Abstract
We previously cloned three endoglucanase genes, rce1, rce2, and rce3, that were isolated from Rhizopus oryzae as the first cellulase genes from a member of the subdivision Zygomycota. In this study, two cDNAs homologous to the rce1 gene, designated the mce1 and mce2 cDNAs, were cloned from Mucor circinelloides, a member of the subdivision Zygomycota. The mce1 cDNA encoded an endoglucanase (family 45 glycoside hydrolase) having one carbohydrate-binding module (CBM), designated MCE1, and the mce2 cDNA encoded the same endoglucanase having two tandem repeated CBMs, designated MCE2. The two cDNAs contained the same sequences but with a 147-bp insertion. The corresponding genomic mce gene consisted of four exons. The mce1 cDNA was created from exons 1, 3, and 4, and the mce2 cDNA was created from exons 1, 2, 3, and 4. These results indicate that the mce1 and mce2 cDNAs were created from one genomic mce gene by alternative splicing. MCE1 and MCE2, purified to apparent homogeneity from the culture supernatant of M. circinelloides, had molecular masses of 43 and 47 kDa, respectively. The carboxymethyl cellulase specific activity of MCE2 was almost the same as that of MCE1, whereas the Avicelase specific activity of MCE2 was two times higher than that of MCE1. Furthermore, MCE2, whose two tandem CBMs might be more effective for degradation of crystalline cellulose than one CBM, was secreted only at an early culture stage when crystalline cellulose was abundant.
Cellulose is the major plant cell wall polysaccharide and is degraded by cellulases. This degradation is considered to be due to the synergistic action of three types of cellulase components: endoglucanases (EC 3.2.1.4; endo-β-d-1,4-glucanases), cellobiohydrolases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21) (11).
The endoglucanases, a large, ubiquitous family of enzymes that hydrolyze β-1,4 linkages adjacent to unsubstituted glucose residues (10), are produced by a broad range of organisms, including fungi (23), bacteria (2), plants (17), and insects (termites) (12). In the textile and detergent industries, endoglucanases have been used for removing microfibrils from the surface of cellulosic fabrics, which enhances the softness and brightness of the cellulosic fabrics (1, 3, 4, 8). Because of the importance of the endoglucanases in these industries, many fungal endoglucanase genes from members of the subdivision Deuteromycotina, such as Aspergillus sp., Fusarium sp., Humicola sp., Penicillium sp., and Trichoderma sp., have been cloned and characterized (21). However, there were no reports of the isolation of endoglucanase genes from members of the subdivision Zygomycota until we isolated two endoglucanases, RCE1 and RCE2, from Rhizopus oryzae as the first cellulases from a member of the subdivision Zygomycota (16) and cloned the encoding genes, rce1, rce2, and rce3 (15). We also cloned the pce1 gene, which is homologous to the codon usage-optimized rce1 gene, from Phycomyces nitens, a member of the same subdivision, and we isolated and characterized endoglucanase PCE1, which is encoded by the pce1 gene (19).
In this study, two cDNAs homologous to rce1, designated the mce1 and mce2 cDNAs, were cloned from Mucor circinelloides. The mce1 cDNA encoded an endoglucanase (family 45 glycoside hydrolase) having one carbohydrate-binding module (CBM), designated MCE1, and the mce2 cDNA encoded the same endoglucanase having two tandem repeated CBMs, designated MCE2. Furthermore, the mce1 and mce2 cDNAs were created from one genomic mce gene by alternative splicing. Therefore, to elucidate the function of the two tandem CBMs, we compared the enzymatic characteristics of MCE1 and MCE2. Also, we compared the amino acid sequences of family 45 glycoside hydrolases from Rhizopus, Phycomyces, and Mucor, which are members of the subdivision Zygomycota.
MATERIALS AND METHODS
Bacterial strains, cultural conditions, and plasmids.
To obtain a chromosomal DNA preparation, M. circinelloides FERM BP-6890 was cultured aerobically at 28°C for 40 h in a medium containing 1.0% yeast extract (Difco), 2.0% polypeptone (Wako), and 2.0% glucose. Plasmid pUC119 (Takara) was used for DNA cloning, and Escherichia coli JM109 was used as the host for the recombinant plasmid. The genomic library of M. circinelloides was constructed in the lambda DASH II vector (Stratagene). E. coli NM514 was used as the host for the genomic DNA library of M. circinelloides. To produce uracil-containing single-stranded DNA, E. coli CJ236 was used as a host and M13KO7 was used as a helper phage. The recombinant E. coli strains were cultured in Luria-Bertani medium containing 50 μg of ampicillin per ml.
Isolation and identification of M. circinelloides.
A small amount of soil from Sakado-shi, Saitama, Japan, was suspended in sterilized water and spread on Mucor agar medium (4% glucose, 0.2% asparagine, 0.05% KH2PO4, 0.025% MgSO4, 0.0005% thiamine, 1.5% agarose), a selective medium for Zygomycota. The plates were incubated at 28°C for a suitable period. Isolated colonies were transferred as inocula into a cellulase production medium (3.0% corn steep liquor, 2.4% potato dextrose, 2.0% sucrose, 0.5% yeast extract), and the cultures were incubated at 28°C with continuous shaking. The carboxymethyl cellulase (CMCase) activity in the culture supernatant was then assayed.
The fungus was classified on the basis of the cultural and morphological characteristics of the colonies. The fungus grew well on potato dextrose agar at 25°C but grew poorly at 37°C. The mycelia were light brown on both sides of a plate and formed a wool-like colony that was over 85 mm in diameter. Colorless sporangiophores that were 10 to 20 μm in diameter arose directly from bifurcated rhizoids. The sporangia were globose or subglobose, dark brown, and 20 to 50 μm in diameter with colorless, globose, or subglobose columella that were12.5 to 30 μm in diameter. The sporangiospores were colorless and subglobose or oval, had smooth walls, and were 3.5 to 5 μm in diameter or 3.5 to 5 by 6 to 8 μm. Based on these observations, we identified this fungus as M. circinelloides (24). This strain has been deposited in the Fermentation Research Institute, Agency of Industrial Science and Technology of Japan, Tsukuba-shi, Ibaraki, Japan, under accession number FERM BP-6890.
Protein and enzyme assays.
To assay CMCase activity, reaction mixtures containing 0.32 μg of MCE1 or MCE2 and 10 mg of carboxymethyl cellulose in 1.0 ml of 50 mM sodium phosphate buffer (pH 7.0) were incubated for 15 min at 45°C (MCE1) or 50°C (MCE2). The reducing sugars liberated were then measured by determining the d-glucose equivalents by the dinitrosalicylic acid assay (14). Avicelase activity was assayed by measuring the reducing sugars liberated by using reaction mixtures containing 10 μg of MCE1 or MCE2 and 10 mg of Avicel in 1.0 ml of 50 mM sodium phosphate buffer (pH 7.0), which were incubated for 120 min at 45°C (MCE1) or 50°C (MCE2). One unit of activity was defined as the amount of enzyme that released 1 μmol of reducing sugar per min.
The CMCase activities of MCE1 and MCE2 were measured under several different pH conditions by using 50 mM sodium citrate (pH 3 to 4), sodium acetate (pH 4 to 6), sodium phosphate (pH 6 to 8), or glycine-NaOH (pH 8 to 10) buffer. The effect of temperature was examined under standard conditions by varying the temperature (20 to 70°C). Protein concentrations were determined by Bradford's method by using a protein assay kit with bovine serum albumin as the standard (7).
Enzyme purification. (i) Step 1.
M. circinelloides FERM BP-6890 was shake cultured at 28°C for 18 h in a liquid medium (3.0% corn steep liquor, 2.4% potato dextrose, 2.0% sucrose, 0.5% yeast extract). After incubation for 18 h, the culture was centrifuged at 27,000 × g at 4°C for 30 min. The supernatant (200 ml) was collected and used in the following steps.
(ii) Step 2.
Ammonium sulfate was added to the culture supernatant at a final concentration of 1.5 M, and by using hydrophobic chromatography, the mixture was chromatographed on an Econo-Pac Methyl HIC cartridge column (15 by 144 mm; Bio-Rad) equilibrated with 1.5 M ammonium sulfate. After the column was washed with 4 bed volumes of the same solution, it was eluted with a stepwise gradient of ammonium sulfate (1.5, 1.0, 0.5, and 0 M), followed by distilled water, at a flow rate of 2 ml/min. The CMCase activity was found in two fractions that eluted at 0 and 0.5 M ammonium sulfate. To each active fraction, ammonium sulfate was added to a final concentration of 1.5 M.
(iii) Step 3.
The two active fractions obtained in step 2 were then chromatographed on an Econo-Pac Methyl HIC cartridge column (15 by 144 mm; Bio-Rad) and again equilibrated with 1.5 M ammonium sulfate. The column was eluted with a stepwise gradient of ammonium sulfate (1.5, 0.9, and 0 M) at a flow rate of 2 ml/min. Fractions with CMCase activity eluted with 0 M ammonium sulfate.
(iv) Step 4.
The active fractions obtained in step 3 were diluted 10-fold with 50 mM acetate buffer (pH 4.0) and were separated by cation-exchange chromatography on a Mono S HR 5/5 column (5 by 50 mm; Pharmacia Biotech) equilibrated with the same buffer. After the column was washed with 3 bed volumes of the same buffer, it was eluted with a continuous linear gradient from 50 mM acetate buffer (pH 4.0) to 0.2 M NaCl in 50 mM acetate buffer (pH 5.2) at flow rate of 1 ml/min. The fractions containing MCE1 and MCE2 eluted at NaCl concentrations of 120 and 190 mM, respectively.
SDS-PAGE.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli (13) by using a ready-made 10% polyacrylamide gel (Tefco). After electrophoresis, the gel was stained with 2D-silver stain II DAIICHI (Daiichi Pure Chemicals).
Protein sequencing.
To determine the amino acid sequences of peptide fragments derived from the internal regions of MCE1 and MCE2, the purified MCE1 and MCE2 were lyophilized and dissolved in 50 mM Tris-HCl (pH 8.0). Each enzyme was digested with lysylendopeptidase, which was added at a molar ratio of MCE1 or MCE2 to lysylendopeptidase of 100:1. The reaction mixture was incubated at 37°C for 48 h, and the peptides generated were separated with a model 172 micropreparative high-performance liquid chromatography system (Applied Biosystems). N-terminal amino acid sequencing of the purified MCE1 and MCE2 proteins and determination of the amino acid sequences of peptide fragments derived from internal regions of MCE1 and MCE2 were performed by using an ABI model 491 protein sequencer (Applied Biosystems).
Western blot analysis.
M. circinelloides FERM BP-6890 was shake cultured at 28°C in a liquid medium containing 3.0% corn steep liquor, 2.4% potato dextrose, 2.0% sucrose, and 0.5% yeast extract. After cultivation for 16, 42, and 90 h, each culture was centrifuged, and the supernatant was subjected to SDS-PAGE as described above. After electrophoresis, the gel was immunostained, and the proteins in the gel were transferred to a membrane (Immobilon-P; Millipre) by using an AE6677 horizon blot (Atto). MCE1 and MCE2 were detected on the membrane by using antibodies from a rabbit immunized with purified RCE1 in combination with anti-rabbit (goat) antibodies conjugated with horseradish peroxidase as secondary antibodies.
Cloning of the genomic mce gene.
For Southern blot analysis, genomic DNA of M. circinelloides was digested with SphI, AccI, EcoRI, or XhoI or with XhoI and AccI. The digested fragments were separated by agarose gel electrophoresis and transferred onto a nylon membrane (Hybond-N+). Chromosomal DNA isolated from M. circinelloides was digested with EcoRI and separated on a 0.8% agarose gel. The 3.0- to 6.5-kbp DNA fragments were collected and ligated into EcoRI-restricted lambda gt10 vector (Stratagene). This vector was packaged in vitro into bacteriophage λ particles by using an in vitro packaging kit (Giga Pack II packaging kit; Stratagene), and the resulting phages were used to infect E. coli NM514. The genomic library was screened by plaque hybridization with the rce1 gene fragment as a probe after labeling with an ECL direct DNA/RNA labeling detection system (Amersham). The sequence reaction was performed by the dideoxy chain termination method by using an A.L.F. DNA Sequencer II (Pharmacia Biotech).
Isolation of mce cDNAs by RT-PCR.
M. circinelloides FERM BP-6890 was grown at 28°C for 18 h with shaking in a medium containing 3.0% corn steep liquor, 2.0% sucrose, 0.5% yeast extract, and 2.4% potato dextrose broth. The resulting cells were lyophilized and finely crushed. Total RNA was isolated by using Isogen (Wako Pure Chemical Industry). Then mRNA was isolated with an mRNA isolation kit (Stratagene). cDNA of the mce gene was prepared from the mRNA by reverse transcription (RT)-PCR by using a Takara RNA PCR kit (avian myeloblastosis virus) (version 2.1; Takara). Oligonucleotide primers were prepared as primers for the N and C termini deduced from the previously determined genomic mce sequence (mce-CN [5′-GCGAATTCATGAAGTTCACCGTTGCTATT-3′] and mce-CC [5′-GCGAATTCTTACTTTCTTTCGCAACCTG-3′]), and cDNA of the mce gene was then amplified by PCR by using the mRNA prepared earlier as the template. The cDNA fragments were collected and ligated into EcoRI-restricted vector pUC118. The DNA sequence was determined by the dideoxy chain termination method as described previously.
Nucleotide sequence accession numbers.
The nucleotide sequences of the mce1 and mce2 cDNAs of M. circinelloides FERM BP-6890 have been deposited in the DDBJ database under accession numbers AB175927 and AB175928, respectively.
RESULTS AND DISCUSSION
Cloning and sequence analysis of the mce gene.
When M. circinelloides genomic DNA digested with EcoRI was analyzed with the rce1 gene as a probe in a Southern blot, one strong signal at about 4.5 kbp appeared. This result suggests the presence of an rce1 homologous gene. Therefore, the rce1 homologous gene, about a 4.5-kbp DNA fragment, was designated the mce gene.
To clone the mce gene, plaque hybridization was performed with the rce1 gene as a probe against the genomic library by using 3- to 6.6-kbp DNA fragments from the M. circinelloides genomic DNA digested with EcoRI. Six positive phages were obtained from about 10,000 phage plaques. These positive phages were considered to contain the mce gene as detected by Southern hybridization. The DNA extracted from the positive phages was digested with EcoRI, and the resultant 4.5-kbp fragment was subcloned into the EcoRI site of pUC119. Nucleotide sequencing of the 4.5-kbp fragment indicated the presence of a single open reading frame encoding a predicted family 45 glycoside hydrolase. However, it was presumed that this sequence contained introns; therefore, cDNA of the mce gene was isolated by RT-PCR.
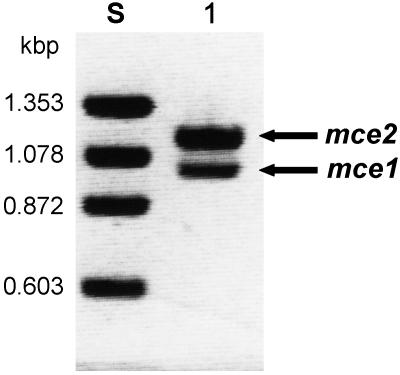
As a result of the RT-PCR, two amplified fragments, about 1.0 and 1.1 kbp long, were detected after agarose gel electrophoresis (Fig. 1). Therefore, the 1.0- and 1.1-kbp DNA fragments were designated the mce1 and mce2 cDNAs, respectively. The mce1 and mce2 cDNAs were subcloned into the EcoRI site of pUC118, and nucleotide sequencing indicated the presence of a single open reading frame encoding predicted 316- and 387-residue proteins (Fig. 2).
FIG. 1.
Agarose gel electrophoresis analysis of cDNA fragments of the mce gene amplified by RT-PCR. Lane S, standard DNAs; lane 1, cDNA fragments of the mce gene amplified by RT-PCR.
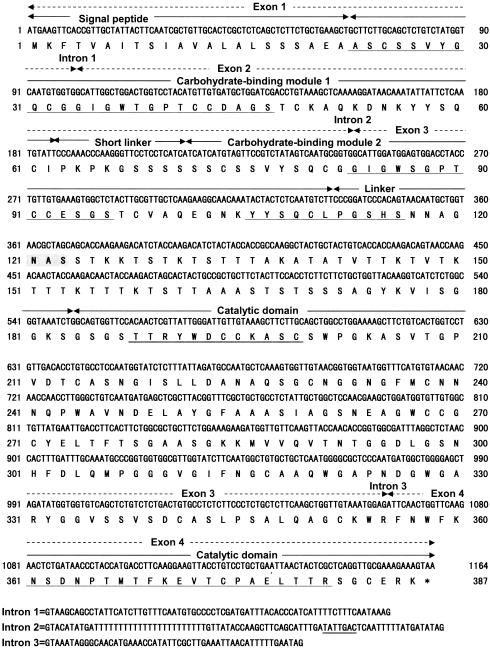
FIG. 2.
Nucleotide and deduced amino acid sequences of the genomic mce gene from M. circinelloides. The mce1 and mce2 cDNAs consist of exons 1, 3, and 4 and of exons 1, 2, 3, and 4, respectively. Arrows indicate the borders of each domain (signal peptide, carbohydrate-binding module 1, short linker, carbohydrate-binding module 2, linker, and catalytic domain). The N-terminal and internal amino acid sequences of purified MCE1 and MCE2 from M. circinelloides are underlined. The consensus amino acid residues of family 45 glycoside hydrolase are double underlined. Potential N-linked glycosylation sites (N-X-S/T) are indicated by shading. The branch point consensus sequences in the intron are double underlined.
Structural characterization of the mce1 and mce 2 cDNAs.
The consensus amino acid residues of family 45 glycoside hydrolases, (S, T, or A)-T-R-Y-(F, Y, or W)-D-X-X-X-X-X-(C or A) (9), were conserved in the amino acid sequences (residues 188 to 199) deduced from the mce1 and mce2 cDNAs (Fig. 2). Therefore, the enzymes (MCE1 and MCE2) encoded by the mce1 and mce2 cDNAs were considered to belong to glycoside hydrolase family 45. The consensus amino acid residues of CBM in family 1 (6) were conserved in the amino acid sequences deduced from the mce1 and mce2 cDNAs (Fig. 2). The amino acid sequences of MCE1 and MCE2 contained one potential N-linked glycosylation site (N-X-S/T) at position 123.
MCE1 and MCE2 consisted of three distinct domains: an N-terminal CBM (family 1), a linker, and a C-terminal catalytic domain (family 45) (Fig. 2). MCE1 had one CBM, and MCE2 had two CBMs at the N terminus. The latter two CBMs were connected by a short linker consisting of nine amino acids. Of the various family 45 endoglucanases, only MCE1, MCE2, RCE1, RCE2, and RCE3 from R. oryzae and PCE1 from P. nitens had the CBM at the N terminus; thus, this structural feature seems to be unique to endoglucanases from members of the subdivision Zygomycota.
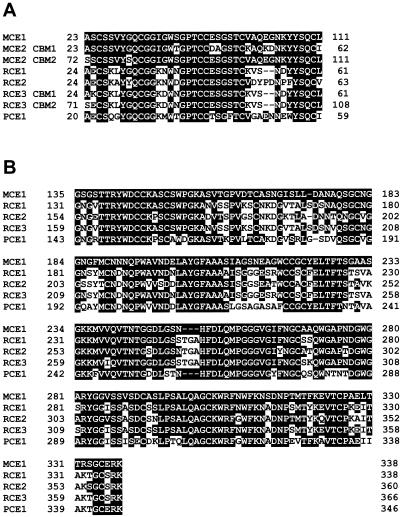
The amino acid sequences of the CBMs of MCE1 and MCE2 were very similar to those of RCE1, RCE2, RCE3, and PCE1 (Fig. 3A). Phylogenetic tree analysis of the amino acid sequences of CBMs from fungal cellulases suggested that the CBMs of MCE1 and MCE2 are evolutionarily closer to those of RCE2 and PCE1 than to those of RCE1 and RCE3 (Fig. 4). The amino acid sequence of the catalytic domain of MCE1 (residues 143 to 346) was very similar to the amino acid sequences of the catalytic domains of RCE1 (75%), RCE2 (73%), RCE3 (75%), and PCE1 (69%) (Fig. 3B). The catalytic domain of MCE1 had lower levels of similarity to the catalytic domains of the other family 45 glycoside hydrolases, indicating that the catalytic domains of family 45 glycoside hydrolases from members of the subdivision Zygomycota might be evolutionarily closer to each other than to the catalytic domains of the other family 45 glycoside hydrolases. Among the members of the subdivision Zygomycota, the catalytic domains of MCE1 and MCE2 exhibited higher levels of similarity to the catalytic domains of RCE1, RCE2, and RCE3 than to the catalytic domain of PCE1. This result indicates that the catalytic domains of family 45 glycoside hydrolases from members of the genus Mucor are evolutionarily closer to those from members of the genus Rhizopus than to those from members of the genus Phycomyces. It might indicate that Mucor and Rhizopus species are closer evolutionarily than the other Zygomycota species in the 18S and 28S rRNA gene tree topology (22).
FIG. 3.
Alignment of the amino acid sequences of the CBMs (A) and catalytic domains (B) of endoglucanases (MCEs, RCEs, and PCE). Amino acids that are identical to amino acids in the CBM and catalytic domain of MCE1 are indicated by white letters on a black background.
FIG. 4.
Phylogenetic tree of the CBMs from fungal cellulases. The analysis was done based on a comparison of amino acid sequence data by using the Clustal W program (version 1.82). Bar = 1% amino acid substitutions.
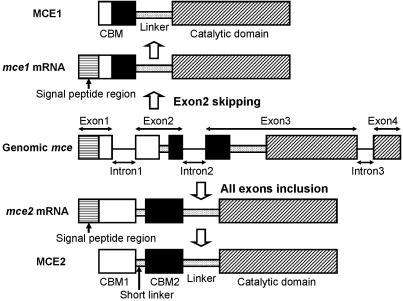
Surprisingly, the sequences of the mce1 and mce2 cDNAs coincided perfectly except for a 147-bp insertion, suggesting that the mce1 and mce2 cDNAs were created from one genomic mce gene by alternative splicing. When genomic DNA from M. circinelloides digested with SphI, AccI, EcoRI, or XhoI or with XhoI and AccI was probed with the mce1 cDNA in a Southern blot, there was one strong signal in each lane (data not shown). This result suggests that there is only one mce1 homologous cDNA in the genome. In fact, the genomic mce gene consisted of four exons, and the mce1 and mce2 cDNAs were created from exons 1, 3, and 4 and from exons 1, 2, 3, and 4, respectively, by alternative splicing. Consequently, MCE1 having one CBM and MCE2 having two tandem CBMs may be secreted as active enzymes (Fig. 5).
FIG. 5.
Schematic representation of the production of MCE1 and MCE2 by alternative splicing of four exons from the genomic mce gene.
Purification and identification of active endoglucanases MCE1 and MCE2.
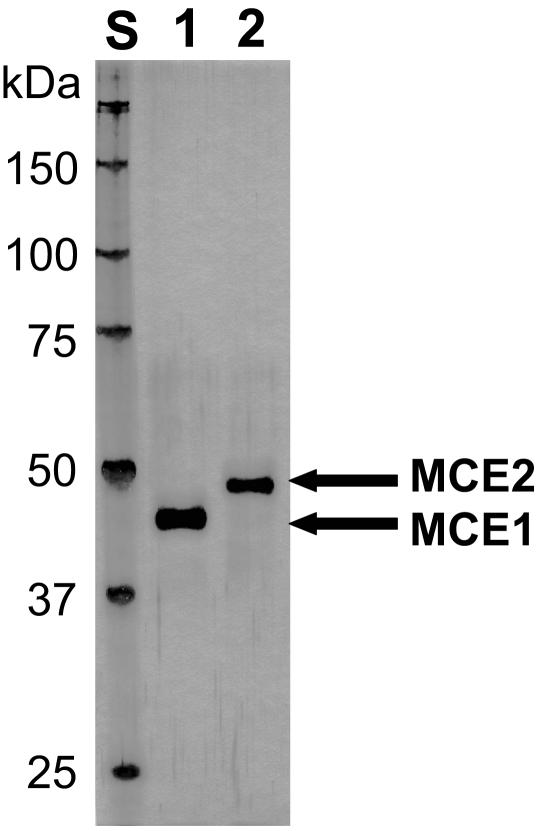
To confirm that endoglucanases encoded by the mce1 and mce2 cDNAs created by alternative splicing are secreted as active enzymes, MCE1 and MCE2 were each purified from culture supernatant of M. circinelloides (Table 1). By using three-step chromatographic fractionation, MCE1 and MCE2 were purified to apparent homogeneity as single bands by SDS-PAGE, and the mobilities corresponded to apparent molecular masses of 43 and 47 kDa, respectively (Fig. 6). MCE1 was purified 69-fold with a level of recovery of 8.9% of the initial activity, and the specific activity was 256 U/mg of protein. MCE2 was purified 60-fold with a level of recovery of 2.4% of the initial activity, and the specific activity was 220 U/mg of protein.
TABLE 1.
Purification of MCE1 and MCE2 from M. circinelloides
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Culture supernatant | 112.0 | 30.180 | 3.7 | 1.0 | 100 |
| First Methyl HIC column | |||||
| 0.5 M ammonium sulfate (MCE1) | 34.9 | 2.412 | 14.4 | 3.9 | 31.2 |
| 0 M ammonium sulfate (MCE2) | 11.4 | 0.304 | 37.4 | 10.1 | 10.2 |
| Second Methyl HIC column | |||||
| 0 M ammonium sulfate (MCE1) | 29.1 | 1.750 | 16.6 | 4.5 | 26.0 |
| 0 M ammonium sulfate (MCE2) | 8.9 | 0.213 | 41.8 | 11.2 | 7.9 |
| Mono S column | |||||
| 120 mM NaCl (MCE1) | 9.97 | 0.039 | 255.6 | 69.1 | 8.9 |
| 190 mM NaCl (MCE2) | 2.64 | 0.012 | 220.0 | 59.5 | 2.4 |
FIG. 6.
SDS-PAGE of purified MCE1 and MCE2 from M. circinelloides. Lane S, standard proteins; lanes 1 and 2, purified MCE1 and MCE2, respectively, obtained by three-step chromatographic fractionation.
The N-terminal amino acid sequences of the purified MCE1 and MCE2 that were determined were as follows: MCE1-N, ASCSSVYGQCGGIGWSGPTCCESGS; and MCE2-N, ASCSSVYGQCGGIGWTGPTCCDAGS. Purified MCE1 and MCE2 were digested with lysylendopeptidase, and each peptide fragment obtained (MCE1a to MCE1c and MCE2a to MCE2c) was sequenced. The amino acid sequences of these peptide fragments were as follows: MCE1a, YYSQCLPGSHS; MCE1b, NSDNPTMTFK; MCE1c, EVTCPAELTTR; MCE2a, YYSQCLPGSH; MCE2b, NSDNPTMTFK; and MCE2c, EVTCPAELTT.
The N-terminal and internal amino acid sequences of purified MCE1 perfectly coincided with the amino acid residues (residues 23 to 33, 83 to 96, 106 to 116, 361 to 370, and 371 to 381) deduced from the mce1 cDNA (Fig. 2). Also, the N-terminal and internal amino acid sequences of purified MCE2 perfectly coincided with the amino acid residues (residues 23 to 47, 106 to 115, 361 to 370, and 371 to 380) deduced from the mce2 cDNA (Fig. 2). These results indicate that the mce1 and mce2 cDNAs coded for purified MCE1 and MCE2, respectively, and these enzymes were secreted in an active form.
Because residues 1 to 22 comprise a signal peptide, the calculated molecular masses of mature MCE1 and MCE2 were 32,168 and 37,194 Da, respectively.
Characterization of endoglucanases MCE1 and MCE2.
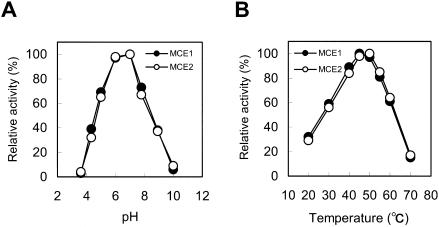
Because MCE1 and MCE2 have the same amino acid sequence except for the CBM sequence, we compared the characteristics of MCE1 and MCE2 to obtain insight into the function of the two tandem CBMs. First, pH and temperature profiles of the CMCase activities of MCE1 and MCE2 were examined (Fig. 7). The optimal pH for activity of MCE1 and MCE2 was 7.0, and the optimum temperature for MCE1and MCE2 was 45 to 50°C. These results indicate that the pH and temperature profiles of MCE1 are almost the same as those of MCE2.
FIG. 7.
pH (A) and temperature (B) profiles of the CMCase activities of purified MCE1 and MCE2.
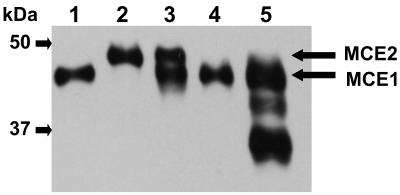
Next, the specific CMCase and Avicelase activities of MCE1 and MCE2 were examined. The CMCase specific activity of MCE2 was 220 ± 5 U/mg, which was almost the same as that of MCE1 (256 ± 6 U/mg), whereas the Avicelase specific activity of MCE2 was 0.161 ± 0.009 U/mg, which was twofold higher than that of MCE1 (0.079 ± 0.005 U/mg). This result indicates that two tandem CBMs might be more effective than one CBM for degrading crystalline cellulose. As shown by the Western blot analysis, both MCE1 and MCE2 were secreted at an early-stage (16-h) culture of M. circinelloides, whereas MCE2 was not secreted at a later stage (42 to 90 h) (Fig. 8). This result indicates that MCE2 might be secreted only during an early stage of culture, when crystalline cellulose is abundant.
FIG. 8.
Western blot analysis of MCE1 and MCE2 secreted by M. circinelloides. Lane 1, purified MCE1; lane 2, purified MCE2; lane 3, 16-h culture supernatant of M. circinelloides; lane 4, 42-h culture supernatant of M. circinelloides; lane 5, 90-h culture supernatant of M. circinelloides.
The mce2 cDNA was created from exons 1, 2, 3, and 4, and introns 1, 2, and 3 were spliced. On the other hand, the mce1 cDNA was created from exons 1, 3, and 4, and exon 2 was skipped. The 5′ splice site of intron 1 (GTAA) and the 3′ splice site of intron 2 (TAG) were strong consensus sequences, whereas the 3′ splice site of intron 1 (AAG) and the 5′ splice site of intron 2 (GTAC) were weak consensus sequences (Fig. 2). Furthermore, the branch point consensus sequences ([C or T]-[G, C, T, or A]-[C or T]-U-[G or A]-A-C) (18) were conserved in intron 2. These results indicate that exon 2 is easily skipped and the mce1 cDNA is constantly created. Since MCE2 was secreted only at an early stage of culture when crystalline cellulose was abundant, the 3′ splicing of intron 1 and the 5′ splicing of intron 2 might be specifically regulated by cultural conditions.
There have been only two previous reports on alternative splicing in the fungal cellulase (5, 20). Birch et al. have shown differential splicing of an intron in the cbhI.1 and cbhI.2 genes encoding cellobiohydrolases from Phanerochaete chrysosporium. The alternative splicing produced two cellulases with a CBM lacking the intron and with a CBM containing the intron and thus, presumably, an altered cellulose binding affinity. Furthermore, Avicel induced the synthesis of both cellulases. In contrast, carboxymethyl cellulose predominantly induced the synthesis of cellulase with the CBM lacking the intron. Although this phenomenon could not clearly explain the role of alternative splicing, it seems likely that alternative splicing creates the diversity of CBMs of fungal cellulases.
REFERENCES
- 1.Azevedo, H., D. Bishop, and A. Cavaco-Paulo. 2000. Effects of agitation level on the adsorption, desorption, and activities on cotton fabrics of full length and core domains of EGV (Humicola insolens) and CenA (Cellulomonas fimi). Enzyme Microb. Technol. 27:325-329. [DOI] [PubMed] [Google Scholar]
- 2.Beguin, P., J. Millet, S. Chauvaux, S. Salamitou, K. Tokatlidis, J. Navas, T. Fujino, M. Lemaire, O. Raynaud, M. K. Daniel, and J. P. Aubert. 1992. Bacterial cellulases. Biochem. Soc. Trans. 20:42-46. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, M. K., and S. Bhat. 1997. Cellulose degrading enzymes and their potential industrial applications. Biotechnol. Adv. 15:583-620. [DOI] [PubMed] [Google Scholar]
- 4.Bhat, M. K. 2000. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 18:355-383. [DOI] [PubMed] [Google Scholar]
- 5.Birch, P. R. J., P. F. G. Sims, and P. Broda. 1995. Substrate-dependent differential splicing of introns in the regions encoding the cellulose binding domains of two exocellobiohydrolase I-like genes in Phanerochaete chrysosporium. Appl. Environ. Microbiol. 61:3741-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boraston, A. B., D. N. Bolam, H. J. Gilbert, and G. J. Davies. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Cavaco-Paulo, A. 1998. Mechanism of cellulase action in textile processes. Carbohydr. Polym. 37:273-277. [Google Scholar]
- 9.Coutinho, P. M., and B. Henrissat. 1999. Carbohydrate-active enzymes server. [Online.] http://afmb.cnrs-mrs.fr/CAZY/GH_45.html.
- 10.Henrissat, B., M. Claeyssens, P. Tomme, L. Lemesle, and J. P. Mornon. 1989. Cellulase families revealed by hydrophobic cluster analysis. Gene 81:83-95. [DOI] [PubMed] [Google Scholar]
- 11.Henrissat, B., H. Driguez, C. Viet, and M. Schulein. 1985. Synergism of cellulases from Trichoderma reesei in the degradation of cellulose. Bio/Technology 3:722-726. [Google Scholar]
- 12.Inoue, T., K. Murashima, J. Azuma, A. Sugimoto, and M. Slaytor. 1997. Cellulose and xylan utilization in the lower termite Reticulitermes speratus. J. Insect Physiol. 43:235-242. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Miller, G. L. 1959. Use of dinitrosalicyclic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 15.Moriya, T., K. Murashima, A. Nakane, K. Yanai, N. Sumida, J. Koga, T. Murakami, and T. Kono. 2003. Molecular cloning of endo-β-d-1,4-glucanase genes, rce1, rce2, and rce3, from Rhizopus oryzae. J. Bacteriol. 185:1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murashima, K., T. Nishimura, Y. Nakamura, J. Koga, T. Moriya, N. Sumida, T. Yaguchi, and T. Kono. 2002. Purification and characterization of new endo-1,4-β-d-glucanases from Rhizopus oryzae. Enzyme Microb. Technol. 30:319-326. [Google Scholar]
- 17.Ohmiya, Y., T. Takeda, S. Nakamura, F. Sakai, and T. Hayashi. 1995. Purification and properties of wall-bound endo-1,4-beta-glucanase from suspension-cultured poplar cells. Plant Cell Physiol. 36:607-614. [PubMed] [Google Scholar]
- 18.Reed, R., and T. Maniatis. 1988. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 2:1268-1276. [DOI] [PubMed] [Google Scholar]
- 19.Shimonaka, A., Y. Baba, J. Koga, A. Nakane, H. Kubota, and T. Kono. 2004. Molecular cloning of a gene encoding endo-β-d-1,4-glucanase PCE1 from Phycomyces nitens. Biosci. Biotechnol. Biochem. 68:2299-2305. [DOI] [PubMed] [Google Scholar]
- 20.Sims, P. F. G., M. S. Soares-Felipe, Q. Wang, M. E. Gent, C. Tempelaars, and P. Broda. 1994. Differential expression of multiple exo-cellobiohydrolase I-like genes in the lignin-degrading fungus Phanerochaete chrysosporium. Mol. Microbiol. 12:209-216. [DOI] [PubMed] [Google Scholar]
- 21.Tomme, P., R. A. Warren, and N. R. Gilkes. 1995. Cellulose hydrolysis by bacteria and fungi. Adv. Microb. Physiol. 37:1-81. [DOI] [PubMed] [Google Scholar]
- 22.Voigt, K., E. Cigelnik, and K. O'Donnell. 1999. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal-DNA sequence data. J. Clin. Microbiol. 37:3957-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood, T. M. 1992. Fungal cellulases. Biochem. Soc. Trans. 20:46-53. [DOI] [PubMed] [Google Scholar]
- 24.Zycha, H., R. Siepmann, and G. Linnemann. 1969. Eine Beschreibung aller Gattungen und Arten dieser Pilzgruppe, p. 313-346. In Mucorales. Verlag von J. Cramer, Weinheim, Germany.