Abstract
Bacillus anthracis Sterne cured of the pXO1 plasmid had enhanced secreted protease activity during the postexponential phase but no change in hemolytic or lecithinase activities. A zymogen profile revealed at least six proteases, including serine, metal, and perhaps cysteine types. There were similar amounts of protease secreted by the closely related species Bacillus cereus and Bacillus thuringiensis, but the patterns differed. Among the pXO1 plasmid-encoded proteins, there is a tetratricopeptide protein designated Cot43 that is related to the Rap proteins of Bacillus subtilis and the PlcR pleiotropic regulator of secreted enzymes and toxins in B. thuringiensis. A disruption of the cot43 gene resulted in overproduction of several proteases to a somewhat greater extent than in the plasmid-cured strain. Transformation of either of these strains with a clone of the cot43 gene resulted in the inhibition of accumulation of some of the proteases and induction of at least one. On the basis of lacZ fusions, transcription of the cot43 gene increased in late exponential cells at the time of protease accumulation. The expression of lacZ fusions to the upstream regions of two B. anthracis extracellular protease genes was greater in the strain with the disruption of cot43 than in the Sterne strain, indicating regulation at the level of transcription. In B. anthracis, a pXO1 plasmid-encoded protein directly modulates or indirectly regulates the transcription of genes for several chromosomally encoded extracellular proteases.
Members of the Bacillus cereus group secrete a variety of pathogenic factors during sporulation, including toxins, lecithinases, phospholipases, hemolysins, and proteases (2, 18, 24). These are important for the pathogenicity of Bacillus thuringiensis for insect larvae (25) and possibly for various types of B. cereus infections. There is a pleiotropic regulator, PlcR, which is responsible for controlling the production of many of these factors as well as of other proteins (2, 18). In Bacillus anthracis, however, there is very little secretion of many of these proteins, which is attributable to a nonsense codon within the plcR gene. Introduction of an intact plcR gene from B. thuringiensis into a plasmid-free derivative of B. anthracis restored synthesis of many of these factors. This change did not, however, enhance the pathogenicity of the transformant for mice (19).
It has been speculated that the absence of a functional plcR gene in B. anthracis is the first step in the ultimate removal of the genes encoding the extracellular factors that are no longer needed for pathogenicity (19). One scenario is that B. anthracis was derived from a B. cereus isolate, a so-called transitional strain (22) containing a functional PlcR regulon, by acquiring the two plasmids, pXO1 and pXO2, necessary for pathogenicity. These plasmids encode critical pathogenicity factors and some key regulators (8, 12). One of these, AtxA, is involved in the regulation of both the capsule and the toxin genes, among others, and appears to be antagonistic to PlcR, at least for sporulation (19). This is one explanation for the nonfunctional plcR gene in B. anthracis although the genome sequence of an atypical B. cereus isolate that contains a pXO1 plasmid has both intact atxA and plcR genes (13).
While the synthesis of many of these pathogenic factors is substantially reduced in B. anthracis, there is an array of genes encoding extracellular proteases (NCBI protein database for B. anthracis Ames, accession no. AE017031.1). Some of these may have a function in pathogenesis, such as the InhA2 metalloprotease in B. thuringiensis (9, 11), while others could play a role by providing nutrients for postexponential survival and sporulation. If so, there are likely to be regulatory protein(s) other than PlcR.
In the course of examining the spore coat profile of B. anthracis, a novel protein of 43 kDa, designated Cot43, was found which is a Rap-like protein encoded on the pXO1 plasmid (21). Rap proteins are members of the tetratricopeptide repeat family (6) which, together with an interacting pentapeptide encoded within a small downstream open reading frame (ORF), serves to modulate the phosphorelay initiating sporulation in Bacillus subtilis (23). Surprisingly, the PlcR pleiotropic regulator is also a member of this tetratricopeptide family and is activated by a pentapeptide (27) that is processed from a secreted oligopeptide of 48 amino acids and then taken up by an oligopeptide permease (10). It was thus conceivable that Cot43, in addition to its presence in spore coats, served a similar regulatory function, especially given the absence of a functional PlcR in B. anthracis. This possibility has been explored by examining the pattern of extracellular enzymes secreted by strains lacking or containing the cot43 gene. The results are consistent with regulation by Cot43 of the pattern and amounts of several extracellular proteases.
MATERIALS AND METHODS
Bacterial strains and growth.
B. anthracis Sterne was the vaccine strain, 34F2. A plasmid-cured derivative, strain St-13, was obtained by growth at 40°C for >40 generations and by screening individual colonies by PCR for the absence of the pag and atxA genes. Cells from these colonies were also examined for the absence of the pXO1 plasmid by agarose gel electrophoresis. B. anthracis Weymouth (UM44) and a derivative with a disruption of the atxA gene (UT53) were obtained from T. Koehler (12). B. cereus T and Bacillus thuringiensis subsp. kurstaki HD1 were laboratory stocks. The Sterne strain with a disruption of the pXO1-encoded cot43 gene, strain d2, was isolated as described below. Strains containing clones of the cot43 gene or cot43 plus a downstream ORF encoding a polypeptide of 46 amino acids (see below) were introduced by electroporation (15) with selection on Luria-Bertani agar plus 25 μg of erythromycin ml−1.
Cells were grown in 30 ml nutrient sporulation medium (NSM) (26), brain heart infusion broth (BHI), or the latter containing 0.5 M sorbitol. B. cereus and B. thuringiensis were also grown in a yeast extract-glucose medium (G-Tris) (4). Growth was in sidearm flasks and the density was monitored in a Klett colorimeter (660-nm filter [K600]). One-milliliter aliquots were removed and centrifuged in an Eppendorf microcentrifuge for 8 min, and the supernatants were passed through 0.2-μm-pore-size Nalgene syringe filters. In some cases the supernatants were dried in a lyophilizer and dissolved in water adjusting the volumes to equate K660 values. Alternatively, the sample volumes for protease assays or for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were adjusted to compensate for differences in the K660 values.
Cloning.
The Cot43 protein was originally identified as a band in SDS-15% PAGE gels of spore coat extracts of the Sterne strain but not of B. cereus. The band was excised and partially sequenced (MIVSVKGNEQITKMLND?YI), and this sequence was aligned with the amino terminal end of Orf136 in the pXO1 plasmid (21). For expression, a primer for a region 246 bp upstream of the start of translation (5′-CTCCTTTCACGCAGACGTGACATAATCCG) and one for the terminus (5′-TTTTCTTCCTTTTAAAGCCTCCTTTTC) were used. In order to include the downstream ORF of 46 amino acids (an alternative reading frame), the second primer was replaced with 5′-GTTTATTTATGATCCGCCAGTATGGCC. The PCR products were cloned into pCRT7/CT-TOPO (Invitrogen), and the inserts were sequenced. Each was excised with XbaI and HincII and subcloned into the E. coli/Bacillus shuttle vector, pHT315 (3) digested with XbaI and SmaI (referred to as cot43 and cot43P, respectively).
For disruption of the cot43 gene, primers 500 bp up- and downstream of the coding region were used: 5′-CGTTATCCAATAGGTTAAGGTCTCCC and 5′-GCTTTCCATGACTGGAAACTAGCACTG. The PCR product was cloned as described above, and a neomycin resistance gene cassette excised with SmaI from plasmid pBEST501 (14) was inserted into the unique EcoRV site within the cot43 gene. The entire disrupted gene region was then excised with XbaI and HindIII and cloned into the shuttle vector, pUTE29 (17). Electroporation and screening for neomycin-resistant (Neor), tetracycline-sensitive (Tcs) colonies were done as described previously (15). Clones were checked by PCR for the Neor insert in cot43 and by immunoblotting of spores and extracts from postexponential cells for the absence of Cot43 antigen.
A lacZ fusion to 246 bp of the region upstream plus 50 bp into the coding region of the cot43 gene was constructed by PCR amplification with primer 5′-CTCCTTTCCTGCAGACGTGACATAATCCG containing a PstI site and primer 5′-GTCTGGCACGTATCTCTAGATACCAGTC containing an XbaI site. Following purification with a QIAGEN kit, the PCR product was digested with these enzymes and ligated into an identically digested vector, pHT304-18Z, containing a promoterless lacZ gene (1).
The upstream regions (130 to 140 bp) of genes encoding two extracellular proteases were cloned by PCR. For a serine protease (NP846139), primers 5′-GCTGATTACTATAATCTCTGCAGTAGAAAGG and 5′-CTCTAGACGCACACATTTTTTGAAAC were used. These primers contained a PstI or XbaI site, respectively, and the resulting oligonucleotide (144 bp) was cloned into PHT305-18Z as described above. The same strategy was used to clone the upstream region of an immune inhibitor protease (NP843199) by employing primers 5′-GCCTTTTTATATAGGGACTGCAGAAAG and 5′-GACGATAACACTTCTAGAGGCGC.
The three plasmids containing lacZ fusions were electroporated into the Sterne strain, and the two protease fusion plasmids were electroporated into the d2 strain as well. Transformants were grown in 30 ml of NSM plus 25 μg of erythromycin ml−1 in sidearm flasks. One-milliliter duplicate samples were removed periodically and centrifuged for 6 min in an Eppendorf microcentrifuge. The pellets were frozen at −80°C and subsequently assayed for β-galactosidase activity (7). The specific activities are the averages of duplicate samples, and plots show the ranges for one standard deviation.
Extracellular enzyme assays.
The initial screening was done by spotting cells onto BHI agar containing 5% low-fat milk (Parmalat), 5% sheep red blood cells, or 5% egg yolk (Difco) and incubating at 37°C for 16 to 36 h (19). Total protease in cell supernatants was measured by using succinylated-casein (QuantiCleave protease assay kit; Pierce) under the conditions specified in the notes to Table 1. Reaction conditions were selected to ensure a linear increase of color with time (over 60 min) and proportional increases with quantities of supernatants (2.5 to 10 μl). Zymogen profiles were determined in SDS-8% PAGE containing Parmalat 2% milk at a 5% (vol/vol) final concentration. Gelatin (Difco) at 2% (w/vol) was also used, but there were fewer bands. Stock solutions (0.01 M) of the protease inhibitors O-phenanthroline and phenylmethylsulfonyl fluoride (PMSF) were dissolved in ethanol; EGTA (Sigma) and E-64 (Sigma), an inhibitor of cysteine proteases, were dissolved in deionized water. Inhibitors were added to the supernatants to a 2.5 mM final concentration, and the solutions were incubated at 27°C for 5 min before loading on gels. A control with an equal concentration of ethanol was also included. After electrophoresis, the gels were treated as described previously (16). A purified protease of 36 kDa from Bacillus polymyxa (Sigma) served as a molecular weight marker.
TABLE 1.
Protease activity in supernatants of Bacillus speciesa
| Strain | A450 |
|---|---|
| B. anthracis Sterne | 0.62 |
| B. anthracis d2 (Δcot43) | 1.60 |
| B. anthracis d2/cot43 | 0.55 |
| B. anthracis St-13 | 1.35 |
| B. anthracis St-13/cot43 | 0.88 |
| B. anthracis St-13/cot43P | 0.40 |
| B. cereus Tb | 0.65 |
| B. cereus T/cot43Pb | 0.45 |
| B. thuringiensisb | 0.65 |
Activity was measured with succinylated casein (Pierce QuantiCleave Protease Assay Kit). Incubation was at 27°C for 30min (values still increasing at a constant rate), followed by 20 min with the colorimetric reagent. Assays were performed with 5 μl of supernatants (values with 10 μl were twice as high) from cells grown in NSM or G-tris at 37°C until 50% contained phase-white endospores. The initial volumes of the supernatants were adjusted so that all were from cells at equivalent Klett units. Values are the averages of three experiments with a standard deviation of ± 5%.
Grown in G-tris medium.
RESULTS
Total protease assays.
The patterns of protease, hemolysin, and lecithinase accumulation were compared between B. anthracis Sterne and the plasmid-cured derivative (strain St-13). On the basis of a plate assay measuring diameters of zones of clearing, there was about a twofold increase in total protease secreted by postexponential cells of the plasmid-cured strain (unpublished results). There were no comparable changes in either hemolytic activity or lecithinase, which were barely detectable in either strain. There was a two- to threefold higher level of protease activity in strain St-13 than in the Sterne strain in an assay with azocasein (Table 1). Total protease activity was comparable for the three related species, B. anthracis, B. cereus, and B. thuringiensis (Table 1).
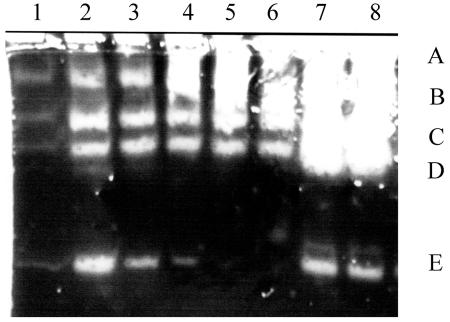
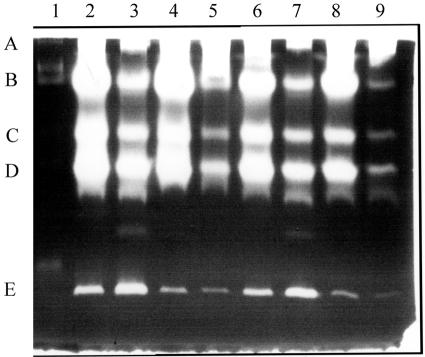
Zymogen profiles of the two B. anthracis strains were compared with sampling during growth and at various stages of sporulation (Fig. 1). Five protease activities were resolved (Fig. 1, bands A to E). For all of the relative sampling times, protease bands A to D in the plasmid-cured strain were more intense than the comparable bands in the Sterne strain. For the latter, bands A to D increased to late sporulation, whereas band E peaked in early sporulation. There was another protease activity of ca. 40 kDa found predominantly in exponentially growing cells (see Fig. 6 and the band just above band E). On the basis of the effects of protease inhibitors (Fig. 2), band A and perhaps bands C and D, which were less intense in the presence of PMSF (lane 3), are likely to be serine proteases. Bands B to D were not detected in the presence of EGTA (lane 2), and so are probably calcium-dependent proteases. Band E was less intense in the presence of the cysteine protease inhibitor, E64, when the bands in lanes 4 and 5 were compared, but its intensity varied with the time of sampling (Fig. 1), and so there is less certainty as to the type of protease.
FIG. 1.
Zymogen profiles of a gel containing low-fat milk. Lanes 1 to 5 are supernatants from the Sterne strain grown in NSM at 37°C with samples removed at 60-min intervals, commencing at the end of growth. Phase-white endospores were present in >80% of the cells by sample 4. Lanes 6 to 8 are for the plasmid-free strain, St-13, sampled at the same time as lanes 2, 4, and 5. Bands labeled A to E are discussed in the text (Fig. 2). In all cases, 5 μl of the supernatants was loaded from cultures grown to a Klett value of 230 or the equivalent (see Materials and Methods).
FIG. 6.
A clone of the cot43 gene regulates the intensity of the protease bands. Supernatants of strain St-13 (lanes 1, 4, and 7), strain St-13/cot43 (lanes 2, 5, and 8), and strain St-13/cot43P (lanes 3, 6, and 9) were used. Samples were removed during late exponential growth (lanes 1 to 3), at the end of growth (lanes 4 to 6), and 90 min later (lanes 7 to 9). Amounts were loaded as described in the legend to Fig. 1.
FIG. 2.
Effect of inhibitors on extracellular proteases. Lane 1, 5 μl of the supernatant from the Sterne strain grown in NSM at 37°C and harvested as described for lane 4, Fig. 1; lane 2, addition of EGTA to a concentration of 2.5 mM prior to loading; lane 3, 2.5 mM PMSF; lane 4, 2.5 mM O-phenanthroline; lane 5, 2.5 mM E64. A molecular weight marker was a metalloprotease of 36 kDa from B. polymyxa (Sigma P-6141) which migrated a little bit more slowly than band E (Fig. 5).
Differences with medium, temperature, and species.
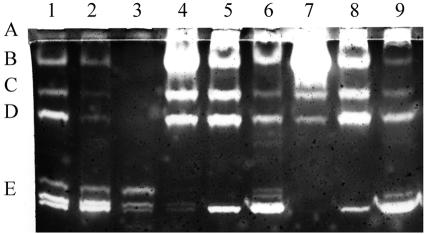
The extracellular protease profile for B. cereus differed somewhat from that of the Sterne strain (Fig. 3). There was more protease in the supernatants of B. cereus grown at 37 versus 30°C (lanes 1 and 3, respectively), but this was not so apparent for the Sterne strain (lanes 5 and 7, respectively). There also appears to be more produced in G-Tris medium than in NSM (lanes 1 to 4). Protease activity in BHI with 0.5 M sorbitol (used for electroporation) was much less, probably due to poorer sporulation in this medium. The zymogen pattern for B. thuringiensis subsp. kurstaki HD1 was similar in profile and amount (Table 1) to that of B. cereus T.
FIG. 3.
Zymogen profiles with variations in strains, media, and temperature of growth. Lane 1, B. cereus T grown in G-tris medium at 30°C; lane 2, B. cereus T grown in NSM at 30°C; lane 3, B. cereus T grown at 37°C in G-tris; lane 4, B. cereus T grown at 37°C in NSM; lane 5, Sterne strain grown at 30°C in NSM; lane 6, Sterne strain grown at 30°C in BHI plus 0.5 M sorbitol; lane 7, Sterne strain grown at 37°C in NSM; lane 8, Sterne strain grown at 37°C in BHI plus 0.5 M sorbitol; lanes 9 and 10, Sterne strain grown at 37°C in NSM and samples incubated at 27°C for 30 min prior to loading (lane 9 supernatant from cells sampled 90 min earlier than the samples in the other lanes). Supernatants in lanes 1 to 8 and 10 are from samples taken 3 h after the end of growth (20 to 30% phase-dull spores in each). See legend to Fig. 1 for the amounts loaded.
Regulation by Cot43.
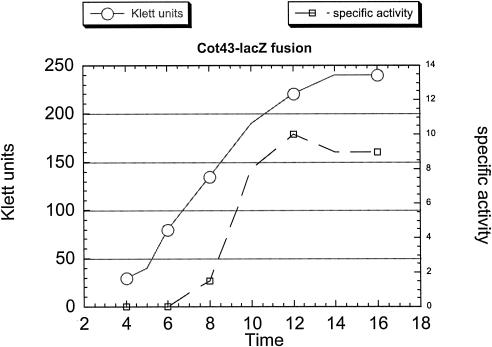
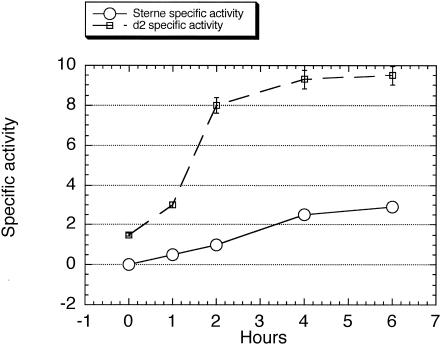
As mentioned above, a possible pXO1-encoded regulator is the Rap-like protein Cot43. This protein was initially isolated from spore coat extracts; so if it were also to function as a regulator, it must be accessible at the time of protease synthesis. Expression of a lacZ fusion to 246 bp upstream of the cot43 coding region was determined (Fig. 4). Transcription increased during growth, and while the specific activity leveled off at the end of growth, Cot43 antigen increased in cell extracts until about stage II of sporulation and was still present when >90% of the cells contained phase-white endospores (unpublished results).
FIG. 4.
Time course of the increase in the specific activity of β-galactosidase in the Sterne strain containing a lacZ fusion to the cot43 upstream region. Sampling and analysis were performed as described in Materials and Methods.
The absence of Cot43 due either to the loss of the pXO1 plasmid (strain St-13) or to a disruption of the gene (strain d2) resulted in higher levels of protease bands A to D in the zymograms (Fig. 5). The values for strain d2 are somewhat greater than for strain St-13 (Table 1). A clone of the cot43 gene with and without the downstream ORF (46 amino acids) was introduced into strain d2, and the resulting transformant produced less protease (Table 1) and a pattern similar to that of the Sterne strain. Introduction of these clones into strain St-13 resulted in some inhibition of proteases A to D for the transformant containing the cot43 clone, and more extensive inhibition if cot43 plus the downstream ORF were introduced (Table 1 and Fig. 6). In contrast, the ca. 36-kDa band E, which is a doublet in late exponential cells, subsequently disappeared in strain St-13 but increased during sporulation in the transformant containing the cot43P clone. The more extensive inhibition of proteases A to D in strain St-13/cot43P than in strain St-13/cot43 (Fig. 6) implies that regulation by Cot43 involves the downstream ORF perhaps as a pentapeptide as for the PlcR regulator in B. thuringiensis (23). The presence of the cot43P clone in B. cereus T did result in some inhibition of several protease bands but less than that found for strain St-13/cot43P (Table 1).
FIG. 5.
Enhanced protease in strains lacking Cot43. Lane 1, B. polymyxa metalloprotease (the lower band is 36 kDa); lane 2, Sterne strain with a disruption of the cot43 gene (strain d2); lane 3, strain UT44; lane 4, strain St-13; lane 5, Sterne strain; Lanes 6 to 9, the same strains used in lanes 2 to 5, respectively, but with cells harvested 90 min earlier. Strains were grown at 37°C in NSM until 90 min after the end of growth (lanes 6 to 9) or 180 min after the end of growth (lanes 2 to 5). Bands labeled A to E are as discussed in the legend to Fig. 2 and the text. Amounts loaded are as described in the legend to Fig. 1.
The AtxA regulator is involved.
The inhibition in strain St-13/cot43P of bands A to D was greater than in the Sterne strain sampled at comparable times (Fig. 1, lanes 2 and 3, versus Fig. 6, lane 9), suggesting that the pXO1 plasmid contains another gene product which is involved in this regulation. The possibility of a plasmid-encoded protease “inducer” prompted an examination of a role for the AtxA regulator (14). The supernatant of a strain with a disruption of the atxA gene, strain UT53, had a somewhat less intense zymogen profile for several of the bands when compared to the parental strain, UM44 (Fig. 7, lanes 1 and 2). This profile in strain UT53 was substantially reduced by introduction of the cot43 clone (lane 3).
FIG. 7.
Differential effects of the presence of Cot43 on the zymogen profiles of a mutant with a disruption of the atxA gene. Lane 1, strain UM44; lane 2, strain UT53; lane 3, strain UT53/cot43P. All strains were grown at 37°C in NSM until 1 h after the end of exponential growth. Amounts were loaded as described in the legend of Fig 1.
LacZ fusions to promoters of extracellular proteases are regulated by Cot43.
Fusions to lacZ of ca. 100 bp of the region upstream of the coding sequences of a serine and an immune inhibitor protease were constructed as described in Materials and Methods and electroporated into the Sterne and d2 strains. As shown in Fig. 8, β-galactosidase-specific activity increased from the end of growth until about stage II to III of sporulation and was severalfold higher in the d2 strain than in the Sterne strain. Similar results were obtained with the fusion to the upstream region of the serine protease gene. Regulation by Cot43 (directly or indirectly) for at least these two proteases is at the level of transcription.
FIG. 8.
Specific activities of β-galactosidase of a fusion of the upstream region of the immune inhibitor protease to lacZ in the Sterne strain and in mutant d2. Time zero is the end of growth, with phase-dull endospores present in >70% of the cells by 4 h.
DISCUSSION
As previously reported, B. anthracis secretes only small amounts of the pathogenic factors which are regulated by the pleiotropic regulator PlcR, despite the presence of intact structural genes for several factors including lysolecithinases, hemolysins, phospholipases, and at least some proteases. This deficiency is attributable to a missense mutation in the plcR gene (19). Even with this defect, B. anthracis does secrete a variety of proteases. Six were resolved in zymogens, and these appeared to be distinct proteases with differing sizes and inhibition patterns. The overall zymogen pattern did not change markedly with the time of sampling, medium, temperature of cell growth, or strain used. These patterns were relatively stable as they were unaltered after incubation of the supernatants at 27°C for 30 min prior to loading, indicating that the lower molecular weight bands were not degradation products. The pattern does depend upon the substrate included in the gel, the resolving power of the gel, and probably the extent of renaturation of the activity after electrophoresis in SDS-PAGE.
An examination of the ORFs in the Ames genome (entrez protein search of the B. anthracis Ames genome, accession no. AE017031.1) indicates that there are ca. 10 proteases with potential signal sequences (5) ranging in size from about 40 to 100 kDa. Nothing is known about the expression or secretion patterns of these enzymes. Two appear to be similar to the InhA protease secreted by B. thuringiensis which may function to destroy host immune proteins and thus aid in establishing infection (9, 11). None of these protease genes contained the consensus palindrome recognized by the PlcR regulator (19), at least within 50 bp of the start codon. There was in the upstream regions, however, a consensus sequence present among six of these protease genes. Its role, if any, in regulation by Cot43 is being examined.
Among the six protease bands identified here, four were present primarily in postexponential cells and at least three were down-regulated in the Sterne strain relative to a derivative devoid of the pXO1 plasmid (Fig. 5). It was noted that a pXO1 gene designated cot43 encoded a tetratricopeptide repeat protein and that there was a potential downstream ORF of 46 amino acids. Cot43 is similar in sequence to the Rap proteins in B. subtilis, many of which also contain a small downstream ORF (23). In addition, the PlcR regulator from B. thuringiensis is a member of this family, and thus the possibility that Cot43 had a regulatory function was explored. While this protein was originally identified in spore coat extracts of the Sterne strain, disruption of the cot43 gene resulted in overexpression of the proteases in a pattern similar to that in the plasmid-free strain. Expression of Cot43 on a plasmid resulted in extensive inhibition of protease activity, especially if the clone contained a downstream orf encoding a 46-amino-acid peptide. Variations in the level of expression of the cot43 operon could thus modulate extracellular postexponential protease synthesis. It appears that Cot43 has a dual function: as part of the B. anthracis spore coat where it modulates germination (A. I. Aronson and H. Hu, unpublished data) and directly or indirectly as a transcriptional regulator of several extracellular proteases.
A small but significant enhanced expression was found for the cot43 mutant strain relative to the plasmid-free strain (Table 1 and Fig. 5), implying that another pXO1 gene was involved as an inducer. One candidate would be the AtxA regulator of pathogenic factors (8, 12), and a strain with a disruption of this gene produced somewhat less protease than the parental strain (Fig. 7). The two plasmid-encoded proteins, Cot43 and AtxA, may serve to modulate extracellular protease synthesis, at least for the six proteases resolved in this analysis.
It is intriguing that chromosomally encoded proteases are regulated by pXO1 factors. A similar regulatory pattern was found for the S-layer proteins that are encoded on the chromosome and regulated by AtxA (20). It is tempting to speculate that in both cases, there is some relation of this pattern of regulation to pathogenicity. This interplay implies that pathogenic B. anthracis strains have evolved beyond a B. cereus“transition strain” containing pXO1 and pXO2 as autonomously functioning plasmids.
Acknowledgments
This research was supported by NIH grant AI48505.
REFERENCES
- 1.Agaisse, H., and D. Lereclus. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13:97-107. [DOI] [PubMed] [Google Scholar]
- 2.Agaisse H., M. Gominet, O. A. Okstad, A.-B. Kolsto, and D. Lereclus. 1999. PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32:1043-1053. [DOI] [PubMed] [Google Scholar]
- 3.Arantes, O., and D. Lereclus. 1991. Construction of cloning vectors for Bacillus thuringiensis. Gene 108:115-119. [DOI] [PubMed] [Google Scholar]
- 4.Aronson, A. I., N. Angelo, and S. C. Holt. 1971. Regulation of extracellular protease production in Bacillus cereus T: characterization of mutants producing altered amounts of protease. J. Bacteriol. 106:1016-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendtsen, J. D., H. Nielsen, G. von Heinje, and S. Brunek. 2004. Improved prediction of signal peptides: signal IP30. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 6.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, P., L. Wu, Y. Ziniu, and A. Aronson. 1999. Subspecies-dependent regulation of Bacillus thuringiensis protoxin genes. Appl. Environ. Microbiol. 65:1849-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, Z., J.-C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 9.Fedhila, S., P. Nel, and D. Lereclus. 2002. The InhA2 metalloprotease of Bacillus thuringiensis strain 407 is required for pathogenicity in insects infected via the oral route. J. Bacteriol. 184:3296-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 11.Grandvalet, C., M. Gominet, and D. Lereclus. 2001. Identification of genes involved in the activation of the Bacillus thuringiensis inhA metalloprotease gene at the onset of sporulation. Microbiology 147:1805-1813. [DOI] [PubMed] [Google Scholar]
- 12.Hoffmaster, A. R., and T. M. Koehler. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. k. Marston, B. K. de, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. J. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. R. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax: public health challenges in the age of bioterrorism. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itaya, M., K. Kondo, and T. Tanaka. 1989. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acid Res. 17:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, H.-S., D. Sherman, F. Johnson, and A. I. Aronson. 2004. Characterization of a major Bacillus anthracis spore coat protein and its role in spore inactivation. J. Bacteriol. 186:2413-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleiner, E. D., and W. D. Stetler-Stevenson. 1993. Quantitative zymography: detection of picogram quantities of gelatinases. Anal. Biochem. 218:325-329. [DOI] [PubMed] [Google Scholar]
- 17.Koehler, T. M., Z. Dai, and M. K. Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lereclus, D., H. Agaisse, M. Gominet, S. Salamitou, and V. Sanchis. 1996. Identification of a Bacillus thuringiensis gene that positively regulates transcription of the phosphatidylinositol-specific phospholipase C gene at the onset of the stationary phase. J. Bacteriol. 178:2853-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mignot, T., M. Mock, D. Robichon, A. Landier, D. Lereclus, and A. Fouet. 2001. The incompatibility between the PlcR-and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Mol. Microbiol. 42:1189-1198. [DOI] [PubMed] [Google Scholar]
- 20.Mignot, T., M. Mock, and A. Fouet. 2003. A plasmid-encoded regulator couples the synthesis of toxins and surface structures in Bacillus anthracis. Mol. Microbiol. 47:917-927. [DOI] [PubMed] [Google Scholar]
- 21.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Ricke, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patra, G., J. Vaissaire, M. Weber-Levy, C. Le Doujet, and M. Mock. 1998. Molecular characterization of Bacillus strains involved in outbreaks of anthrax in France in 1997. J. Clin. Microbiol. 36:3412-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perego, M. 1999. Self-signaling by Phr peptides modulates Bacillus subtilis development, p. 243-258. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. American Society for Microbiology, Washington, D. C.
- 24.Pomerantsev, A. P., K. V. Kalnin, M. Osorio, and S. H. Leppla. 2003. Phosphatidylcholine-specific phospholipase C and shingomyelinase activites in bacteria of the Bacillus cereus group. Infect. Immun. 71:6591-6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salamitou, S., F. Ramisse, M. Brehelin, D. Bourguet, N. Gilois, M. Gominet, E. Hernandez, and D. Lereclus. 2000. The plcR regulon is involved in the opportunistic properties of Bacillus thuringiensis and Bacillus cereus in mice and insects. Microbiology 146:2825-2832. [DOI] [PubMed] [Google Scholar]
- 26.Schaeffer, P., H. Ionesco, A. Ryter, and G. Balassa. 1963. La sporulation de Bacillus subtilis: etude genetique et physiologique. Colloq. Int. CNRS 124:553-563. [Google Scholar]
- 27.Slamti, L., and D. Lereclus. 2002. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21:4550-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]