Abstract
Virulent lactococcal prolate (or c2-like) phages are the second most common phage group that causes fermentation failure in the dairy industry. We have mapped two host range determinants in two lactococcal prolate phages, c2 and 923, for the host strains MG1363 and 112. Each phage replicates on only one of the two host strains: c2 on MG1363 and 923 on 112. Phage-phage recombinants that replicated on both strains were isolated by a new method that does not require direct selection but rather employs an enrichment protocol. After initial mixed infection of strain 112, two rotations, the first of which was carried out on strain MG1363 and the second on 112, permitted continuous amplification of double-plating recombinants while rendering one of the parent phages unamplified in each of the two rotations. Mapping of the recombination endpoints showed that the presence of the N-terminal two-thirds of the tail protein L10 of phage c2 and a 1,562-bp cosR-terminal fragment of phage 923 genome overcame blocks of infection in strains MG1363 and 112, respectively. Both infection inhibition mechanisms act at the stage of DNA entry; in strain MG1363, the infection block acts early, before phage DNA enters the cytoplasm, and in strain 112, it acts late, after most of the DNA has entered the cell but before it undergoes cos-end ligation. These are the first reported host range determinants in bacteriophage of lactic acid bacteria required for overcoming inhibition of infection at the stage of DNA entry and cos-end ligation.
Acquisition and exchange of functional modules by recombination has been proposed as the major mechanism of viral evolution, based on analyses of arrangements of conserved and nonconserved segments along the genomes of lambdoid phage and functional analysis of the recombinants (hybrids) between distant members of the family comprising phage P22 of Salmonella enterica serovar Typhimurium and phage λ of Escherichia coli K12 (3, 5, 22, 23, 55). The selective pressure for generating phage-phage recombinants in nature is most likely the ability to replicate on an increased number of hosts.
Bacteriophages of Lactococcus lactis, a lactic acid bacterium (LAB) used in dairy fermentation, are numerous and diverse. The most frequently encountered are two groups of small isometric-headed phages named 936 and P335 and one group with prolate-headed phage named c2 (28, 38). Recent large-scale sequence analyses of bacteriophages of LAB indicate that exchange of functional modules by recombination is widespread among these phages (2, 6, 10, 11, 33, 37).
Prolate-headed (or prolate) phages are the secondmost common group isolated after fermentation failure in New Zealand dairy plants. This is a relatively conserved group with an exclusively lytic life style. The two prolate phages (c2 and bIL67) whose genome sequences have been determined to date have an overall identity of 80% (36, 54). Both 20- to 22-kbp double-stranded DNA (dsDNA) genomes code for two blocks of divergently oriented open reading frames (ORFs), transcribed early and late, separated by a noncoding region that serves as an origin of replication (51, 53, 61). The early ORFs are detectable 2 min postinfection. They code for products whose function has not yet been determined. Late ORF expression begins approximately 8 min postinfection, and the products are virion proteins and proteins that carry out assembly and release of the virions from the host cells (34, 36). The termini of the genome carry cos ends, which are ligated in the host but appear linear when DNA is extracted from the virion (35, 36, 47).
A given prolate phage typically replicates on only 5 to 10 L. lactis strains among hundreds (21). The molecular basis for this tropism is largely unknown, except for one phage/strain pair. The determinant of the tropism of phage CHL92 for host strain CAa120 has recently been revealed by in vivo phage-plasmid recombination to be gL15 (58). The product of gL15, a minor tail protein, has been proposed to be an adsorption protein. It is not yet clear how host range determination by variants of gL15 relates to currently recognized primary and secondary receptors for prolate phage infection, namely, rhamnose residues of the cell wall (44, 60) and the integral membrane protein phage infection protein (PIP) (19, 44). The protein coded by gene gL10, located at the tip of the tail, was proposed to be an adsorption protein based on its location and presence of a binding motif for Ca2+, an ion required for prolate phage infection (35).
Several nonprolate LAB phages have been subjected to in vivo recombination experiments to reveal strain-specific host range determinants that are capable, when acquired by a given phage, of overcoming adsorption and several other defense mechanism barriers (4, 13, 14, 42, 58). In these experiments, recombination with engineered homologous segments on plasmids or with chromosomally located prophages was demonstrated. Very recently, recombination between two lactococcal virulent phages of the 936 (small isometric) species was reported, with direct selection of recombinants resistant to two defense systems (cloned on a plasmid and transformed into a host strain) (12).
Phage-phage recombination in the environment is clearly necessary for phage evolution. However, no reports are yet available that feature a design relevant for generation of virulent phage-phage recombinants in the environment, without use of engineered hosts. In this study, we applied a novel procedure that consisted of unmodified host strains and phage and used an enrichment rather than a direct selection protocol to isolate the recombinants. This approach was used to map the host range determinants of two prolate phages, c2 and 923, for two L. lactis strains, MG1363 and 112, respectively.
MATERIALS AND METHODS
Strains, media, and culture conditions.
L. lactis strains used in this report were MG1363, a plasmid-free strain and prophage-cured derivative of NCDO 712 (18); AJ376 and MG1363/pAJ376 (35); AJ510 and MG1363/pAJ510 (30); IL-1403, a plasmid-free strain (7); C6 (50); 112, a New Zealand (NZ) dairy plant isolate; and 2282, a NZ dairy plant isolate. Phages used in this report were c2 (32, 40, 48); bIL67 (54); c6A (32, 50); 923, a NZ dairy plant isolate (26); 943, a NZ dairy plant isolate; 5440, a NZ dairy plant isolate; 5447, a NZ dairy plant isolate; 5469, a NZ dairy plant isolate; Rc1 through Rc12, recombinants between 923 and c2 phage (this work); and Rc13 through Rc16, recombinants between 923 phage and plasmid pAJ376 (this work).
Plasmids used in this work were pAJ510 and pSA3 (9), carrying the 7.5-kbp EcoRI fragment of c2 phage (nucleotide [nt] 15681-cos through 633 of the c2 genomic sequence), and Emr (35) and pAJ376 and pSA3, carrying the 4,230-bp EcoRI fragment of c2 (nt 11451 through 15680 of the c2 genomic sequence), and Emr (30). L. lactis strains were grown in M17 medium supplemented with 0.5% (wt/vol) glucose at 30°C (59) unless otherwise stated (Table 1). For selection of L. lactis carrying plasmids pAJ376 and pAJ510, 5 μg/ml erythromycin (Ery) was added to the medium.
TABLE 1.
Host range of the phage
| Phagea | Efficiency of plating on bacterial strain:
|
L10 type | |||||
|---|---|---|---|---|---|---|---|
| MG1363
|
IL1403, 30°C | C6, 30°C | 112, 30°C | 2282, 30°C | |||
| 30°C | 37°C | ||||||
| c2 | 1 | 0.63 | 0.055 | <9.1 × 10−10 | <9.1 × 10−10 | <9.1 × 10−10 | Long |
| bIL67 | <9.1 × 10−9 | <9.1 × 10−9 | 1 | <9.1 × 10−9 | <9.1 × 10−9 | <9.1 × 10−9 | Short |
| c6A | <1.9 × 10−9 | <1.9 × 10−9 | 0.0028 | 1 | <1.9 × 10−9 | <1.9 × 10−9 | Short |
| 923 | <2.4 × 10−9 | 0.0046b | 0.022 | <2.4 × 10−9 | 1 | 3.0 × 10−7 | Short |
| 943 | <2.3 × 10−9 | 0.0024b | <2.3 × 10−9 | <2.3 × 10−9 | 1 | 2.0 × 10−7 | Short |
| 5440 | <3.6 × 10−9 | <3.6 × 109 | <3.6 × 10−9 | <3.6 × 10−9 | <3.6 × 10−9 | 1 | Short |
| 5447 | <4.8 × 10−9 | 0.0012b | <4.8 × 10−9 | <4.8 × 10−9 | 0.015 | 1 | Short |
| 5469 | 0.35 | 0.37 | 9.8 × 10−8 | <1.9 × 10−9 | 0.72 | 1 | Long |
Phage stocks were prepared on standard permissive host strains (efficiency of plating underlined).
Phages from the plaques could not be propagated further at 37°C.
Bacteriophage propagation and isolation.
Preparation of phage stocks was carried out in liquid medium or by plate lysis in the presence of 5 mM CaCl2 (26). When required, phages were concentrated by centrifugation at 40,000 × g for 2 h in a Sorvall centrifuge or by precipitation with polyethylene glycol (26).
Recombinant DNA methods.
Purification of DNA fragments from agarose gels was carried out using commercial DNA purification kits from Roche Molecular Biochemicals and QIAGEN according to the manufacturers' instructions. DNA from polyethylene glycol-precipitated phages was isolated using the QIAGEN λ phage DNA purification kit (Midi), according to the manufacturer's instructions. DNA from phage-infected L. lactis was isolated as described previously (24). Transformation of L. lactis was carried out by electroporation (49).
Sequencing strategy of L10 and recombinants.
For sequencing of the gL10 gene, genomic fragments equivalent to the c2 sequence from coordinates 13127 to 15263 (36) were amplified using primers complementary to the sequences conserved in c2 and bIL67 phage genomes, JR127 (5′ CAATGGCTAAAGAAAAATATGTC 3′) and JR128 (5′ GCTTTTAGTATAAATCAACGCTT 3′) in six prolate phages, c6A, 923, 943, 5440, 5447, and 5449, using DNA from phage particles as templates.
To sequence the fragment in phages 923 and Rc13 through Rc16 equivalent to the c2 EcoRI fragment from coordinates 11451 to 15681 (insert in plasmid pAJ376 carrying ORFs L7 through L11), gL10 and the regions upstream and downstream of it were amplified separately in three PCRs. The amplified products were sequenced using primer walking. The sequence of the genome fragment from gL14 to the cosR end (c2 coordinates 18601 through 22163) and further to the cosL intergenic (IG) fragment (c2 coordinates 1 through 176) in phages 923, Rc5, and Rc6 was determined from three overlapping PCR fragments.
To minimize the error rate, proofreading polymerase Pwo (Roche Molecular Biochemicals) was used in all amplifications. As an additional precaution, each preparative PCR was divided in four PCR tubes prior to cycling. After the PCR, the four reactions were combined for use in subsequent purification and sequencing. The sequencing was carried out using the BigDye mix (Applied Biosystems) at the Allan Wilson Centre DNA Analysis Services (Institute of Molecular Biosciences, Massey University). Both strands were sequenced by primer walking, each with an at least twofold redundancy. Sequences were assembled and analyzed using the GeneWorks program (Accelrys; Oxford Molecular), BioEdit (Tom Hall, North Carolina State University, Raleigh, NC), and Vector NTI (Informax-Invitrogen).
Bacteriophage assays.
A phage adsorption assay was carried out in the linear adsorption range (multiplicity of infection [MOI] = 0.001) at 30°C for 10 min as described previously (27). Transduction of strains MG1363 and 112 was carried out using phage lysates of strain MG1363 containing a plasmid carrying the 7.5-kbp EcoRI fragment that spans the cos ends of phage c2. These lysates were obtained from the infection of the strain AJ510 (MG1363 transformed with pAJ510, which carries c2 phage cos ends ([35]) with phage c2 or Rc15. Bacterial cells were removed from the lysate by filtering them through a 0.22-μm-pore-size filter, and the phage particles in the lysates were titered on MG1363. Transduction experiments were carried out by infecting exponentially growing strain MG1363 or 112 in liquid medium at an MOI of 0.1. The mixtures of phages and bacteria were diluted and plated on erythromycin-containing plates to determine the number of cells that acquired plasmid from the transducing particles. The transduction efficiency was calculated as a ratio of the number of Eryr transductants per ml of lysate to phage titer in the lysate (number of PFU per ml).
Membrane inactivation assay.
Membrane protein extract was prepared and the inactivation assay carried out as described previously (44). Each assay was carried out using 10 μg of protein extract (in a volume of 5 μl) and 5 × 105 phages (in a volume of 5 μl). Inactivation was determined relative to the buffer control. For each experiment, a total of 600 to 900 plaques on three plates were counted; triplicates of each experiment were carried out.
cos ligation assay.
To monitor cos-end ligation, total DNA was isolated from 112 and MG1363 host cells 5 and 20 min after infection with phages c2, 923, Rc16, and Rc5. DNA extracted from cells that had been infected on ice was used as a 0-min time point. Isolation of replicating phage DNA was carried out by the rapid phage purification method (24). DNA was further purified with a QIEX kit (QIAGEN) and digested with appropriate restriction enzymes: BstUI for phages c2 and Rc5 and EcoRI for 923 and Rc16. The digested DNA was separated by agarose electrophoresis and Southern blotted onto a nitrocellulose filter, and the fragments carrying cos ends were detected using a PCR-generated probe spanning the cos ends. To generate the probe, primers JR150 (forward, 5′-CAGTAGTAGTTAGTCATCTGTATAA-3′; nucleotides 21933 through 21967 of the c2 sequence) and JR151 (reverse, 5′-CCTCCTATATAATACCCCTTTAA-3′; nucleotides 154 through 176 of the c2 sequence) were used. Two probes were created. The first probe was amplified from c2 DNA as a template and used for the detection of phage c2, and the second was amplified from 923 DNA as a template and used for detection of phage 923, Rc16, and Rc5 cos ends.
Phage-phage recombination.
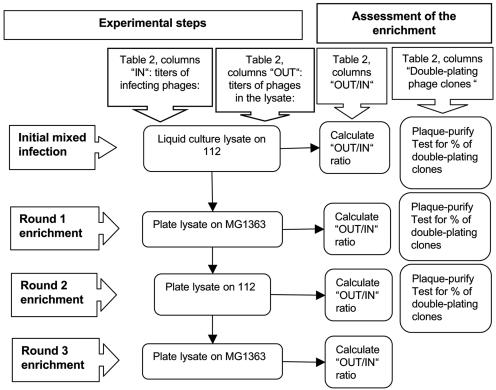
Recombinants between phages 923 and c2 were isolated from mixed infections of one host (MG1363 or 112), followed by alternate plating on each host to enrich for recombinants, which plate on both strains, as opposed to parental phage, which each plate on only one of the two hosts (Table 2). The experiment is outlined by a flowchart diagram (Fig. 1). In the Table 2 and Fig. 1, “IN” and “OUT” represent the total number of PFU used in the infection and obtained after the lysis, respectively, and “OUT/IN” represents the ratio of the two values. The initial mixed infection of 112 was carried out at 30°C and that of MG1363 at 37°C. In brief, to exponentially growing cells (optical density at 600 nm of 0.1; 0.1 ml), CaCl2 was added to 5 mM. After 10 min, both phages were added to the growing cells, 923 at an MOI of 11 and c2 at an MOI of 60, and incubated for 10 min. To remove the majority of unadsorbed phage particles, cells were pelleted by centrifugation, medium was removed, and cells were resuspended in 1 ml of fresh medium. Lysate was collected after 1 h, time required for lysis of the host cells. Bacterial cells were removed by filtering through a 0.22-μm-pore-size filter. The subsequent rounds of enrichment were carried out on plates by using 105 PFU of the phage (as titered on the strain on which the following round of enrichment was to be carried out). To recover the phages, 5 ml of dilution buffer (M17 medium diluted 10-fold with distilled water) was pipetted onto the plate, followed by slow shaking for 1 h at room temperature. The dilution buffer containing phages was filtered through a 0.22-μm-pore-size filter to eliminate bacteria.
TABLE 2.
Phage-phage recombination experiment
| Step of the expt | INa
|
OUTb
|
OUT/INc
|
No. of double-plating phage clones (%)d
|
||||
|---|---|---|---|---|---|---|---|---|
| On 112 | On MG1363 | On 112 | On MG1363 | On 112 | On MG1363 | 112e | MG1363e | |
| Initial mixed infection of 112f | 1.1 × 108 | 6.0 × 108 | 2.3 × 109 | 5.3 × 107 | 2.1 × 101 | 8.8 × 10−2 | <2 | <2 |
| Round 1g on MG1363 | 4.3 × 106 | 1.0 × 105 | 7.5 × 107 | 1.8 × 1010 | 1.7 × 101 | 1.8 × 105 | 96 | 2 |
| Round 2g on 112 | 1.0 × 103 | 2.5 × 108 | 9.7 × 109 | 1.4 × 1010 | 9.7 × 104 | 5.6 × 101 | 92 | 90 |
| Round 3g on MG1363 | 1.0 × 103 | 1.4 × 105 | 1.2 × 1010 | 5.3 × 1010 | 1.2 × 105 | 3.8 × 105 | NDh | NDh |
Total PFU used in the infection.
Total PFU in the lysate.
The ratio of the number of PFU in the lysate to the number of PFU used in the infection. Data are in boldface as explained in Results.
Lysates were plated on 112 or MG1363 at a density of 300 to 400 PFU per plate. Single plaques were purified through two passages by streaking on the strain from which they had been picked. To test whether the phages from the isolated plaques were single or double plating, phages from the purified plaques were streaked on 112 and MG1363.
Strain on which the clones were plaque purified.
The total number of cells was 107 in a volume of 0.1 ml. Note that this is one-cycle phage growth in the liquid medium.
The enrichment rounds 1, 2, and 3 were plate lysates.
ND, not determined.
FIG. 1.
Flowchart diagram for isolation of phage-phage recombinants by an enrichment strategy.
After each round of plating, the enrichment was monitored in a sample of 50 plaques obtained by plating the mixed lysate on each host by using the following procedure. The lysate was first plated at low density on each host (MG1363 and 112) to obtain individual plaques. Fifty plaques from each host plate were passaged twice by streaking onto a new plate with the same host. The purpose of this passage was to make master plates and eliminate the residual nongrowing parental phages that may have colocalized near the original plaques. To detect recombinants, phages from each master plate were streaked on both hosts, and the phages that formed plaques on both MG1363 and 112 were scored as recombinants. A total of 12 recombinant plaques were purified and analyzed further.
Restriction mapping of phage-phage recombinants.
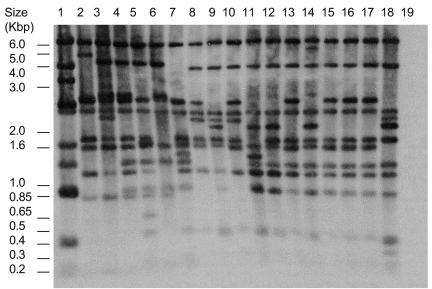
The sequence of phage c2 has been determined; therefore, its restriction map is known (36). Phage 923 and 12 recombinants from the phage-phage recombination experiment (Rc1 through Rc12) were mapped using EcoRI and BstUI restriction enzymes. Phage DNA was isolated from infected cells by the rapid method of Hill et al. (24). This method, rather than phage DNA isolation from lysates, was used because of its rapidity. The isolated phage DNA was probed with several probes sufficient for coverage of the whole genome. The restriction maps were constructed based on cumulative data from Southern blots with all probes. Figure 2 shows a representative Southern blot of the BstUI-digested DNA of parent phages and recombinants Rc1 through Rc16 and the digest probed with the full-length phage c2 DNA isolated from the phage lysate.
FIG. 2.
Restriction digest patterns of the recombinant phages. BstUI digest of the phage DNA isolated from the cells after 20 min of infection, probed with the full-length c2 genomic DNA isolated from the phage lysate. The host strain of c2 was MG1363, and that of all other phages was 112. Lanes: 1, c2; 2, 923; 3, Rc13; 4, Rc14; 5, Rc15; 6, Rc16; 7, Rc1; 8, Rc2, 9, Rc3; 10, Rc4; 11, Rc5; 12, Rc6; 13, Rc7; 14, Rc8; 15, Rc9; 16, Rc10; 17, Rc11; 18, Rc12; 19, DNA from uninfected strain 112.
Phage-plasmid recombination.
A culture of AJ376 grown at 37°C to an OD600 of 0.1 was infected with phage 923 at an MOI of 1 for 10 min at 37°C. Cells were then centrifuged to remove unadsorbed phage, and the culture was incubated for 1 h at 37°C. To determine the presence and frequency of recombinants, the lysate was titered on strain MG1363 at 30°C (a temperature at which the parental phage 923 does not form plaques on that strain) and on 112, the permissive host strain for phage 923.
Alternatively, 104 PFU of phage 923 was used to infect AJ376 at 37°C and plated on AJ376 on solid medium. Recombinants gave clear plaques on AJ376 at 37°C, as opposed to parent phage 923, which gave small, turbid plaques. Four clear plaques were isolated and passaged by streaking onto MG1363-containing plates at 30°C to eliminate parental phage 923. A single plaque from each streak was picked into medium, and residual bacteria were removed by filtering through a 0.22-μm-pore-size filter. The dissolved plaques were used to obtain plate lysates of recombinants on strain 112 for further analysis.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences were as follows: for gL10, AY569312 for 943, AY569313 for 5440, AY569314 for 5447, AY569315 for 5469, and AY569311 for c6A; for phage 923 sequences, AY570983 for L6-L12, AY572847 for L14-cosL, AY570984 for Rc13 and L6-L12, AY570985 for Rc14 and L6-L12, AY570986 for Rc15 and L6-L12, AY570987 for Rc16 and L6-L12, AY572848 for Rc5 and L14-cosL, and AY572849 for Rc6 and L14-cosL.
RESULTS
Generation of phage-phage recombinants.
To examine the host ranges of a group of laboratory and industrial phages, a plating matrix of eight phages on five host strains was carried out (Table 1). We focused on identification of the host range determinants of two phages, c2 and 923, on their respective standard permissive strains, MG1363 and 112, by in vivo phage-phage recombination. Phage c2 was chosen because it is the best characterized prolate phage and its host is the most commonly used L. lactis laboratory strain, MG1363. Phage 923 is a New Zealand industrial isolate from 1975. It was chosen because its host, strain 112, does not restrict phages passaged on strain MG1363 (data not shown). Neither phage can form plaques on the other phage's standard permissive host at 30°C, the optimal growth temperature for L. lactis (Table 1). At 37°C, strain MG1363 is slightly less resistant to infection with phages 923, 943, and 5447, permitting low-efficiency formation of small and turbid plaques that could not be propagated further (Table 1 and data not shown). The influence of growth temperature on the stringency of phage resistance mechanisms is well documented in L. lactis (for examples, see references 29 and 52). In the group of strains tested in Table 1, decreased stringency of phage resistance at 37°C was also observed for IL-1403 and C6 (data not shown). For strains 112 and 2282, incubation at 37°C was lethal; therefore, the influence of increased temperature on their phage resistance mechanisms could not be examined.
Mixed infection of strain 112 with phages c2 and 923 was performed in an attempt to obtain recombinants between the two phages. Direct selection of such recombinants was not possible because each parent phage and the recombinants were expected to form plaques on one of the two host strains, MG1363 or 112. Indeed, in the lysate obtained after the initial mixed infection of strain 112, the standard permissive strain for phage 923, the titer of this phage was 2.3 × 109/ml. Moreover, plating 103 of these phages on a mixed lawn of strains 112 and MG1363 in an effort to detect recombinants as clear plaque formers gave a negative result. The titer of the phages from the same lysate on strain MG1363 (host of c2 phage) was 5.3 × 107/ml. This population should consist of the residual unadsorbed phage from the c2 stock used to infect the host cells, which form plaques only on strain MG1363 and recombinants which should plate on both strains (MG1363 and 112). To search for recombinants, 50 plaques from strain MG1363 (host of c2) were purified and tested for their ability to form plaques on both hosts but did not reveal any recombinants. Hence, the frequency of recombinants was too low for any rapid method of detection in the lysate after the initial mixed infection. We therefore devised an enrichment strategy whereby the putative recombinants in the lysate obtained after the mixed infection stage would be enriched by alternate plating (rotation) on the two host strains. Each round would prevent amplification of one of the parent phages while supporting amplification of double-plating recombinants. The method is described in detail by the flowchart diagram (Fig. 1). The total numbers of PFU before and after initial mixed infection and at every step of enrichment are given in Table 2 (columns “IN” and “OUT”). The OUT/IN ratio was used as an indicator of enrichment. In particular, the OUT/IN ratios on the host other than that from which the lysate was obtained were expected to be very low in the absence of recombination. Therefore, this ratio (Table 2, OUT/IN column pair, bold font) was used as an indicator of the appearance and enrichment of recombinants. To confirm that double-plating recombinant clones were present in the lysates and to determine what fraction of the lysate they represented, 100 single plaques from each round (50 from a plate of each host strain) were purified and tested for double-plating (Table 2, data columns 7 and 8).
In the initial mixed lysate obtained from infecting strain 112, the OUT/IN PFU ratio, as determined on MG1363, was 8.8 × 10−2 (Table 2, “Initial mixed infection” row, OUT/IN column, bold). An identical OUT/IN ratio was obtained in the parallel control experiment in which only c2 was used for infection of strain 112 (data not shown), suggesting that there was no complementation between phages c2 and 923 in the mixed infection.
In the first round of enrichment, 105 PFU (as titered on strain MG1363) from the initial mixed-infection lysate was used to infect MG1363 (Table 2, “Round 1” row, data column 2). The OUT/IN PFU ratio of this enrichment round, as determined on strain 112, was 1.7 × 101 (“Round 1” row, OUT/IN column, bold), suggesting that there was a population of strain 112-plating phages that had been amplified on MG1363. Moreover, testing for double-plating phages among the plaques on strain 112 showed that 96% were double-plating recombinants and 4% parent phage 923 (Table 2, “Round 1” row, data column 7). Therefore, this rotation was very successful in preventing replication of the 923 parent phages and allowing replication of c2 phages and recombinants. To enrich for recombinants relative to c2, a second rotation, this time on strain 112, was carried out. After this rotation, as determined by a double-plating assay, most of the plaques purified from either of the host strains were recombinants (Table 2, “Round 2” row, data columns 7 and 8). To confirm equal amplification of the double-plating recombinant phages on both hosts, a third round of plating was carried out. The OUT/IN ratio on both hosts was over 105, suggesting that the recombinants were amplifying equally well on both hosts (Table 2, “Round 3” row, columns 5 and 6).
The reciprocal experiment was also carried out, where MG1363 was the primary host of the mixed infection, with similar results. In order to obtain recombinants in this experiment, the initial mixed infection of MG1363 had to be carried out at 37°C.
Restriction analysis of recombinants and mapping of the host range determinants.
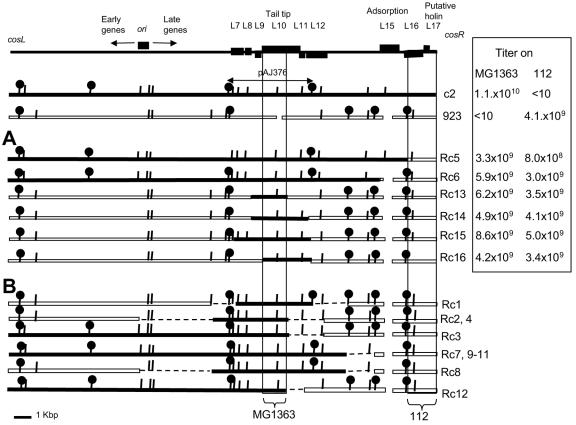
Twelve recombinants isolated from the two recombination experiments were purified and analyzed further. Some of the recombinants isolated from the same recombination experiments were expected to be clonal (derived from the same original recombinant phage), but the analysis was pursued to determine the number of distinct recombinants in each of the two experiments.
Restriction mapping of recombinants was carried out by Southern blotting of BstUI and EcoRI digests of phage DNA isolated from infected cells with a series of overlapping probes that covered the whole phage genome. Figure 2 shows a representative Southern blot in which full-length c2 phage DNA isolated from the virions was used as a probe. Mapping was facilitated by availability of the genome sequence (and hence the precise restriction map) of phage c2. Restriction mapping revealed that there were most likely five and three different recombinants, respectively, in the two experiments in which 112 and MG1363 were the primary hosts (Fig. 3). Interestingly, no recombination endpoints were found within the block of early genes (Fig. 3).
FIG. 3.
Comparison of parental and recombinant phages. (Top) Schematic map of the prolate phages with the relevant genes indicated. First two lines, restriction maps of the parent phages. Arrow indicates the insert in plasmid pAJ376. Panels A and B show restriction maps of the recombinants. Black box, c2-derived sequences; white box, 923-derived sequences. Restriction sites: BstUI (vertical lines), EcoRI (lollipops). (A) Hybrid phages whose recombination endpoints were determined by sequencing. The exchanged DNA fragment sequences have been deposited in GenBank. (See Material and Methods section for the accession numbers.) Table contains the titers of each phage on strains MG1363 and 112. (B) Hybrid phages Rc1 through Rc4 and Rc7 through Rc12, whose recombination endpoints were determined by restriction mapping only. Stippled lines denote the fragments where the crossover points were expected to be based on the restriction mapping. The titers of these phages were similar to those of the phages in panel A (data not shown).
Infection in nonpermissive phage/host pairs is inhibited at the step of DNA entry or cos-end ligation.
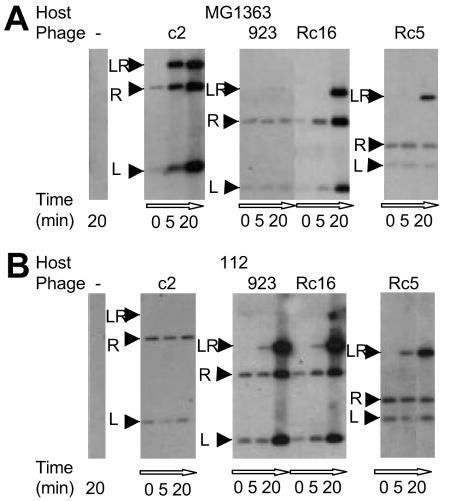
To reveal at what step of the phage life cycle the infection was inhibited in nonpermissive phage/host pairs, 923/MG1363 and c2/112, assays for adsorption, entry, and DNA replication were carried out. Assays of adsorption and inactivation of the virions by membrane protein extracts showed that, for the nonpermissive phage/host strain pairs, the efficiencies of these two steps were within the range reported to be sufficient for infection (Table 3) (1, 44, 58). Therefore, neither adsorption nor inactivation of virions by membrane proteins is inhibited in these nonpermissive phage/host pairs. To resolve whether infection was inhibited early or late, a time course of phage DNA replication during infection was examined by Southern blotting of DNA isolated from infected cells. No replication was detected within the first 20 min of infection, suggesting an early block of infection. This experiment also examined whether the cos ends of the phage DNA in the nonpermissive phage/host pairs had undergone ligation, using as a probe a phage genome fragment that spanned the cos ends (Fig. 4). This experiment showed that the cos-end ligation did not occur in nonpermissive phage/host pairs within the first 20 min of infection and that the amount of recovered phage DNA remained constant during that time (Fig. 4A, phage 923 infection of strain MG1363, and B, phage c2 infection of strain 112). Because of the ability of phage to adsorb to the nonpermissive host cells, phage DNA was isolated even when phages were mixed with prechilled bacteria on ice and subjected to DNA extraction without warming the mix (Fig. 4, time point, 0 min), the conditions that prevent phage DNA entry. Therefore, this assay could not distinguish whether or not DNA entered the cytoplasm. Consequently, it was not possible to resolve whether the failure of cos ligation in the nonpermissive infections was due to the inhibition of DNA entry or direct inhibition of ligation of the cos ends of the genome. In contrast to the nonpermissive host strain infection, the cos-end ligation in the permissive strain infection occurred within the first 5 min of infection and it was followed by replication, as demonstrated by the increase in the amount of phage DNA (Fig. 4A, phage c2 infection of strain MG1363, and B, phage 923 infection of strain 112).
TABLE 3.
Adsorption and membrane inactivation assaysa
| Phage | Adsorption by:
|
Membrane inactivation by:
|
||
|---|---|---|---|---|
| MG1363 | 112 | MG1363 | 112 | |
| c2 | 96 ± 2% | 60 ± 2% | 92 ± 3% | 31 ± 3% |
| 923 | 42 ± 3% | 92 ± 2% | 49 ± 3% | 49 ± 9% |
Each value was obtained from three independent experiments.
FIG. 4.
cos-end ligation assay. The assay was performed as described in Materials and Methods. LR, ligated cosL and cosR ends; R, unligated cosR; L, unligated cosL. For the zero time point, phages were added to an aliquot of a chilled culture and incubated for 20 min on ice before DNA isolation. Hence, the bands on the Southern blots of the 0-min time points are derived from the adsorbed phages, the DNA of which has not entered the host cells.
A cosR fragment of phage 923 is the host range determinant for strain 112.
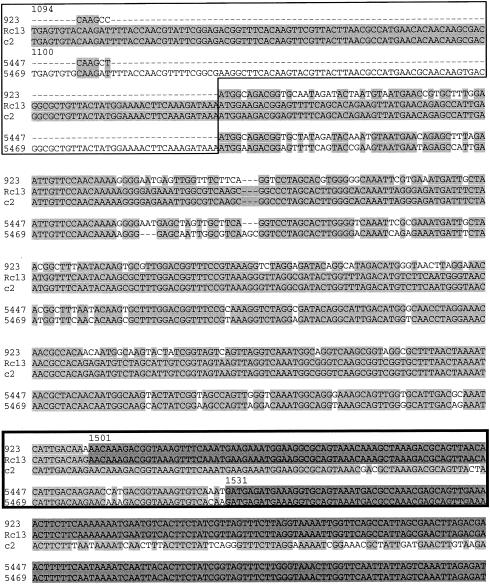
Restriction mapping of phage-phage recombinants showed that recombinant Rc5 had acquired the shortest segment of phage 923 DNA (Fig. 3). Sequence analysis showed that Rc5 carried 1,562 bp from the cosR end of the phage 923 genome, comprising the 3′ 93% of gL16, the complete gL17, and the cosR portion of the cos IG region (GenBank accession number AY572848) (Fig. 5). Therefore, gL16, gL17, or the cosR IG DNA of 923 carried by the exchanged fragment are required for propagation on strain 112.
FIG. 5.
The 5′ and 3′ ends of the phage 923-derived fragment in the Rc5 recombinant. Alignment of c2, 923, and Rc5 DNA sequences around the recombination endpoints. Shaded background indicates identities. Darker-shaded background indicates the parental phage from which a particular portion of the sequence is acquired by the recombinant. The ends of the Rc5 fragment acquired from 923 are indicated by arrows above the sequence. (A) Alignment beginning with the ATG codon of gL16. Box, sequence identical in all three phages within which the crossover has occurred. (B) Alignment of the sequences of cosR and cosL termini. The alignment was broken at the cos-end nonamer (white font) to indicate ends of the genome. Assuming that recombination occurred between the linear c2 and 923 genomes (before the cos-end ligation), the cos nonamer should be the end of the Rc5 sequence acquired from the phage 923. Underlining, the low-identity segment of the cosR IG sequence.
From the cos-end ligation experiment, it was clear that the recombinant Rc5 had overcome inhibition in the nonpermissive strain 112 (Fig. 4). However, it was not clear whether the inhibition was acting at the step of DNA injection or cos-end ligation. To distinguish between these two possibilities, it would have been necessary to examine whether bypassing infection by direct transfection of c2 phage DNA into host 112 would overcome the inhibition. For the c2/112 nonpermissive phage/host pair, however, this was not possible because of the very poor transformability of strain 112. Instead, an indirect approach was taken. It was based on transduction of the plasmid pAJ510 that carries phage c2 cos-end DNA. This plasmid is packaged into the c2 virions upon infection of strain AJ510 (MG1363 carrying pAJ510) (35). The transduction efficiency of the nonpermissive host strain 112 was 10−4 that of the permissive strain MG1363 (Table 4). This confirms the block in DNA injection or recircularization by cos-end ligation, two steps upon which the transduction efficiency depends. To investigate whether the strain 112 inhibitory mechanism was acting on c2-derived virion proteins or c2 cos-end DNA, plasmid pAJ510 was encapsidated into the recombinant Rc15 virion (which carries phage 923-derived virion proteins) and tested for the transduction of strains 112 and MG1363. The transduction efficiency of these chimeric particles into the nonpermissive strain 112 was as low as that of the particles consisting of the phage c2 virion proteins (Table 4), suggesting that virion proteins from phage 923 could not bypass the inhibition of infection in 112. This in turn indicates that the transduction efficiency is low in these particles because the c2 cosR DNA is a target of an inhibition mechanism in strain 112. A postentry blocking mechanism is also indicated by the fact that, in the phage-phage recombination experiment, phage c2 DNA had to enter into the cytoplasm of the nonpermissive host 112 in order to recombine with the phage 923 DNA. Therefore, in strain 112, the cosR DNA of phage c2 is likely the target of a blocking mechanism, a consequence of which is the failure of the phage c2 cos-end ligation.
TABLE 4.
Transduction of the cos-end-carrying plasmid pAJ510
| Transducing lysate | Recipient | Frequency of transductantsa | Relative transduction frequency (112/MG1363) |
|---|---|---|---|
| Rc15(pAJ510) | MG1363 | 5.4 × 10−3 | |
| Rc15(pAJ510) | 112 | 2.1 × 10−7 | 2.6 × 10−4 |
| c2(pAJ510) | MG1363 | 1.5 × 10−3 | |
| c2(pAJ510) | 112 | 1.4 × 10−7 | 9.3 × 10−5 |
Relative to the phage titer in the lysate.
With the premise that the least-conserved portion of the exchanged cosR fragment may be a candidate for the target of the blocking mechanism, the conservation of the coding and IG portions of this fragment between the c2 and 923 phages was examined. The least-conserved segment was a portion of the cosR IG sequence of 98 bp immediately downstream of the L17 stop codons (49% identity). This sequence was followed by highly conserved 44 nt that included the absolutely conserved cohesive-ends nonamer (Fig. 5B). The gL16 and gL17 coding sequences conservation (90% and 85% identity, respectively) matches that of the average prolate phage genome conservation. Therefore, the low-identity 98-bp fragment is a candidate for a target of the cos-end ligation inhibitory mechanism in strain 112.
The gL10 of c2 phage is a host range determinant for strain MG1363.
When restriction maps of phage-phage recombinants were aligned, they all carried a common fragment from phage c2 comprising a set of late genes from gL6 to gL11, suggesting that this fragment of phage c2 carries the host range determinant for plating on strain MG1363. To define more precisely the host range determinant of c2 required for plating on MG1363, phage-plasmid recombination was carried out using plasmid pAJ376, which carries a 4,230-bp EcoRI fragment of c2 containing genes gL7 through gL11 (30). Strain AJ376 (MG1363 carrying pAJ376) was infected with phage 923 at 37°C. (The recombination could not be carried out in the industrial strain 112, which is 100% permissive for 923, because this strain was very poorly transformable, and pAJ376 transformants were not obtained even after several attempts.)
The recombinants in the resulting lysate were detected by their ability to propagate on MG1363 at 30°C, the temperature at which the parental phage 923 does not form plaques on this strain. After 1 h of infection, 16% of the phages in the lysate were recombinants. The entire 4.2 kbp corresponding to pAJ376 was sequenced in 4 independent recombinants to determine the portion of the c2 genome acquired from the plasmid. The minimum common c2 sequence in all four recombinants spanned from nt −25 to + 1500 of gL10c2 and included a 300-bp (100-amino acid) insert which is present in c2 but absent in 923 gL10 (Fig. 3A, Rc13 through Rc16). Therefore, the 5′ portion of gL10 of c2 is a host range determinant for plating on the laboratory strain MG1363.
The cos-end ligation experiment could not distinguish between inhibition of DNA entry and direct blocking of cos-end ligation in the 923/MG1363 nonpermissive phage/host pair. Hence, further experiments were performed to distinguish between these two possibilities. This could be resolved directly by taking advantage of the relatively high transformation efficiency of the laboratory strain MG1363. Transfection of purified phage 923 DNA bypassed the adsorption and entry steps and allowed the ability of the phage DNA to replicate following introduction into the host cytoplasm by electroporation to be determined. The burst size of transfected phage 923 DNA was 25% that of the c2 DNA control, suggesting that when phage 923 DNA is introduced into the nonpermissive host, it completes the life cycle, including cos-end ligation. In contrast, infectious center formation after conventional infection of MG1363 with phage 923 occurred at much lower efficiency, only 1% that of c2. An infection inhibition that prevents phage DNA entry was also suggested by the inability to obtain phage-phage recombinants after mixed infection of strain MG1363 at 30°C. All these findings suggest that the major block of MG1363 infection by phage 923 is at the step of DNA injection, before cos-end ligation.
The “long” gL10 correlated with the replication of prolate phages on strain MG1363 in the sample of eight phages (Table 1). Among those, two industrial phage isolates from the same cheesemaking “season” (1995), named 5649 and 5447, carried “long” and “short” gL10 genes, respectively (GenBank accession numbers AY569314 and AY569315). Interestingly, the 5′ 1530 nucleotides (carrying the “insert”) were 66% identical, while the remaining 603 residues were 100% identical between the two phages. This pattern of nucleotide conservation within gL10 can be rationalized by a recombination event that had created a hybrid “long” gL10 with the recombination endpoint located only 30 nucleotides downstream of the one found in our experimentally derived c2/923 recombinant Rc13 (Fig. 6). Phage 5469 has a broader host range than does 5447 (Table 1); therefore, the putative recombination event has created a more promiscuous hybrid phage. Phage 5469 does not have identical host range to that of c2 with respect to the other tested strains (Table 1), most likely because of differences in other host range determinants, e.g., cosR (this work) or gL15 (58).
FIG. 6.
Comparison of the gL10 recombination endpoints in the laboratory recombinant Rc13 and industrial isolate 5469. Top alignment, c2, 923, and Rc13; bottom alignment, 5447 and 5469. The numbering in the alignments starts from the A residues of the ATG codons of gL10 of c2 and 5469, respectively. Thin-line box indicates a fragment of the “insert” in the “long” gL10 genes of phages c2 and 5469. Shaded background indicates identities in each of the alignments. Darker-shaded background indicates sequences of the recombinants Rc13 and 5469 that match the short-L10 parent phages 923 and 5447, respectively. Sequences 5′ to these blocks in Rc13 and 9469 match the long-L10 parent phage c2 and an unknown parent phage, respectively. Thick-line box indicates the rows of the two alignments where the recombination endpoints are located.
DISCUSSION
Among the large number of prolate phages that infect numerous strains of L. lactis, the host range determinant, gL15, for only one phage/host strain pair, CHL92/CAa120, has been revealed to date (58). The detection of host range determinants in the lactococcal bacteriophages were impeded by the poor transformation capacity of many industry lactococcal strains, lack of appropriate genetic and molecular biology tools, and toxicity of significant portions of prolate phage gene products for E. coli and L. lactis (31; J. Rakonjac and M. W. Lubbers, unpublished data). We have used an approach with minimal requirement for genetic manipulation, in which phage-phage recombinants were isolated by enrichment through strain rotation, to reveal two new host range determinants for two phage/host pairs, c2/MG1363 and 923/112, respectively, which overcame strain-specific barriers of phage DNA entry and cos-end ligation. Previous reports have identified the host range determinants of LAB phages acting at the stage of phage adsorption (4, 13, 14, 42, 58). This report reveals the first two LAB phage host range determinants involved in phage DNA entry and possibly cos ligation.
In the nonpermissive phage/host pair 923/MG1363, phage DNA injection was inhibited at a stage after the virion inactivation step but before DNA had entered the host cell. Recombinant phages that completely overcame the infection barrier had all acquired a minimal common fragment of c2 carrying the 5′ two-thirds of gL10, from nt −25 to + 1500. The gL10 genes of the two parent phages, c2 and 923, were 70% identical, and the portion of c2 required for plating on strain MG1363 carried a 300-bp “insert” relative to the 923 gL10 gene.
Our findings suggest that gL10 of prolate phages is involved in DNA entry into the host cells. This is interesting because pL10 has been shown to be located at the tip of the virion tail and proposed to be an adsorption protein (36). In the set of strains that are the subject of the present study, pL10 did not determine the host specificity at the step of adsorption. However, it is possible that this protein provides conserved functions for adsorption (such as Ca2+ binding), while the receptor recognition resides in other minor tail proteins, such as pL15, which determines the host specificity of adsorption of prolate phage CHL92 to host strain CaA120 (58). Interestingly, in the two phage/host pairs in which we mapped the host range determinants c2/MG1363 and 923/112, pL15 did not determine the host specificity or adsorption efficiency despite the low conservation between the phages c2 and 923 (34% identity) (data not shown).
The questions remain as to which host proteins interact with pL10 to mediate DNA entry and whether variants of these proteins determine the host range of the prolate phages. The lactococcal membrane protein PIP is required for infection by many prolate phages, and its presence correlates with the ability of host cells to inactivate the virions (1, 19, 44). Phage 923 is inactivated by the MG1363 membrane protein extract with 42% efficiency, sufficiently high for infection (44). Therefore, the inhibition of infection in MG1363 either is not dependent on PIP or depends on a PIP function different from its role in the inactivation of the virion. Alternatively, by analogy with the model cos-end-containing phage λ, which is inactivated by the surface (outer membrane) protein LamB but still requires another, inner membrane protein Pel (or MalY) for injection, additional host protein(s) may participate in prolate phage DNA injection. Provided this protein is variable in different L. lactis strains, only certain variants of the host protein would interact with the matching variants of phage protein pL10 (17). The absence of such protein in certain strains could also be responsible for variations in host range. Another possibility is that an active injection inhibition mechanism could be mediated by a putative prophage-encoded protein present in MG1363 and counteracted by long gL10. An example of prophage-encoded injection inhibition protein in L. lactis is Sie2009 (39), which inhibits the injection of a large number of species 936 phages (but not prolate phages). MG1363 is a plasmid-free laboratory strain used to test various phage defense mechanisms. Considering that in our sample, only one-quarter of the tested prolate phages could propagate on MG1363, it would be of interest to identify and, if possible, eliminate putative inhibitory proteins in order to broaden this strain's use for the expression and testing of phage defense mechanisms for an increased number of phages.
The second host range determinant, that for growth on strain 112, overcomes an infection barrier of a very late step of infection, when most of the phage DNA has entered the cytoplasm, and/or at the step of cos-end ligation. Transduction experiments showed that the target of the inhibition mechanism was the cos-end DNA, consistent with this portion of the genome of phage 923 being the host range determinant required for replication of the recombinant phages in strain 112.
A major portion of the cosR IG sequence (except for the cos-end nonamer and adjacent 35 bp) is poorly conserved between phages c2 and 923; therefore, it could be hypothesized that a putative DNA-binding inhibitory protein could distinguish between the cosR IG sequences of phages c2 and 923. In λ phage, an asymmetric mutation of the cosN site prevents injection-coupled cos-end ligation. If mutant phage DNA is introduced into host cells by CaCl2-dependent transformation, the cos-end ligation proceeds normally (62). A possible explanation for this observation could be that a host protein required for injection (e.g., Pel) may bind to the cos end of the genome that is injected last into the host cell and, under certain circumstances, fails to release it, thus preventing the cos-end ligation. It can be speculated that this type of interference might be active in strain 112, perhaps as a consequence of putative membrane protein variability among lactococcal strains, where the variant present in strain 112 may prevent the release of the cosR end of c2 but not that of phage 923.
In this work, virulent phage-phage recombinants were obtained with ease by a protocol that does not involve mutation or genetic modification of the phages and the host strains. This represents the situation in the natural environment and suggests how it is possible for virulent phage-phage recombinants to arise in nature.
The divergence between the prolate phage genomes is about 20%, thus the recombination would be expected to occur with a low efficiency due to the action of host repair systems (16, 57). Consequently, the recombinants should not be obtainable without direct selection. The unexpectedly high recombination efficiency between the prolate phages could be due to the presence of a phage-encoded recombinase capable of bypassing the host repair systems. A candidate recombinase in prolate phage is the product of ORF E15, which shares 37% identity and 57% similarity to the ERF recombinase of phage P22 of Salmonella enterica serovar Typhimurium (36, 45, 54). The ERF recombinase mediates high-efficiency recombination and mutagenesis by a mechanism similar to lambda Redβ, and it belongs to the same protein superfamily (25, 46, 63, 64). Further work will be carried out to determine whether the product of ORF E15 is a recombinase.
Strain rotation protocol presented in this paper is related to a common practice of changing or rotating strains in the dairy fermentation to avoid phage attack (8, 15, 56). Thus, the strain rotation practice in dairy fermentation is very likely a factor that stimulates LAB phage evolution by horizontal gene transfer, plausibly contributing to the observed genetic mosaicism (2, 6, 10, 11, 33, 37). With respect to managing the risks of phage-mediated fermentation failure in dairy industry, our work suggests that an undesirable long-term outcome of the strain rotation practice may actually be the appearance of highly promiscuous hybrid “super-phages” with an expanded host range, a phenomenon encountered in the industry (20, 29, 43).
Phage therapy is an alternative to antibiotic treatment of bacterial infection (41). One of the limitations of phage therapy is the narrow host range of bacteriophage. Our approach to obtain recombinants with broadened host range without in vitro genetic modifications can be used to increase the number of bacterial targets of one phage. It may therefore help decrease the costs of phage production and cut expenses associated with obtaining approvals for usage of therapeutic phages by regulatory authorities such as the Food and Drug Administration.
Acknowledgments
We thank Marjorie Russel, John Tweedie, and Anja Schiemann for critical reading of the manuscript and suggestions, Lawrence Ward and Howard Heap for generously providing the prolate phage isolates, and Qing Deng for excellent technical help.
This work was supported by the Marsden Fund of the Royal Society of New Zealand (grant number MAU803) to P. W. O'Toole and M. W. Lubbers and by the Institute of Molecular BioSciences start-up fund to J. Rakonjac.
REFERENCES
- 1.Babu, K. S., W. S. Spence, M. R. Monteville, and B. L. Geller. 1995. Characterization of a cloned gene (pip) from Lactococcus lactis required for phage infection. Dev. Biol. Stand. 85:569-575. [PubMed] [Google Scholar]
- 2.Blatny, J. M., L. Godager, M. Lunde, and I. F. Nes. 2004. Complete genome sequence of the Lactococcus lactis temperate phage phiLC3: comparative analysis of phiLC3 and its relatives in lactococci and streptococci. Virology 318:231-244. [DOI] [PubMed] [Google Scholar]
- 3.Botstein, D., and I. Herskowitz. 1974. Properties of hybrids between Salmonella phage P22 and coliphage lambda. Nature 251:584-589. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, A., and D. Botstein. 1983. Evolution of the lambdoid phages, p. 365-380. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 6.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 8.Cogan, T. M., N. Peitersen, and R. L. Sellars. 1991. Starter systems, p. 16-23. Bulletin of the International Dairy Federation, vol. 263/1991. International Dairy Federation, Brussels, Belgium.
- 9.Dao, M. L., and J. J. Ferretti. 1985. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl. Environ. Microbiol. 49:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desiere, F., S. Lucchini, C. Canchaya, M. Ventura, and H. Brussow. 2002. Comparative genomics of phages and prophages in lactic acid bacteria. Antonie Leeuwenhoek 82:73-91. [PubMed] [Google Scholar]
- 11.Desiere, F., C. Mahanivong, A. J. Hillier, P. S. Chandry, B. E. Davidson, and H. Brussow. 2001. Comparative genomics of lactococcal phages: insight from the complete genome sequence of Lactococcus lactis phage BK5-T. Virology 283:240-252. [DOI] [PubMed] [Google Scholar]
- 12.Domingues, S., A. Chopin, S. D. Ehrlich, and M.-C. Chopin. 2004. A phage protein confers resistance to the lactococcal abortive infection mechanism AbiP. J. Bacteriol. 186:3278-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont, K., F. K. Vogensen, H. Neve, J. Bresciani, and J. Josephsen. 2004. Identification of the receptor-binding protein in 936-species lactococcal bacteriophages. Appl. Environ. Microbiol. 70:5818-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durmaz, E., and T. R. Klaenhammer. 1995. A starter culture rotation strategy incorporating paired restriction/modification and abortive infection bacteriophage defenses in a single Lactococcus lactis strain. Appl. Environ. Microbiol. 61:1266-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duwat, P., A. Cochu, S. D. Ehrlich, and A. Gruss. 1997. Characterization of Lactococcus lactis UV-sensitive mutants obtained by ISS1 transposition. J. Bacteriol. 179:4473-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elliott, J., and W. Arber. 1978. E. coli K-12 pel mutants, which block phage lambda DNA injection, coincide with ptsM, which determines a component of a sugar transport system. Mol. Gen. Genet. 161:1-8. [DOI] [PubMed] [Google Scholar]
- 18.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geller, B. L., R. G. Ivey, J. E. Trempy, and B. Hettinger-Smith. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heap, H. A., and J. T. Harnett. 2003. Bacteriophage in the dairy industry, p. 136-142. In H. Roginski, J. W. Fuquay, and P. F. Fox (ed.), Encyclopedia of dairy sciences, vol. 1. Academic Press, Amsterdam, The Netherlands.
- 21.Heap, H. A., and A. W. Jarvis. 1980. A comparison of prolate and isometric-headed lactic streptococcal bacteriophages. New Zealand J. Dairy Sci. Technol. 15:75-81. [Google Scholar]
- 22.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Highton, P. J., Y. Chang, and R. J. Myers. 1990. Evidence for the exchange of segments between genomes during the evolution of lambdoid bacteriophages. Mol. Microbiol. 4:1329-1340. [DOI] [PubMed] [Google Scholar]
- 24.Hill, C., I. J. Massey, and T. R. Klaenhammer. 1991. Rapid method to characterize lactococcal bacteriophage genomes. Appl. Environ. Microbiol. 57:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyer, L. M., E. V. Koonin, and L. Aravind. 2002. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics 3:8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jarvis, A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis, A. W. 1978. Serological studies of a host range mutant of a lactic streptococcal bacteriophage. Appl. Environ. Microbiol. 36:785-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 29.Jarvis, A. W., H. A. Heap, and G. K. Y. Limsowtin. 1989. Resistance against industrial bacteriophages conferred on lactococci by plasmid pAJ1106 and related plasmids. Appl. Environ. Microbiol. 55:1537-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarvis, A. W., M. W. Lubbers, T. P. Beresford, L. J. Ward, N. R. Waterfield, L. J. Collins, and B. D. Jarvis. 1995. Molecular biology of lactococcal bacteriophage c2. Dev. Biol. Stand. 85:561-567. [PubMed] [Google Scholar]
- 31.Jarvis, A. W., M. W. Lubbers, N. R. Waterfield, L. J. Collins, and K. M. Polzin. 1995. Sequencing and analysis of the genome of lactococcal phage c2. Int. Dairy J. 5:963-976. [Google Scholar]
- 32.Keogh, B. P. 1973. Adsorption, latent period and burst size of phages of some strains of lactic streptococci. J. Dairy Res. 40:303-309. [Google Scholar]
- 33.Labrie, S., and S. Moineau. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308-320. [DOI] [PubMed] [Google Scholar]
- 34.Lubbers, M. W., K. Schofield, N. R. Waterfield, and K. M. Polzin. 1998. Transcription analysis of the prolate-headed lactococcal bacteriophage c2. J. Bacteriol. 180:4487-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubbers, M. W., L. J. Ward, T. P. Beresford, B. D. Jarvis, and A. W. Jarvis. 1994. Sequencing and analysis of the cos region of the lactococcal bacteriophage c2. Mol. Gen. Genet. 245:160-166. [DOI] [PubMed] [Google Scholar]
- 36.Lubbers, M. W., N. R. Waterfield, T. P. Beresford, R. W. Le Page, and A. W. Jarvis. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lucchini, S., F. Desiere, and H. Brüssow. 1999. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 73:8647-8656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maniloff, J., and H. W. Ackermann. 1998. Taxonomy of bacterial viruses: establishment of tailed virus genera and the order Caudovirales. Arch. Virol. 143:2051-2063. [DOI] [PubMed] [Google Scholar]
- 39.McGrath, S., G. F. Fitzgerald, and D. van Sinderen. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509-520. [DOI] [PubMed] [Google Scholar]
- 40.McKay, L. L., and K. A. Baldwin. 1984. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl. Environ. Microbiol. 47:68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merril, C. R., D. Scholl, and S. L. Adhya. 2003. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2:489-497. [DOI] [PubMed] [Google Scholar]
- 42.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1994. Evolution of a lytic bacteriophage via DNA acquisition from the Lactococcus lactis chromosome. Appl. Environ. Microbiol. 60:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moineau, S., D. Tremblay, and S. Labrie. 2002. Phages of lactic acid bacteria. ASM News 68:388-393. [Google Scholar]
- 44.Monteville, M. R., B. Ardestani, and B. L. Geller. 1994. Lactococcal bacteriophages require a host cell wall carbohydrate and a plasma membrane protein for adsorption and ejection of DNA. Appl. Environ. Microbiol. 60:3204-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy, K. C., L. Casey, N. Yannoutsos, A. R. Poteete, and R. W. Hendrix. 1987. Localization of a DNA-binding determinant in the bacteriophage P22 Erf protein. J. Mol. Biol. 194:105-117. [DOI] [PubMed] [Google Scholar]
- 46.Passy, S. I., X. Yu, Z. Li, C. M. Radding, and E. H. Egelman. 1999. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc. Natl. Acad. Sci. USA 96:4279-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perrin, R., P. Billard, and C. Branlant. 1997. Comparative analysis of the genomic DNA terminal regions of the lactococcal bacteriophages from species c2. Res. Microbiol. 148:573-583. [DOI] [PubMed] [Google Scholar]
- 48.Pillidge, C. J., and A. W. Jarvis. 1988. DNA restriction maps and classification of the lactococcal bacteriophages c2 and sk1. New Zealand Dairy Sci. Technol. 23:411-416. [Google Scholar]
- 49.Powell, I. B., M. G. Achen, A. J. Hillier, and B. E. Davidson. 1988. A simple and rapid method for genetic transformation of lactic streptococci by electroporation. Appl. Environ. Microbiol. 54:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Powell, I. B., and B. E. Davidson. 1985. Characterization of streptococcal bacteriophage c6A. J. Gen. Virol. 66:2737-2741. [DOI] [PubMed] [Google Scholar]
- 51.Rakonjac, J., L. J. Ward, A. H. Schiemann, P. P. Gardner, M. W. Lubbers, and P. W. O'Toole. 2003. Sequence diversity and functional conservation of the origin of replication in lactococcal prolate phages. Appl. Environ. Microbiol. 69:5104-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanders, M. E., and T. R. Klaenhammer. 1984. Phage resistance in a phage-insensitive strain of Streptococcus lactis - temperature-dependent phage development and host-controlled phage replication. Appl. Environ. Microbiol. 47:979-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiemann, A. H., J. Rakonjac, M. Callanan, J. Gordon, K. Polzin, M. W. Lubbers, and P. W. O'Toole. 2004. Essentiality of the early transcript in the replication origin of the lactococcal prolate phage c2. J. Bacteriol 186:8010-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schouler, C., S. D. Ehrlich, and M. C. Chopin. 1994. Sequence and organization of the lactococcal prolate-headed bIL67 phage genome. Microbiology 140:3061-3069. [DOI] [PubMed] [Google Scholar]
- 55.Simon, M. N., R. W. Davis, and N. Davidson. 1983. Heteroduplexes of DNA molecules of lambdoid phages: physical mapping of their base sequence relationship by electron microscopy, p. 313-328. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 56.Sing, W. D., and T. R. Klaenhammer. 1993. A strategy for rotation of different bacteriophage defenses in a lactococcal single-strain starter culture system. Appl. Environ. Microbiol. 59:365-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stambuk, S., and M. Radman. 1998. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics 150:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuer-Lauridsen, B., T. Janzen, J. Schnabl, and E. Johansen. 2003. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 309:10-17. [DOI] [PubMed] [Google Scholar]
- 59.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valyasevi, R., W. E. Sandine, and B. L. Geller. 1990. The bacteriophage kh receptor of Lactococcus lactis subsp. cremoris KH is the rhamnose of the extracellular wall polysaccharide. Appl. Environ. Microbiol. 56:1882-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterfield, N. R., M. W. Lubbers, K. M. Polzin, R. W. Le Page, and A. W. Jarvis. 1996. An origin of DNA replication from Lactococcus lactis bacteriophage c2. Appl. Environ. Microbiol. 62:1452-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, S. Y., and M. Feiss. 1991. The last duplex base-pair of the phage lambda chromosome. Involvement in packaging, ejection and routing of lambda DNA. J. Mol. Biol. 220:293-306. [DOI] [PubMed] [Google Scholar]
- 63.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., J. P. Muyrers, J. Rientjes, and A. F. Stewart. 2003. Phage annealing proteins promote oligonucleotide-directed mutagenesis in Escherichia coli and mouse ES cells. BMC Mol. Biol. 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]