutants defective in this trait cannot replicate inside host cells and are thus unable to cause disease (24, 26). One large class of proteins that allow L. pneumophila to alter the endocytic pathway is encoded by the dot/icm genes (3, 5, 37). To date, over two dozen dot/icm genes have been identified and are clustered in two areas of the L. pneumophila chromosome (region I and region II) (63). Based on the similarity of the Dot/Icm proteins to proteins involved in conjugative DNA transfer, and the fact that the Dot/Icm system can transfer the mobilizeable plasmid RSF1010, it was proposed that the dot/icm genes of L. pneumophila encode a type IV secretion system (31, 50, 63).
Type IV secretion systems are able to export DNA and/or proteins out of the bacterial cell and include plasmid transfer systems (e.g., the tra and trb genes of the plasmid RP4), as well as systems involved in the delivery of virulence factors (10, 46, 66). The canonical type IV secretion system is encoded by the virB operon of the plant pathogen Agrobacterium tumefaciens (66). A number of other pathogens, including Bartonella tribocorum, Bordetella pertussis, Brucella abortus, Helicobacter pylori, and Rickettsia prowazekii, contain orthologues to the VirB proteins, and some of these systems have been shown to export proteins essential for virulence (10). In contrast to these type IV systems, the L. pneumophila Dot/Icm proteins have limited sequence similarity to the VirB proteins. Instead, the Dot/Icm proteins show high similarity to the transfer proteins from IncI plasmids (e.g., R64 and ColIb-P9) and compose a type IVB secretion system (31, 57).
As with most conjugative transfer systems, little is known about the specific function of many of the L. pneumophila Dot/Icm proteins. DotB was recently shown to possess ATPase activity and likely provides energy to the secretion apparatus (56). A second Dot protein, DotL, also contains a nucleotide binding motif and shows extensive sequence similarity to the conjugal transfer protein TrbC from IncI plasmids (19, 31). DotL also has detectable sequence similarity to a family of proposed ATPases known as TraG-like or type IV secretion system coupling proteins (T4CPs). The more notable members of the T4CP family include TraG (RP4 plasmid), TrwB (R388 plasmid), TraD (F plasmid), and the A. tumefaciens VirD4 protein (8, 18, 33).
The term “coupling protein” was proposed for this family because its members are believed to target, or couple, exported substrates to the secretion apparatus (8, 9, 15, 22, 23, 32, 61). This proposal was initially based on the phenotype of RP4 traG mutants, which were still able to process plasmid DNA into a secretion-competent intermediate and assemble a functional pilus but were unable to transfer the plasmid. This indicated that TraG plays a role in linking the two processes (9). Consistent with the idea of T4CPs linking substrates to the secretion apparatus, a number of T4CPs have been shown to interact with both exported substrates and with components of the secretion apparatus (2, 15, 20, 35, 61). Although T4CPs are absolutely required for export of substrates, their specific molecular function remains unknown (22).
We demonstrate here that a T4CP homologue, the DotL protein, is not only required for growth of L. pneumophila inside macrophages but is also essential for viability of certain strains on bacteriological media. The lethality caused by loss of dotL in those strains can be suppressed by mutations that inactivate the Dot/Icm complex, which is consistent with a DotL role in regulating the activity of this type IV secreton.
MATERIALS AND METHODS
Bacterial strains and media.
All L. pneumophila strains used in the present study are derived from Lp02 (hsdR rpsL thyA) or JR32 (hsdR rpsL), two separate isolates of L. pneumophila Philadelphia-1 (3, 7, 38) (Table 1). L. pneumophila strains were cultured on N-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered charcoal yeast extract agar (CYET) or ACES-buffered yeast extract broth (AYET) supplemented with thymidine (100 μg/ml). Salt sensitivity was assayed on CYET plates containing 0.65% sodium chloride (11, 45, 64). Antibiotics (kanamycin, 20 μg/ml; chloramphenicol, 5 μg/ml; streptomycin, 50 μg/ml; gentamicin, 5 μg/ml) and sucrose (5%) were added as needed. Escherichia coli strains were cultured on Luria-Bertani medium, and antibiotics (kanamycin, 20 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 17 μg/ml) were added as needed. Replication-competent plasmids were propagated in the E. coli strain XL1-Blue. In order to propagate suicide plasmids containing the R6K origin of replication, a strain expressing the R6K π protein, E. coli strain DH5α(λpir), was used (30, 67).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH5α | endA1 hsdR17 (rK− mK+) glnV44 thi-1 recA1 gyrA (Nalr) relA1 (ΔlacIZYA-argF)U169 deoR (φ80dlac ΔlacZ) M15 | 67 |
| DH5α(λpir) | DH5α(λpir) tet::Mu | 30 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| L. pneumophila strains | ||
| Philadelphia-1 | Wild-type strain | 3,38 |
| Lp01 | Philadelphia-1 rpsL hsdR | 3 |
| Lp02 | Philadelphia-1 rpsL hsdR thyA mutant | 3 |
| Lp03 | Lp02 dotA mutant | 3 |
| JR32 | Philadelphia-1 rpsL hsdR | 38 |
| JV1001 | Lp02 dotL+/ΔdotL | This study |
| JV1003 | Lp02 dotL+/ΔdotL::Cmr | This study |
| JV1067 | JV1003 + pJB1014 | This study |
| JV1630 | JR32 ΔlvhB | This study |
| JV1631 | JR32 ΔlvhB | This study |
| JV2114 | JR32 ΔdotL | This study |
| JV2348 | JR32 ΔdotL + pJB1079 | This study |
| JV2349 | JR32 ΔdotL + pJB1081 | This study |
| JV2097 | Lp02 dotM+/ΔdotM::Cmr | This study |
| JV2100 | Lp02 dotN+/ΔdotN::Cmr | This study |
| JV2114 | JR32 ΔlvhB dotL+/ΔdotL::Cmr | This study |
| JV2116 | JR32 ΔlvhB dotL+/ΔdotL::Cmr | This study |
| JV2118 | JR32 ΔlvhB dotL+/ΔdotL::Cmr | This study |
| JV2153 | JR32 ΔdotB | This study |
| JV2238 | Lp02 dotM+/ΔdotM::Cmr ΔdotA | This study |
| JV2240 | Lp02 dotN+/ΔdotN::Cmr ΔdotA | This study |
| JV2256 | Lp02 ΔdotV dotL+/ΔdotL::Cmr | This study |
| JV2258 | Lp02 ΔdotO dotL+/ΔdotL::Cmr | This study |
| JV2260 | Lp02 ΔdotP dotL+/ΔdotL::Cmr | This study |
| JV2262 | Lp02 ΔdotE dotL+/ΔdotL::Cmr | This study |
| JV2264 | Lp02 ΔicmQ dotL+/ΔdotL::Cmr | This study |
| JV2274 | Lp02 ΔicmX dotL+/ΔdotL::Cmr | This study |
| JV2276 | Lp02 ΔdotA dotL+/ΔdotL::Cmr | This study |
| JV2282 | Lp02 ΔicmS dotL+/ΔdotL::Cmr | This study |
| JV2284 | Lp02 ΔicmR dotL+/ΔdotL::Cmr | This study |
| JV2286 | Lp02 ΔdotB dotL+/ΔdotL::Cmr | This study |
| JV2290 | Lp02 ΔdotU dotL+/ΔdotL::Cmr | This study |
| JV2292 | Lp02 ΔicmF dotL+/ΔdotL::Cmr | This study |
| JV2294 | Lp02 ΔcitA dotL+/ΔdotL::Cmr | This study |
| JV2465 | Lp02 dotM+/ΔdotM::Cmr + pdotL+ | This study |
| JV3748 | Lp02 ΔdotH dotL+/ΔdotL::Cmr | This study |
| JV3750 | Lp02 ΔicmT dotL+/ΔdotL::Cmr | This study |
| JV3752 | Lp02 ΔicmW dotL+/ΔdotL::Cmr | This study |
| JV3754 | Lp02 ΔdotI dotL+/ΔdotL::Cmr | This study |
| JV3756 | Lp02 ΔdotJ dotL+/ΔdotL::Cmr | This study |
| JV3758 | Lp02 ΔdotK dotL+/ΔdotL::Cmr | This study |
| JV3760 | Lp02 ΔdotD dotL+/ΔdotL::Cmr | This study |
| JV3762 | Lp02 ΔdotC dotL+/ΔdotL::Cmr | This study |
| JV3765 | Lp02 ΔicmV dotl+/ΔdotL::Cmr | This study |
| JV3767 | Lp02 ΔdotG dotL+/ΔdotL::Cmr | This study |
| JV3769 | Lp02 ΔdotF dotL+/ΔdotL::Cmr | This study |
| Plasmids | ||
| pJB908 | pKB5 ΔoriT | 56 |
| pJB921 | ΔdotB in pSR47S | 55 |
| pJB1001 | ΔdotL in pSR47S | This study |
| pJB1005 | ΔdotL::Cmr in pSR47S | This study |
| pJB1010 | His-tagged DotL in pQE-32 | This study |
| pJB1014 | pJB908 + dotL+ | This study |
| pJB1079 | pJB1014 + Genr cassette | This study |
| pJB1081 | pJB908 + Genr cassette | This study |
| pJB1242 | ΔlvhB in pSR47S | This study |
| pJB1300 | pKB5 with NotI site | This study |
| pJB1304 | lvhB operon in pJB1300 | This study |
| pJB3046 | ΔdotN in pSR47S | This study |
| pJB3050 | ΔdotM in pSR47S | This study |
| pKB5 | RSF1010 cloning vector | 3 |
| pSR47S | oriR6K oriTRP4 kan sacB | 40 |
Nalr, nalidixic acid resistant; Tetr, tetracycline resistant; Genr, gentamicin resistant.
Plasmid construction.
To make the ΔdotL suicide plasmid, pJB1001, two PCR-amplified fragments were cloned into the NotI/SalI sites of pSR47S (40). Fragment 1 was amplified by using the primers 5′-CCCAAACGGCCGCCAAACGAGTATTTACCATGC(JVP201 with the EagI site underlined) and 5′-CCCAAAGGATCCCGCATCATGGCTCTAATTCC(JVP202 with the BamHI site underlined). Fragment 2 was amplified by using the primers 5′-CCCAAAGGATCCGCTATTGGGCATGAAGAGAGC(JVP203 with the BamHI site underlined) and 5′-CCCAAAGTCGACCCTACTGATGCAACTTTAATCC(JVP204 with the SalI site underlined). Plasmid pJB1005 was constructed by inserting a gene encoding chloramphenicol acetyltransferase that was amplified from pKRP10 by using the primers 5′-CCCAAAGGATCCGAGGTTCCAACTTTCACC(JVP206 with the BamHI site underlined) and 5′-CCCAAAGGATCCCTGCCTTAAAAAAATTACGC(JVP207 with the BamHI site underlined) into the BamHI site of plasmid pJB1001.
To make the ΔdotN suicide plasmid, pJB3046, two PCR-amplified fragments were cloned into the NotI/SalI sites of pSR47S (40). Fragment 1 was amplified by using the primers 5′-CCCGCGGCCGCGGTGTATCGTTAGGTAAAATGG(JVP289 with the NotI site underlined) and 5′-CCCGGATCCCGCCATAGTTTGGTTCACATTCAGTC(JVP903 with the BamHI site underlined). Fragment 2 was amplified by using the primers 5′-CCCGGATCCGAGAAATGGGCTGCCAGTGC(JVP904 with the BamHI site underlined) and 5′-CCCGTCGACGCAGCTTTTAACTGATCGC(JVP286 with the SalI site underlined).
To make the ΔdotM suicide plasmid, pJB3050, two PCR-amplified fragments were cloned into the NotI/SalI sites of pSR47S (40). Fragment 1 was amplified by using the primers 5′-CCCGCGGCCGCGAAGCAATCTTCAGTCCTGG(JVP297 with the NotI site underlined) and 5′-CCCGGATCCCTGCTGTTGTTGTGCCATCTC(JVP901 with the BamHI site underlined). Fragment 2 was amplified by using the primers 5′-CCCGGATCCGATGAAGCGATTAGAGCTCTGG(JVP902 with the BamHI site underlined) and 5′-CCCGTCGACGCATACAGAGAGTTATCTCC(JVP294 with the SalI site underlined).
pJB1010, the His-tagged version of DotL, was constructed by amplifying the dotL open reading frame (ORF) using plasmid pJB359 and the primers 5′-GACATGCATGCGATGGGGTTGACTAATTAAGG (JVP217 with the SphI site underlined) and 5′-GACATGCATGCCCCGAAAGCAAAAGTTGCC(JVP218 with the SphI site underlined). The PCR product was digested with SphI and cloned into the SphI site of pQE-32 (Qiagen). The final construct can be used to express a fusion protein containing six histidines fused to amino acids 72 through 783 of DotL.
The dotL complementing clone, pJB1014, was constructed by first amplifying the dotL ORF from Lp02 chromosomal DNA by using the primers 5′-GGGGTACCGGAATTAGAGCCATGATGCG(JVP227 with the KpnI site underlined) and 5′-GACATGCATGCGATGGGGTTGACTAATTAAGG(JVP217 with the SphI site underlined). The resulting product was digested with KpnI and SphI and ligated into KpnI/SphI-digested pJB908. pJB908, a derivative of the plasmid pKB5, has the following features: (i) an RSF1010 origin to permit replication in L. pneumophila, (ii) an ΔoriT mutation to prevent inhibition of growth in macrophages, and (iii) a tac promoter driving DotL expression (3). Constitutive expression from pJB1014 is able to rescue a dotL deletion strain for viability on plates and in macrophages and expresses similar levels of DotL compared to a wild-type strain.
pJB1242, the ΔlvhB suicide plasmid, was constructed by cloning two PCR-amplified fragments into the SalI and NotI sites of pSR47S. Fragment 1 was amplified by using the primers 5′-CCCGTCGACGTTTGGAGAAGTCAGTTTAAGG(JVP342 with the SalI site underlined) and 5′-CCCGGATCCTCATGGCGCCACCTTTTGC(JVP343 with the BamHI site underlined). Fragment 2 was amplified with the primers 5′-CCCGGATCCGAAGCACTCGAACTATAAACC(JVP344 with the BamHI site underlined) and 5′-CCCGCGGCCGCGTTTCGCCATTGTATCCC(JVP345 with the NotI site underlined).
pJB1304, containing the lvhB operon, was constructed by first amplifying the lvhB operon from JR32 chromosomal DNA by using the primers 5′-CCCGTCGACGTTTGGAGAAGTCAGTTTAAGG(JVP342 with the SalI site underlined) and 5′-CCCGCGGCCGCGTTTCGCCATTGTATCCC(JVP345 with the NotI site underlined). The resulting product was digested with SalI and NotI and ligated into SalI/NotI-digested pJB1300. pJB1300, a derivative of the plasmid pKB5 (3), has the HindIII site in the polylinker replaced with a unique NotI site.
Antibody production.
pJB1010, a polyhistidine-tagged version of DotL in which the amino-terminal signal sequence of DotL was replaced with six histidines, was purified by using Ni-nitrilotriacetic acid chromatography (Qiagen). The purified His6-DotL fusion protein was injected into rabbits to raise polyclonal antibodies against DotL (Cocalico). The serum recognized a single protein from wild-type L. pneumophila extracts that was absent in extracts from an E. coli strain and a L. pneumophila strain lacking the dotL gene.
Fractionation and Western analysis.
L. pneumophila was fractionated as previously described (55). Briefly, a culture of Lp02 was grown to mid-exponential phase, and the cells were pelleted and resuspended in 50 mM Tris-HCl (pH 8.0), 0.5 M sucrose, 5 mM EDTA, and 0.1 mg of lysozyme/ml. The cell suspension was incubated on ice for 1 h, MgSO4 was added to a final concentration of 20 mM, and spheroplasts were collected by centrifugation at 5,000 × g. The pellet was resuspended in 50 mM Tris-HCl (pH 8.0), sonicated, and then centrifuged at 5,000 × g to collect any unlysed cells. The supernatant was then centrifuged at 100,000 × g for 1 h at 4°C to obtain a total membrane fraction. The supernatant was removed, centrifuged at 100,000 × g, and saved as the cytoplasmic sample. The pellet was washed and resuspended in 50 mM Tris-HCl (pH 8.0). The inner membranes were solubilized by the addition of Triton X-100, and the outer membranes were collected by centrifugation at 100,000 × g. Fractions were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and either Coomassie blue stained for total protein or transferred to a membrane and probed with the anti-DotL serum (1:5,000).
Cell culture.
The histiocytic cell line U937 (American Type Culture Collection) was cultured in RPMI 1640 media (BioWhittaker) containing 10% fetal bovine serum (BioWhittaker). Cells were differentiated with 12-O-tetradecanoylphorbol-13-acetate (TPA; Sigma) as described previously (3). Differentiated U937 cells were plated as a confluent monolayer in 24-well plates, with each well containing ca. 2 × 106 cells per well.
Southern blot analysis.
L. pneumophila chromosomal DNA was isolated by a combination of a high-salt precipitation to eliminate contaminating proteins, followed by isopropanol precipitation of the DNA. Chromosomal DNA was digested with 10 U of HaeII restriction enzyme overnight at 37°C. Southern blots were performed according to the ECL Southern hybridization kit (Amersham), with probes specific to regions flanking dotL (from pJB1001) or dotB (pJB921).
Transposon mutagenesis of L. pneumophila.
L. pneumophila was mutagenized by using the transposon delivery system encoded on pJK211-2 (13). pJK211-2 contains a temperature-sensitive origin that is not permissive for replication at 37°C, an altered sites transposase that increases the randomness of insertion, and a mini-Tn10 transposon containing a kanamycin cassette (KanR) and a conditional origin from plasmid R6K later used to recover the transposon insertions in E. coli strain DH5α(λpir). A pool of insertions was placed on sucrose chloramphenicol plates to select for recombinants (sucrose to select for recombinants and chloramphenicol to select for the ΔdotL::Cmr). Chloramphenicol-resistant (Cmr), kanamycin-sensitive, sucrose-resistant (Sucr) colonies were colony purified and scored for loss of the plasmid-encoded resistance cassette to ensure they had resolved the integrated plasmid. Insertions were recovered as previously described (13). The site of insertion was identified by sequencing by using the primers JVP348 (GGATCTGGTACCGGATCC) or JVP349 (TCAACAGGTTGAACTGCGGATC).
Screen for suppression of the ΔdotL lethality.
Plasmids pJB1001 and pJB1005 were transferred into L. pneumophila strains by using an RP4 conjugation system encoded on pRK600 (14). L. pneumophila strains containing the integrated plasmid were selected by plating on CYET containing kanamycin and streptomycin. Resulting merodiploid strains that had a second crossover event were selected by plating on CYET plates containing 5% sucrose. Resolution of the integrated plasmid was confirmed by loss of kanamycin resistance. In the case of strains containing the ΔdotL::Cmr cassette, sucrose-resistant colonies were streaked onto chloramphenicol to screen for the wild-type or mutant dotL alleles.
Replication of L. pneumophila strains in U937 cells.
L. pneumophila strains were resuspended in phosphate-buffered saline to an optical density at 600 nm of 1. The bacterial suspensions were then diluted 1:1,000 in RPMI 1640 containing 10% fetal bovine serum, and 2 mM glutamine. A monolayer of TPA-treated U937 cells were infected with various L. pneumophila strains at a multiplicity of infection of one for 1 h. The monolayers were washed with fresh RPMI and then incubated in RPMI 1640 containing 10% fetal bovine serum and 2 mM glutamine at 37°C and 5% CO2. Thymidine was added when appropriate. At 1, 24, 48, and 72 h postinfection, cells were lysed in sterile ddH2O and dilutions were plated on CYET. Plates were incubated for 4 days at 37°C, and viable counts were determined.
Accession numbers.
GenBank accession numbers for submitted sequences are as follows: DotU is AF533658 and DotV is AF533657.
RESULTS
DotL is an inner membrane protein with homology to T4CPs.
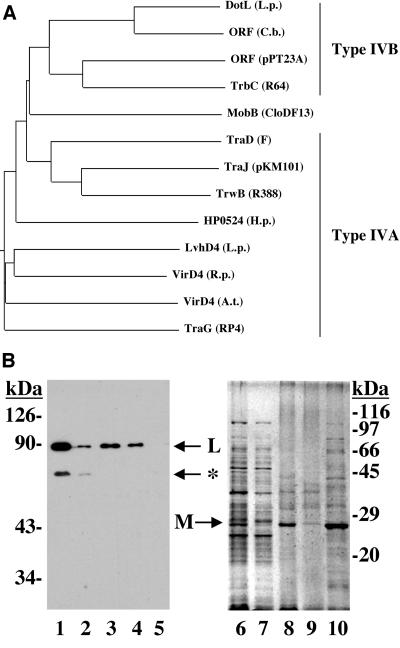
The L. pneumophila DotL protein has extensive similarity to several proteins found in GenBank (Fig. 1A), with the highest degree of similarity (56% identity) to an uncharacterized ORF found in Coxiella burnetii that has been proposed to be part of a type IV secretion system (57). DotL also has similarity to TrbC (27% identity), a protein required for the transfer of the IncI plasmids R64 and ColIb-P9 (Fig. 1, top) (19, 31), and to an ORF on a plasmid found in Pseudomonas syringae strains that may be part of the conjugative transfer apparatus for this plasmid (57). DotL also has sequence similarity, extending primarily over the Walker A box, to members of the type IV coupling protein family, most notably TraD, TraG, TrwB, and VirD4 (Fig. 1A). In addition, DotL shares a number of characteristics with members of the T4CP family. These include a predicted size of 86 kDa, the presence of a potential nucleotide binding motif (a Walker A box), and an amino-terminal hydrophobic sequence that would likely target the protein to the bacterial inner membrane (52, 65). These characteristics, combined with its homology, suggest DotL may be a T4CP.
FIG. 1.
(Top) DotL shows sequence similarity to members of the T4CP family. DotL has extensive similarity to a number of putative type IVB secretion system ATPases, including an uncharacterized ORF in Coxiella burnetii [ORF (C.b.)], the TrbC protein of the IncI plasmid R64 [TrbC (R64)], and a TrbC orthologue on the pPT23A plasmid of Pseudomonas syringae strains [ORF (pPT23A)]. DotL has similarity to plasmid T4CPs (MobB, TraD, TraJ, TrwB, and TraG from the plasmids CloDF13, F, pKM101, R388, and RP4, respectively) and to T4CPs from adapted conjugation systems found in pathogens (HP0524 from Helicobacter pylori, VirD4 from the Ti plasmid of Agrobacterium tumefaciens, and a VirD4 orthologue from Rickettsia prowazekii). Most strains of L. pneumophila contain at least one additional T4CP, LvhD4, which is part of a second type IV secretion system (51). The dendrogram was generated by using CLUSTAL W alignment. (Bottom) The DotL protein is localized to the inner membrane of L. pneumophila. Extracts of wild-type L. pneumophila were separated into cytoplasmic and membrane fractions by high-speed centrifugation. The membrane fractions were then further separated into inner membrane versus outer membrane fractions by extraction with the detergent Triton X-100. Duplicate samples were run on two 7.5% acrylamide gels; the first gel was transferred to a polyvinylidene difluoride membrane and probed with anti-DotL serum (lanes 1 to 5), whereas the second gel was stained with Coomassie blue for total protein (lanes 6 to 10). Lanes 1 and 6 are total cell lysates, lanes 2 and 7 are soluble cytoplasmic fractions, lanes 3 and 8 are total membrane, lanes 4 and 9 are Triton X-100 soluble (inner membrane), and lanes 5 and 10 are Triton X-100 insoluble (outer membrane). All samples were loaded proportionally except for lanes 8, 9, and 10, which were overloaded in order to detect the protein profile (lane 8 is 3-fold, lane 9 is 2-fold, and lane 10 is 25-fold overloaded relative to lanes 1 to 7). The quality of the fractionation procedure can be determined by monitoring the localization of the major outer membrane protein, MOMP, on the Coomassie blue-stained gel (lane 10) (44). A DotL breakdown product detected by Western analysis is indicated with an asterisk (lane 2).
To confirm the subcellular localization of DotL, L. pneumophila extracts were prepared, fractionated, and the protein was detected by Western analysis with a DotL specific antibody. The DotL protein was primarily localized to the membrane fraction (Fig. 1B, lane 3). Moreover, the majority of the protein was Triton X-100 soluble, indicating it was likely to be in the inner membrane of the bacterial cell (Fig. 1B, lane 4) (48). In addition, a smaller cross-reacting species of ca. 75 kDa could be detected that localized completely to the cytoplasmic fraction (Fig. 1B, lane 2) and is consistent with a DotL breakdown product lacking the hydrophobic amino-terminal transmembrane domains.
DotL is essential for viability on bacteriological media.
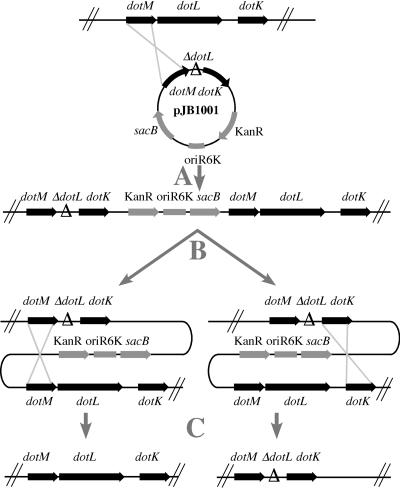
To investigate the function of the DotL protein, we attempted to delete the dotL gene from the chromosome of Lp02, a strain of L. pneumophila with an intact dot/icm system (3, 63). Previous attempts to delete dot/icm genes have been uniformly successful, indicating that the Dot/Icm complex is not required for viability on bacteriological media (1, 3, 63). To construct an in-frame deletion of the dotL gene, ca. 500 bp of DNA upstream and downstream adjacent to the dotL gene was cloned into the suicide vector pSR47S, generating plasmid pJB1001 (Fig. 2). The dotL deletion plasmid was electroporated into strain Lp02 and introduced onto the chromosome by selecting for a single crossover event generating a dotL/ΔdotL merodiploid strain. Merodiploids that had resolved were selected by plating on sucrose, a toxic compound for gram-negative organisms containing the counterselectable marker sacB (4). Resolution of the merodiploid should result in an equal proportion of strains containing either the wild-type copy of dotL or ΔdotL on the chromosome (Fig. 2).
FIG. 2.
Assay for ability of L. pneumophila to tolerate the ΔdotL mutation. A merodiploid consisting of a wild-type copy of dotL and a dotL deletion was constructed by integration of the suicide plasmid pJB1001. This plasmid contains an origin, from the R6K plasmid, that is unable to replicate in L. pneumophila strains lacking the replication protein π (30). pSR47S also contains the selectable marker, Kanr, and a counterselectable marker, sacB, which confers sensitivity to sucrose. The kanamycin marker was used to select for a single crossover generating a merodiploid strain containing both dotL+ and ΔdotL (step A). Recombination between duplicated sequences in the heterozygote was selected by growth on 5% sucrose (step B). If dotL is a nonessential gene, both dotL+ and the ΔdotL will be obtained (step C). If dotL is an essential gene, then only wild-type dotL will be recovered.
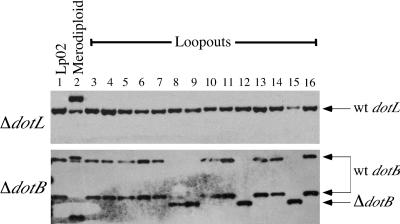
Examination of 14 independent sucrose resistant recombinants derived from the dotL/ΔdotL merodiploid strain revealed no strains that lacked the wild-type copy of dotL (Fig. 3, top panel). In contrast, sucrose resistant recombinants derived from a similarly constructed dotB/ΔdotB merodiploid (55) re sulted in ten strains containing wild-type dotB and four strains containing ΔdotB (Fig. 3, bottom panel). To ensure that the recombination event in the dotL/ΔdotL merodiploid strain was not theoretically impossible, recombinants were selected in a merodiploid containing the dotL+ plasmid pJB1014. In this situation, chromosomal dotL deletions were recovered, indicating that a ΔdotL could be obtained in a strain exogenously expressing DotL (data shown below). Finally, the ΔdotL::Cmr reporter plasmid pJB1005 could not be introduced directly onto the chromosome of the L. pneumophila strain Lp02 by using natural transformation (56), confirming the difficulty of constructing a dotL deletion. These results indicated a strong bias against deleting the wild-type version of dotL and suggested that loss of dotL may result in lethality of L. pneumophila on bacteriological media.
FIG. 3.
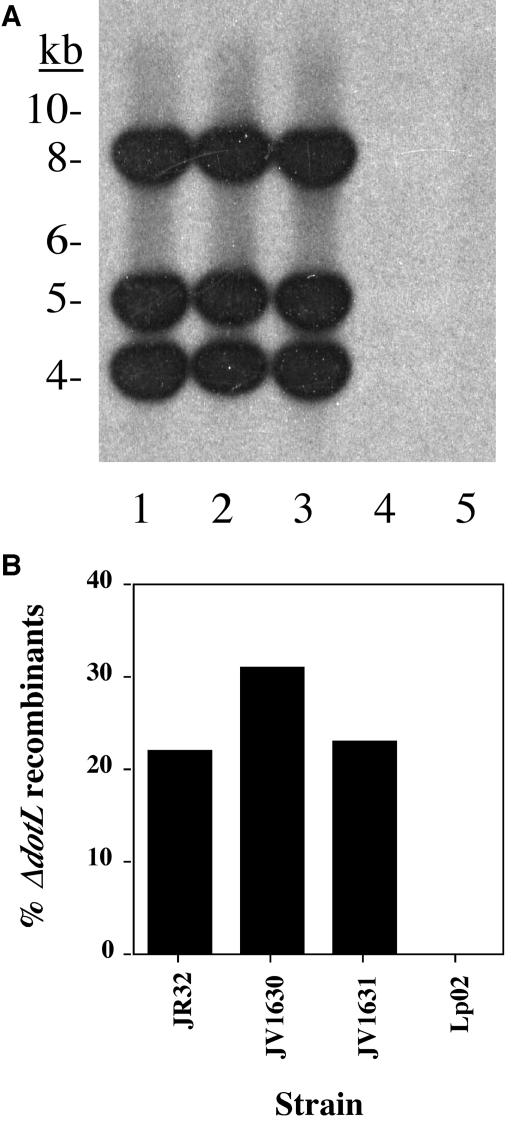
dotL is an essential gene on bacteriological media. dotL and dotB merodiploids were constructed and recombinants selected as described in Fig. 2 were analyzed by Southern analysis (as described in Materials and Methods). The top panel shows a Southern blot of recombinants derived from a parental dotL/ΔdotL merodiploid strain probed with a 700-bp SalI fragment of DNA adjacent to the dotL gene. Lane 1 is the wild-type strain Lp02; lane 2 is JV1003, a ΔdotL/dotL merodiploid; and lanes 3 to 16 are JV1003 plated on CYET plus 5% sucrose. All 14 strains that were selected for sucrose resistance in this fashion retained the wild-type version of dotL. In contrast, the bottom panel is a similar experiment in which sucrose resistant recombinants derived from a dotB/ΔdotB merodiploid strain were probed with a dotB-region specific probe. Lane 1 is the wild-type strain Lp02; lane 2 is JV941, a dotB/ΔdotB merodiploid; and lanes 3 to 16 are JV941 selected on CYET plus 5% sucrose. In this case, two distinct types of recombinants are observed, a finding consistent with either dotB or ΔdotB, indicating that dotB is not an essential gene.
Although it appeared not to be feasible to isolate a strain lacking dotL, it was possible that an insufficient number of events were examined in order to identify such a strain. To screen a larger number of recombination events, the deletion strategy was repeated with a chloramphenicol-marked version of the dotL deletion. A dotL/ΔdotL::Cmr merodiploid was subjected to selection on sucrose, in the absence of chloramphenicol, and the presence of the ΔdotL::Cmr cassette was subsequently screened by plating sucrose resistant recombinants on medium containing chloramphenicol. Examination of a larger number of sucrose-resistant strains still failed to detect a recombinant that contained just the ΔdotL::Cmr allele (0 of 753 events scored). Based on these results, we conclude that dotL is required for the viability of the L. pneumophila strain Lp02 on bacteriological media.
Isolation of suppressors of ΔdotL.
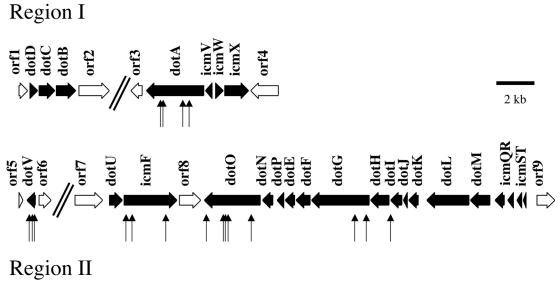
In order to determine whether it was possible to suppress the lethality caused by loss of dotL, we plated an even greater number of the dotL/ΔdotL::Cmr merodiploid on plates containing sucrose and chloramphenicol, thereby directly selecting for loss of dotL. Rare sucrose-resistant, chloramphenicol-resistant recombinants were isolated at a rate of ∼10−6. This was consistent with dotL being an essential gene, with the chloramphenicol-resistant colonies that arose being pseudorevertants due to spontaneous mutations in other genes. To identify the nature of the pseudorevertants, random transposon insertions were generated in the dotL/ΔdotL::Cmr merodiploid strain background by using a mini-Tn10 transposon, and the insertion pool was plated on sucrose and chloramphenicol to select for strains that could tolerate loss of dotL. Thirty-three such insertions were isolated from independent pools. These strains were first analyzed by Southern blot to ensure that they had only one insertion. To confirm that the phenotype was linked to the transposon insertion, the strains were recreated by transforming the transposon and flanking chromosomal DNA into the original, unmutagenized merodiploid strain by using natural transformation (56). Examination of the 33 strains by this assay demonstrated that, in each case, the phenotype was linked to the transposon insertion. Finally, the transposons and flanking DNA were recovered on a plasmid, and the sites of the transposon insertions on the L. pneumophila chromosome were identified by sequencing off the end of each transposon.
Surprisingly, approximately one-half of the insertions (16 of 33) were in other dot/icm genes. This included four insertions in dotA, two in dotG, one in dotI, five in dotO, three in icmF, and one in icmX (Fig. 4). In most cases, the phenotype appeared to be due to inactivation of the gene the transposon was inserted in, because the insertions were in terminal genes of proposed operons (e.g., dotA, dotO, icmF, and icmX). Among the insertions that were not in known dot/icm genes, three mutants (JV1308, JV1343, and JV1499) were defective for intracellular growth of L. pneumophila when the insertions were separated from the ΔdotL (data not shown). The three mutants each contained an insertion in a different site of the same gene, which is located ca. 20 kb from region II (Fig. 4) (63). This gene codes for a small protein of 180 amino acids that has extensive homology to DotE (40% amino acid identity over 171 amino acids). We have designated this gene dotV because it is required for proper targeting of the L. pneumophila phagosome and for intracellular growth (unpublished results) (accession no. AF533657). Finally, the remaining insertions were not in known dot/icm genes or homologous genes and, when separated from the dotL deletion, caused the corresponding strains to exhibit various degrees of growth inhibition inside host cells (data not shown).
FIG. 4.
Mini-Tn10 insertions in multiple dot/icm genes suppress the lethality caused by loss of dotL. The dotL/ΔdotL merodiploid strain, JV1003, was mutagenized with mini-Tn10, and viable strains harboring ΔdotL were directly selected on sucrose-chloramphenicol-containing plates. Shown are the L. pneumophila dot/icm regions I and II (63). dot/icm genes are indicated with filled arrows, whereas flanking genes that are not required for intracellular growth are designated by open arrows (ORFs 1 to 9). Region I contains an 8-kb intervening region, which contains apparent housekeeping genes, separating the two dot/icm loci. dotV is separated from the rest of the dot/icm genes in region II by 20 kb. Mini-Tn10 insertions that suppressed the ΔdotL lethality were found in dotA, dotG, dotI, dotO, dotV, and icmF, and the sites of insertions are indicated with vertical arrows.
Other dot/icm mutations suppress loss of dotL.
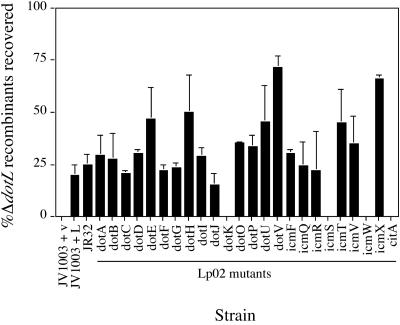
To confirm that loss of a specific dot gene could suppress a ΔdotL, we attempted to delete dotL in a strain containing an in-frame deletion of the dotA gene. In contrast to the previous attempt to delete dotL (Fig. 3), both dotL and ΔdotL loopouts were obtained from the ΔdotA dotL/ΔdotL merodiploid, demonstrating that loss of a single dot gene could allow the isolation of the ΔdotL mutation (Fig. 5). Because the ΔdotL suppressor hunt identified only a subset of dot/icm genes, we investigated whether they were the only dot/icm genes that, when inactivated, could suppress the ΔdotL lethality. In-frame deletions were constructed in 23 of the 26 dot/icm genes, and the ΔdotL suicide plasmid was integrated into each strain to assay for the ability to tolerate loss of dotL. Remarkably, dotL could be deleted in almost all of the strains containing different dot/icm mutations (Fig. 6). This suppression was specific in that dotL could not be deleted in a strain lacking a housekeeping gene found in region II, citA, which is not required for intracellular growth (Fig. 6) (42).
FIG. 5.
The dotL gene can be deleted in a strain lacking dotA. Southern blot analysis of ΔdotL recombinants in a ΔdotA background. A 700-bp SalI fragment from pJB1001 encoding DNA flanking dotL on the chromosome was used as a probe to determine the status of dotL in these strains. Lane 1 is the wild-type strain Lp02; lane 2 is JV1005, a ΔdotL/dotL merodiploid in a dotA mutant background; and lanes 3 to 16 are JV1005 resolved on CYET plus 5% sucrose.
FIG. 6.
Deletion of most dot/icm genes can suppress the lethality caused by deletion of dotL. A dotL/ΔdotL::CmR merodiploid was constructed in a variety of different dot/icm backgrounds. Sucrose-resistant recombinants were selected and then screened for the ΔdotL allele by resistance to chloramphenicol. ΔdotL recombinants could not be obtained from the dotL/ΔdotL::Cmr merodiploid strain JV1003. The presence of a wild-type copy of dotL on a low-copy vector allowed the isolation of ΔdotL recombinants (JV1003 plus dotL). Inactivation of 20 of 23 dot/icm genes suppressed the loss of dotL. In addition, deletion of citA/tphA, a housekeeping gene found near the dot/icm genes, did not allow loss of dotL (42). In contrast, dotL is not essential in a related L. pneumophila strain, JR32. The data shown reflects the average number of ΔdotL recombinants recovered from scoring 50 events from four independent experiments.
In contrast, inactivation of three dot/icm genes, dotK, icmS, and icmW, did not suppress loss of dotL (Fig. 6). dotK encodes an outer membrane protein with homology to OmpA and is only partially required for growth in amoebae (53). icmS and icmW encode two small, acidic, cytoplasmic proteins that have been proposed to function as secretion chaperones (12). Similar to dotK, icmS and icmW are not absolutely required for the growth of L. pneumophila in permissive cell lines such as U937s and HL60s (12, 53). Therefore, it was possible that inactivation of these genes failed to suppress loss of dotL simply because they are not absolutely required for intracellular growth. However, inactivation of three other dot/icm genes—icmF, dotU, and icmR—did suppress loss of dotL (Fig. 6), even though loss of these genes caused only a partial inhibition of growth in permissive hosts (12, 53, 54, 62, 68). These results indicate that, although inactivation of the majority of the dot/icm genes can suppress the lethality caused by loss of dotL, there is specificity to the suppression.
dotL is not essential in all L. pneumophila strain backgrounds.
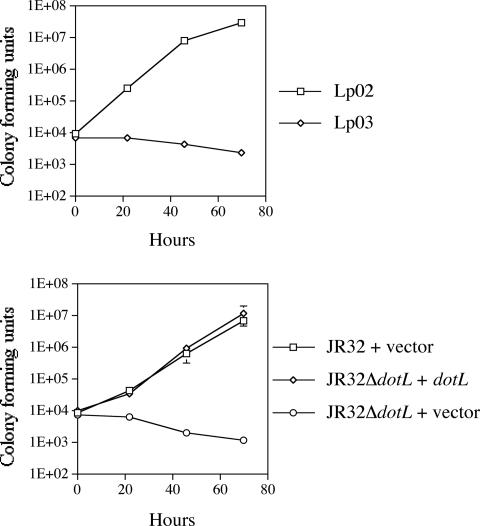
dotL, also known as icmO, is essential for viability in the Lp02 background. However, it has been previously published that loss of dotL in another L. pneumophila strain, JR32, is not a lethal event (52). Lp02 and JR32 were independently derived from L. pneumophila Philadelphia-1, an organism isolated from the original Legionnaires' disease outbreak in 1976 (3, 38). Each strain was individually selected to be streptomycin resistant and to lack host restriction, and Lp02 was then also selected to be a thymidine auxotroph. Due to the relatedness of these two strains, it was surprising that dotL/icmO was essential for viability in Lp02 but was dispensable in JR32. To confirm that dotL was not an essential gene in JR32, a clean ΔdotL was constructed in the JR32 background and was indeed found to be viable on buffered CYE plates (data not shown). To examine intracellular growth, monolayers of U937 macrophages were challenged with wild-type L. pneumophila strains Lp02 and JR32. Both strains were able to multiply >1,000-fold in 3 days (Fig. 7). In contrast, an Lp02 strain lacking a functional dotA gene, Lp03, was unable to replicate inside macrophages. As previously shown, deletion of dotL in JR32 prevented the strain from replicating in U937 macrophages, and this defect could be complemented by the addition of a plasmid containing dotL+ (Fig. 7) (52).
FIG. 7.
dotL is required for growth of the L. pneumophila strain JR32 in U937 cells. A number of L. pneumophila strains were assayed for their ability to replicate inside U937 cells over 3 days. The top panel includes Lp02 (wild type) and Lp03 (a dotA mutant derivative of Lp02) as controls. The bottom panel includes JR32 containing the vector pJB908, a JR32ΔdotL strain containing the dotL+ complementing clone pJB1014, and a JR32ΔdotL strain containing the vector pJB908. The data shown are the average of triplicate samples and are representative of two independent experiments.
Although the JR32 ΔdotL strain was viable, closer examination revealed that it displayed a key difference from other dot/icm mutants (Table 2). Wild-type L. pneumophila stains, such as Lp02 and JR32, exhibit a significantly decreased plating efficiency on buffered CYE plates containing a low amount of sodium chloride (0.65%) compared to growth on plates lacking sodium chloride (11, 45, 64). Most dot/icm deletions (e.g., ΔdotA) exhibit an increased plating efficiency on plates supplemented with salt (Table 2). However, the JR32 ΔdotL strain was even more sensitive to sodium chloride than JR32 (Table 2), suggesting that the physiology of the JR32 ΔdotL is perturbed. These data suggest that loss of dotL in either the Lp02 strain or the JR32 strain is detrimental to the cell.
TABLE 2.
Deletion of dotL in JR32 confers an enhanced salt sensitivity to the strain
| Strain | Genotype | Plating efficiencya (%) | Phenotype |
|---|---|---|---|
| Lp02 | Wild type | 0.09 | Salt sensitive |
| Lp03 | Lp02 dotA | 13 | Salt resistant |
| JR32 | Wild type | 0.05 | Salt sensitive |
| JV2153 | JR32 ΔdotB | 9.0 | Salt resistant |
| JV2114 | JR32 ΔdotL | 0.002 | Hyper-salt sensitive |
The plating efficiency was calculated as the number of colonies on a buffered CYE plate containing 0.65% sodium chloride divided by the number of colonies on a buffered CYE plate × 100.
Mutation of lvhD4 does not suppress the lethality caused by loss of dotL.
Since the sequences of the dot/icm genes are identical between Lp02 and JR32, it is likely that there is an additional genetic difference between the two strains responsible for the more severe effect of deleting dotL in Lp02. For example, a gene may have been inactivated or lost during the derivation of JR32 that allows the JR32 ΔdotL strain to survive. Alternatively, a gene may be absent in Lp02 that is normally able to suppress the lethality caused by loss of dotL. In fact, a number of differences have been reported between various L. pneumophila serogroup I isolates including Lp01, the progenitor strain of Lp02, and JR32 (Table 1) (6, 36, 47). One potential candidate is lvhD4, which is present in JR32 but not in Lp01 (47). lvhD4 is encoded in the lvhB1-11/lvhD operon and is a component of a second type IV secretion system found in L. pneumophila strains such as JR32 (51). lvhD4 encodes a protein with similarity to T4CPs, most specifically to the A. tumefaciens VirD4 (51), and could in theory functionally substitute for DotL.
To confirm that the JR32 and Lp02 isolates we were working with contained and lacked lvhD4, respectively, we performed Southern analysis with a probe specific to lvhD4 (Fig. 8, top). Consistent with previous reports, Lp01 and Lp02 lacked lvhD4, whereas JR32 and Philadelphia-1, the progenitor strain for both Lp02 and JR32, both contained it (Fig. 8A). Lp01 and Lp02 may have lost the lvhB-lvhD region during their derivation to become restriction minus, since a number of restriction or modification genes are located adjacent to the lvhB/lvhD4 system (47). To determine whether there was a connection between the presence of lvhD4 and ΔdotL lethality, we deleted lvhD4 from JR32 or added it back to Lp02 and then assayed the consequence of deleting dotL. dotL could still be deleted in a JR32 strain lacking the lvhB-lvhD4 region, indicating that lvhD4 was not responsible for the viability of the JR32 ΔdotL strain (Fig. 8B). Likewise, the addition of the lvhB-lvhD4 region from JR32 to the Lp02 strain did not suppress loss of dotL. Therefore, lvhD4 does not appear to be responsible for the altered requirements of dotL in these two L. pneumophila strains.
FIG. 8.
The lvhB operon is not responsible for viability of the JR32 ΔdotL strain. (Top) The presence of the lvhB/lvhD operon in a variety of L. pneumophila strains was assayed by Southern analysis with a probe that contains the entire operon: lane 1 is the Philadelphia-1 progenitor of JR32, lane 2 is JR32, lane 3 is the Philadelphia-1 progenitor of Lp01 and Lp02, lane 4 is Lp01, and lane 5 is Lp02. (Bottom) A dotL/ΔdotL merodiploid of JR32 can be resolved to the ΔdotL, indicating it is not an essential gene. Two independently derived JR32 strains lacking the lvhB operon, JV1630 and JV1631, still allow deletion of dotL, whereas dotL is essential for viability in Lp02. The data shown reflect the average number of ΔdotL recombinants recovered from scoring 50 events from four independent experiments.
In addition to the dot/icm and the lvhB systems, certain L. pneumophila strains contain an additional type IV secretion system (6). This system is encoded in an ca. 65-kb locus, LpPI-1, that bears the hallmarks of a pathogenicity island. It contains homologues to a type IV secretion system that resembles the F plasmid, including a T4CP that resembles the F plasmid TraD protein, mobile genetic elements, and several putative virulence factors (6). In contrast to lvhD4, LpPI-1 is present in Lp02 but is absent from JR32. However, deletion of the TraD-like protein from Lp02 did not suppress the lethality caused by loss of dotL (C. Vincent and J. P. Vogel, unpublished data). Thus, some additional, as-yet-uncharacterized, mutation must exist in one of these strains that is responsible for the differential requirement of dotL for viability.
dotM and dotN are also essential for viability in the Lp02 background.
While constructing a collection of dot/icm deletions, we were able to generate in-frame deletions in 23 of the 26 known dot/icm genes. However, similar to the dotL deletion, we could not construct a deletion in dotM, the gene upstream of dotL (Table 3). dotM, also known as icmP, codes for a predicted inner membrane protein with similarity to TrbA of the IncI plasmids R64 and ColIb-P9 (24% amino acid identity). Since dotL and dotM are likely to be cotranscribed in a two gene operon, dotML, it was possible that the lethality of the dotM deletion was due solely to polarity on the downstream dotL gene (Fig. 4). However, the ΔdotM mutation could not be obtained in the presence of a complementing clone containing a wild-type version of dotL, suggesting that the dotM lethality was not due to polarity but reflected the essentiality of dotM (Table 3). Moreover, insertions in dotM were obtained in a screen for genes that resemble dotL, i.e., genes that are essential in the presence of a functional Dot/Icm complex (13).
TABLE 3.
dotM and dotN are required for the viability of Lp02
| Strain | Genotype | No. of ΔdotM or ΔdoctN recombinants/totala |
|---|---|---|
| JV2097 | dotM+/ΔdotM::Cmr | 0/600 |
| JV2465 | dotM+/ΔdotM::Cmr + pdotL+ | 0/280 |
| JV2238 | dotM+/ΔdotM::Cmr ΔdotA | 172/400 |
| JV2100 | dotN+/ΔdotN::Cmr | 0/700 |
| JV2240 | dotN+/ΔdotN::Cmr ΔdotA | 158/400 |
This value was determined as the number of events in which the merodiploid resolved to either ΔdotM::Cmr or to ΔdotN::Cmr.
A third gene, dotN, also proved difficult to delete from the L. pneumophila chromosome. dotN, also known as icmJ, is located ca. 12 kb downstream of the proposed dotML operon and is the first gene of another predicted operon, dotNO (Fig. 4). dotN codes for a small protein of 208 amino acids that contains a high proportion of cysteines (4.3%). The lethality of the ΔdotN could not be due to simple polarity on the downstream gene dotO because deletions could easily be made in the dotO gene. Similar to dotL, both dotM and dotN could each be deleted in strains lacking a functional dot complex (Table 3). These results indicate that three dot genes, dotL, dotM, and dotN are each essential for viability on bacteriological media in the Lp02 background and in each case, the lethality can be suppressed by inactivation of the Dot/Icm complex.
DISCUSSION
The dot/icm genes are required for the intracellular replication of L. pneumophila and encode a type IVB secretion system that appears to have evolved from the conjugation apparatus of an IncI plasmid. We have demonstrated here that three dot genes, dotL, dotM, and dotN, are essential for growth of L. pneumophila strain Lp02 on bacteriological media. This is in direct contrast to the established paradigm that the dot/icm genes are dispensable under the laboratory conditions of growth on plates (3, 37). In addition, we were able to isolate a large collection of suppressors of the ΔdotL lethality and have shown that the majority of these map to other dot/icm genes. However, inactivation of several dot/icm genes (dotK, icmS, and icmW) did not suppress loss of dotL, indicating specificity to the suppression.
DotL has limited homology to the T4CP family of proteins. T4CPs have been proposed to play a central role in type IV secretion systems (34). They have been shown to bind substrates synthesized in the cytoplasm and target them to the secretion apparatus in the inner membrane (2, 15, 61). T4CPs have also been shown to interact with other components of the secretion apparatus, namely, the VirB10-family of proteins (20, 35). Finally, T4CPs are absolutely required for export of substrates (22). Based on their homology to Escherichia coli FtsK and Bacillus subtilis SpoIIIE, and their ability to bind DNA, T4CPs have been proposed to function as molecular pumps, driving export of substrates via hydrolysis of ATP (22). In consideration of these traits, T4CPs would appear to be likely candidates to function as regulators of the type IV secretion complexes.
Based on the similarity of DotL to T4CPs, it is surprising that inactivation of the dotL gene in strain Lp02 is lethal. No other known T4CP is essential for viability. Moreover, the only proteins associated with conjugative transfer that that are required for bacterial viability are inhibitors of plasmid toxin segregation factors (43). DotL, however, shows no sequence similarity to such factors. In addition, if DotL functioned as an inhibitor of a plasmid segregation toxin, then the ΔdotL lethality suppressors would be predicted to map to the toxin. In contrast, many of the ΔdotL suppressors are components of the Dot/Icm machinery, and the non-dot/icm suppressors do not have homology to any known toxin inhibitors.
To explain these overall observations regarding toxicity induced by loss of dotL, we propose that loss of the DotL protein results in the accumulation of a toxic structure consisting of a portion of the Dot/Icm complex (Fig. 9). This partial Dot/Icm complex could be deleterious for a number of different reasons. First, a partial Dot/Icm complex could misassemble or misfold in the absence of DotL, disrupting the membrane in some fashion. Alternatively, loss of dotL could be toxic because the type IV secretion system forms an unregulated pore in the membrane in the absence of DotL (Fig. 9). In this model, DotL would play the role of a regulator of the complex, controlling the opening and closing of the pore.
FIG. 9.
Model for potential DotL regulation of the Dot/Icm translocator. (A) In wild-type L. pneumophila strains, the Dot/Icm proteins form a secretion apparatus in the membrane, which is used to export substrate(s). DotL is shown interacting with the complex as a hexameric gate based on homology to the hexameric T4CP, TrwB (21). Translocated substrates would be exported through the complex after interacting with DotL on the cytoplasmic face of the inner membrane. (B) During conditions in which L. pneumophila is not actively secreting substrates, the export apparatus would be closed via DotL and potentially substrates such as LidA (indicated as a ball) (13). (C) In the absence of DotL, the secretion pore might remain constitutively open and the cell would die, possibly due to cell lysis. (D) Inactivation of the Dot/Icm complex would suppress the ΔdotL lethality since an unregulated pore would no longer exist.
We favor the unregulated pore model for the following reasons. First, if a misfolded subcomplex were the cause of the lethality one would not anticipate that inactivation of the majority of dot/icm genes (20 of 23) would suppress the loss of dotL. Second, the JR32 ΔdotL phenotype, increased sensitivity to sodium relative to a wild-type strain, is much more consistent with an unregulated pore. Although the sodium sensitivity of wild-type L. pneumophila strains is not well understood, it is believed to result from leakage of sodium ions through the Dot/Icm secretion apparatus (11, 64). This model is supported by the observation that strains resistant to sodium chloride often contain mutations in dot/icm genes (63). Taken in this context, loss of a regulator of the secretion pore is predicted to enhance the effect of exogenous sodium and is consistent with the hypersensitivity of the JR32 ΔdotL. Finally, there is precedence in the literature of an example in which loss of a protein resulted in an unregulated pore that can be lethal under certain circumstances. Inactivation of Yersinia pestis lcrG results in an unregulated type III secretion pore under certain conditions and has led to the model where LcrG forms a plug at the base of the apparatus (39, 58).
Based on the phenotype of a strain lacking dotL, mutations that cause lowered viability in the presence of an intact Dot/Icm apparatus were previously isolated (13). lidA was shown to encode a protein exported by the Dot/Icm system that may interact directly with DotL (13). Other lid genes may encode proteins necessary for proper assembly of the Dot/Icm complex, particularly a subcomplex consisting of DotL, DotM, and DotN. For example, three Lid proteins are involved in disulfide bond metabolism and, since the DotN protein is rich in cysteine residues, it may be that mutations affecting the formation of disulfide bonds could disrupt folding of DotN (13).
The ΔdotL lethality phenotype in Lp02 has proven to be useful for several additional reasons. First, it has provided a convenient plate selection for additional dot/icm mutants. This is noteworthy because many of the dot/icm genes were identified by labor-intensive screens that have never been performed to saturation (1, 3, 45). The only selection for dot/icm mutants previously available was based on the phenomenon that sodium-resistant L. pneumophila strains were often avirulent, although this phenomenon is poorly understood and may be mutagenic (11, 64). The benefit of our new selection is amply demonstrated since we have already identified an additional dot/icm gene, dotV, by this procedure.
The ΔdotL lethality phenotype also provides information about existing Dot/Icm proteins. A number of Dot/Icm proteins that appear to be primarily cytoplasmic and not membrane associated were still able to suppress the loss of DotL when their genes were inactivated. For example, IcmQ and IcmR have been shown to be soluble proteins in the cytoplasm of L. pneumophila where IcmR appears to function as a chaperone for IcmQ (16). Although the specific function of IcmQ remains unknown, the fact that ΔicmQ and ΔicmR were able to suppress the lethality caused by the ΔdotL suggests that they are directly required for the assembly or activity of the Dot/Icm complex. Another example is the DotB ATPase (55). Although DotB does not appear to be an integral component of the Dot/Icm membrane complex, it is required for expression of the ΔdotL lethality trait, thus indicating that the protein plays a role in the assembly and/or function of the apparatus.
In contrast, inactivation of icmS or icmW did not suppress loss of dotL. Since icmS and icmW are predicted to encode cytoplasmic proteins and have been proposed to function as chaperones for secreted substrates (12), their failure to suppress is consistent with our model. Moreover, inactivation of a secreted substrate ralF (41) also failed to suppress loss of dotL (unpublished results). One additional Dot/Icm protein, the putative lipoprotein DotK, was also not required for ΔdotL lethality. Combined with the observation that a ΔdotK strain shows only mild defects for intracellular growth (53), this suggests that DotK is not essential for the formation of the Dot/Icm complex. Further examination of how various dot/icm mutants are able to suppress loss of dotL may reveal information on which components are key to formation of the secretion pore.
A third interesting observation that resulted from our analysis of the ΔdotL lethality involved dotM and dotN. Similar to dotL, we discovered that dotM and dotN are also essential for viability in the Lp02 background and are not essential for the viability of JR32 on bacteriological media but are required for growth of JR32 inside macrophages (50). Since all three proteins appear to code for inner membrane components of the secretion apparatus, it is possible that DotM and DotN interact with DotL and regulate its activity, perhaps by modulating its proposed nucleotide hydrolysis capability. In fact, we have recently shown that DotM can be coimmunoprecipitated by using DotL specific antibodies (Vincent and Vogel, unpublished).
It is interesting that deleting dotL in two very closely related strains results in very different phenotypes: death versus life. This is likely to be due to a genetic difference between the two strains acquired during their derivation. The JR32 strain may have acquired a suppressor mutation or Lp02 may have lost a gene that prevents ΔdotL lethality. One difference between these strains is that Lp02 lacks the second type IV secretion system encoded by the lvhB operon (47). However, deletion of the lvhB operon in JR32 did not cause the dotL deletion to be lethal, and therefore the identity of the suppressor(s) remains to be discovered. Nevertheless, the difference between these two strains may not be as profound as it initially appeared, since the JR32 ΔdotL strain is less fit than a wild-type strain, as demonstrated by its hyper-NaCl sensitivity. It is possible that the difference in phenotypes between the two strains is more a matter of degrees of sensitivity to loss of dotL rather than JR32 being impervious to its loss.
The ΔdotL phenotype described here is consistent with the proposal that T4CPs function as inner membrane gates for exported substrates (49). Further characterization of this interesting phenomenon should shed light not only on the function of DotL and other T4CPs but also on the L. pneumophila Dot/Icm complex and other type IV secretion systems.
Acknowledgments
We thank James Kirby for the generous gift of pJK211-2. We also thank Jessica Sexton and Carr Vincent for suggestions and critical reading of the manuscript.
G.M.C. was supported by training program J32AI1007422. This study was funded by the Whitaker Foundation (J.P.V.), the American Lung Association (J.P.V.), and NIH grant AI48052-01A2 (J.P.V.) and by funding from the Howard Hughes Medical Institute to R.R.I.
REFERENCES
- 1.Andrews, H. L., J. P. Vogel, and R. R. Isberg. 1998. Identification of linked Legionella pneumophila genes essential for intracellular growth and evasion of the endocytic pathway. Infect. Immun. 66:950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmakuri, K., Z. Ding, and P. J. Christie. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol. Microbiol. 49:1699-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, K. H., and R. R. Isberg. 1993. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol. Microbiol. 7:7-19. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisenstein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 5.Brand, B. C., A. B. Sadosky, and H. A. Shuman. 1994. The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol. Microbiol. 14:797-808. [DOI] [PubMed] [Google Scholar]
- 6.Brassinga, A. K., M. F. Hiltz, G. R. Sisson, M. G. Morash, N. Hill, E. Garduno, P. H. Edelstein, R. A. Garduno, and P. S. Hoffman. 2003. A 65-kilobase pathogenicity island is unique to Philadelphia-1 strains of Legionella pneumophila. J. Bacteriol. 185:4630-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner, D. J., A. G. Steigerwalt, and J. E. McDade. 1979. Classification of the Legionnaires' disease bacterium: Legionella pneumophila, genus novum, species nova, of the family Legionellaceae, familia nova. Ann. Intern. Med. 90:656-658. [DOI] [PubMed] [Google Scholar]
- 8.Cabezon, E., E. Lanka, and F. de la Cruz. 1994. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J. Bacteriol. 176:4455-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabezon, E., J. I. Sastre, and F. de la Cruz. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400-406. [DOI] [PubMed] [Google Scholar]
- 10.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catrenich, C. E., and W. Johnson. 1989. Characterization of the selective inhibition of growth of virulent Legionella pneumophila by supplemented Mueller-Hinton medium. Infect. Immun. 57:1862-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coers, J., J. C. Kagan, M. Matthews, H. Nagai, D. M. Zuckman, and C. R. Roy. 2000. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol. Microbiol. 38:719-736. [DOI] [PubMed] [Google Scholar]
- 13.Conover, G. M., I. Derre, J. P. Vogel, and R. R. Isberg. 2002. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 48:305-321. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 15.Disque-Kochem, C., and B. Dreiseikelmann. 1997. The cytoplasmic DNA-binding protein TraM binds to the inner membrane protein TraD in vitro. J. Bacteriol. 179:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dumenil, G., and R. R. Isberg. 2001. The Legionella pneumophila IcmR protein exhibits chaperone activity for IcmQ by preventing its participation in high-molecular-weight complexes. Mol. Microbiol. 40:1113-1127. [DOI] [PubMed] [Google Scholar]
- 17.Fields, B. S. 1996. The molecular ecology of legionellae. Trends Microbiol. 4:286-290. [DOI] [PubMed] [Google Scholar]
- 18.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuya, N., and T. Komano. 1996. Nucleotide sequence and characterization of the trbABC region of the IncI1 plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J. Bacteriol. 178:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilmour, M. W., J. E. Gunton, T. D. Lawley, and D. E. Taylor. 2003. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 49:105-116. [DOI] [PubMed] [Google Scholar]
- 21.Gomis-Ruth, F. X., G. Moncalian, F. de la Cruz, and M. Coll. 2002. Conjugative plasmid protein TrwB, an integral membrane type IV secretion system coupling protein. Detailed structural features and mapping of the active site cleft. J. Biol. Chem. 277:7556-7566. [DOI] [PubMed] [Google Scholar]
- 22.Gomis-Ruth, F. X., M. Sola, F. de la Cruz, and M. Coll. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr. Pharm. Des. 10:1551-1565. [DOI] [PubMed] [Google Scholar]
- 23.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz, M. A. 1987. Characterization of avirulent mutant Legionella pneumophila that survive but do not multiply within human monocytes. J. Exp. Med. 166:1310-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz, M. A. 1983. Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J. Exp. Med. 158:1319-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horwitz, M. A. 1983. The Legionnaires' disease bacterium (Legionella pneumophila) inhibits phagosome-lysosome fusion in human monocytes. J. Exp. Med. 158:2108-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz, M. A., and S. C. Silverstein. 1980. Legionnaires′ disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J. Clin. Investig. 66:441-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagan, J. C., and C. R. Roy. 2002. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell. Biol. 4:945-954. [DOI] [PubMed] [Google Scholar]
- 30.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 31.Komano, T., T. Yoshida, K. Narahara, and N. Furuya. 2000. The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol. Microbiol. 35:1348-1359. [DOI] [PubMed] [Google Scholar]
- 32.Lessl, M., D. Balzer, K. Weyrauch, and E. Lanka. 1993. The mating pair formation system of plasmid RP4 defined by RSF1010 mobilization and donor-specific phage propagation. J. Bacteriol. 175:6415-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lessl, M., W. Pansegrau, and E. Lanka. 1992. Relationship of DNA-transfer-systems: essential transfer factors of plasmids RP4, Ti and F share common sequences. Nucleic Acids Res. 20:6099-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz Fd. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 35.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luneberg, E., B. Mayer, N. Daryab, O. Kooistra, U. Zahringer, M. Rohde, J. Swanson, and M. Frosch. 2001. Chromosomal insertion and excision of a 30 kb unstable genetic element is responsible for phase variation of lipopolysaccharide and other virulence determinants in Legionella pneumophila. Mol. Microbiol. 39:1259-1271. [DOI] [PubMed] [Google Scholar]
- 37.Marra, A., S. J. Blander, M. A. Horwitz, and H. A. Shuman. 1992. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc. Natl. Acad. Sci. USA 89:9607-9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marra, A., and H. A. Shuman. 1989. Isolation of a Legionella pneumophila restriction mutant with increased ability to act as a recipient in heterospecific matings. J. Bacteriol. 171:2238-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matson, J. S., and M. L. Nilles. 2001. LcrG-LcrV interaction is required for control of Yops secretion in Yersinia pestis. J. Bacteriol. 183:5082-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagai, H., J. C. Kagan, X. Zhu, R. A. Kahn, and C. R. Roy. 2002. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295:679-682. [DOI] [PubMed] [Google Scholar]
- 42.Purcell, M., and H. A. Shuman. 1998. The Legionella pneumophila icmGCDJBF genes are required for killing of human macrophages. Infect. Immun. 66:2245-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rawlings, D. E. 1999. Proteic toxin-antitoxin, bacterial plasmid addiction systems and their evolution with special reference to the pas system of pTF-FC2. FEMS Microbiol. Lett. 176:269-277. [DOI] [PubMed] [Google Scholar]
- 44.Roy, C. R., and R. R. Isberg. 1997. Topology of Legionella pneumophila DotA: an inner membrane protein required for replication in macrophages. Infect. Immun. 65:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sadosky, A. B., L. A. Wiater, and H. A. Shuman. 1993. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect. Immun. 61:5361-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmond, G. P. C. 1994. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32:181-200. [Google Scholar]
- 47.Samrakandi, M. M., S. L. Cirillo, D. A. Ridenour, L. E. Bermudez, and J. D. Cirillo. 2002. Genetic and phenotypic differences between Legionella pneumophila strains. J. Clin. Microbiol. 40:1352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schnaitman, C. A. 1971. Solubilization of the cytoplasmic membrane of Escherichia coli by Triton X-100. J. Bacteriol. 108:545-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schroder, G., S. Krause, E. L. Zechner, B. Traxler, H. J. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Segal, G., M. Purcell, and H. A. Shuman. 1998. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl. Acad. Sci. USA 95:1669-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 52.Segal, G., and H. A. Shuman. 1997. Characterization of a new region required for macrophage killing by Legionella pneumophila. Infect. Immun. 65:5057-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segal, G., and H. A. Shuman. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72:5983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sexton, J. A., J. S. Pinkner, R. Roth, J. E. Heuser, S. J. Hultgren, and J. P. Vogel. 2004. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J. Bacteriol. 186:1658-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sexton, J. A., and J. P. Vogel. 2004. Regulation of hypercompetence in Legionella pneumophila. J. Bacteriol. 186:3814-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sexton, J. A., and J. P. Vogel. 2002. Type IVB secretion by intracellular pathogens. Traffic 3:178-185. [DOI] [PubMed] [Google Scholar]
- 58.Skryzpek, E., and S. C. Straley. 1993. LcrG, a secreted protein involved in negative regulation of the low- calcium response in Yersinia pestis. J. Bacteriol. 175:3520-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Swanson, M. S., and R. R. Isberg. 1995. Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infect. Immun. 63:3609-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Szpirer, C. Y., M. Faelen, and M. Couturier. 2000. Interaction between the RP4 coupling protein TraG and the pBHR1 mobilization protein Mob. Mol. Microbiol. 37:1283-1292. [DOI] [PubMed] [Google Scholar]
- 62.VanRheenen, S. M., G. Dumenil, and R. R. Isberg. 2004. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect. Immun. 72:5972-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vogel, J. P., H. L. Andrews, S. K. Wong, and R. R. Isberg. 1998. Conjugative transfer by the virulence system of Legionella pneumophila. Science 279:873-876. [DOI] [PubMed] [Google Scholar]
- 64.Vogel, J. P., C. Roy, and R. R. Isberg. 1996. Use of salt to isolate Legionella pneumophila mutants unable to replicate in macrophages. Ann. N. Y. Acad. Sci. 797:271-272. [DOI] [PubMed] [Google Scholar]
- 65.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zusman, T., M. Feldman, E. Halperin, and G. Segal. 2004. Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect. Immun. 72:3398-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]